Abstract
A general method for the dynamic control of single gene expression in eukaryotes, with no off-target effects, is a long-sought tool for molecular and systems biologists. We engineered two artificial transcription factors (ATFs) that contain Cys2His2 zinc-finger DNA-binding domains of either the mouse transcription factor Zif268 (9 bp of specificity) or a rationally designed array of four zinc fingers (12 bp of specificity). These domains were expressed as fusions to the human estrogen receptor and VP16 activation domain. The ATFs can rapidly induce a single gene driven by a synthetic promoter in response to introduction of an otherwise inert hormone with no detectable off-target effects. In the absence of inducer, the synthetic promoter is inactive and the regulated gene product is not detected. Following addition of inducer, transcripts are induced >50-fold within 15 min. We present a quantitative characterization of these ATFs and provide constructs for making their implementation straightforward. These new tools allow for the elucidation of regulatory network elements dynamically, which we demonstrate with a major metabolic regulator, Gcn4p.
INTRODUCTION
A current goal of synthetic biology is the development of modular tools for programming genetic circuits. This includes the ability to control gene expression in a fast, quantitative fashion without otherwise affecting cellular physiology. Rapid perturbation of a protein’s abundance followed by monitoring of the genome-wide transcriptional response has previously been used to dissect the architecture of complex regulatory networks in vivo (1,2). This kinetic information can directly inform quantitative models of gene regulation and elucidate novel design principles.
In yeast, the most commonly used expression systems rely on nutritional perturbations such as the addition of copper, sugar or methionine to modulate levels of a target gene’s expression (3,4). These nutritional perturbations can be quite severe. For example, a gene whose expression is driven by the MET25 promoter can be selectively repressed, but this requires the addition of 1 mM extracellular methionine (5). For overexpression of a target gene, the transcription factor (TF) Gal4p is commonly used. A target gene placed under the control of a promoter containing Gal4p recognition sequences (UASGAL) can be selectively induced in the presence of galactose (5). Since Gal4p activity is attenuated in glucose, this approach requires growing cells on alternative carbon sources like raffinose or glycerol.
Improvements to expression systems using otherwise inert inducers such as the tetracycline analog doxycycline or the mammalian hormone β-estradiol have reduced the perturbations of the physiological cell state due to introduction of the inducer (6–8). Although these previous systems have provided useful tools, demonstration of an expression system that satisfies the criteria of being (i) fast-acting, (ii) tightly regulated, (iii) nearly gratuitous (i.e. relatively few off-target effects) and (iv) graded over a range of inducer concentrations was only recently published for yeast (7).
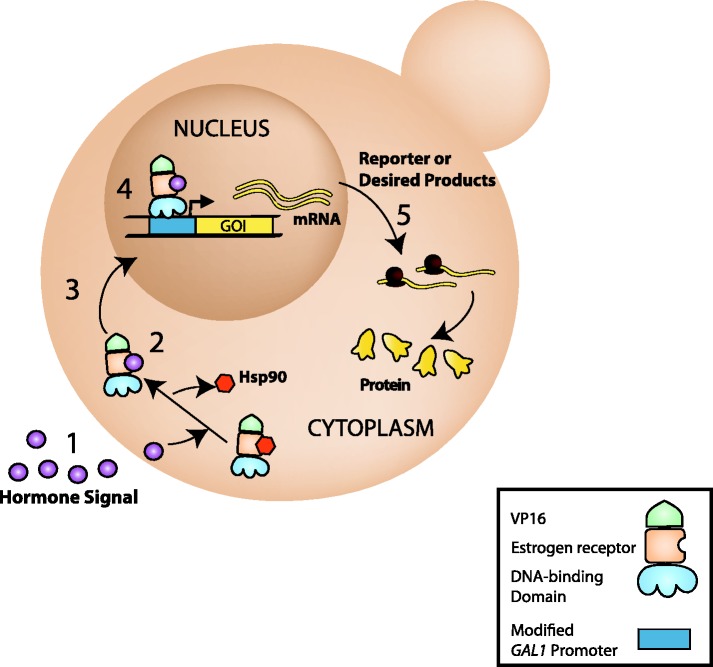
Here, we provide a full characterization of a next-generation β-estradiol inducible expression system for yeast that displays true single-gene precision today as well as the potential for multiplex regulation in the future. This system utilizes a constitutively expressed artificial transcription factor (ATF) from the ACT1 promoter. The ATF consists of a DNA-binding domain (DBD), the human estrogen receptor (ER) and the VP16 activation domain (Figure 1). In the absence of inducer, the ER interacts with the Hsp90 chaperone complex, sequestering the ATF to the cytoplasm (9). Introduction of β-estradiol displaces Hsp90, revealing a nuclear localization signal, and the ATF translocates to the nucleus (Figure 1). The ATF as described provides a strong transcriptional activator that is dependent on the presence of β-estradiol. By using synthetic DBDs that bind only a cognate DNA sequence in place of the Gal4p DBD, residual off-target effects have been completely eliminated.
Figure 1.
Schematic of hormone-based gene expression system. ATFs contain a DNA-binding zinc-finger array, the ligand binding domain of the human estrogen receptor and the VP16 activation domain. In the presence of β-estradiol (1), ATFs dissociate from Hsp90 (2), translocate to the nucleus (3) and activate transcription of a gene of interest (GOI) (4). Once produced (5) the gene products can be detected using a variety of methods.
Previously, the DBD from the yeast transcriptional activator Gal4p was used in the chimeric activator Gal4dbd.ER.VP16 (GEV) (7,10). Yet, as a DBD from a yeast TF, and a factor with only 6 bp of specificity, this DBD has many potential off-target sites in the yeast genome. DBDs of bacterial TFs (e.g. LacI and tetR) have been repurposed for use in the development of gene expression systems that are orthogonal to the native eukaryotic regulatory machinery. However, as discussed elsewhere (11), bacterial DBDs have numerous limitations (oligomerization issues, cooperative binding, etc.) that restrict their use for engineering novel TFs for use in eukaryotes.
We constructed a set of ATFs that utilize the modular and designable Cys2His2 zinc-finger DBD. A single zinc-finger domain contains ∼30 amino acids and will offer three to four bases of specificity, while multiple domains can be assembled as zinc-finger arrays where each additional finger adds three bases of specificity [reviewed in (12)]. This domain has previously shown to be engineer-able to take on novel specificity and function in a somewhat modular fashion while high-quality zinc-finger arrays have been selected by multiple methods (13–15). Through decades of research some basic rules of specificity have been gleaned and specificity can sometimes be designed (16,17). ATFs have previously been constructed by fusing a designed Cys2His2 zinc-finger DBD to a hormone-responsive domain and transcriptional activation domain and were functional in mammalian cells (18,19). A repressive ATF utilizing an engineered Cys2His2 zinc-finger DBD has been reported to achieve single-target precision (also in mammalian cells) (20).
In this study, we replaced the Gal4dbd of GEV with either the three-fingered DBD of the mouse TF Zif268 or a rationally designed four-fingered zinc-finger array (Z4). We refer to these constructs as Z3EV and Z4EV, respectively. The design of the Z4 array leaned heavily on previously defined design principles and targets that have been shown to be most effective (i.e. GNN fingers). The Z4 array was designed to target not only a sequence that is not found in the yeast genome but also a sequence that accommodated a likely to be successful assembly of GNN-binding zinc fingers.
We demonstrate that the ATFs Z3EV and Z4EV are fast-acting and achieve a graded output response over a range of inducer concentrations, as did the GEV system. Using gene expression microarrays, we find that while GEV activation results in the induction or repression of several hundred genes within 30 min, these new ATFs have no significant off-target expression response. As a result, these ATFs eliminate the growth defect incurred by strong induction of GEV. Finally, we demonstrate the power of these new expression systems by inducing GCN4 and following the genome-wide expression responses dynamically.
MATERIALS AND METHODS
Media and growth conditions
For batch culture experiments, cells were grown in either synthetic complete medium lacking uracil or YPD (1% yeast extract, 2% bacto-peptone and 2% dextrose), unless noted otherwise. Induction of GEV, Z3EV or Z4EV by β-estradiol (Tocris Biosciences, Ellisville, MO, USA) was performed in cells during log-phase growth (culture absorbance = 50–100 Klett units). Chemostat cultures were maintained in phosphate-limited growth medium (20 mg/l potassium phosphate). When needed to complement an auxotrophy, phosphate-limited media was supplemented with excess l-Leucine (200 mg/l). The components of chemostat medium are carefully prepared to mimic the composition of standard yeast nitrogen base medium and have been detailed previously (21).
Strain and plasmid construction
Transformations were performed with a standard lithium acetate method. To construct Z3EV and Z4EV strains, the Gal4dbd of GEV was deleted by PCR-mediated disruption with URA3. Zif268 and Z4 DNA-binding domains with homology to the ER and ACT1 promoter were then transformed into cells and selected via 5-Fluoroorotic Acid (5-FOA) counter selection of URA3. Single colonies were isolated and sequenced to verify the presence of the proper DNA-binding domain (Supplementary Figure S1A).
The reporter plasmid was created from the base plasmid pRS416 (gift from Megan McClean), a CEN plasmid containing the URA3 selectable marker. The GAL1 promoter region was amplified from genomic DNA. Overlap-extension PCR was used to add the restriction enzyme sites for XbaI and NotI, respectively, on either side of the region of the three continuous Gal4p-binding sites 5′-CGG-N11-CCG-3′ (22). This promoter fragment was then cloned into pRS416 in front of green fluorescent protein (GFP) using the restriction enzymes NheI and XmaI. Before zinc-finger-binding sites were added the XbaI site in GFP was re-coded using a silent mutation to remove this restriction site. Finally, the three canonical Gal4p-binding sites were removed via digestion with XbaI and NotI and triplets of dimeric Z3EV and Z4EV-binding sites were cloned into respective plasmids (Supplementary Data and Supplementary Figure S1B).
To construct the inducible GCN4 allele, KanMX-Z4EVpr was amplified from pMN10 with the primers 5′-caatttgtctgctcaagaaaataaattaaatacaaataaaCGCACTTAACTTCGCATCTG-3′ and 5′-tggatttaaagcaaataaacttggctgatattcggacatTATAGTTTTTTCTCCTTGACG-3′ and transformed into Z4EV-containing parent strain. The uppercase portions of the sequences share homology with the KanMX-Z4EVpr cassette on the plasmid pMN10.
RNA extraction, labeling and hybridization
RNA extraction, labeling and hybridization were performed as described previously (7). Five milliliters of cells were harvested from chemostat cultures by vacuum filtration onto 0.45-µm nylon membranes (Millipore, HNWP02500) and flash frozen in liquid nitrogen. Crude RNA was extracted with a standard acid-phenol method and subsequently cleaned with RNeasy (QIAGEN, Valencia, CA, USA). Cleaned RNA was labeled using the Agilent Quick-Amp Labeling Kit (#5190-0447). Reference RNA was extracted from a laboratory wild-type strain (DBY12001) grown to steady state in phosphate-limited growth medium with a doubling time of 3.9 h. Microarrays were hybridized for 17 h at 65°C on a rotisserie at 20 rpm. Hybridized microarrays were washed, scanned and raw data were extracted with Agilent Feature Extraction Software version 9.5.
Microarray analysis
Sample and reference channel intensities were first floored to a value of 350 (23,24). Once log2 ratios were computed between samples and reference, the data were time-zero transformed. Genes that were flagged by Agilent Feature Extraction Software or equal to 0 at every time point were removed in R (25). Data were hierarchically clustered in the Cluster 3.0 software package (26) with average linkage using the Pearson correlation distance as the metric of similarity between genes (27). K-means clustering and figures of merit analysis were carried out with the MeV software suite (28,29). Enrichment of TF binding to promoters in each cluster was determined with Fisher’s exact test. P-values were Bonferroni-corrected to account for multiple comparisons.
Quantitative real-time PCR
RNA was converted to cDNA with MultiScribe™ Reverse Transciptase using random hexamer priming (Applied Biosystems). Quantitative real-time PCR (qRT–PCR) of 100 ng cDNA was performed in triplicate with SYBR® Green PCR Master Mix on an ABI 7900HT series PCR machine. PCR conditions were 10 min at 95°C (one cycle) followed by 15 s at 95°C and 1 min at 60°C (40 cycles). Analysis was performed using SDS2.3 software. CT values were computed using the standard curve method. GFP expression levels were normalized to TDH2 expression. Primers for amplification from GFP were 5′-TTTCTGTCTCCGGTGAAGGT-3′ and 5′-GACTAAGGTTGGCCATGGAA-3′. Primers for amplification from the housekeeping gene TDH2 were 5′-TGACTCCACTGGTGTTTTCAAG-3′ and 5′-ACCTTCTTCAATACCGAAAGCA-3′.
Flow cytometry
Approximately 107 cells were harvested by centrifugation and subsequently washed and resuspended in DPBS + 0.1% Tween-20. Measurements of GFP fluorescence were performed with a BD LSRII Multi-Laser Analyzer with HTS (BD Biosciences, Sparks, MD, USA). Mean fluorescence values were determined from at least 50 000 cells.
In Figure 3, dose response curves were fit to a Hill function of the form 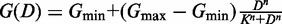 , where G is the level of GFP, D is the amount of β-estradiol, n is the Hill coefficient and K is the dose yielding half of
, where G is the level of GFP, D is the amount of β-estradiol, n is the Hill coefficient and K is the dose yielding half of 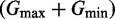 . Gmin and Gmax are constants that are the minimum and maximum mean GFP levels from the data. Data were fit using least-squares regression with the cftool function in MATLAB (MathWorks, Natick, MA, USA).
. Gmin and Gmax are constants that are the minimum and maximum mean GFP levels from the data. Data were fit using least-squares regression with the cftool function in MATLAB (MathWorks, Natick, MA, USA).
Figure 3.
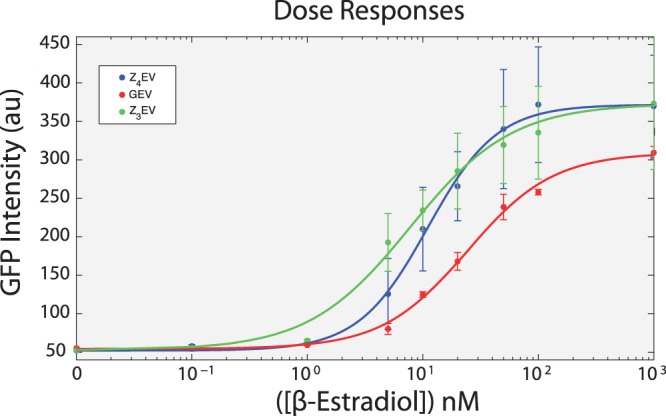
Dose response curves. Strains yMN5 (GEV), yMN7 (Z3EV) and yMN14 (Z4EV) were grown to early log-phase and then different amounts of β-estradiol were added to the growth medium. GFP was quantified following 12 h of induction by flow cytometry. Error bars represent ±1 SD from three independent cultures.
Zinc-finger array design
The Z4 array was designed to bind a sequence that does not exist in the yeast genome while keeping design principles biased toward GNN sequences that have been shown to be more functional and modular (30,31). The zinc fingers were designed with four primary contacts that have been described: Arg’s at position −1 and 6 of each recognition helix to offer G specificity, Asp at position 3 of the helix to offer C specificity and Asn at position 3 to offer A specificity. To avoid high affinity that may be offered by cross-strand contacts, and therefore bias binding by a subset of zinc fingers as opposed to the whole four-fingered array, small amino acids such as Ala and Gly were employed at position 2 of each helix. At the more flexible positions 1 and 5, residues were either chosen because of size or charge. Furthermore, as a four-fingered protein, the backbone of one finger of the three-fingered Zif268 was duplicated and recoded. We chose to repeat finger 2 and extend the linker between the second and third fingers (TGSQKP) as has been described (32).
The final assembly used the backbones of Zif268 fingers 1–2–2–3. As a result, the modest design principles noted above were used to design the recognition helices on each of the four fingers, while the backbone residues were copied from the Zif268 fingers in a finger 1–2–2–3 pattern (the exact coding sequence can be found in the Supplementary Data). Similar finger helices to those designed here have been shown to be functional on these targets when engineered with the OPEN system, giving us confidence that this would be a functional finger array with the desired specificity (31).
Bacterial one-hybrid assay
Characterization of the DNA-binding specificity of the Z4 protein was performed by bacterial one-hybrid assay as previously described (33,34). The protein was assembled by overlapping PCR using the Zif268 scaffold as a template and cloned into the pB1H2w2 plasmid (34). Using this system, functional binding sites are found by selecting for sequences that are able to activate the HIS3 reporter gene in bacteria from a 28-bp library of randomized DNA sequences upstream of the promoter (Supplementary Figure S2). Functional binding sites were recovered by sequencing this promoter region from surviving clones (see Supplementary Data for sequences) and analyzed by MEME to discover an over-represented motif that represents the DNA-binding specificity of the protein (Supplementary Figure S2) (33).
RESULTS
Identifying non-yeast DNA-binding domains with few potential genomic-binding sites
In order to produce a synthetic system that has no off-target induction, we sought synthetic target sequences extremely rare or absent from the yeast genome. The Gal4p DNA-binding domain of GEV dimerizes and binds to the sequence 5′-CGG-N11-GCC-3′ (Figure 2). Scanning the Saccharomyces cerevisiae genome for instances of this motif, we identified more than 500 potential binding sites. The canonical motif recognized by Zif268, the zinc-finger domain used in Z3EV, is GCGTGGGCG (Figure 2), for which we only find 11 predicted binding sites. The motif recognized by the rationally designed zinc-finger array used in Z4EV is GCGGCGGAGGAG (Figure 2). The Z4 zinc-finger array was specifically designed to target this sequence because it is not present in the yeast genome. While the motifs for Gal4p and Zif268 have been published previously, we confirmed the DNA specificity of the Z4 array with a bacterial one-hybrid system (33,34).
Figure 2.
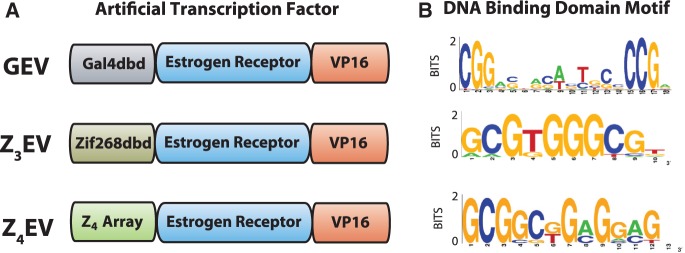
(A) The structure of the ATFs Z4EV, GEV and Z3EV. (B) The DNA-binding motifs of Gal4p, Zif268 and the Z4 array.
Measuring transcriptional output
To characterize the responsiveness of Z3EV and Z4EV, we constructed synthetic promoters based on the zinc-finger DNA-binding specificity (see Supplementary Data for sequences). Each promoter mimics the GEV promoter, containing six monomeric binding sites, which allows for three dimerizations of the ER as one would see in the GEV system. These promoters are referred to as Z3EVpr and Z4EVpr, respectively. Gal4p recognition sequences used in the previous GEV system were replaced with either the Z3EV or Z4EV targeting sequences within the GAL1 promoter (Supplementary Data and Supplementary Figure S1B). Control experiments confirmed that the correct combination of DBD and promoter, as well as introduction of β-estradiol, were required for activation of a target gene (Supplementary Figure S3). When the UASGAL sequences are removed, Gal4p can no longer induce transcription (Supplementary Figure S4). Furthermore, with the appropriate DBD–promoter combinations, the dose response of each construct was characterized (Figure 3). No induction of GFP was observed for any ATF at concentrations of β-estradiol <1 nM. Interestingly, a similar amount of GFP intensity is achieved with ∼10-fold less induction of Z3EV or Z4EV in comparison to GEV (Figure 3). We determined that the Hill coefficient for each ATF-promoter pair is ∼1, consistent with non-cooperative activation of transcription.
The effect of ATF induction on gene expression
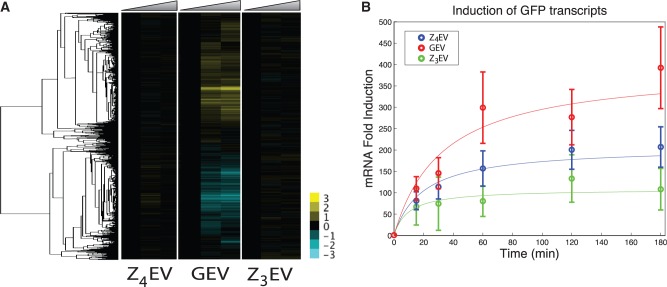
To assess off-target expression directly, we measured genome-wide gene expression of cells grown to steady state in chemostats containing GEV, Z3EV or Z4EV and cognate reporter constructs (Figure 4A), and assessed global gene expression 0, 0.5 and 3 h after addition of 1 µM β-estradiol to the steady-state cultures. In response to GEV induction, 155 genes are induced >2-fold and 263 genes are repressed >2-fold 3 h following β-estradiol addition. In contrast only one gene is induced >2-fold in response to Z3EV activation at 3 h. This gene is YOR343C, which is a dubious open reading frame (ORF) unlikely to encode a protein according to the Saccharomyces Genome Database (35). Similarly, zero genes are induced >2-fold in response to Z4EV activation 3 h following β-estradiol addition; however, two genes, RPS8A (a ribosomal protein) and YDR133C (a dubious ORF), were induced 2-fold in the Z4EV time course at 30 min, but returned to 0-fold at 3 h.
Figure 4.
(A) Hierarchically clustered gene expression responses of continuous cultures containing GEV, Z3EV or Z4EV upon addition of 1 µM β-estradiol (data are time-zero normalized in each experiment). Time points in each experiment are 0, 0.5 and 3 h. Each culture was maintained at a doubling time of 4.07 h. (B) Induction of GFP reporters in cultures from (A) measured by qRT–PCR. Error bars represent ±1 SEM of three technical replicates. Strains are yMN5 (GEV), yMN7 (Z3EV) and yMN14 (Z4EV).
Each of the chimeric activators induced a GFP reporter over the course of the experiment. Cells were visualized by microscopy to confirm GFP expression (data not shown), and the level of GFP transcript was measured by qRT–PCR, revealing that each of the chimeric activators induced GFP mRNA >50-fold by 15 min following addition of β-estradiol (Figure 4B). The maximum level of induction for the chimeric activators ranges from ∼100 - to 300-fold.
The effect of ATF induction on cell growth
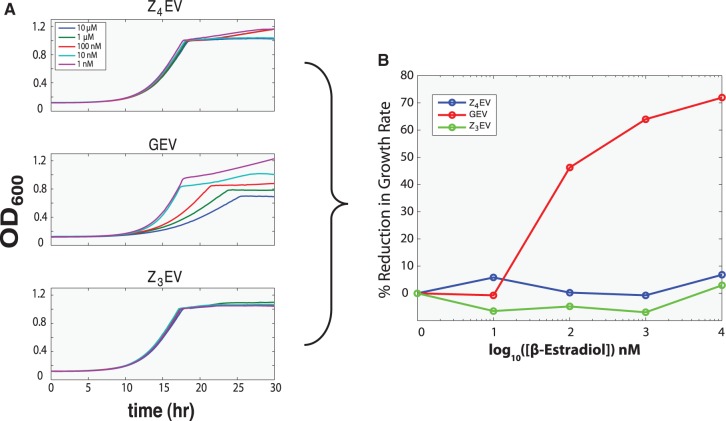
We next examined whether these differences in gene expression might be related to the growth defect we previously reported to be conferred by the GEV system. We compared growth between Z4EV-, GEV- and Z3EV-containing strains in the presence of different amounts of β-estradiol (Figure 5A). Between 10 nM and 10 µM β-estradiol, the growth rate of GEV-containing cells decreased ∼70% in rich medium (Figure 5B). In contrast, induction of either Z3EV or Z4EV with 10 nM–10 µM β-estradiol has no effect on growth (Figure 5B).
Figure 5.
Non-yeast DNA-binding domains eliminate growth defect conferred by GEV induction. (A) Growth curves of GEV-, Z3EV- or Z4EV-containing strains in the presence of different amounts of β-estradiol. (B) The level of growth reduction conferred by ATFs.
Tuning the transcriptional output of Z3EV
The above results demonstrate the precision with which the Z3EV or Z4EV systems are able to activate a single target. However, they do little to indicate how sensitive the system is to the strength of the protein–DNA interaction. To this end, we tested the Z3EV system against four targets with known differences in binding affinity in comparison to the consensus target of Zif268 (36). Within a 9-fold range of dissociation constants, relative to the consensus sequence, activation of GFP is correlated to the strength of the protein–DNA interaction (Figure 6). Furthermore, a binding-site that offers a 20-fold reduction in affinity is not above background after 6 h of induction. These results illustrate that our system is tunable and is unlikely to function on binding sites that offer >20-fold reduction of binding affinity. Future research might investigate the perturbation of networks with finely tuned levels of transcription based on the use of one of these or other characterized binding sites.
Figure 6.
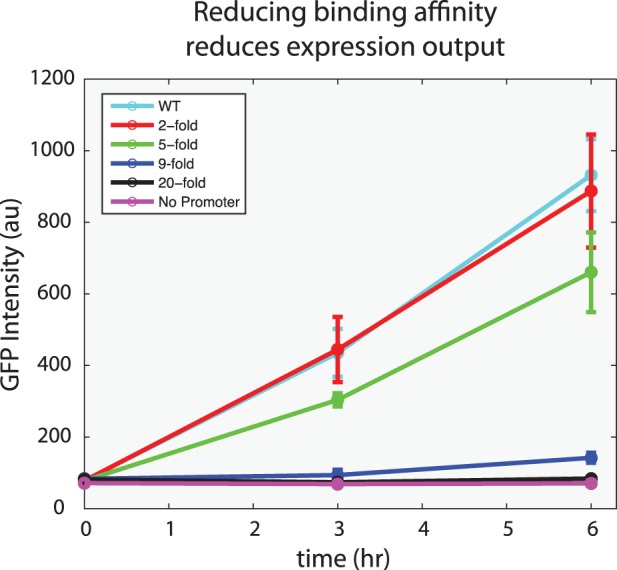
Induction of GFP reporters driven by promoters of different affinity by Z3EV following addition of 1 µM β-estradiol. Error bars represent ±1 SD of three independent cultures.
Using Z4EV to probe the Gcn4p transcriptional regulatory network
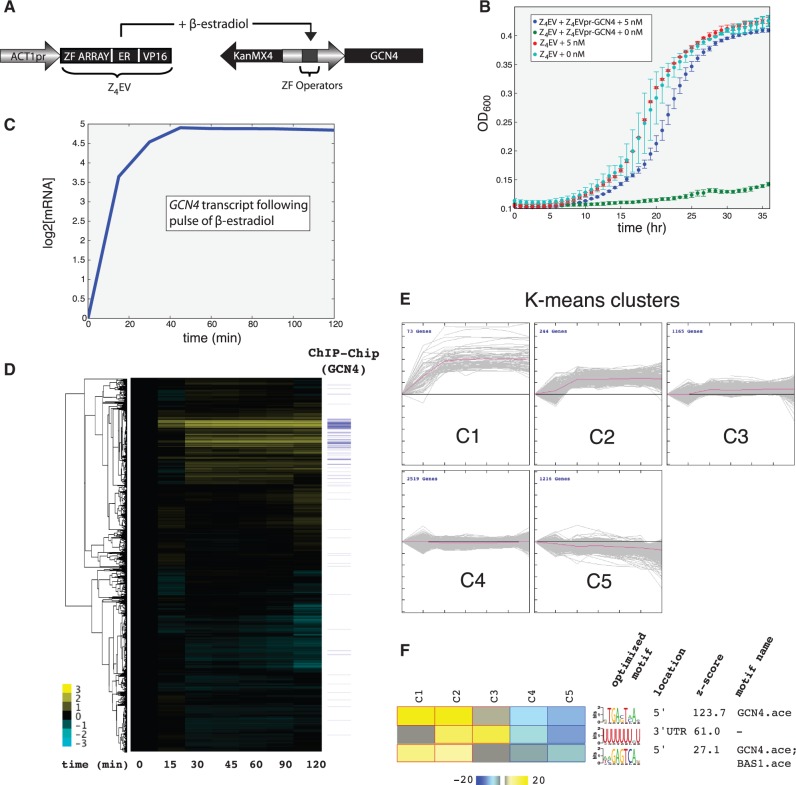
To test whether this system is able to provide single-gene precision on a genomic target, we decided to target GCN4. To simplify constructing inducible genomic alleles, we created plasmids (pMN9 and pMN10) with the KanMX cassette, which confers resistance to G418, fused to either Z3EVpr or Z4EVpr (template sequences can be found in the Supplementary Data). These synthetic promoters can be inserted into the genome by homologous recombination to make an allele that is conditionally expressed in the presence of β-estradiol (Figure 7A). Gcn4p is a strong transcriptional activator of a wide range of enzymes required for the production of amino acids (Supplementary Figures S5–S14). Cells lacking GCN4 grow slowly in minimal medium (37), a phenotype we were able to recapitulate in a strain in which GCN4 is under the control of KanMX-Z4EVpr (Figure 7B). This growth defect is fully alleviated by adding 5 nM β-estradiol (Figure 7B).
Figure 7.
(A) Schematic of strains used for inducing genomic GCN4 allele. Z4EV is driven by the ACT1 promoter. In the presence of β-estradiol, Z4EV activates transcription of the native GCN4 allele driven by the synthetic promoter Z4EVpr. KanMX is expressed in the reverse orientation. (B) Measuring growth Z4EV-containing haploids containing GCN4 or Z4EVpr-GCN4 in the presence or absence of 5 nM β-estradiol. Cells were grown in low phosphate chemostat medium. (C) The transcriptional response of Z4EVpr-GCN4 (strain = DBY12423) in response to 1 µM β-estradiol in a continuous controlled maintained under phosphate limitation. The culture was maintained at a doubling time of 6.3 h. (D) The global transcriptional response of cells in (C) measured out to 2 h. We denote the presence of Gcn4p binding to a promoter by a blue line [(38) P-value <0.001]. (E) Expression data from (D) were divided into five expression clusters using K-means clustering with the Euclidean distance similarity metric. The optimal number of clusters over-represented and under-represented motifs within these clusters was determined using the FIRE algorithm (F).
GCN4 is rapidly induced by Z4EV in response to 1 µM β-estradiol (Figure 7C) and in continuous cultures we were able to quantify the dynamic transcriptional impact of GCN4 expression on cells. By 2 h, induction of GCN4 resulted in >2-fold induction of 327 genes (∼5% of the genome; Figure 7D and Supplementary Dataset S1), which are strongly enriched for cellular amino acid biosynthesis processes (corrected P-value = 1.81 × 10−50). There are 255 genes repressed >2-fold. These genes are enriched for processes involved with cytoplasmic translation (corrected P-value = 1.41 × 10−11), ATP synthesis coupled proton transport (corrected P-value = 6.1 × 10−4) and nucleotide biosynthesis (corrected P-value = 3 × 10−3) (Figure 7D and Supplementary Dataset S1). The presence of Gcn4p binding based on previously published ChIP–Chip data (38) is noted next to the hierarchically-clustered gene expression data (Figure 7D), and clearly correlates with induced expression. Based on ChIP–Chip, of the induced genes, 116 have previously been shown to be direct Gcn4p targets. Only five of the repressed genes (DLD3, ICY1, MET3, RFA3 and TIM10) (Supplementary Figure S15) have been shown to be direct targets. Repression is typically mediated at late time points, consistent with an indirect response to Gcn4p production.
To perform an unbiased search for enriched regulatory motifs corresponding to particular expression responses (determined via K-means clustering) we used the FIRE algorithm (39). The number of K-means clusters was determined using figures of merit analysis (Supplementary Figure S16). Two clusters were identified as having been strongly induced by Gcn4p (clusters 1 and 2; Figure 7E). Based on FIRE, promoters of genes in these clusters are enriched for the presence of Gcn4p-binding sequences (Figure 7F; z-scores = 123.7 and 27.1, respectively). Furthermore, cluster 1 genes are enriched for Gcn4p, Rtg3p, Yap7p, Gln3p and Leu3p binding based on ChIP–Chip (corrected P-values = 1.47 × 10−55, 4.77× 10−5, 0.006, 0.031 and 0.06, respectively; Fisher’s Exact Test). Cluster 2 genes are enriched only for Gcn4p binding (corrected P-value = 1.50 × 10−30; Fisher’s Exact Test). The remaining clusters show no enrichment for Gcn4p binding.
These results, in combination with those detailed above, demonstrate the utility of these expression systems for dissecting complex regulatory networks in vivo. They offer the ability to maintain precise levels of an individual gene’s expression. We plan in the future to adapt other DBDs, target sequences or hormone receptors to make it possible to control several inputs simultaneously or in sequence with the same level of precision.
DISCUSSION
The synthetic biology and broader experimental communities have been hampered by a lack of quantitative expression systems for both engineering novel and probing native regulatory circuitry in vivo. In this article, we developed quantitative expression systems for yeast that address this problem. These expression systems achieve single-gene specificity: in an appropriately modified strain a single gene of interest can be selectively activated in an inducer-dependent fashion. Output expression can be tuned to different levels by either changing the amount of inducer or the identity of the binding sites upstream of a target gene. We have taken advantage of the functionality of hormone receptors as effective post-translational switches in S. cerevisiae. With this methodology, we anticipate that by utilizing multiple hormone receptors with orthogonal-binding domains a combinatorial library of tightly-regulated, inducible expression systems can be used to design and program more sophisticated regulatory functions. These may also be combined with IPTG- and doxycycline-based expression systems, the most recent of which have been characterized in (11).
Here, we have used the Cys2 His2 zinc-finger DBD to provide the necessary affinity and specificity to site-specifically activate the target of interest. However, transcription activation-like (TAL) effector domains have emerged as promising tools for engineering DBDs in eukaryotes. TAL effectors are virulence factors utilized by pathogenic bacteria in the genus Xanthomonas (40). Specificity is determined by repeat variable di-residues (RVDs) at positions 12 and 13 while RVD regions that recognize each of the four nucleotides have been identified (41,42). Their modularity and relative simplicity suggests that TAL domains can be stitched together to form novel DNA-binding domains that recognize any DNA sequence of interest. Synthetic TAL effectors have been constructed that function as activators or repressors of eukaryotic transcription (43). Therefore, if applications would benefit from the use of multiple inducers used simultaneously, a conversion to TAL-hormone receptor system might be beneficial. Still, limitations in the TAL assembly may result in the continued use of zinc fingers (size, repetitive coding sequence, etc.). Moreover, as we have demonstrated in this manuscript, extensive biochemical characterization such as knowledge of Cys2His2 DNA-binding affinity can be used to predictably tune output expression of distinct targeting sequences. The wealth of literature on the zinc-finger protein–DNA interaction may prove advantageous toward this end.
Different inputs can be used to stimulate a network and reveal its relevant components and architecture. A ‘transfer function’ relates a prescribed input to a measured output. In the described case, we provided a step increase of Gcn4p to identify and quantify the responsive outputs of Gcn4p activation. It is, however, often desirable to provide alternative stimuli. For example, to determine the bandwidth of a signaling pathway, the pathway input can be varied at different frequencies and the output can be measured using fluorescently tagged reporters. This approach has been used previously to show that the Hog1p MAPK-signaling cascade acts as a low-pass filter in response to fluctuations in extracellular salt concentration and quantify its bandwidth (44). Future work may investigate how to directly provide cells with varying amounts of protein at desired frequencies. While varying extracellular concentrations of stimulus is relatively straightforward with microfluidic-based approaches, varying intracellular levels of protein over time presents a significant technical challenge. One solution may be the implementation of light-responsive expression systems, which can induce expression of a target gene in the presence of a particular frequency of light, and ablate expression in its absence (45).
We previously demonstrated that by inducing the TEV protease with GEV, we could rapidly degrade an N-degron-tagged protein with a half-life of <15 min (7). By combining two orthogonal induction systems (one for inducing an N-degron-tagged allele and a second for inducing TEV) one could provide cells with different size and/or length protein pulses. In response to the first inducer the protein would be synthesized, and in response to the second inducer, the protein would be destroyed by the protease. This approach could also utilize the TIR1 ubiquitin ligase from Arabidopsis thaliana, which is post-translationally induced by the auxin indole-3-acetic acid and is functional in yeast (46). This may be advantageous because TIR1 can be expressed constitutively and induced by auxin without having to be first transcribed and translated as in the case of TEV.
In conclusion, synthetic approaches can aid in the understanding of native regulatory machinery and provide the toolkit to design and implement rationally designed genetic programs. We demonstrated the utility of a perturbative approach in quantifying the downstream effects of a major transcriptional regulator of amino acid biosynthetic genes. More broadly our work provides a new set of expression systems for a variety of experimental and potentially industrial applications. These systems serve as a flexible platform that can be used to tune, integrate and re-purpose eukaryote functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–16 and Supplementary Dataset 1.
FUNDING
The NSF Graduate Research Fellowship (to R.S.M.), Lewis-Sigler Fellowship, the endowed gift of Peter Lewis (to M.B.N.), National Institutes of Health grant [GM046406 to D.B.] and NIGMS Center for Quantitative Biology [GM071508 to D.B.]. Funding for open access charge: Lewis-Sigler Fellowship, endowment of Peter Lewis.
Conflict of interest statement. McIsaac, Noyes, and Botstein (with others) have submitted part of this system as a Patent Disclosure.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the great undergraduate students of the QCB301 course, in particular Brian Hsueh and Catalina Hwang, with whom they initiated this project, Dr Megan McClean for her gift of primary plasmids and advice while developing many of the tools in this project. Also, they would like to thank the anonymous reviewers, whose constructive remarks greatly improved the manuscript.
REFERENCES
- 1.McIsaac RS, Petti AA, Bussemaker HJ, Botstein D. Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway. Mol. Biol. Cell. 2012;23:2993–3007. doi: 10.1091/mbc.E12-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickman MJ, Petti AA, Ho-Shing O, Silverman SJ, McIsaac RS, Lee TA, Botstein D. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol. Biol. Cell. 2011;22:4192–4204. doi: 10.1091/mbc.E11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romanos MA, Scorer CA, Clare JJ. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 4.Labbe S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Expr. Recombin. Genes Eukary. Sys. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 5.Ronicke V, Graulich W, Mumberg D, Muller R, Funk M. Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Cell Cycle Control. 1997;283:313–322. doi: 10.1016/s0076-6879(97)83025-x. [DOI] [PubMed] [Google Scholar]
- 6.Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIsaac RS, Silverman SJ, McClean MN, Gibney PA, Macinskas J, Hickman MJ, Petti AA, Botstein D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell. 2011;22:4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander RD, Barrass JD, Dichtl B, Kos M, Obtulowicz T, Robert MC, Koper M, Karkusiewicz I, Mariconti L, Tollervey D, et al. RiboSys, a high-resolution, quantitative approach to measure the in vivo kinetics of pre-mRNA splicing and 3′-end processing in Saccharomyces cerevisiae. RNA. 2010;16:2570–2580. doi: 10.1261/rna.2162610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fliss AE, Benzeno S, Rao J, Caplan AJ. Control of estrogen receptor ligand binding by Hsp90. J. Steroid Biochem. Mol. Biol. 2000;72:223–230. doi: 10.1016/s0960-0760(00)00037-6. [DOI] [PubMed] [Google Scholar]
- 10.Louvion JF, Havaux-Copf B, Picard D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 11.Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Quart. Rev. Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 13.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 14.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl Acad. Sci. USA. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 17.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 18.Beerli RR, Schopfer U, Dreier B, Barbas CF., 3rd Chemically regulated zinc finger transcription factors. J. Biol. Chem. 2000;275:32617–32627. doi: 10.1074/jbc.M005108200. [DOI] [PubMed] [Google Scholar]
- 19.Xu L, Zerby D, Huang Y, Ji H, Nyanguile OF, de los Angeles JE, Kadan MJ. A versatile framework for the design of ligand-dependent, transgene-specific transcription factors. Mol. Ther. 2001;3:262–273. doi: 10.1006/mthe.2000.0254. [DOI] [PubMed] [Google Scholar]
- 20.Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y, Zhang HS, Howes K, McNamara AR, Lai A, et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc. Natl Acad. Sci. USA. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protocols. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 23.Petti AA, McIsaac RS, Ho-Shing O, Bussemaker HJ, Botstein D. Combinatorial control of diverse metabolic and physiological functions by transcriptional regulators of the yeast sulfur assimilation pathway. Mol. Biol. Cell. 2012;23:3008–3024. doi: 10.1091/mbc.E12-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petti AA, Crutchfield CA, Rabinowitz JD, Botstein D. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc. Natl Acad. Sci. USA. 2011;108:E1089–E1098. doi: 10.1073/pnas.1101494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Development Core Team R. R Foundation for Statistical Computing. Vienna, Austria: Vienna, Austria; 2009. [Google Scholar]
- 26.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 27.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- 29.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 33.Noyes MB. Analysis of specific protein-DNA interactions by bacterial one-hybrid assay. Methods Mol. Biol. 2012;786:79–95. doi: 10.1007/978-1-61779-292-2_5. [DOI] [PubMed] [Google Scholar]
- 34.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JC, Pabo CO. Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition. J. Mol. Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 37.Hinnebusch AG, Fink GR. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1983;80:5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 41.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 42.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 43.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc. Natl Acad. Sci. USA. 2008;105:7165–7170. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat. Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.