Abstract
We used a novel single-cell strategy to examine the fate of histones during G2-phase. Consistent with previous results, we find that in G2-phase, the majority of nuclear histones are assembled into chromatin, whereas a small fraction comprises an unassembled pool. Small increases in the amount of histones within the free pool affect the extent of exchange, suggesting that the free pool is in dynamic equilibrium with chromatin proteins. Unexpectedly, acetylated H4 is preferentially partitioned to the unassembled pool. Although an increase in global histone acetylation did not affect overall nucleosome dynamics, an H4 containing lysine to glutamine substitutions as mimics of acetylation significantly increased the rate of exchange, but did not affect the acetylation state of neighbouring nucleosomes. Interestingly, transcribed regions are particularly predisposed to exchange on incorporation of H4 acetylation mimics compared with surrounding regions. Our results support a model whereby histone acetylation on K8 and K16 specifically marks nucleosomes for eviction, with histones being rapidly deacetylated on reassembly.
INTRODUCTION
The dynamic nature of chromatin structure ensures accessibility of the genetic information to trans-acting factors. A critical process in this regard is the disassembly/reassembly of nucleosomes manifest as the exchange of core histones into and out of chromatin (1). However, many aspects regarding the extent of histone exchange and the factors that modulate this process remain undefined. A current model proposes that specific epigenetic marks designate regions of chromatin for more or less dynamic exposure of the underlying DNA. Transcriptionally active regions of the genome are associated with specific histone modifications, such as acetylation [for review (2)]. However, acetylation has little effect on the salt or thermal stability of individual nucleosomes and only marginally increases the probability of DNA unwrapping and exposure of internal sites in nucleosome DNA (3,4). Characterization of the role of histone acetylation in transcription has led to the idea that this post-translational modification facilitates the binding of transcription activators containing bromo-domains with affinity for acetylated histone tails and also destabilizes repressive higher-order chromatin structures (5,6). Although these studies provide a mechanistic link between histone acetylation and transcription, whether this modification is directly involved in targeting nucleosomes for disassembly/reassembly is unclear. Early analyses of the acetylation dynamics have shown a rapid turn over of the histone modification at active loci (7,8). Interestingly, genome-wide mapping analyses of histone acetyltransferases (HATs) and histone deacetylases (HDACs) in primary human cells revealed that both activities are co-localized in the vicinity of active genes, rather than HATs associated with active and HDACs with inactive genes (9). The co-localization of these antagonist enzymes is consistent with a high turn over of this histone modification associated with active genes (10,11).
Nucleosome eviction and histone turn-over is also evident by replacement of canonical histones with histone variants. Mapping the sites of incorporation of the variant H3.3 within the genome shows an enrichment of this variant in vicinity of the regulator elements and across active genes (12,13). Interestingly, H3.3 is enriched in post-translational modifications associated with transcriptionally active chromatin (14,15). However, genetic depletion of this histone variant failed to exhibit a phenotype related to an alteration of transcription regulation (16,17). Although the role of H3.3 in predisposing nucleosomes to turn over is unclear, histone variant-containing nucleosomes border nucleosome-free regions of transcription regulatory regions and H2AZ/H3.3-containing nucleosomes have been reported to exhibit a lower stability than canonical histones (18,19).
To examine the role of histone acetylation in nucleosome disassembly/reassembly in vivo, we developed a novel single-cell biochemical approach that allows histone exchange to be assessed at specific times throughout the cell cycle. This approach exploits the natural synchrony of millions of nuclei within a single cell of Physarum polycephalum. We find nuclear histones are partitioned into two distinct histone pools, wherein exchange occurs between a chromatin pool representing the majority of the nuclear histones and a small unassembled histone pool. Using the unique ability of Physarum cells to internalize exogenous histone complexes, we show that the amount of histone within the unassembled histone pool affects the nucleosome exchange pattern, suggesting that the free pool is in dynamic equilibrium with chromatin proteins. Surprisingly, we found that during G2-phase acetylated H4 is preferentially located within the unassembled histone pool, with acetylation at lysines 8 and 16 preferentially appearing in the free pool. Moreover, we found that nucleosomes containing mimics of H4 acetylation are more rapidly displaced from chromatin than those containing unmodified H4. These results support a model wherein H4 acetylation signals prompt nucleosome disassembly and reassembly with histones from the unassembled histone pool.
MATERIALS AND METHODS
Physarum cultures
Physarum polycephalum strain TU291 was maintained in liquid culture. Naturally synchronous macroplasmodia were prepared as previously described (20). Onset of the second synchronous mitosis was determined by phase-contrast microscopy observations of small explants. All the experiments were carried out during the third synchronous cell cycle between M2 and M3.
Incorporation of exogenous proteins into macroplasmodia
The histones used for the incorporation experiments were expressed in Escherichia coli BL21 transformed with pET3a plasmid bearing a gene encoding for Flag H4 (FH4), Flag H4-K4Q (FH4-K4Q) and H3-C110A. For all our experiments of incorporation, we used H3-C110A to ensure the absence of in vitro formation disulphide bridge between two H3s. The different histone complexes were purified as described (21). Defined amounts of tagged H3/FH4 complexes were spread onto the upper cellular surface of macroplasmodia fragments at three time points throughout S-phase similarly to (22). Time course experiments of nucleosome exchange were carried out using a single cell at least in duplicate. The cell fragments were harvested as depicted in the figures.
Analyses of the subnuclear fractions of histones
The analyses of the subnuclear localization of histones were carried out on isolated nuclei prepared by percoll gradient as reported (21). The suspensions of nuclei (50%, 20 μl) were incubated on ice for 30 min in 1 ml of phosphate-buffered saline (PBS) (control) and in 1 ml of PBS supplemented with 0.1% Triton X-100 and the salt concentration indicated in the figures. Chromatin-unbound histones were extracted by treating nuclei with 0.1% Triton to prevent salt-dependent destabilization of nucleoprotein complexes. The fractions were resolved in sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and analysed by western blotting with anti-H3 (Abcam), anti-H3.3 (Abcam), anti-Flag (Sigma) and anti-acetyl H4 (site specific or not) (Active Motif) antibodies as indicated in the figures. For the analyses of nucleosomes, nuclei were isolated, chromatin was digested with MNase followed by fractionation of 5–20% sucrose gradient as described (21,22).
Chromatin immunoprecipitation and quantitative polymerase chain reaction
For chromatin immuno-precipitation (ChIP) experiments, the cell fragments were incubated for 8 min in 1% formaldehyde to allow cross-linking. Nuclear fractions were prepared by homogenization in swelling buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 1% NP40), followed by centrifugation at 700 g for 5 min. The nuclear pellet was resuspended in 200 µl of lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM ethylenediaminetetraacetic acid, 1% SDS) and sonicated five times for 6 sec with 30% output in a Branson sonifier. Debris were pelleted by centrifugation at 13 000 g for 10 min, and chromatin was transferred into a new tube. ChIP analyses were carried out using M2 anti-Flag antibody coupled to agarose beads (Sigma). For anti-K27 methyl (Active Motif) ChIP, the immunocomplexes were purified with protein A/G magnetic beads (Ademtech). The ChIP experiments were performed at least in duplicate. The real-time polymerase chain reactions (PCRs) were performed using appropriate primer sets (Supplementary Table S1) and Maxima SYBR green quantitative PCR (qPCR) master mix (Fermentas) according to manufacturer instructions and the opticon monitor 3 system (BioRad). For a given region, the value was calculated as the log2 of the ratio between the IP signal and the respective input DNA. Each PCR was performed in duplicate for each biological duplicate.
In vitro reconstitution of oligonucleosomes and analyses
Oligonucleosome arrays containing wt H2A/H2B and increasing ratios of wt H3/H4 : H3/H4K4Q [ratios: 100:0 (0% H3/H4KQ4), 80:20 (20%), 60:40 (40%), 40:60 (60%), 20:80 (80%), 0:100 (100%)]—were reconstituted onto a 2.5 kb 5S 12mer DNA template via standard salt dialysis as described in (23). Arrays were then incubated with increasing concentrations of MgCl2 at room temperature for 10 min then centrifuged at 12 500g in a benchtop centrifuge for 10 min. The resulting supernatants were then electrophoresed briefly on 0.8% SDS–agarose gels. The fraction of arrays in the supernatant was quantified and plotted versus MgCl2 concentration.
RESULTS
Nuclear histones are partitioned into chromatin and unassembled pools
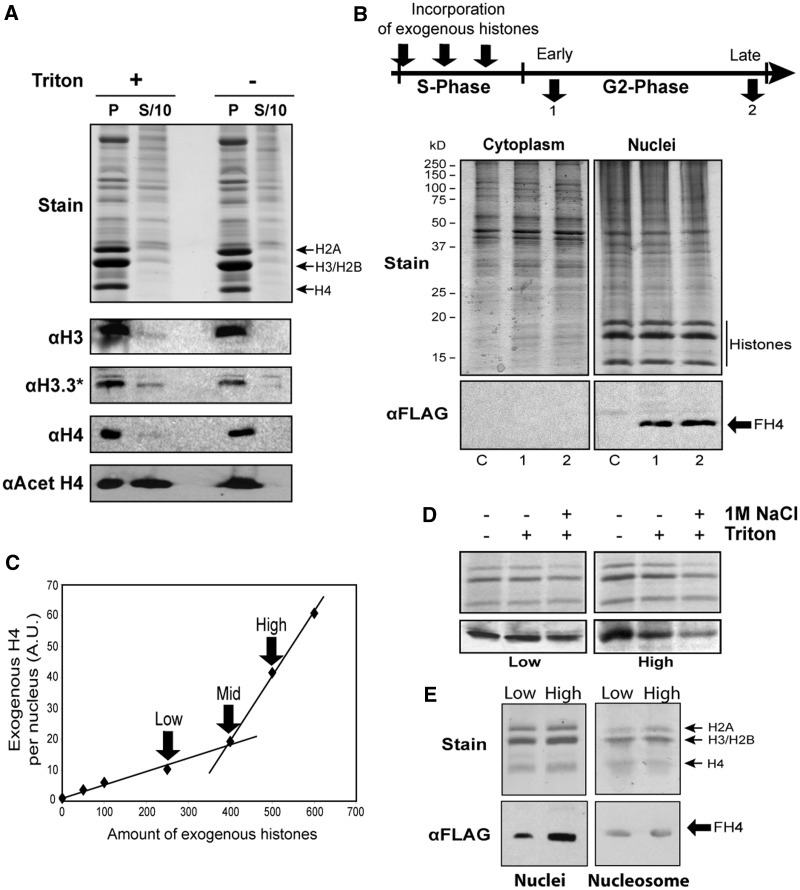
Selective extraction of nuclei from asynchronous mammalian cell cultures indicates that a small fraction of nuclear histones exist within a soluble pool that is not assembled with DNA (24,25). First, we wanted to determine whether a free pool of histones exists outside S-phase. We used cells of the slime mould Physarum polycephalum, which contain several million nuclei that proceed with complete synchrony throughout the cell cycle. Nuclei were isolated from a cell in G2-phase, and half of the nuclear pellet was extracted with PBS containing 0.1% Triton to solubilize nuclear proteins not assembled into chromatin, and the second half was incubated in PBS and used as control (26) (Figure 1A). Western blotting analyses of the soluble fraction and nuclear pellet from the Triton-treated nuclei and the control revealed that a small fraction of total H3 was extracted from the nuclei by the detergent treatment, and thus resident with a pool of nuclear core histones not incorporated into chromatin in G2-phase. Interestingly, blotting with an anti-H3.3 antibody showed that a somewhat greater fraction of total H3.3 was present in the soluble pool [note the antibody to H3.3 exhibited some cross-reactivity with bulk H3 (Supplementary Figure S1)]. Similarly to the H3 data, we found that only a small fraction of nuclear histone H4 is present within the unincorporated pool (Figure 1A). In striking contrast, we observed that acetylated H4 was highly enriched in the soluble nuclear histone pool. Thus, consistent with higher eukaryotes, we find that the nuclear histones are partitioned into chromatin and unassembled pools during G2-phase of the cell cycle.
Figure 1.
Nuclear histone partitioning is experimentally manipulated by the incorporation of different amount of exogenous histones into Physarum macroplasmodia. (A) Endogenous Physarum nuclear histones are partitioned into a chromatin pool and an unassembled pool. Nuclei from fragments of a single Physarum macroplasmodium were incubated in PBS in presence of 0.1% Triton (+) and in absence of detergent (−). Nuclei (P) and supernatants (S/10) were analysed by SDS–PAGE (Stain) and western blotting using antibodies to H3 (αH3), to H3.3 (αH3.3*) (note that this antibody also reacts with canonical H3; Supplementary Figure S1A), to H4 (αH4) and to acetylated H4 (αAcet H4), respectively. (B) Exogenous histones internalized in S-phase are stably recovered in nuclei throughout the G2-phase. Exogenous H3/FH4 complex was spread onto a Physarum cell as depicted on the scheme. One hour after S-phase (Early, 1) and 30 min before mitosis (Late, 2), the cell fragments were harvested and fractionated. Cytoplasmic (Cytoplasm) and nuclear (Nuclei) fractions from control (C), 1 and 2, respectively, were resolved in SDS–PAGE (Stain) and analysed by western blotting with anti-Flag antibody (αFLAG). (C) Titration of exogenous histones internalized into Physarum nuclei. A range of concentration of H3/FH4 was incorporated into Physarum in S-phase as in (B). The nuclei were isolated by percoll gradient in early G2-phase and subjected to western blotting analyses with no further treatments. The immune reactions with anti-Flag antibodies were quantified with ImageQuant. The arrows point out the different concentration used in subsequent experiments, low, mid and high, respectively. (D) Partitioning of the exogenous histones within the two pools depends on the amount of exogenous H3/FH4 internalized during S-phase. Cell fragments were treated as in (B and C) with low amount (low) and high amount (High) of exogenous histones. Nuclei were isolated, analysed by SDS–PAGE and western blotted after different incubation in PBS, PBS + 0.1% Triton and PBS + 0.1% Triton + 1 M NaCl, respectively. (E) The assembly of the exogenous histones in chromatin was examined by conventional sucrose gradient purification of nucleosome prepared from nuclei digested with MNase. The nuclei and nucleosome fraction prepared from cell fragments treated with ‘low’ and ‘high’ amount of exogenous were analysed by SDS–PAGE and western blot.
We and others have previously established that the Physarum giant cell is capable of taking up exogenous proteins and using them in cellular metabolism (20–22,27–30). Thus, we decided to take advantage of this ability to examine whether exchange between the chromatin-assembled histones and the free pool occurs, and the extent to which H4 acetylation influenced such exchange in G2-phase. It is well known that in Physarum, like other eukaryotes, the majority of histone synthesis occurs in S-phase, when replication of chromatin takes place (31,32). We have demonstrated that H3/H4 complexes prepared from recombinant proteins and incorporated into Physarum cells during S-phase are rapidly transported into the nuclei and assembled in chromatin (22,33). As a prelude to G2 exchange experiments, we thus wanted to determine the extent to which exogenous histones incorporated during S-phase persisted in the nuclear fraction during the subsequent G2-phase. Trace amounts of Flag-tagged exogenous canonical H3/H4 complex (H3/FH4) were incorporated into a cell throughout S-phase, to randomly integrate these proteins into the genome (Supplementary Figure S1B), and the cell fragments were harvested in early G2-phase and in late G2-phase. To verify the cellular localization of the exogenous proteins throughout the G2-phase, the cell fragments were fractionated into cytoplasmic and nuclear fractions, and proteins were analysed by western blotting (Figure 1B). Clearly, the Flag-tagged exogenous histones were found only in the nuclear fractions and at a level that was unchanged throughout the 6 h G2-phase, demonstrating that the exogenous histones are not significantly degraded during this period.
Given that our experimental strategy relies on the incorporation of exogenous proteins into the cell rather than transgene expression, it is possible to precisely control the amount of exogenous histones introduced. Consequently, we examined whether the amount of exogenous H3/FH4 introduced into the cell affected the extent of incorporation into nuclei. Cell fragments were treated with different amounts of exogenous histones during S-phase; nuclei were isolated in early G2-phase; and the amount of exogenous Flag-tagged histone was determined by western blotting (Figure 1C). Interestingly, the total amount of exogenous histones per nucleus varied as a function of protein introduced, with a relationship described by two adjacent linear profiles with distinct slopes. These results revealed that although throughout the range of incorporation, the amount of exogenous histones was at trace levels [<1/1000 as estimated by comparing the total endogenous histone amount with the incorporated amount (Supplementary Figure S1B)], the quantity of histones spread onto the cell influenced the amount that accumulated in the nuclei. The biphasic behaviour may be related to the relative ratios of exogenous and endogenous cytoplasmic histones and the abundance of endogenous histone chaperones. Moreover, the inflection in the dependence curve implies that a threshold amount with regard to endogenous histone supply had been reached, possibly because of competition with endogenous cytoplasmic histones for chaperones. These results suggested that histone supply and demand in Physarum cell is as precisely balanced as it is in other organisms (25,34).
Given our finding that the level of H3/FH4 within nuclei depends on the amount introduced into the cell, we next wanted to determine how the different amounts of incorporated exogenous histones were distributed into the chromatin and soluble nuclear histone pools in G2-phase. One half of a cell was incorporated in S-phase with a ‘low’ amount of exogenous H3/FH4, and the other was treated with a ‘high’ amount (Figure 1C), and nuclei were isolated in early G2-phase. One fraction of the nuclei was incubated in PBS as a control, a second was incubated in PBS with Triton to extract the histones in the free pool, and a third was incubated in PBS with Triton and 1 M NaCl to ensure the release of core histones that were not stably assembled into chromatin. The nuclear pellets were then analysed for the amount of exogenous histone remaining in the chromatin-associated nuclear fractions by western blotting (Figure 1D). Surprisingly, the exogenous histones from the two cell fragments exhibited different distributions at beginning of G2-phase. Although the exogenous histones from cells receiving the low amount of tagged H3/FH4 complex were primarily stably incorporated into chromatin and insensitive to detergent and salt extraction (∼85%), a much greater fraction of the Flag-tagged histones from cell fragments treated with the higher amount of exogenous histones was present in the labile, unassembled nuclear fraction. To confirm that the exogenous histones were assembled into nucleosomes, we prepared the nucleosomal fractions by MNase digestion and sucrose gradients. Analyses of the nuclei and nucleosome fractions revealed that, consistent with the previous analysis, the amount incorporated into nuclei depended on the amount of histones spread onto the cellular surface, and that exogenous histones were indeed assembled into nucleosomes (Figure 1E). Importantly, the comparative western blot revealed that regardless of the amount of exogenous histones (low or high) introduced into cells, similar quantities of exogenous histones were incorporated into chromatin (Figure 1E, right). These results suggest that the chromatin assembly machinery is saturated at lower amounts of histones than nuclear import and histone storage machineries. Therefore, the partitioning of the exogenous histones into the different nuclear histone pools was directly related to the amount of exogenous proteins introduced into the Physarum cell fragments.
Apparent exchange of exogenous histones in G2-phase is dependent on the amount of incorporation
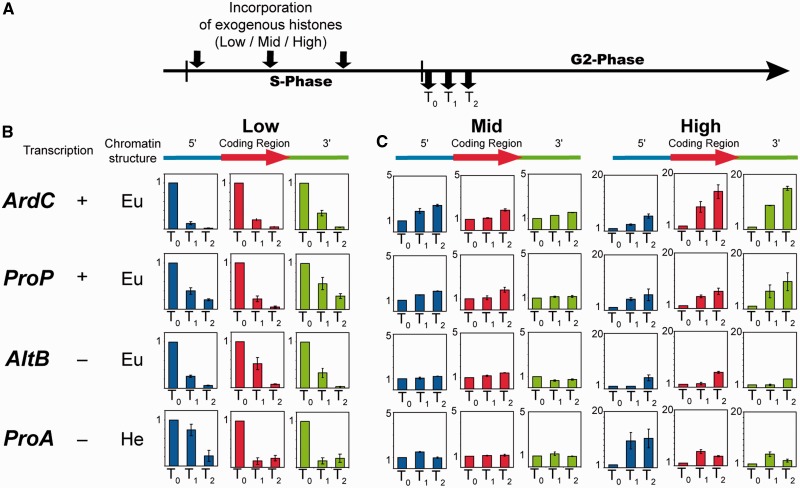
Next, we examined the relationship between histones in the free nuclear pool and histone assembled into chromatin by monitoring the fate of exogenous histones introduced into Physarum cells in S-phase during the subsequent G2-phase. Given that incorporation of different amounts of exogenous histones led to distinct distributions of the tagged histones in the incorporated and free nuclear histone pools, we first monitored histones associated with specific DNA sequences by ChIP, using the ‘low’ amount of exogenous histone, as in these conditions the exogenous histones were almost entirely assembled into chromatin at the beginning of G2-phase (Figure 2A). To carry out these analyses, loci were chosen for their different representative chromatin structures and transcription regulation during the cell cycle (22,35,36) (Supplementary Figure S2A and B). The amount of tagged protein associated with the 5′-region, coding region and 3′-region of each locus at three time points during G2-phase was determined by ChIP and qPCR analyses (Figure 2B and Supplementary Figure S3A). We found that exogenous H3/FH4 is rapidly displaced from chromatin. To verify that the loss of exogenous was caused by the exchange of nucleosomes rather than a simple eviction of the exogenous histones from chromatin, we examined the apparent exchange under conditions (mid and high) where incorporation resulted in exogenous proteins within both chromatin and free pools (Figure 2C and D). The ChIP analyses revealed that when exogenous histones are incorporated at levels corresponding to the inflection point (Figure 1C), almost no apparent exchange was detected, likely because of a relative balance between eviction of tagged proteins from chromatin and assembly of tagged proteins from the soluble pool into chromatin (Figure 2C, ‘mid’). In contrast, when the highest amounts of exogenous histones were incorporated in S-phase, resulting in the greatest fraction of tagged histones within the free pool, the amount of tagged proteins associated with the loci actually increased over the course of the experiment, with the greatest increases in active loci (Figure 2C, ‘high’). These results suggest that the exogenous histones were both evicted from and underwent replication-independent assembly into chromatin in G2-phase. Our findings support a model in which some fraction of bulk H3/H4 is in a constant flux between the assembled and unassembled pools in early G2-phase.
Figure 2.
The profile of nucleosome exchange depends on the amount of histones within the unassembled pool. (A) The scheme on the top represents the experimental diagram with the incorporation of exogenous histones at three time points throughout the S-phase and in the three concentrations defined in Figure 1C, low, mid, and high, respectively. The cell fragments were harvested in early G2-phase, and the association of exogenous histones with specific sequences was determined by ChIP/qPCR (see ‘Materials and Methods’ section) at three time points, T0, T0 + 30 min (T1) and T0 + 60 min (T2), respectively. (B and C) The different loci examined by ChIP are indicated on the left (ArdC: Actin; ProP: plasmodial profilin; AltB: α-tubulin; ProA: amoebal profilin), the relative transcription level and the chromatin structure (Eu: euchromatin and He: heterochromatin) were determined by reverse transcriptase–PCR (Supplementary Figure S2A) and by methyl-K27 of H3 ChIP (Supplementary Figure S2B), respectively. The exogenous histone recovery within the 5′-region (blue bars), coding region (red bars) and 3′-region (green bars) of each locus was determined for each incorporation condition and at the three time points in early G2-phase. The graphs plot the log2 of the ratio IP to input DNA, and the deviation was calculated for biological and PCR duplicates.
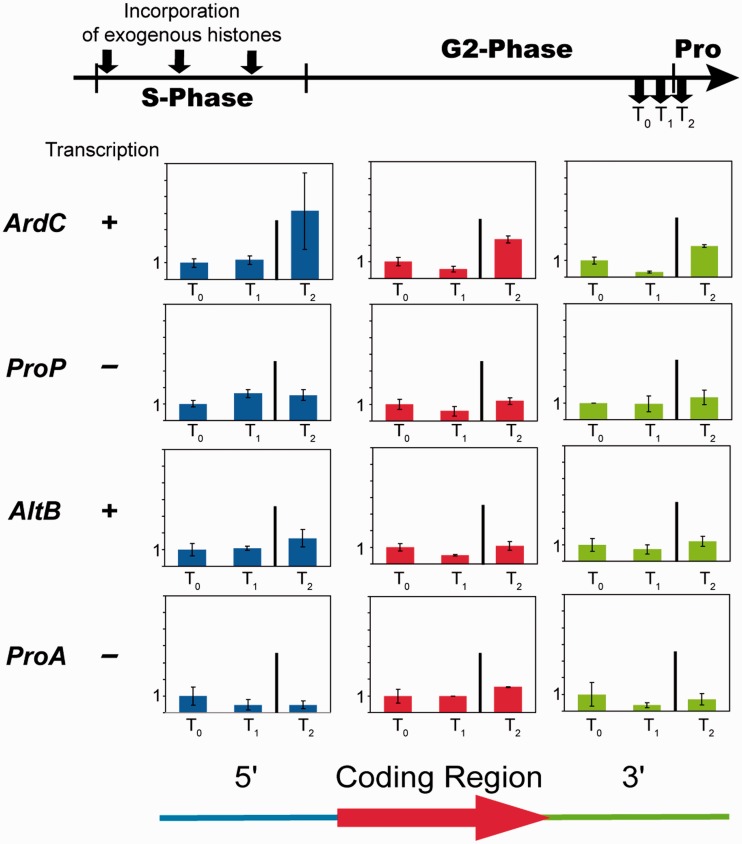
To determine whether the flux of histones into/out of chromatin persists later in G2-phase, low amounts of exogenous histones were incorporated in S-phase and histone exchange examined in late G2-phase and at the G2/M transition (1 h, 30 min and 5 min before mitosis, respectively) (Figure 3). Consistent with previous results (22), we found that the amount of tagged histones associated with each locus examined remained at a constant level, indicating little or no apparent turn over of tagged histones in late G2-phase. However, ChIP experiments performed after the G2/M transition, which corresponds to the formation of mitotic chromosomes, revealed an increase in exogenous histones associated with the highly transcriptionally active ArdC locus, suggesting that in some regions an additional fraction of the unassembled histone pool is deposited into chromatin during mitotic chromosome formation, possibly to completely fill in nucleosome-depleted regions (Figure 3, T2; Supplementary Figure S2C). It is likely that the newly assembled histones come from the nuclear free pool, as Physarum mitosis does not involve the complete disassembly of the nuclear envelope and incorporation of exogenous histones in S-phase does not result in detectable amounts of cytoplasmic histones (Figure 1B) (33).
Figure 3.
Replication-independent nucleosome exchange is a homeostatic equilibrium between assembled and unassembled histone pools. The top scheme represents the experimental diagram of exogenous histone incorporation throughout the S-phase and the harvest of the cell fragments in late G2-phase and at G2/M transition point. The different loci (see the legend of Figure 2) and their relative transcription state in late G2-phase are indicated on the left. The ChIP analyses were carried out as in Figure 2 at specific time points, T0 (mitosis −1 h), T1 (mitosis −30 min) and T2 (mitosis −5 min), respectively. The graphs plot the log2 of the ratio IP to input DNA, and the deviation was calculated for biological and PCR duplicates.
Acetylation stimulates histone exchange
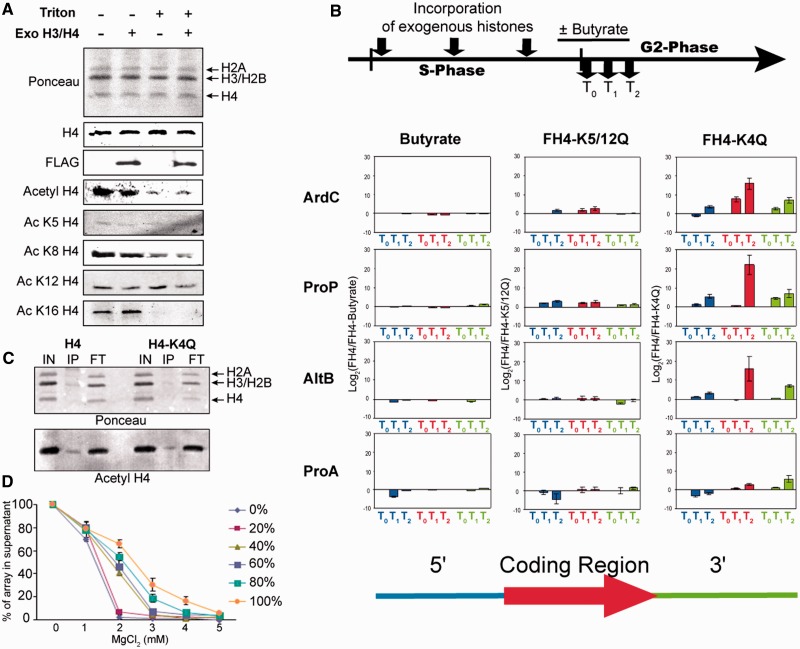
As gene transcription correlates with histone acetylation, we asked whether this modification is associated with the observed histone exchange. We first determined the disposition of acetylated H4 within Physarum nuclei. Half a Physarum cell was treated with the ‘low’ amount of exogenous histones throughout the S-phase, whereas the other half was examined as an untreated control. In early G2-phase, nuclei from the two halves were isolated at ∼15 min beyond S/G2, and the amount of acetylated and unacetylated H4 was estimated by western blotting of purified nuclei and of nuclei subjected to Triton extraction to remove the free pool of histones (Figure 4A). These analyses allowed us to not only determine the nuclear distribution of H4 but also know whether the incorporation of exogenous histones affected the distribution. As expected, the vast majority of endogenous bulk H4 was assembled in chromatin and stable against Triton extraction (Figure 4A, H4 blot, compare lanes 1 and 3). Likewise, almost all of the Flag-tagged exogenous H4 was stably assembled into chromatin and insensitive to extraction with Triton (Figure 4A, lanes 2 and 4). We also found that in both cell fragments (exogenous histone-treated fragment and control fragment), the majority of endogenous acetylated H4 was extracted by Triton; therefore, it was primarily partitioned to the unassembled pool (Figure 4A, compare lanes 1 and 2 with 3 and 4), as revealed with an antibody that reacts with all forms of acetylated H4 (37). Thus, these results showed that the incorporation of ‘low’ amounts of exogenous did not affect the distribution of endogenous histones. We then examined different sites of H4 acetylation using specific antisera. The results of the western blots revealed two distinct patterns of acetylated H4. We found H4 acetylated on K5 and K12 are stably associated within chromatin (Figure 4A, Ac K5 and Ac K12). In striking contrast, H4 acetylated at K8 and especially K16 showed a remarkable decrease after detergent treatment, suggesting these acetylated isoforms are preferentially partitioned to the soluble, unassembled pool (Ac K8 H4 and Ac K16 H4, compare lane 1, 2 and 3, 4).
Figure 4.
Chromosomal H4 acetylation enhances the nucleosome dynamics within euchromatin active loci. (A) Unassembled histone pool is enriched in acetylated H4. Physarum cell fragments were treated and untreated with exogenous histones (Exo H3/H4 +/−) as indicated and harvested in early G2-phase. Nuclei were purified and either loaded on SDS–PAGE directly or extracted with 0.1% Triton to remove the soluble pool of histones before loading (Triton −/+). The nuclear pellets were analysed by western blotting with anti-H4 (H4), anti-Flag (FLAG), antibodies recognizing any acetylated H4 (Acetyl H4) and individual anti-acetylated K5 H4 (Ac K5 H4), anti-acetylated K8 H4 (Ac K8 H4), anti-acetylated K12 H4 (Ac K8 H4) and anti-acetylated K16 H4 (Ac K16 H4) antibodies, respectively. (B) Alteration of the nucleosome exchange by the histone acetylation. The scheme represents the experimental diagram. The timing of incorporation of exogenous proteins and the harvests of cell fragments were identical to Figure 2, except for the butyrate treatment, 30 min before the end of the 3-h S-phase, the cell fragments were cultured in presence of 5 mM sodium butyrate. The ChIP experiments were carried out at least twice on a half cell treated with exogenous histones and corresponding to the experimental conditions, H3/FH4 + butyrate (Butyrate), H3/FH4-K4Q (FH4-K4Q) and H3/FH4-K5-12Q (FH4-K5-12Q), respectively, and the other half used as control and treated with H3/FH4. The regions analysed by ChIP are the same as in Figures 2 and 3. (C) Acetyl mimic did not induce a raise of chromatin acetylation. Chromatin from cells treated with H3/FH4 (H4) and H3/FH4-K4Q (H4-K4Q) during S-phase was prepared by MNase digestion. The soluble chromatin fractions were immunoprecipitated with anti-Flag antibodies coupled to agarose beads. The input (IN), bound (IP) and unbound (FT) fractions were analysed by western blot with a mix of anti-acetyl H4 (Acetyl H4) as in Figure 4A. (D) A threshold level of ∼30% acetylated H4 is required to alter salt-dependent chromatin condensation. Self-association assays were performed as described in the ‘Materials and Methods’ section on arrays reconstituted with increasing percents of H4-K4Q over native H4 and the results plotted.
To further examine the effect of acetylation on histone dynamics, cell fragments were spread with low amounts of exogenous H3/FH4 throughout S-phase, and half the fragments were cultured in presence of butyrate in late S-phase followed by ChIP and qPCR analyses in early G2-phase (Figure 4B). The HDAC inhibitor butyrate globally increased the level of histone acetylation of all nuclear histones (unassembled pool and chromatin pool) (Supplementary Figure S3B). Interestingly, the amount of exogenous histones associated with specific chromatin loci in cells treated with butyrate was similar to that in control fragments, suggesting that butyrate-induced global acetylation did not significantly alter rates of histone exchange for all loci examined (Figure 4B, Butyrate).
We next asked whether levels of histone acetylation beyond that achievable by butyrate treatment can affect histone exchange by incorporating low amounts of exogenous histones containing K→Q substitutions as mimics of acetylation. We first incorporated H3/FH4 containing K→Q substitutions at residues 5 and 12, mimicking the highly conserved deposition pattern of acetylation (33,38). Previous work showed this complex is efficiently assembled into Physarum chromatin when introduced during S-phase (33). ChIP analyses revealed minimal exchange of nucleosomes containing the diacetylated H4 mimic compared with the unacetylated control in the active ArdC and ProP loci and no significant effect on exchange in the inactive loci. These results were consistent with the western blotting analyses of specific acetyl-lysines.
To test whether the exchange is dependent on acetylation at other sites in the H4 tail, we incorporated exogenous H3/FH4, wherein all four acetylable lysines at position 5, 8, 12 and 16 were substituted with glutamine. These histone complexes were efficiently assembled into chromatin during S-phase (Supplementary Figure S3C). We found that chromatin-assembled H3/FH4-K4Q exhibited a significantly greater rate of exchange with the free pool than unacetylated H3/FH4 in early G2-phase, with the greatest levels of exchange occurring in the coding regions of the ProP, ArdC and AltB genes (Figure 4B, FH4-K4Q). Therefore, maximal levels of histone exchange are induced by H4 tetraacetylation, but not diacetylation at lysines 5 and 12, suggesting that nucleosome disassembly requires the modification of the four acetylable lysines of H4 or, minimally, acetylation at H4 lysines 8 and 16. Unfortunately, this hypothesis could not be directly tested, as previous work has shown that the FH4-K8Q/K16Q/H3 histone complex is not imported into Physarum nuclei (failure of nuclear import was also observed for the individual substitutions K8Q and K16Q, data not shown), possibly because of the inhibition of HAT1 acetylation of K5 and K12 (33,39). Nevertheless, these results of incorporation of exogenous histones together with the partitioning of acetylated H4 into the nuclear pools (Figure 4A) strongly suggest an important role for the acetylation of H4 K8 and K16 in nucleosome eviction, as these modifications can occur independently in vivo.
To gain insight into the mechanism whereby acetylation increases histone exchange, we examined whether nucleosomes containing FH4-K4Q induced acetylation, and thus potentially altered the exchange of neighbouring nucleosomes. We immunoprecipitated oligonucleosomes from cell fragments treated with Flag-tagged wild-type H3/FH4 and H3/FH4-K4Q and examined by western blotting the histone acetylation within the oligonucleosomes (Figure 4C). We found that the acetylation status in chromatin surrounding the exogenous histone-containing nucleosome was unaffected by the exogenous FH4-K4Q (Figure 4C, compare IP lanes). Thus, we conclude that the incorporation of the exogenous acetylated H4 mimic does not induce recruitment of HATs to acetylate to nearby nucleosomes. To estimate local chromatin structure alterations induced by acetylated H4 mimic, we carried out chromatin condensation assays with reconstituted nucleosome arrays, wherein the range of H4-K4Q varied from 0 to 100% (Figure 4D) (23). Interestingly, our in vitro analyses revealed that the presence of H4-K4Q diminished the folding of the array when the acetylated H4 mimic-containing nucleosome represented ∼30% of the nucleosomal array. This concentration was obviously higher than the possible concentration found in Physarum cells, as the exogenous/endogenous ratio of histones in our experiments was <1/1000. These results suggest that acetylation-induced nucleosome exchange is not because of alteration of chromatin structure in regions where H4-K4Q is assembled into chromatin.
DISCUSSION
In this work, we document a surprisingly rapid and wide-spread exchange of histones H3/H4 between nucleosomes and unassembled, free histone pools during early G2-phase and show that this exchange likely requires specific acetylation of H4. Histones acetylated at lysines 8 and 16 are preferentially located in the free pool and installation of acetylation mimics at these positions in H4 significantly enhances exchange of exogenous proteins. These results point to a model whereby specific H4 acetylation marks histones for rapid exchange out of nucleosomes as a mechanism for increasing accessibility of specific DNA sequences (Figure 5).
Figure 5.
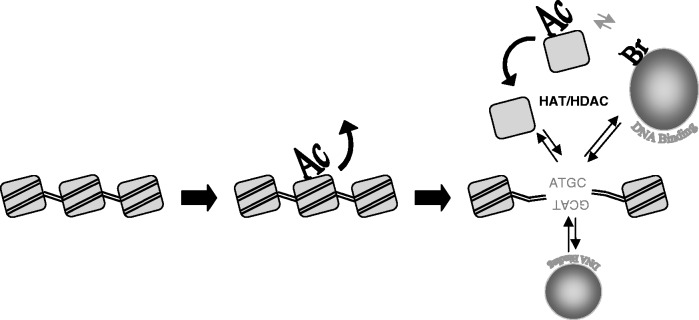
Model of the acetylation-dependent nucleosome turn over. The model proposes that histone acetylation by HATs within euchromatin induces the displacement of the nucleosome from chromatin, making accessible the genetic information. The unassembled acetylated nucleosome might favour the recruitment of factors, like bromo-domain containing proteins (40). The vicinity of HATs and HDACs reported by (9) facilitates the histone deacetylation and the nucleosome recycling.
Like the distribution found in human cell cultures, our experiments show that Physarum nuclear histones are partitioned into two pools; the vast majority of nuclear histones are assembled into chromatin, whereas a small fraction exists in an unassembled pool (Figures 1 and 4A). These results are consistent with FRAP analyses of human cells wherein a subpopulation of H3 (<16%) was found to diffuse freely (41), and the finding of a small fraction of nuclear core histones is associated with chaperones and not assembled into chromatin (40,42,43). Moreover, we find that despite introduction of trace quantities of exogenous histones, small increases in the amounts introduced into the cell result in the exogenous histones being preferentially partitioned to the unassembled nuclear pool. These results suggest that the amount of histone produced by the cell closely matches the capacity for assembly into chromatin during S-phase. Hence, a fine balance between histone supply and demand is maintained during S-phase (25,44,45). Thus, we are able to control the amount of H3/FH4 in the free pool by modest increases in the amount of proteins applied to the cell during S-phase.
In absence of obvious degradation of the exogenous histones throughout the G2-phase, we observed that the apparent exchange in early G2-phase depended on the amounts of the exogenous histone introduced into the cells and their partitioning in the nucleus. Indeed, introduction of the lowest amount of exogenous histones we used, resulted in their near-complete assembly into chromatin during S-phase. In this case, we detected the eviction of chromatin-associated tagged proteins in early G2-phase. In contrast, when higher amounts of exogenous histones were introduced, a greater fraction was partitioned to the free pool, and the amount of tagged protein associated with chromatin actually increased during G2 under some conditions. These results suggest that the two pools of nuclear histones are in constant flux, and the apparent replication-independent exchange is affected by the amount of tagged histones available in the free pool. We previously observed rapid H2A/H2B dimer exchange associated with actively transcribed regions in Physarum, whereas H3/H4 exchanged with proteins in the free pool at much slower rate (22). Thus, our finding that levels of H3/H4 associated with chromatin remain constant when ‘mid’ amounts of histones are introduced (Figure 2) may be because of equilibration of tagged exogenous protein between the two nuclear histone pools. This steady state may also be reached after extended periods when the constant flux of histone leads to the equilibrium of target histone within the two pools (Figure 3, T0 and T1). Alternatively exchange may be a more active and prevalent feature in early G2 (Figure 2). In this model, nucleosomes early G2 still retain deposition-related acetylation or are present in ‘open’ chromatin regions, more exposed to exchange machinery (46,47), whereas mature chromatin in late G2 exhibits much lower levels of exchange (Figure 3).
Importantly, inspection of the acetylation state of nuclear proteins indicated that the vast excess of acetylated H4 was present within the Triton-extractable, free pool. As the pan-acetyl H4 antibody reacts most strongly with hyper-acetylated protein, we tested acetylation at individual sites with specific antibodies. Interestingly, we find that while acetylation at lysines 5 or 12 is associated with H4 stably assembled into chromatin, acetylation of lysines 8 and 16 is preferentially represented in the free histone pool (Figure 4A). These results are consistent with a role in chromatin assembly for H4 acetylated at lysines 5 and 12 (38). Interestingly, in yeast substitution of H4 lysine 16 with arginine indicates a critical function in acetylation at this position in transcription of chromatin (48). This H4 lysine 16 acetylation plays an important role in Drosophila dosage compensation, as this specific acetylation mark is concomitant with the RNA polymerase II recruitment (49). In these experiments, the H4 acetylation has been examined by ChIP, which provides information on the location of the modification within the genome, but did not examine the relative abundance of the acetylation mark within the assembled and free pools of histones. Our analyses revealed that the lysine 16 acetylation of H4 is mainly found in the free pool, suggesting that this modification plays a key role in eviction of core histones. Consistently, it has shown that H4 acetylation at lysine 8 and 16 facilitates H2A/H2B dimer exchange, although in these experiments, H3/H4 tetramer displacement was not investigated and could also be exchanged (50). Moreover, we find that installation of glutamine substitutions as mimics of acetylated lysine at all four positions of the H4-tail domain greatly stimulated the rate and extent of histone exchange compared with the native proteins, whereas H4-containing mimics of acetylation at K5 and K12 induced only low levels of exchange (Figure 4B). These results indicate that the stimulation in histone exchange observed with the tetraacetyl mimic is because of additional acetylation on K8 and/or K16 (40). Together these results indicate that specific acetylation at K8/K16 within the H4 tail marks nucleosomes for rapid exchange. This model predicts that incorporation of a mutant H4 in which the four acetylable lysines are substituted for arginine would result in a protein that undergoes much less frequent exchange. Unfortunately, this hypothesis cannot be tested in our system, as H3/FH4 K5,8,12,16Q tetramers are not efficiently transported into Physarum nuclei and assembled into chromatin, likely because of the role of H4 K5,12 acetylation in these processes (33).
It is well-established that transcription activity coincides with histone acetylation. Interestingly, our results showed that epigenetic marks of transcription, such as the histone variant H3.3 and acetylated H4, are preferentially recovered within the unassembled pool of histones (Figures 1A and 4A). Moreover, HATs and HDACs are preferentially co-localized at active regions within yeast and human genomes (9). Thus, we propose that a recycling mechanism occurs, wherein specific acetylation induces histone displacement, whereas rapid deacetylation occurs after re-deposition of histones into chromatin from the free pool (Figure 5). Recent genome-wide analyses in yeast and Drosophila revealed that histone acetylation promotes nucleosome turn over, suggesting that this model is likely conserved throughout eukaryotes (49,51). It will be interesting to determine whether other reversible histone modifications within the nucleosome influence rates of nucleosome eviction and histone deposition.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and Supplementary Figures 1–3.
FUNDING
CNRS, ANR and La Ligue contre le Cancer [41, 44, 49 and 86 to C.T.]; CNRS post-doctoral fellowship (to G.E.); NIH [GM52426 to J.J.H.]. Funding for open access charge: La Ligue contre le Cancer.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Thiriet Laboratory for helpful discussions.
REFERENCES
- 1.Mito Y, Henikoff J, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Anderson JD, Lowary PT, Widom J. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2001;307:977–985. doi: 10.1006/jmbi.2001.4528. [DOI] [PubMed] [Google Scholar]
- 4.Ausio J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 5.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 6.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 7.Zhang DE, Nelson DA. Histone acetylation in chicken erythrocytes. Rates of deacetylation in immature and mature red blood cells. Biochem. J. 1988;250:241–245. doi: 10.1042/bj2500241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendzel MJ, Delcuve GP, Davie JR. Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem. 1991;266:21936–21942. [PubMed] [Google Scholar]
- 9.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterborg JH. Dynamics of histone acetylation in Saccharomyces cerevisiae. Biochemistry. 2001;40:2599–2605. doi: 10.1021/bi002480c. [DOI] [PubMed] [Google Scholar]
- 11.Waterborg JH. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem. Cell Biol. 2002;80:363–378. doi: 10.1139/o02-080. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 13.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 14.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 16.Cui B, Liu Y, Gorovsky MA. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol. Cell Biol. 2006;26:7719–7730. doi: 10.1128/MCB.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr. Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 18.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiriet C, Hayes JJ. Histone proteins in vivo: cell-cycle-dependent physiological effects of exogenous linker histones incorporated into Physarum polycephalum. Methods. 1999;17:140–150. doi: 10.1006/meth.1998.0725. [DOI] [PubMed] [Google Scholar]
- 21.Thiriet C. Analysis of chromatin assembled in vivo using exogenous histones in Physarum polycephalum. Methods. 2004;33:86–92. doi: 10.1016/j.ymeth.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell. 2011;44:918–927. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 27.Bradbury EM, Inglis RJ, Matthews HR, Langan TA. Molecular basis of control of mitotic cell division in eukaryotes. Nature. 1974;249:553–556. doi: 10.1038/249553a0. [DOI] [PubMed] [Google Scholar]
- 28.Prior CP, Cantor CR, Johnson EM, Allfrey VG. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980;20:597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- 29.Prior CP, Cantor CR, Johnson EM, Littau VC, Allfrey VG. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983;34:1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- 30.Thiriet C, Hayes JJ. A novel labeling technique reveals a function for histone H2A/H2B dimer tail domains in chromatin assembly in vivo. Genes Dev. 2001;15:2048–2053. doi: 10.1101/gad.910201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loidl P, Grobner P. Histone synthesis during the cell cycle of Physarum polycephalum. Synthesis of different histone species is not under a common regulatory control. J. Biol. Chem. 1987;262:10195–10199. [PubMed] [Google Scholar]
- 32.Annunziato AT, Hansen JC. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol. Biol. Cell. 2011;22:245–255. doi: 10.1091/mbc.E10-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 35.Pierron G, Benard M, Puvion E, Flanagan R, Sauer HW, Pallotta D. Replication timing of 10 developmentally regulated genes in Physarum polycephalum. Nucleic Acids Res. 1989;17:553–566. doi: 10.1093/nar/17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maric C, Benard M, Pierron G. Developmentally regulated usage of Physarum DNA replication origins. EMBO Rep. 2003;4:474–478. doi: 10.1038/sj.embor.embor822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin R, Leone JW, Cook RG, Allis CD. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J. Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makowski AM, Dutnall RN, Annunziato AT. Effects of acetylation of histone H4 at lysines 8 and 16 on activity of the Hat1 histone acetyltransferase. J. Biol. Chem. 2001;276:43499–43502. doi: 10.1074/jbc.C100549200. [DOI] [PubMed] [Google Scholar]
- 40.Akai Y, Adachi N, Hayashi Y, Eitoku M, Sano N, Natsume R, Kudo N, Tanokura M, Senda T, Horikoshi M. Structure of the histone chaperone CIA/ASF1-double bromodomain complex linking histone modifications and site-specific histone eviction. Proc. Natl Acad. Sci. USA. 2010;107:8153–8158. doi: 10.1073/pnas.0912509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell. Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 43.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunjan A, Paik J, Verreault A. Regulation of histone synthesis and nucleosome assembly. Biochimie. 2005;87:625–635. doi: 10.1016/j.biochi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 46.Cusick ME, Lee KS, DePamphilis ML, Wassarman PM. Structure of chromatin at deoxyribonucleic acid replication forks: nuclease hypersensitivity results from both prenucleosomal deoxyribonucleic acid and an immature chromatin structure. Biochemistry. 1983;22:3873–3884. doi: 10.1021/bi00285a024. [DOI] [PubMed] [Google Scholar]
- 47.Annunziato AT, Seale RL. Maturation of nucleosomal and nonnucleosomal components of nascent chromatin: differential requirements for concurrent protein synthesis. Biochemistry. 1982;21:5431–5438. doi: 10.1021/bi00265a008. [DOI] [PubMed] [Google Scholar]
- 48.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl Acad. Sci. USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conrad T, Cavalli FM, Vaquerizas JM, Luscombe NM, Akhtar A. Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters. Science. 2012;337:742–746. doi: 10.1126/science.1221428. [DOI] [PubMed] [Google Scholar]
- 50.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 51.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.