Abstract
The mitochondrial genome exists in numerous structural conformations, complicating the study of mitochondrial DNA (mtDNA) metabolism. Here, we describe the development of 2D intact mtDNA agarose gel electrophoresis (2D-IMAGE) for the separation and detection of approximately two-dozen distinct topoisomers. Although the major topoisomers were well conserved across many cell and tissue types, unique differences in certain cells and tissues were also observed. RNase treatment revealed that partially hybridized RNAs associated primarily with covalently closed circular DNA, consistent with this structure being the template for transcription. Circular structures composed of RNA:DNA hybrids contained only heavy-strand DNA sequences, implicating them as lagging-strand replication intermediates. During recovery from replicative arrest, 2D-IMAGE showed changes in both template selection and replication products. These studies suggest that discrete topoisomers are associated with specific mtDNA-directed processes. Because of the increased resolution, 2D-IMAGE has the potential to identify novel mtDNA intermediates involved in replication or transcription, or pathology including oxidative linearization, deletions or depletion.
INTRODUCTION
The mammalian mitochondrial genome is present in hundreds to thousands of copies per cell and encodes essential components of oxidative phosphorylation (OXPHOS) complexes I, III, IV and V. Mitochondrial DNA (mtDNA) is located within the mitochondrial matrix in single or multi-genome protein–DNA structures termed nucleoids (1–6), often associated with the inner membrane (7,8). Total mtDNA copy number is under tight cellular control and responds to changes in demand for mitochondrial respiration [reviewed in (9)]. The maintenance and integrity of this genome is vital, as alterations such as point mutations, insertions and deletions, or low copy number (depletion) impair mitochondrial respiration and result in adenosine triphosphate insufficiency that can cause a wide spectrum of heterogeneous multi-systemic disorders (10–13). Consistent with their causative role in disease, mtDNA abnormalities are increasingly associated with a range of human pathologies, including neurodegenerative disorders (14,15), type 2 diabetes mellitus (16–18), cancer (19–21) and aging (22,23).
Mitochondrial function and mtDNA integrity are maintained by a dynamic balance among replication, transcription, repair and degradation. It is not yet known how this balance is regulated, or how specific mtDNA nucleoids (or genomes within the nucleoid) are assigned to each process. It is possible that genomes performing different roles may be structurally distinct; however, the inability to isolate mtDNA structures has limited the study of these processes. We expected that the separation and detection of mtDNA based on size, shape, double or single-stranded content and topological conformation would identify molecules involved in specific aspects of mtDNA metabolism. To begin to address this hypothesis, we first explored established methods to separate mtDNA structures based on their shape.
Agarose gel electrophoresis is a robust method for separating plasmid DNA topoisomers, and it was first used to examine mtDNA damage in isolated HeLa cell mitochondria (24,25). Improved 1D separation methods can detect changes in mtDNA topology caused by transient knockdown or induced expression of the mtDNA organizing protein and mitochondrial transcription factor A (TFAM) in mouse cells (26), and they were later used to identify complex tangled structures (27), catenanes and double-length linear molecules (28) present in human heart mitochondria. The addition of a second gel separation step, which uses ethidium bromide (EtBr) intercalation to compact and accelerate topologically confined DNA, can further resolve undigested genomes, but it has only been applied to petite yeast mtDNA (29) and high-molecular weight structures in heart (27). This approach contrasts the classic 2D agarose electrophoresis gels (2D-AGE) used to examine fragments of genomes (30). In the present study, we adapted the concept of resolving undigested (intact) mtDNA to optimize the separation of all major structures, a technique that we call 2D intact mitochondrial DNA agarose gel electrophoresis (2D-IMAGE). We have used a detailed molecular biology approach to describe and characterize the structures revealed by 2D-IMAGE, many of which are associated with replication and possibly transcription.
MATERIALS AND METHODS
Cell culture and treatments
Mouse embryonic fibroblasts (MEFs), C2C12 myoblasts, MIN6, HeLa, HMEC 151p and MDA-MB-231 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 25 mM of glucose, 1 mM of sodium pyruvate, 1× pen/strep and 10% of fetal bovine serum:fetal calf serum (1:1). MIN6 cell medium was further supplemented with 0.001% of 2-mercaptoethanol. SKBR3 cells were cultured in Roswell Park Memorial Institute medium supplemented with 10% of fetal bovine serum. Isolation of cardiomyocytes from neonatal myocardium was performed by the myocyte core of the Penn Cardiovascular Institute at the University of Pennsylvania using standard procedures (31).
EtBr treatment of wild-type MEFs was conducted on subconfluent cells for 24 h in the presence of 1 mM of pyruvate, 0.2 mM of uridine and either 100, 250 or 500 ng/ml EtBr. For EtBr recovery experiments, subconfluent MEFs were grown in 500 ng/ml EtBr plus pyruvate and uridine for 24 h, at which time the medium was aspirated, cells were washed once with phosphate buffered saline without magnesium or calcium (MP Biomedicals) and fresh medium lacking EtBr was added. Cells were harvested at 1, 3 and 6 h after medium change. Cell collection was performed as follows: cells were trypsinized, neutralized with media plus serum, pelleted at 200 RCF for 5 min, washed with phosphate buffered saline, collected at 4°C by centrifugation, flash frozen in liquid nitrogen, then stored at −80°C for later DNA preparation.
Relative mtDNA content by quantitative polymerase chain reaction (qPCR) was conducted using MT9 and MT11 primers (32) for mtDNA and NDUFV1 F and NDUFV1 R primers for nuclear DNA (33) in a RealPlex PCR mastercycler (Eppendorf) with SYBR green mastermix (Applied Biosystems).
DNA isolation, Southern blotting, 1-D and 2-D electrophoresis
Please see Supplementary Methods for description of enrichment of mitochondria, DNA preparation, Southern blotting, 1D and 2D electrophoresis and step-by-step protocols.
DNA treatment
For enzymatic treatment of DNA, samples were diluted into the appropriate buffer provided by each manufacturer and treated with the indicated units of enzyme at the following temperatures and times: topoisomerase I (Invitrogen; 20 U at 37°C for 1 h); topoisomerase II (USB Affymetrix; 40 U at 37°C for 1 h); topoisomerase IV (Inspiralis; 20 U at 37°C for 1 h); DNA gyrase (New England Biolabs; 10 U at 37°C for 1 h); BglII (Fermentas; 4 U at 37°C for 30 min); S1 nuclease (Promega; 0.9 U at 37°C for 30 min); Escherichia coli exonuclease I (New England Biolabs; 20 U at 37°C for 30 min); RNase A (Ambion; 2 µl at 24°C for 1 h, used to generate data in Figure 4 and Supplementary Figures S1 and S2); RNase H (Fermentas; 15 U at 37°C for 30 min).
Figure 4.
2D-IMAGE profiling reveals similar mtDNA structural forms with different distributions among many cell types. (A) Mouse cells: MEF, MIN6, C2C12 and MC3T3-E1. (B) Human cells: HMEC 151p (hTERT), MDA-MB-231, HeLa and SKBR3. MtDNA detected by Southern blot with total mtDNA probe. Black diamond in MIN6 cells indicates unique structure similar to those found in tissues (Figure 5). Abbreviations for structures are from Figure 3.
RESULTS
1D separation of mtDNA reveals major topoisomers
We first examined the qualitative distribution of total mtDNA topoisomers in wild-type MEFs or C2C12 myoblasts by an established 1D agarose gel-based method (26). RNase A-treated total wild-type MEF DNA (10 µg based on OD260) was electrophoresed through a low-percentage agarose gel overnight at low voltage to allow higher molecular weight DNAs to resolve (see Supplementary Methods for details). DNAs were transferred to nylon membranes and Southern blotted for mtDNA with a probe for mouse COXI sequence (Supplementary Figure S1). Catenated circles, nicked circles, linear molecules and supercoiled circles were resolved in a ladder pattern (Figure 1A), typical for this approach (25,26,28,34–37). Preliminary characterizations to confirm the predicted identities of the major bands in each lane included treatment of total MEF DNA with the BglII restriction enzyme, which cleaves mouse mtDNA once, and topoisomerase I, which relaxes supercoiled DNA but does not separate catenated molecules (Figure 1A). DNA molecules sensitive to linearization by BglII treatment are presumed to have a circular form and to be double-stranded (ds) at the restriction site; therefore, the location of the BglII linearization product marks the position of genome-length linear mtDNA in the undigested 1D gel profile. Several other low abundance molecules were detected after restriction digest (each indicated by asterisks) and were addressed in later experiments (see Figure 2). The position of maximally supercoiled mtDNA molecules was revealed by topoisomerase I treatment, which converted them to relaxed circles (Figure 1A and B). To confirm the location of supercoiled molecules, we also treated samples with DNA gyrase, which increases the degree of supercoiling of closed circular DNA (Figure 1B). DNA gyrase treatment did not promote supercoiling of all relaxed circles, suggesting that the relaxed circles were not exclusively a covalently closed population. We next treated total genomic DNA with type II topoisomerases (topoisomerase II and IV), which decatenate or relax supercoiled molecules but leave concatemers (head-to-tail dimers) intact. Both type II topoisomerases relaxed supercoiled molecules and resolved catenated circles (Figure 1B).
Figure 1.
Separation of mtDNA into major topoisomers: catenanes, relaxed circles, linear molecules and supercoiled circles. (A) Treatment of total cellular DNA isolated from MEFs with BglII and topoisomerase I (Topo I). Untreated control (untr.) is shown for reference. Additional structures present after BglII digest are indicated by asterisks. (B) Treatment of total cellular DNA isolated from C2C12 myoblasts with topoisomerases I, II, IV and gyrase. (C) Treatment of total cellular DNA isolated from C2C12 myoblasts with S1 nuclease and E. coli exonuclease I. All gels were probed by Southern blot with COXI mtDNA sequence.
Figure 2.
2D separation of mtDNA resolves previously undetected structural forms. (A–F) Total MEF DNA resolved by 2D-IMAGE and detected by Southern blot analysis with total mtDNA probe: (A) untreated; (B) DNA from isolated mitochondria; (C) topoisomerase I treated; (D) topoisomerase II treated; (E) BglII digested; and (F) S1 nuclease treated. Within panel (A) above 2D-IMAGE profile is a 1D lane in the correct orientation for the second dimension separation for comparison, with the following annotated species: catenanes (cat.); unit length nicked circles (1n NC); two-genome length nicked circle catenane (2n NC); CCCs; unit-length linear molecules (1n L). Arrow in panel A indicates position of CCC bar, which collapses by topoisomerase I or II treatment (black triangle in panels C and D). In panel D, the signal reduction in catenanes is completely recovered in linear molecules, all signal from CCC bar collapses onto relaxed CCC and a small amount of mtDNA at the 2n NC region that is resistant to topoisomerase II treatment is indicated by number sign. In panel E, single asterisks indicate structures potentially also identified in Figure 1 after BglII digestion, whereas open diamonds indicate additional molecules that were undetected in Figure 1. In panel (F), single-stranded species removed by S1 nuclease treatment are indicated by black squares.
In addition to the four major topoisomer species resolved by 1D electrophoresis, we observed significant hybridization to species migrating between each of the major bands within each lane. Some of this signal corresponded to low-level species that were not cleaved by restriction digestion and, therefore, could contain single-stranded (ss) DNA (Figure 1A). The less abundant species, including multiple bands between the relaxed and supercoiled circles, between relaxed circles and 2n catenanes, and above 2n catenanes, have been observed by others (26,34). To test whether any of these structures contained ssDNA, we treated total DNA with two common enzymes that remove unpaired DNA (Figure 1C). Treatment with low levels of S1 nuclease, which digests ssDNA, dsDNA at nicks or gaps and RNA [with 5-fold lower activity than for ssDNA (38)], reduced the signal that was not associated with the major topoisomers and produced more relaxed circles and various sizes of linear mtDNA. Treatment with E. coli exonuclease I produced no clear changes in the resolved molecules, suggesting that the ssDNA did not contain 3′-overhangs. Although the overall pattern in 1D gels agreed with previous findings that mtDNA is present in multiple species (24–26,39), we observed numerous other hybridizing species, including those around relaxed circular, linear and supercoiled mtDNA molecules. These data show that 1D gel electrophoresis is insufficient to separate and identify all mtDNA topological and structural isomers, including those that apparently contain ssDNA.
2D separation of mtDNA reveals ∼25 structures, including ssDNA molecules
To resolve and characterize these additional mtDNA species, we developed 2D-IMAGE. This procedure introduces a second electrophoretic separation step after the 1D gel that uses EtBr to induce supercoiling in covalently closed or topologically constrained DNAs, resulting in their accelerated migration. Furthermore, EtBr enhances the rigidity of DNA, thus exaggerating separation of molecules with differences in extended DNA shape. After electrophoresis in the first dimension, the gel was incubated in EtBr, slices were excised and rotated 90°, embedded in a second gel of the same percentage that contained EtBr and samples electrophoresed under the same conditions as the first dimension (step-by-step method is detailed in Supplementary Methods). Because the agarose concentration is constant in both dimensions, distinct populations of DNA appear in the shape of the well used in the first dimension. The four major topoisomers resolved in 1D gels (catenanes, relaxed circles, linear molecules and maximally supercoiled circles) migrated near the diagonal of the 2D gels, indicating that their mobilities were largely unaffected by EtBr; however, numerous additional species were affected by EtBr and were now separated from these major structures. DNA from enriched mitochondria produced a similar pattern, eliminating the possibility that these structures are hybridization artifacts of using whole-cell DNA preparations (Figure 2A and B).
Among the new DNA structures visible in the 2D-IMAGE profile was a horizontal continuum of signal spanning the region between supercoiled and relaxed circular DNA in the first dimension (Figure 2A, arrow). These molecules are covalently closed circles (CCCs), which are separated in the first dimension by writhe, but have equal migration characteristics in the second dimension because of the presence of EtBr, which drives closed circles into a maximally supercoiled state. These molecules ranged from completely relaxed [left-most molecules that co-migrated with open (i.e. nicked) circles in the first dimension] to maximally supercoiled (right-most molecules corresponding to the species labelled ‘supercoiled’ in the first dimension). The CCCs are clearly separated from unit-length nicked circular DNAs (1n NC), whose migration is EtBr insensitive. The identity of CCCs was confirmed by topoisomerase I treatment (Figure 2C), which converted all supercoiled CCCs (scCCC) to relaxed CCCs (rCCC) that still remained EtBr sensitive in the second dimension (indicated with black triangle). These data reveal a broad population of supercoiled molecules not detected by 1D methods.
To remove catenanes, we treated total genomic DNA with topoisomerase II, which after separation by 2D-IMAGE revealed a small number of remaining molecules that are likely concatemers (Figure 2D, indicated by number sign). The data suggest that the topoisomerase reactions were complete because scCCC was fully converted to rCCC. The increase in linear forms was initially unexpected, but has been observed previously for topoisomerase II (40).
Linear mtDNAs (1n L) were identified by digestion with BglII to collapse all full-length circular dsDNA (Figure 2E). A diagonal trail of signal originating at the linear molecules and proceeding towards lower molecular weights represents linear mtDNAs of decreasing length (these are included in quantitations of all linear molecules in subsequent experiments; see Supplementary Figure S2). As observed in 1D analyses, restriction digestion converted the majority of mtDNA to linear molecules of unit length, yet several species remained. Those structures detected in 1D (Figure 1A) are labelled with an asterisk in Figure 2E, whereas those only detected by 2D-IMAGE are indicated with an open diamond.
To test whether any of the additional species present after BglII digest contained ssDNA, we treated wild-type MEF DNA with S1 nuclease (Figure 2F) and observed a complete disappearance of the two near-vertical series of molecules proceeding downwards from 1n NC and linear molecules and of the haze emanating diagonally from the linear molecules towards the origin, all of which are marked with black squares. We also detected some ‘sharpening’ of the molecules between 1n NC and 2n NC, which we term 1n–2n molecules. The S1-sensitive species were not affected by E. coli exonuclease I treatment (data not shown). Taken together, these results identify 25 mtDNA species (summarized in Figure 3) and suggest that 2D-IMAGE could be applied to the study of the biogenesis and metabolism of these molecules.
Figure 3.
Model for mtDNA structures resolved by 2D-IMAGE. (A) 2D-IMAGE profile of DNA isolated from MEF mitochondria in Figure 2B. Each population of mtDNA is indicated with a number. (B) Diagram of molecules resolved by 2D-IMAGE. The numbers correspond to populations indicated in panel (A). Those numbers with annotations are predicted molecular species based on enzyme sensitivity and electrophoretic characteristics. The four primary topoisomers are indicated in bold. Species 26 is found in other cell types and tissues but not MEFs. Positions of nicks in DNA are arbitrary. Abbreviations include: catenane (cat.); nicked circle (NC); concatemer (CNC); intermediate between NC and cat. (1n-2n); linear (L); covalently closed circles (CCC); relaxed CCC (rCCC); supercoiled CCC (scCCC); circle (C); single-stranded (ss). Lengths are indicated relative to one genome unit length (1n).
Similar mtDNA structural forms exist across human and mouse cell and tissue types
The diversity in mtDNA structures separated by 2D-IMAGE analysis prompted us to determine how well-conserved the mtDNA structural profiles might be in several human and mouse cell lines. We isolated total DNA (treated with RNase A) from four lines each of mouse and human cells listed in Table 1 and subjected 10 µg to 2D-IMAGE analysis (Figure 4). Although the diversity of molecules was shared among the different sources of DNA, some cells demonstrated a specific distribution of the major mtDNA topoisomers. For example, the slowest growing cell line MIN6, a pancreatic insulinoma line, harboured an additional band (Figure 4A, upper right panel, indicated by black diamond) and altered abundance of both ss and dsDNA structures, including low level of species 23 and 24.
Table 1.
Cell lines and tissues used in this study
| Mouse cell lines | |
| MEF | Embryonic fibroblast |
| MIN6 | Pancreatic insulinoma |
| C2C12 | Myoblast |
| MC3T3-E1 | Pre-osteoblast |
| Human cell lines | |
| HMEC 151P | Normal mammary epithelium (hTERT) |
| MDA-MB-231 | Mammary epithelial cancer |
| HeLa | Cervical cancer |
| SKBR3 | Mammary epithelial cancer |
| Mouse tissues | |
| Liver | |
| Heart | |
| Quadriceps | |
| Kidney | |
| Human tissues | |
| Pancreatic islets | |
| Myocardium | |
To determine how 2D-IMAGE profiles varied among tissues, which have a reduced mtDNA replication demand compared with actively growing cell lines, we isolated total DNA from four mouse tissues and two human tissues (Table 1). From mice, we analysed total DNA isolated from liver, heart, kidney and quadriceps (Figure 5A). As with cells, we observed the same major mtDNA topoisomer species, but again there were clear differences in population distribution (Figure 5C). These differences are emphasized when total mtDNA copy number (Figure 5D) is factored into their relative abundance (Figure 5E). In the two human tissues analysed (Figure 5B), pancreatic islets and left ventricle myocardium, we saw a clear laddering of catenated structures. These patterns are similar to results from others that focused on high molecular weight structures in human heart, such as junctional complexes and catenanes (27. 2) D-IMAGE analysis reiterated the high levels of catenated mtDNA molecules in not only human heart but also pancreatic islets. This heavily catenated mtDNA network has been suggested to be the replicating population in heart (28). As an extension of those findings, the simpler catenanes present in the majority of cell and tissue types shown here could be involved in replication or be replication products. Myocardium samples also contained the novel structure observed in MIN6 cells (black diamonds) (Figure 5B), which was also observed in approximately half of the mouse heart samples (Figure 5A). The relative distribution of the major mtDNA topoisomers differed between the two human tissues. Notably, human and mouse tissues showed a limited abundance of structures that contain ssDNA (structure #9, 13, 14, 18, 19, 23, 24 and 25). 2D-IMAGE profiling clearly revealed tissue and cell type-specific distributions of mtDNA topoisomers, suggesting that alterations in these profiles could be used to detect qualitative changes in mtDNA metabolism.
Figure 5.
Mouse and human tissues share many common structures, but differ in relative abundance and additional structures. (A) 2D-IMAGE analysis of mouse tissues: liver, heart, kidney and quadriceps. (B) 2D-IMAGE analysis of human tissues: pancreatic islets and non-failed myocardium. Total DNA from indicated tissue was resolved by 2D-IMAGE and detected by Southern blot with total mouse mtDNA probe (panel A) or mixed human mtDNA probe (panel B). Black diamond indicates structure similar to that found in MIN6 cells in Figure 8. (C) Relative distribution of major topoisomers in four mouse tissues shown with standard deviation of biological replicates (± standard error of the mean) (n = 3). Asterisk indicates P < 0.05; double asterisk indicates P < 0.01; triple asterisk indicates P < 0.001. (D) MtDNA copy number in mouse liver, heart, kidney and quadriceps as determined by Southern blot. Data from biological replicates (n = 4) are shown with standard deviation (± standard error of the mean). Liver mtDNA levels were used for normalization. Asterisk indicates same P-values as in panel (C). (E) MtDNA copy number-adjusted distribution of major topoisomers in tissues expressed in arbitrary units (A.U.).
Fully hybridized and partially hybridized RNA associated with discrete mtDNA structures in 2D-IMAGE
RNA has long been known to be associated with the mtDNA nucleoid, playing roles in linking mtDNA to the membrane (41,8,7), DNA replication priming (42–44) and mtDNA conformation (37,45). To test whether RNA might be associated with specific DNA structures, we examined the effects of several well-characterized RNAses on the 2D-IMAGE profile. RNase T1, which cleaves after unpaired G residues in RNA, and RNase V1, which cleaves base-paired RNA, had no overt effect on the 2D-IMAGE profiles relative to the untreated sample (Supplementary Figure S3). We were, however, able to detect changes in 2D-IMAGE profiles induced by treatment with RNase A, which digests single and double-stranded RNA after U and C bases in low salt (46), and with RNase H, which cleaves the RNA portion of a fully hybridized RNA–DNA duplex (Figure 6: dashed and solid arrows indicate decreased or increased signal after treatment, respectively). Relative to RNase A treatment (Figure 6C), untreated samples showed significantly stronger hybridization, indicating the presence of more nucleic acid and with new hybridizing populations (Figure 6A: labelled a–e). Treatment with RNase H specifically removed one of these populations [Figure 6B (a), dashed arrow] to produce several additional populations (corresponding to Figure 3, #23–25). These RNase H-sensitive species may represent replication intermediates and were characterized further in subsequent experiments (see following section). These effects on the distribution of signal were also shared by RNase A treatment (Figure 6C). Those structures generated by both RNase A and H were removed by S1 nuclease treatment in sequential digestion experiments, indicating that they contain ssDNA (Figure 2F and Supplementary Figure S4). Our data show that RNase H-sensitive species are a subset of RNAse A (from several manufacturers) sensitive RNAs, with those RNA species sensitive only to RNase A likely being partially hybridized to DNA.
Figure 6.
Mitochondrial DNA topoisomers are associated with RNA in 2D-IMAGE profiles. (A) 2D-IMAGE profile of wild-type MEF DNA without RNAse treatment. New topoisomers relative to those in Figure 3 are assigned letters. (B) RNase H treatment, which digests RNA:DNA hybrids, reveals the vertical spikes of DNA that are sensitive to S1 nuclease (23–25). Molecules that decrease on digestion are indicated with dashed arrows, whereas those that increase are indicated with solid arrows. Numbers correspond to topoisomers in Figure 3. (C) RNase A treatment, which digests heterogeneous RNA after U and C bases, reveals the typical 2D-IMAGE pattern shown in Figure 2. Dashed arrows indicate a reduction in signal, and solid arrows indicate increased signal. (D) Quantitation of change in abundance of topoisomers shown in panels B and C relative to untreated.
In untreated samples, signal surrounding the 2n catenanes was much less distinct than in RNase A-treated samples [Figure 6A (b)], with additional horizontal extensions observed to the right of 1n NC (c) and at the lower left of 1n NC (d). Hybridization within (e) and to the left of the CCC molecules ( f) was significantly higher without RNase A treatment, suggesting that denaturation before transfer was not sufficient to remove all RNAse A-sensitive RNA. This qualitative pattern was seen in multiple independent preparations (data not shown). The signal within the diagonal region between 1n L and 1n NC species is strongly enhanced by the RNase A treatment (Figure 6C: #14). Assignments and signal intensity are plotted as fold change in Figure 6D. The majority of RNAs partially hybridized to DNA are associated with CCC mtDNA, which is the preferred substrate for transcription in bacteria (47). These data suggest that CCC molecules are a potential substrate for mitochondrial transcription.
Discrete topoisomers contain heavy strand-specific ssDNA
RNase H can remove RNA primers from replicating DNAs (48), implicating the RNase H-dependent structures in replication (49). Although these and other structures were sensitive to S1 nuclease treatment, such sensitivity could be because of hairpin structures, nicks or abasic sites. To determine whether there were extensive single-stranded regions in the S1 nuclease-sensitive structures, we examined the 2D-IMAGE hybridization pattern of non-denatured DNA (Figure 7). Alkaline denaturation before transfer is required for hybridization of double-stranded nucleic acids with a single-stranded radiolabelled probe. Omission of this step has been used to characterize ssDNA-dependent replication of hypersuppressive petite genomes in yeast (50). To this end, we resolved RNase A-treated DNA isolated from mitochondria by 2D-IMAGE in duplicate and transferred samples with or without alkaline denaturation (Figure 7A and B). RNAse A is required to eliminate RNA completely from the profile to reveal the ssDNA species being tested (see Figure 6). In this system, the only detectable mtDNA structures after non-denaturing transfer were the 1n–2n molecules (Figure 3: #9), the lower termini of the vertical spikes of ssDNA emanating from 1n NC and 1n L molecules (Figure 3: #23, 24), a small portion of the lower part of the linear ssDNA vertical spike (Figure 3: #19) and less than unit-length single-stranded linear DNA (<1n ssL; Figure 3: #25). Taken together with the S1 nuclease sensitivity results, we conclude that these structures harbour significant ssDNA content, with an increase in dsDNA content as 18 and 19 approach 1n NC and 1n L. Notably, these structures are most abundant in fast growing cell types in which mtDNA is rapidly duplicating.
Figure 7.

Strand-specific single-stranded mtDNA species resolved by 2D-IMAGE. (A) Standard 2D-IMAGE profile of total wild-type (WT) MEF mtDNA transferred under denaturing (alkaline) conditions. The 1n–2n molecules are indicated, as are the vertical series of S1-sensitive signal emanating from 1n NC and linear molecules. (B) 2D-IMAGE profile of WT MEF mtDNA from isolated mitochondria transferred under non-denaturing conditions. Only the indicated molecules adhere to the positively charged membrane without previous denaturation (ssC, single-stranded circle; ssL, single-stranded linear). (C–H) 2D-IMAGE profiles of total WT MEF mtDNA transferred under denaturing conditions and probed with three sets of oligo probes corresponding to either the heavy (B, C, D, D2, E, F) or light strand (B′, C′, D′, D2′, E′, F′). Only heavy strand probes hybridized to the ssDNA spikes emanating downwards from 1n NC and 1nL molecules.
To determine whether these ssDNAs comprised a specific strand of mtDNA, RNase A-treated DNA isolated from mitochondria was resolved by 2D-IMAGE, transferred with the denaturation step and probed with three sets of strand-specific oligonucleotides corresponding to different regions of the genome (Figure 7C–H and Supplementary Figure S1). The inclusion of RNase A ensured the removal of RNA and electrophoretically focused ssDNA into structures 18, 19, 23 and 24. As expected, the catenanes, 1n NC, 1n-2n, 1n L, and CCC molecules were detected by both heavy strand-specific and light strand-specific probe sets, as these structures are composed of dsDNA (Figure 7C–H). The heavy-strand probe sets (Figure 7C, E and G), but not the light strand sets (Figure 7D, F and H), detected 1n ssC (#23) and 1n ssL (#24), thus indicating that these structures comprise heavy-strand sequence. Light strand probes sets detected 18 and 19 near 1n NC and 1n L, confirming that these structures become progressively more double stranded as they approach their fully double-stranded counterparts.
Recovery from EtBr replication arrest identifies replication templates, intermediates and products
Intact mtDNAs that contain extensive ss regions could be replication intermediates. To test this possibility, we perturbed mtDNA replication by EtBr arrest and release procedures. EtBr intercalates between base-paired nucleotides to disrupt the processivity of mitochondrial DNA polymerase, without significantly affecting nuclear DNA replication (51–53). We noted that 2D-IMAGE profiles of mtDNA isolated from wild-type MEF cells treated for 24 h with 100, 250 or 500 ng/ml of EtBr showed a significant decrease in total hybridization signal in response to increasing EtBr concentration (Supplementary Figure S5A–D). Cells exposed to 500 ng/ml EtBr contained <10% of the mtDNA signal present in untreated cells by qPCR or total counts in 2D-IMAGE (Supplementary Figure S5E), with all mtDNA structures affected (Supplementary Figure S5F and G).
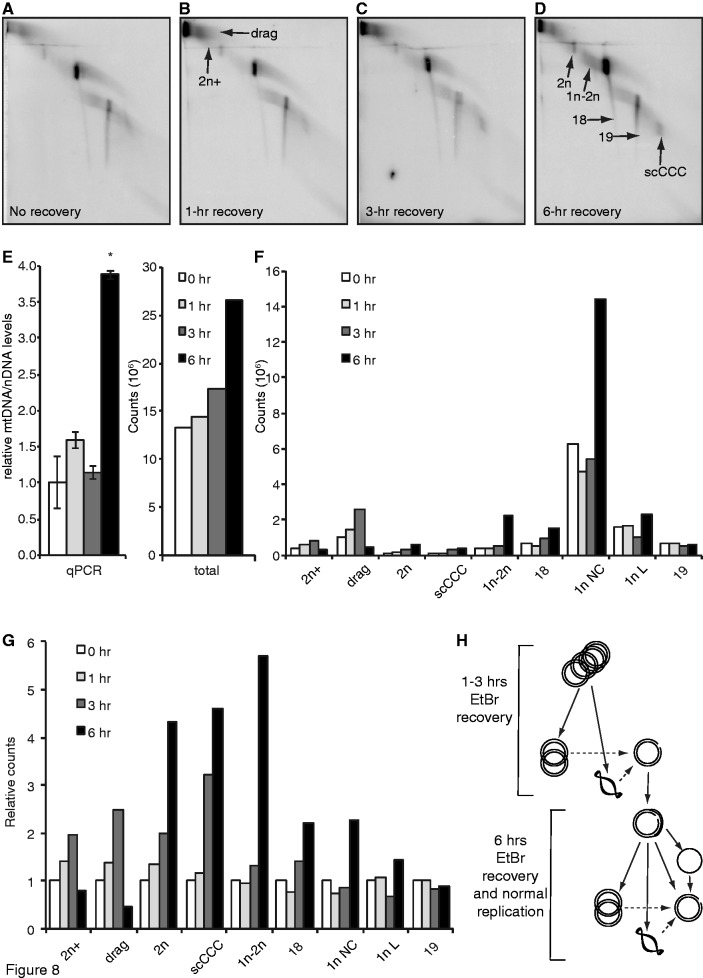
To restart mtDNA replication, we first exposed MEFs to 500 ng/ml of EtBr for 24 h (no recovery; Figure 8A), and then released cells from arrest by washing out the EtBr with fresh medium. Cells recovered for 1, 3 or 6 h (Figure 8B–D) and mtDNA content were monitored by qPCR or total hybridization counts (Figure 8E). The 2D-IMAGE profile of the 6-hour time point is similar to that of unexposed cells actively growing in culture. At this time point, elevation in mtDNA content was statistically significant (Figure 8E). Similar timing of the mtDNA increase was observed by qPCR in an independent experiment (Supplementary Figure S6). The majority of the radioactive signal arose from 1n NC molecules (Figure 8F). We assessed the order of events during EtBr recovery by establishing changes in the relative abundance of various intact mtDNA structures (Figure 8G). At the early time point (1 h), increases in hybridization signal intensity were observed in high molecular weight mtDNA, such as catenanes (2n+ and 2n) and molecules that ‘drag’ out of the well (Figure 8B). These ‘drag’ molecules are not commonly detected in normal cells, and they only appear with 250 and 500 ng/ml of EtBr treatment (Supplementary Figure S5C and D); therefore, these molecules likely represent highly interconnected tangles that cannot migrate into the gel efficiently because of their high molecular weight. In tightly correlated changes (Pearson’s correlation: R = 0.99, P < 0.01), 2n+ and drag molecules increased for the first 3 h of recovery, but dropped to pre-exposure levels at 6 h. These alterations are clearly visible in dark exposures (Supplementary Figure S7). Because replication templates are incorporated into product by semi-conservative replication, the depletion of these high molecular weight structures implicates them as the substrate for replication early in EtBr recovery. Also increasing after washout are 2n and scCCC, which increase 4–5-fold by 6 h washout, suggesting that these structures are replication products during the early stages of EtBr recovery. The increase in 1n NC was positively correlated with 1n–2n abundance (Pearson’s correlation: K = 0.99, P < 0.05), and at 6 h, NCs were present at significant levels. Similar trends were observed in ssDNA molecule #18 at 6 h. In contrast, 1n L and #19, both linear molecules, showed little if any relative increase at the 6-h recovery time point.
Figure 8.
Recovery from EtBr treatment stimulates replication of high-molecular weight mtDNA structures. (A) 2D-IMAGE analysis of wild-type MEFs cultured for 24 h in 500 ng/ml of EtBr. Medium was changed to omit EtBr and cells harvested at 1 (B), 3 (C) or 6 h (D) post-washout. (E) mtDNA content during washout as determined by qPCR (left) or total mtDNA hybridization signal for panels (A–D). (F) Distribution of signal across nine selected topoisomers as mtDNA levels recover. (G) Relative abundance of selected topoisomers from (F) after setting unrecovered sample equal to 1. (H) Model for template–product relationship for early and late recovery from EtBr-mediated mtDNA replication arrest. Solid arrows indicate template–product synthesis, and dashed arrows indicate conversion of one topoisomer to another. Location of nick in 1n NC is arbitrary.
Based on the timing and correlation of changes in the 2D-IMAGE profile, we propose a template–product relationship among several identified structures (Figure 8H). High molecular weight mtDNAs that accumulate during EtBr exposure are the primary template for replication early in the recovery and produce 2n catenanes and scCCC molecules, which could arise from dedicated replication pathways or simply as the result of differences in topology at completion of replication. Because 1n–2n molecules are replication intermediates that originate from the 1n NC structure, these data suggest that late in recovery when these molecules increase–and likely during normal replication–1n NC served as the template for replication. To reach this point, the early replication products (2n catenanes and scCCC) were likely converted to 1n NC species. Continuing replication from 1n NC, the complete replication of both strands produced catenanes and scCCC, whereas a delay in initiation of second-strand synthesis would allow completion of first strand synthesis and displacement of ssC containing heavy-strand sequence (molecule #23). Initiation of second-strand synthesis on ssC formed a gapped circle (molecule #18) becoming progressively more double-stranded during elongation to produce 1n NC.
DISCUSSION
Our 2D-IMAGE technique was built on the successes of 1D separation of mtDNA structures to reveal multiple new forms of double-stranded circular mtDNA, including a range of supercoiled molecules, nicked circles and multiple catenated species, most of which are present in a variety of mammalian cells and tissues. This technique has also identified several forms of mtDNA that contain ssDNA structures, including 1n–2n, 1n ssC and 1n ssL, which are likely replication intermediates. 2D-IMAGE differs from restriction digest-based 2D methods, such as 2D-AGE, which has been successfully used to examine replication and recombination intermediates (27,36,54,55). Although 2D-AGE uses digested DNA to detect low abundance replication or recombination structures, such as X and Y arcs, the topology of the intact genome containing these structures cannot be ascertained using that technique. 2D-IMAGE resolved ∼25 intact mtDNA species of diverse structural content, providing a survey of total mtDNA complexity. Even greater diversity in the mtDNA population can be expected by focusing on further resolution of specific structures [ex. (27)].
RNA in the nucleoid
The assessment of RNA content in the nucleoid may elucidate mitochondrial transcription and genome stability. We found that most partially hybridized RNAs were associated with CCC mtDNA (structure e); given that highly supercoiled DNA plasmids are better transcriptional templates in bacteria (47) and adenovirus (56), our results are consistent with CCC mtDNA being a major template for transcription in vivo. This idea is supported by observations of promoter-specific transcription on supercoiled templates using yeast mitochondrial RNA polymerase (POLRMT) in vitro (57), although mammalian POLRMT does not show promoter specificity (58). This contrast is not yet understood, and it suggests that additional factors may be contributing to transcription of mammalian CCC mtDNA in vivo.
Alternatively, RNAs could play a functional role in the stability of mtDNA by linking the nucleoid to the mitochondrial inner membrane (41). The mtDNA:membrane association (8,7) is likely important for coupled transcription–translation import (59), and by analogy to the bacterial nucleoid (60), has been suggested to be important in mtDNA replication (61), nucleoid division (62), segregation (63–65) and DNA repair (66–68). Furthermore, partially hybridized RNAs have been visualized by atomic force microscopy as bound to DNA, where they increase the apparent DNA writhe (45). In those DNA preparations, RNAs represent ∼25% of the nucleotide content, have short 5′- and 3′-ends and unmodified 3′-ends distinct from total RNA transcripts that suggest a difference in processing and targeting relative to mature transcripts. Additional studies of the sequence and structure of these RNAs may yet reveal essential structural determinants of mtDNA nucleoid content and stability. Further supporting the idea that RNAs are likely part of the nucleoid is the presence of RNA-binding proteins in nucleoid preparations (69–72) and the requirement of some RNA-directed nucleoid proteins or their associated proteins, for mtDNA stability (73–75).
Replicating structures
RNA priming is required for all current proposed models of mtDNA replication: coupled leading and lagging strand synthesis (54); strand-displacement or asynchronous replication (76,77); and RNA incorporation throughout the lagging strand (44). The presence of discrete structures whose electrophoretic characteristics are altered by RNase H and/or S1 nuclease treatment in 2D-IMAGE (structures 18 and 19) suggests the presence of extensive regions of RNA:DNA hybrids. Similar stretches have been visualized on mtDNA by atomic force microscopy after treatment with RNase A and incubation with ssDNA-binding protein (55) and by analysis of replication intermediates in 2D-NAGE (native agarose gel electrophoresis) gels (78).
To determine how these structures respond to changes in mtDNA replication, we depleted mtDNA in MEFs by inducing replication arrest with EtBr, followed by release. Relative to human cell lines [e.g. 143B human sarcoma (54)], a much shorter time of exposure was required to deplete mtDNA levels (24 h), which coincided with a more rapid initiation of replicative recovery after EtBr washout. We observed high molecular weight mtDNA molecules accumulating under conditions of strong mtDNA depletion (500 ng/ml of EtBr), which after release from arrest, began to increase in intensity first to produce 2n and scCCC molecules. At 6 h post-washout, these high-molecular weight structures were diminished, coinciding with strong increases in 1n NC, 1n-2n and structure 18. Based on these observations, we suggest that the early and late replicative mechanisms both produce 2n and scCCC molecules, but vary in template utilization. Specifically, early recovery may use the prevalent >2n molecules as template, whose replication intermediates are not resolved in these gel conditions. Late recovery may use unit-length templates (1n NC) to generate replication intermediates (1n–2n and structure 18) and likely represents the mechanism occurring in unchallenged cells.
New concepts generated by this study may prove important in refining our understanding of mtDNA replication. For example, the significant differences in template utilization during EtBr-induced replication arrest and release have not been considered previously in the literature. Moreover, the evident structural complexity of mtDNA is significant, and the separation of specific structures will provide a clearer description of mtDNA features than will analysing the genome en masse. This latter bulk approach has been taken to describe strand-asynchronous and RNA incorporation throughout the lagging strands replication mechanisms (44), but additional refinement is required to investigate whether one or multiple mechanisms exist specifically or simultaneously, and under what physiological conditions they are regulated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7, Supplementary Methods, Supplementary Protocols and Supplementary References [79–81].
FUNDING
Muscular Dystrophy Association [MDA-69064]; NIH [P30-DK050306] Pilot and Feasibility; Mari Lowe Center for Comparative Oncology Pilot and Feasibility Grant (UPENN). Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank their colleagues for their helpful discussions and review of this manuscript (Dr Joseph Baur, Dr Neal Sondheimer, Dr Brad Johnson, Dr Joel Meyer and Dr Leslie King). Cell and tissue samples were kindly provided by Dr Kenneth Margulies, Dr Chengyang Liu, Dr Ali Naji and Dr Kurt Hankenson.
REFERENCES
- 1.Nass MM. Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J. Mol. Biol. 1969;42:521–528. doi: 10.1016/0022-2836(69)90240-x. [DOI] [PubMed] [Google Scholar]
- 2.Nass MM, NASS S. Intramitochondrial fibers with DNA characteristics. I. fixation and electron staining reactions. J. Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 2011;31:4994–5010. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson N-G, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albring M, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl Acad. Sci. USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl Acad. Sci. USA. 2012;109:6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 10.Zeviani M, Moraes CT, DiMauro S, Nakase H, Bonilla E, Schon EA, Rowland LP. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 11.Wallace DC, Zheng XX, Lott MT, Shoffner JM, Hodge JA, Kelley RI, Epstein CM, Hopkins LC. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 12.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 13.Wong L-JC. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion. 2007;7:45–52. doi: 10.1016/j.mito.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 15.Ikebe S, Tanaka M, Ohno K, Sato W, Hattori K, Kondo T, Mizuno Y, Ozawa T. Increase of deleted mitochondrial DNA in the striatum in Parkinson's disease and senescence. Biochem. Biophys. Res. Commun. 1990;170:1044–1048. doi: 10.1016/0006-291x(90)90497-b. [DOI] [PubMed] [Google Scholar]
- 16.Maassen JA, 'T Hart LM, Van Essen E, Heine RJ, Nijpels G, Jahangir Tafrechi RS, Raap AK, Janssen GMC, Lemkes HHPJ. Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes. 2004;53(Suppl 1):S103–S109. doi: 10.2337/diabetes.53.2007.s103. [DOI] [PubMed] [Google Scholar]
- 17.Gauthier BR, Wiederkehr A, Baquié M, Dai C, Powers AC, Kerr-Conte J, Pattou F, MacDonald RJ, Ferrer J, Wollheim CB. PDX1 deficiency causes mitochondrial dysfunction and defective insulin secretion through TFAM suppression. Cell Metabolism. 2009;10:110–118. doi: 10.1016/j.cmet.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmons RA, Suponitsky-Kroyter I, Selak MA. Progressive accumulation of mitochondrial DNA mutations and decline in mitochondrial function lead to beta-cell failure. J. Biol. Chem. 2005;280:28785–28791. doi: 10.1074/jbc.M505695200. [DOI] [PubMed] [Google Scholar]
- 19.Ye C, Shu X-O, Wen W, Pierce L, Courtney R, Gao Y-T, Zheng W, Cai Q. Quantitative analysis of mitochondrial DNA 4977-bp deletion in sporadic breast cancer and benign breast diseases. Breast Cancer Res. Treat. 2008;108:427–434. doi: 10.1007/s10549-007-9613-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meierhofer D, Mayr JA, Foetschl U, Berger A, Fink K, Schmeller N, Hacker GW, Hauser-Kronberger C, Kofler B, Sperl W. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis. 2004;25:1005–1010. doi: 10.1093/carcin/bgh104. [DOI] [PubMed] [Google Scholar]
- 21.Fan AX-C, Radpour R, Haghighi MM, Kohler C, Xia P, Hahn S, Holzgreve W, Zhong XY. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J. Cancer Res. Clin. Oncol. 2009;135:983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat. Res. 1992;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 23.Baumer A, Zhang C, Linnane AW, Nagley P. Age-related human mtDNA deletions: a heterogeneous set of deletions arising at a single pair of directly repeated sequences. Am. J. Hum. Genet. 1994;54:618–630. [PMC free article] [PubMed] [Google Scholar]
- 24.Singh G, Hauswirth WW, Ross WE, Neims AH. A method for assessing damage to mitochondrial DNA caused by radiation and epichlorohydrin. Mol. Pharmacol. 1985;27:167–170. [PubMed] [Google Scholar]
- 25.Higuchi Y, Linn S. Purification of all forms of HeLa cell mitochondrial DNA and assessment of damage to it caused by hydrogen peroxide treatment of mitochondria or cells. J. Biol. Chem. 1995;270:7950. doi: 10.1074/jbc.270.14.7950. [DOI] [PubMed] [Google Scholar]
- 26.Pohjoismäki JLO, Wanrooij S, Hyvärinen AK, Goffart S, Holt IJ, Spelbrink JN, Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohjoismäki JLO, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohjoismäki JLO, Goffart S, Taylor RW, Turnbull DM, Suomalainen A, Jacobs HT, Karhunen PJ. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS One. 2010;5:e10426. doi: 10.1371/journal.pone.0010426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacAlpine DM, Perlman PS, Butow RA. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 2000;19:767–775. doi: 10.1093/emboj/19.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes A, Yasukawa T, Cluett TJ, Holt IJ. Analysis of mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol. Biol. 2009;554:15–35. doi: 10.1007/978-1-59745-521-3_2. [DOI] [PubMed] [Google Scholar]
- 31.Laugwitz K-L, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin L-Z, Cai C-L, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 33.Amthor H, Macharia R, Navarrete R, Schuelke M, Brown SC, Otto A, Voit T, Muntoni F, Vrbóva G, Partridge T, et al. Lack of myostatin results in excessive muscle growth but impaired force generation. Proc. Natl Acad. Sci. USA. 2007;104:1835–1840. doi: 10.1073/pnas.0604893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2:1007–1012. doi: 10.1093/embo-reports/kve233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M-Y, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 37.Wiesner RJ, Swift H, Zak R. Purification of mitochondrial DNA from total cellular DNA of small tissue samples. Gene. 1991;98:277–281. doi: 10.1016/0378-1119(91)90186-f. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann RI. Endonucleases specific for single-stranded polynucleotides. The Enzymes. 1981;4:193–201. [Google Scholar]
- 39.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burden DA, Froelich-Ammon SJ, Osheroff N. Topoisomerase II-mediated cleavage of plasmid DNA. Methods Mol. Biol. 2001;95:283–289. doi: 10.1385/1-59259-057-8:283. [DOI] [PubMed] [Google Scholar]
- 41.Rickwood D, Chambers JA, Barat M. Isolation and preliminary characterisation of DNA-protein complexes from the mitochondria of Saccharomyces cerevisiae. Exp. Cell Res. 1981;133:1–13. doi: 10.1016/0014-4827(81)90350-5. [DOI] [PubMed] [Google Scholar]
- 42.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl Acad. Sci. USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanrooij S, Fusté JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasukawa T, Reyes A, Cluett TJ, Yang M-Y, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Struhl K. Ribonucleases. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb0313s08. Chapter 3, Unit3.13. [DOI] [PubMed] [Google Scholar]
- 47.Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 48.Leis JP, Berkower I, Hurwitz J. Mechanism of action of ribonuclease H isolated from avian myeloblastosis virus and Escherichia coli. Proc. Natl Acad. Sci. USA. 1973;70:466. doi: 10.1073/pnas.70.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985;4:1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacAlpine DM, Kolesar J, Okamoto K, Butow RA, Perlman PS. Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. EMBO J. 2001;20:1807–1817. doi: 10.1093/emboj/20.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leibowitz RD. The effect of ethidium bromide on mitochondrial DNA synthesis and mitochondrial DNA structure in HeLa cells. J. Cell Biol. 1971;51:116–122. doi: 10.1083/jcb.51.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nass MM. Abnormal DNA patterns in animal mitochondria: ethidium bromide-induced breakdown of closed circular DNA and conditions leading to oligomer accumulation. Proc. Natl Acad. Sci. USA. 1970;67:1926–1933. doi: 10.1073/pnas.67.4.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nass MM. Differential effects of ethidium bromide on mitochondrial and nuclear DNA synthesis in vivo in cultured mammalian cells. Exp. Cell Res. 1972;72:211–222. doi: 10.1016/0014-4827(72)90583-6. [DOI] [PubMed] [Google Scholar]
- 54.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 55.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banerjee S, Spector DJ. Differential effect of DNA supercoiling on transcription of adenovirus genes in vitro. J. Gen. Virol. 1992;73(Pt 10):2631–2638. doi: 10.1099/0022-1317-73-10-2631. [DOI] [PubMed] [Google Scholar]
- 57.Matsunaga M, Jaehning JA. Intrinsic promoter recognition by a ‘core’ RNA polymerase. J. Biol. Chem. 2004;279:44239–44242. doi: 10.1074/jbc.C400384200. [DOI] [PubMed] [Google Scholar]
- 58.Fukuoh A, Ohgaki K, Hatae H, Kuraoka I, Aoki Y, Uchiumi T, Jacobs HT, Kang D. DNA conformation-dependent activities of human mitochondrial RNA polymerase. Genes Cells. 2009;14:1029–1042. doi: 10.1111/j.1365-2443.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 59.Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaimer C, Graumann PL. Players between the worlds: multifunctional DNA translocases. Curr. Opin. Microbiol. 2011;14:719–725. doi: 10.1016/j.mib.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaufman BA, Kolesar JE, Perlman PS, Butow RA. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:457–461. doi: 10.1083/jcb.200306132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boldogh IR, Nowakowski DW, Yang H-C, Chung H, Karmon S, Royes P, Pon LA. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berger KH, Yaffe MP. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 2000;8:508–513. doi: 10.1016/s0966-842x(00)01862-x. [DOI] [PubMed] [Google Scholar]
- 66.Boesch P, Ibrahim N, Dietrich A, Lightowlers RN. Membrane association of mitochondrial DNA facilitates base excision repair in mammalian mitochondria. Nucleic Acids Res. 2010;38:1478–1488. doi: 10.1093/nar/gkp1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, Stevnsner T, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell. Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Bogenhagen DF. Biochemical isolation of mtDNA nucleoids from animal cells. Methods Mol. Biol. 2009;554:3–14. doi: 10.1007/978-1-59745-521-3_1. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 72.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 73.He J, Cooper HM, Reyes A, Di Re M, Kazak L, Wood SR, Mao CC, Fearnley IM, Walker JE, Holt IJ. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012;40:6097–6108. doi: 10.1093/nar/gks257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Molecular Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 75.Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–8047. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 77.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 78.Pohjoismäki JLO, Holmes JB, Wood SR, Yang M-Y, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Rigby RE, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leary SC, Kaufman BA, Pellecchia G, Guercin G-H, Mattman A, Jaksch M, Shoubridge EA. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004;13:1839–1848. doi: 10.1093/hmg/ddh197. [DOI] [PubMed] [Google Scholar]
- 80.Church GM, Gilbert W. Genomic sequencing. Proc. Natl Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martens PA, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: localization and sequence of the light-strand origin of replication. J. Mol. Biol. 1979;135:327–351. doi: 10.1016/0022-2836(79)90440-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.