Abstract
Non-coding RNAs from transposable elements of human genome are gaining prominence in modulating transcriptome dynamics. Alu elements, as exonized, edited and antisense components within same transcripts could create novel regulatory switches in response to different transcriptional cues. We provide the first evidence for co-occurrences of these events at transcriptome-wide scale through integrative analysis of data sets across diverse experimental platforms and tissues. This involved the following: (i) positional anchoring of Alu exonization events in the UTRs and CDS of 4663 transcript isoforms from RefSeq mRNAs and (ii) mapping on to them A→I editing events inferred from ∼7 million ESTs from dbEST and antisense transcripts identified from virtual serial analysis of gene expression tags represented in Cancer Genome Anatomy Project next-generation sequencing data sets across 20 tissues. We observed significant enrichment of these events in the 3′UTR as well as positional preference within the embedded Alus. More than 300 genes had co-occurrence of all these events at the exon level and were significantly enriched in apoptosis and lysosomal processes. Further, we demonstrate functional evidence of such dynamic interactions between Alu-mediated events in a time series data from Integrated Personal Omics Profiling during recovery from a viral infection. Such ‘single transcript—multiple fate’ opportunity facilitated by Alu elements may modulate transcriptional response, especially during stress.
INTRODUCTION
The largely unexplored non-coding regions of the human genome comprising repetitive sequences and other DNA elements can no longer be neglected as ‘junk’ (1–3). These regions harbour a large number of regulatory elements that could govern genome-wide regulation at genetic, epigenetic level and also the transcriptome and proteome diversity (4–8). The primate specific retrotransposon family of Alu repeats is present in >1 million copies and occupy nearly 11% of the human genome. This, with an average size of ∼285 bps translates to almost 108 bps of the human genome (9,10). These elements have been shown to be more abundant in genes related to signalling, metabolism and transport and depleted in genes related to information and structural components (11).
Alus harbour many cryptic splice sites, which potentiates their inclusion in exons, a process referred to as exonization and reported to be common (12,13). These exonized Alus are not present constitutively in all the transcript isoforms and exhibit tissue-specific expression (14–16). In the coding sequence (CDS) region, these result in protein isoforms with different functions and in untranslated regions (UTR), transcripts with differences in stability (17–20). Novel functions through exonization of Alus have also been reported (21–23).
Alus transcribed by RNA pol III also comprise a large fraction of the non-coding RNA. Elevated levels of Alu RNA have been observed in stress, cancer and viral infection (24,25). These have been implicated in diverse functions. For instance, in the nucleus, Alu RNAs can act as transcriptional co-repressors (26), and as the most abundant fraction of the antisense transcriptome, these have the potential to downregulate sense expression both in cis as well as trans (27–29).
A third aspect of the involvement of Alu element in transcriptome has been through its affinity to get A→I edited. Two oppositely oriented Alus in pre-mRNAs can adopt secondary structures, which are the most preferred substrates for double-strand specific adenosine deaminase, ADAR1. A large fraction of A→I editing events in the genome map to Alus (30–32). The edited transcripts have been shown to be preferentially retained in the nucleus, a phenomenon that can regulate the fate of transcripts at the post-transcriptional level. This phenomenon has been implicated in modulating heat shock stress, senescence and stem cell differentiation (33–35).
Expression of a gene having an Alu exonization can be modulated differentially if there are editing events within the exonized regions or if it has an antisense transcript. If either of the events are condition specific, we might anticipate different fates of transcripts from the same gene. Such events at the transcriptome-wide level could have systemic consequences. Although all these events have been reported independently, a possible crosstalk event at the genic level has never been explored.
In this study, through extensive data mining and computational approaches, we demonstrate the possibility of co-existence of Alu exonization, editing and antisense at the transcriptome-wide level (Figure 1). Further, we demonstrate these events to be preferentially localized in the 3′UTR and also a positional preference within Alus. A significantly enriched set of genes from apoptosis pathway have co-occurrence of all the three events at the exon level. Through analysis of RNA-seq data of an individual across subsequent time points following a viral infection, we observe altered dynamics of the sense and antisense transcripts of a subset of genes linked to ubiquitin-mediated proteolysis. This study adds a further dimension to the evolution of novel regulatory networks in primates.
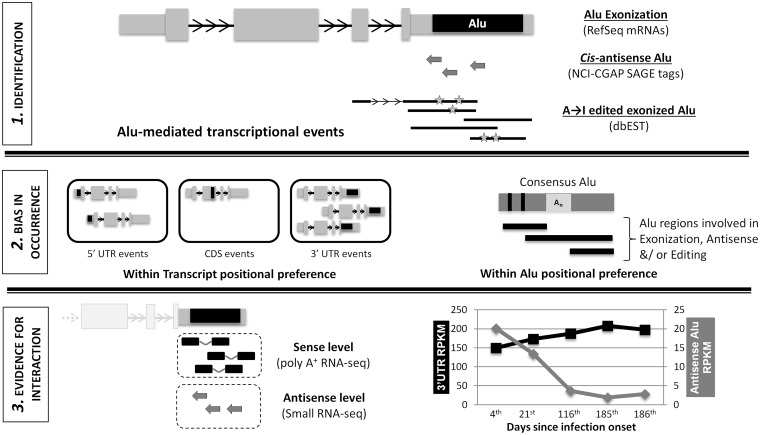
Figure 1.
Overall design of the study for exploring cross-talk between Alu exonization, antisense and editing. Different databases were mined to identify exonization, editing and antisense events mediated by Alu elements. These were mapped to transcript isoform level, their position in the transcripts and within Alu repeats. GO analysis was carried out to see whether these events participate in similar biological processes or pathways.
METHODS
To identify and anchor exonization, editing and antisense events in the transcriptome, an extensive data mining and curation was carried out. The detailed steps for each of them have been provided in Supplementary methods. A brief overview is presented here.
Identification of Alu exonization events in the transcriptome
The comprehensive set of mature mRNA sequences from RefSeq database (release 45, January 2011) (36) was used to identify and map Alu exonization events in the UTR and CDS regions at the transcriptome-wide scale (Supplementary methods). Using the University of California, Santa Cruz (UCSC) table browser (genome build hg18) (37), exon block alignments for 5′UTR, 3′UTR and CDS regions in browser extensible data (BED) format were exported to the Galaxy framework of tools (38–40). Alignments from alternate assemblies (HapMap regions) and unplaced contigs (chr*_random) were filtered out. The Alignment blocks, which had a ≥10% overlap with Alu elements identified through RepeatMasker (version 3.2.7) (http://www.repeatmasker.org) using the Coverage Tool in Galaxy (41,42), were identified. The exonized Alus were mapped back to the gene through the mRNA accession numbers. The number of transcripts (and related genes) in each category, i.e. 5′UTR, 3′UTR and CDS, which contained Alu within exons were documented individually.
Identification of Alu in the antisense transcriptome
There are as of yet no high-throughput experimental platforms, including next-generation sequencing (NGS), that can be used to readily detect, differentiate and map antisense transcripts from repetitive sequences. We explored the potential of serial analysis of gene expression (SAGE) to determine the contribution of Alu elements to antisense transcription (Supplementary methods) (43). Briefly, we initiated our study through a comprehensive search for existence of Alu overlapping transcripts in antisense orientation to the host gene, from all possible transcripts in National Center for Biotechnology Information (NCBI) SAGEMap’s virtual SAGE library data (long SAGE, 17 bp sequences). This data set contains ∼9 million virtual SAGE tags generated from in silico digestion of transcripts by NlaIII from the 3′ end derived from heterogeneous sources like mRNA, cDNA and expressed sequence tags (ESTs) from diverse tissues. We selected database of ESTs (dbEST) (44), containing 7 million EST sequences for identifying not only Alu antisense but also A→I editing events, described in the following section.
As described for Alu exonization, we used the Galaxy tool to identify ESTs that overlapped with Alu elements in exonized transcripts These were mapped with the SAGE tags from the library. The strand information of EST alignment with the genome and the host gene’s orientation were compared to infer the antisense transcripts. These antisense were restricted to only those that target mature mRNA. We ensured that the antisense tags did not match with the Best Tag from National Cancer Institute (NCI)-Cancer Genome Anatomy Project (CGAP) annotation (45,46) and were non-redundant gene-wise. Thus, following an extensive series of filtering criteria, we identified a set of virtual SAGE tags in the transcripts that were potentially derived from Alu and cis-antisense to the genes in the transcriptome. We explored the actual existence of these tags in two NGS-based SAGE data sets (GSE1902 and GSE15314), a part of the CGAP, available from NCBI Gene Expression Omnibus (GEO) (47), which have information on >20 different tissue types across 124 samples. Through this exercise, we identified Alu overlapping transcribed sequences in antisense orientation to the host gene (cis antisense), referred hereafter as Alu antisense.
The Alu antisense identified earlier in the text were anchored to the exonized transcripts to localize these events onto the transcripts with respect to 5′UTR, CDS and 3′UTR regions using the UCSC table browser resource. We anchored the antisense event for only those genes that were present in the RefSeq database and had alignment information available (48).
Identification of Alu editing in the transcriptome
We selected dbEST for profiling A→I editing within Alu repeat using the same set of Alu exonized ESTs that were used for detecting antisense transcripts (Supplementary methods). The editing sites were identified through alignment of EST stretches with corresponding genomic regions and then filtering for A→G mismatches. Briefly, alignment block coordinates for Alu exonized ESTs were retrieved and those that could not be mapped unambiguously or mapped to alternate assemblies in the genome were filtered out. A criteria of a minimum block size of ≥41 bp comprising 16 bp of non-Alu sequence (416 > genome size, the probability of finding a 16-bp sequence stretch more than once exceeds the genome size) and ≥25 bp of Alu sequence was also defined. Global alignment using the Stretcher program from the European Molecular Biology Open Software Suite (EMBOSS) was performed to identify A→G mismatch positions (49). A series of filtering steps were carried out to confirm that these mismatches were a consequence of A→I editing. These included that the positions were within Alu and are not a Single Nucleotide Polymorphism (SNP) (as inferred from frequency information in dbSNP or HapMap validated SNPs (release 129) (50). An additional criterion for the presence of an oppositely oriented Alu proximal to the edited sequence was also used, as the structure formed by such a head-to-tail orientation is a favoured substrate for the dsRNA editing enzyme. The A→G mismatch positions thus identified were termed as possible A→I editing events within Alu elements. These were then mapped back to the 5′UTR, CDS or 3′UTR within exons by overlapping the genomic coordinates of the possible Alu editing events with those of the Alu-containing exon blocks.
Gene-wise mapping of exonization, antisense and editing events
Several genes had more than one transcript isoforms involved in all or either of the three events. Similarly, for any given transcript isoform, either or all of the transcript positions (UTR or CDS) were involved. Also, a given UTR or CDS could have more than one Alu element involved in exonization, editing or antisense. To have a more comprehensive picture of these events, we represented the pattern of these Alu-mediated events across the length of the gene in relation to UTR and CDS positions using Heatmap. Making use of exonization data set as a master set, subsets were created transcript wise for antisense and editing events, and each subclasses were sorted in descending order of Alu counts within exons. The subset were events in 5′UTR only, CDS only, 3′UTR only, all three, 5′UTR and CDS only, CDS and 3′UTR only and finally 5′ and 3′UTR only. The ordered set of gene list was populated for corresponding values in case of antisense and editing, where brighter colour represents higher Alu counts (51).
Enrichment analysis for co-occurrence of Alu-mediated events in 3′UTR
The co-occurrence of Alu-mediated events could be a consequence of a reflection of the predominance of individual events and determined by the probability of Alu exonization in the UTR or CDS regions. The extent of Alu exonization could in turn be determined by the relative genomic length from where the exons were derived. A given gene can have multiple transcript isoforms and also multiple exons in a given region of UTR or CDS. For a normalized base pair count of Alu exonization in a given category, the actual exonized length per gene per category is crucial. Hence, an algorithm was written that would, given a Gene Transfer Format (GTF) file, merge the overlapping exons and calculate the unique length of genomic space covered by the given exons for each gene. Using a GTF file of exonized Alu start-end positions, we calculated unique length for Alu exonization across the categories. Alu exons with incidence of both editing and antisense were selected, and their length summarized to calculate the fraction of exonized Alu with co-occurrence across the categories (Supplementary methods). To infer whether there was positional preference in the transcripts, we carried out a Pearson chi-squared test.
Positional preference within Alu sequence for exonization, antisense and editing events
Apart from studying the positional preference of Alu-mediated events in transcripts, we also attempted to see whether there were positional preferences within Alus. To do this, we first converted the genomic coordinates of the portion of the Alu sequence involved in any of the three phenomena (a stretch in case of exonization and antisense and a single base in case of editing) into the base position within the consensus Alu using RepeatMasker. To make the representation uniform between single base occurrence of Alu editing and stretches of exonization and antisense, all the three were converted into per base position frequency of occurrence.
Intronic region analysis for A→I editing density
We also wanted to compare A→I events in the coding (5′UTR, CDS and 3′UTR) with the intronic regions that housed the exons. Intronic region coordinates for each of the Alu-harbouring transcript identified in our analysis were retrieved using RefSeq database and categorized into 5′UTR, CDS and 3′UTR. We used the DAtabase of RNa EDiting (DARNED) (52) for analysing A→I events within intronic regions. The numbers of events observed in each category were normalized by the total genomic space covered and represented as A→I editing density per Mb of intron length (Supplementary Table S7).
RNA-seq data sets
We explored the co-occurrence of Alu exonization and antisense event in an experimental poly-A+ RNA-seq and small RNA-seq data set from Integrated Personal Omics Profiling (iPOP) resource website (53). We used a five time-points (second, third and fifth to seventh) data set from NCBI GEO (GSE32874).
Annotation sets for RNA-seq analysis
The analysis of interaction between exonization and antisense events in response to viral infection across the time points was restricted to 3′UTR for reasons detailed in results. To compare the expression pattern of Alu-exonized transcripts with that of non-Alu exonized transcripts, two gene lists were created. One set of transcripts had 3′UTR Alu exonization event and the other set used as control had no Alus within transcripts in the RefSeq database. The genomic coordinates (hg19 version) of the 3′UTR exons for both the sets were used to construct GTF files, and 514 genes containing both the transcript isoforms were identified (Supplementary Tables S8 and S9). We used the small RNA-seq data sets, for this selective group of genes for identifying an antisense transcript fragment overlapping an exonized Alu element for specific isoforms across the five time points.
For the 36 base pair cycle of the small RNA-seq data sets, we specified a minimum length of 17 bases. As we observed that majority of Alu exonization events in the 3′UTR were full-length Alus, we searched for antisense reads across the complete length of the exonized Alu and did not limit the search to the exonized fragment only. As such, we retrieved the full-length co-ordinates of 3′UTR exonized Alu and assigned them a strand annotation, which is reverse to that of the host gene. Using this method, we created a BED file for Alu antisense transcripts (Supplementary Table S10). Owing to limitation of depth of coverage for the RNA-seq experiments analysed here, we could not explore the A→I editing events.
Calculation of unique genomic length occupied by 3′UTR
For a normalized expression index of a transcribed region, the actual length calculation for the region is crucial. Multiple transcript isoforms with overlapping 3′UTR exons confound the calculation. Hence, the size of 3′UTR across transcript isoforms was calculated using the algorithm described in methodology section for Enrichment analysis for co-occurrence of Alu mediated events in 3′UTR. Thus, the unique genomic length for 3′UTR exons of the genes in the Alu-exonized and non-Alu-exonized sets were calculated.
Reads/Kb/Million calculation for 3′UTR exons from poly-A+ RNA-seq data sets
Aligned (TopHat-Bowtie pipeline; hg19 genome build) files (binary format, BAM file) available from the GEO webpage for GSE32874 were downloaded for the five time points, (GSM818564 to GSM818568). The alignment statistics were calculated for each of them using bamtools (54). Using the HTSeq package (55) and respective GTF files, strand-specific coverage was calculated separately for Alu-exonized and non-Alu-exonized 3′UTR exons across all the five time points. For each gene, the Reads/Kb/Million (RPKM) for 3′UTR exon was calculated as
RPKM calculation for antisense Alu from small RNA-seq data sets
Aligned files for the small RNA-seq data sets are not available from the GEO webpage, instead read counts are provided. Hence, the raw data (Sequence Read Archive (SRA) files) were downloaded from NCBI SRA database (Accession SRX101440; SRA run files SRR353655 to SRR353659) for the five time points. These SRA files were converted to fastq format using fastq-dump utility of the SRA tool kit (available from NCBI). Using Fastx (56) and FastQC packages (57), the small RNA-seq fastq files were processed for adaptor and primer sequence removal as well as 3′ end trimming, when required, for quality checking. The quality check (QC)-passed reads for each of the time point were then aligned to the hg19 genome build using Bowtie (version 0.12.7, 64 bit) (58). Keeping our question of repetitive Alu element sequence in mind, the Bowtie alignment was performed using the following parameters: fastq quality aware -n mode, only 1 mismatch allowed in seed length of 17, best alignment to be reported only with –best and -k 1 and reads with >5 valid alignments to be rejected with -m 5. The Sequence Alignment/Map (SAM) file obtained from alignment in case of each time point was converted to BAM (binary format, SAM file) file. After calculation of alignment statistics using bamtools, the BAM files were processed by bedtools (59), using the BED file created for antisense transcripts for calculating strand-specific coverage of the antisense Alu coordinates. From the coverage (read counts) data for each antisense Alu coordinate, RPKM value was calculated as
The pattern of RPKM values thus calculated across the five time points for antisense Alu detected in the small RNA-seq data sets were compared with the expression pattern of 3′UTR exons from Alu-exonized genes and that from non-Alu-exonized genes, respectively.
Network analysis of Alu-containing genes responsive to viral recovery
We wanted to perform a network analysis of a set of 59 genes, wherein we detected cis-antisense transcription and a concurrent anti-correlated sense transcription in the iPOP data set. For this, we used the Biological General Repository for Interaction Datasets (BioGRID) interaction database (60), as it is one of the most comprehensive interaction data repository housing curated information from experiments and is also user friendly. We chose BioGRID data set release 3.1.93 for humans to retrieve all interactions for the 36 of 59 genes whose interaction data were present after ensuring that both the source and target instances were from human. These genes along with their different interacting partners resulted in 450 pair-wise interactions. Using Cytoscape (version 2.8.0), this interaction set was plotted using Spring embedded layout algorithm (61,62).
Gene ontology analysis
Gene ontology (GO) analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (63). Briefly, we looked for enriched functional categories using the GO-FAT classification, as this gives specificity during GO classification by filtering out the broadest terms in hierarchy. Also Functional Annotation Clustering tool was used to summarize annotation from, namely UniProt, InterPro, and Kyoto Encyclopedia of Genes and Genomes (KEGG) with GO classification to aid functional interpretation of the gene lists. For GO interpretation of large set of genes (>3000), we used the BiNGO plugin available in the Cytoscape framework (64).
RESULTS
Alus are the most abundant transposons in the transcriptome
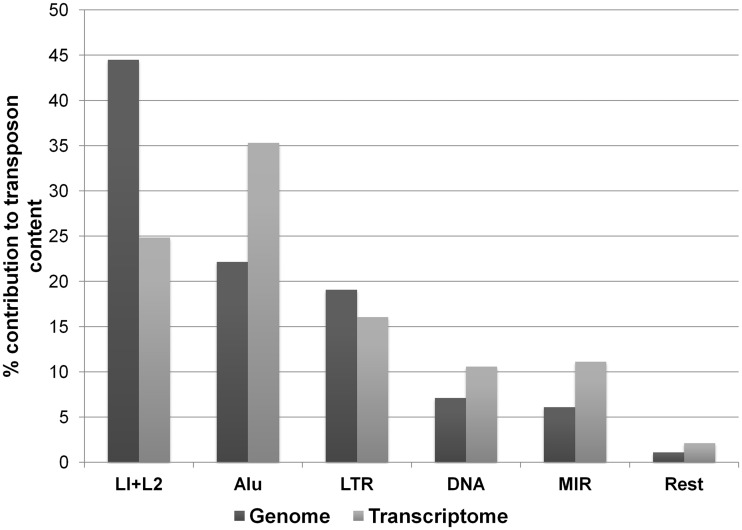
Using the RepeatMasker track from UCSC, we identified all the transposable elements that are present in the RefSeq mRNAs and compared their abundance with respect to their genomic coverage (Figure 2 and Supplementary Table S1). The transposable elements cover nearly 45% of the genome but comprise only 6% of the transcriptome. We observed significant difference (P = 0.05) in the distribution of Long Interspersed Nucleotide Element (LINE) and Short Interspersed Nucleotide Element (SINE), majorly Alu, in the genome and transcriptome. Although LINE was the most abundant in the genome (nearly twice as much as Alu), the Alus were significantly enriched (35%) in the transcriptome amongst all the transposons. Excepting Alu and Mammalian Interspersed Repeats (MIR), which is also a family of SINEs, no other transposon families showed such significant enrichment in the transcriptome.
Figure 2.
Enriched presence of Alu elements in the transcriptome. Though LINEs are most abundant in the genome (in base pairs) and Alu elements are about half in percentage, the ratio nearly flips in the transcriptome. Among all transposons, Alu elements have the highest representation in the transcriptome (in base pairs).
Co-localization of exonization, antisense and editing events in the transcriptome
Alu exonization events in the transcriptome
We carried out analysis on complete RefSeq data set (release 45, January 2011) comprising 36 690 full-length human transcripts. Using the UCSC table browser (hg18), we retrieved the 4 28 976 exon blocks of these transcripts for the 5′UTR, CDS and 3′UTR regions (Supplementary methods). Nearly 1.9% (8457) of these exon blocks was filtered out, as they had ambiguities in their genomic locations, for instance, mapping to alternate genome assemblies or in unplaced chromosomal contigs. Using the UCSC RepeatMasker (version 3.2.7) track, we mapped and retrieved the exons having Alu repeats. These repeats were present in 6695 exon blocks representing 1.56% of the total exons. Of these, 76% had ≥10% overlap with Alu. These were considered to be Alu-exonized events.
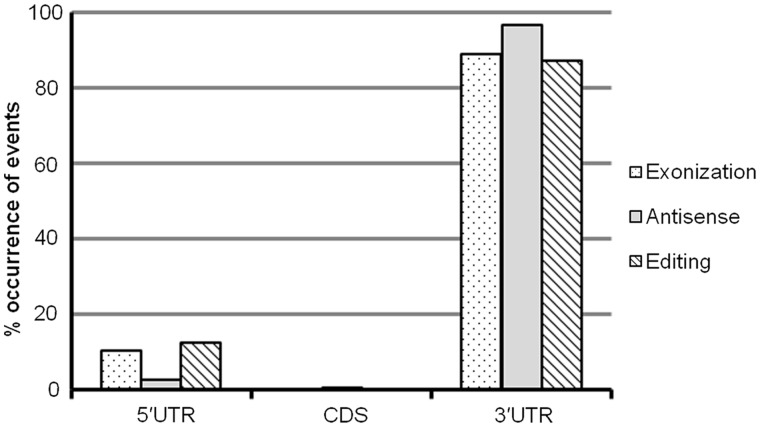
Therefore, of the total 36 690 transcripts, 4663 (12.7%) have Alu exonization mapping to 3177 genes. Of the 3177 genes, 80% (2529 genes) have Alu exonization in 3′UTR. In terms of the base pair coverage, 88% of the Alus are present in the 3′UTR, whereas 5′UTR and CDS have ∼11 and 1%, respectively (Figure 3 and Supplementary Table S2). There are a large number of transcripts, which have Alu exonization in multiple regions of the genes but not necessarily in the same isoform. For example, as evident from the Table 1, 110 genes have Alu exonization in the 5′UTR and 3′UTR, but in terms of the transcripts, the numbers are much smaller. It might be possible that the preponderance of Alus in the 3′UTR could be owing to larger length of this region compared with the remaining transcript. However, we observed >50% of the base pairs to be occupied by CDS and the rest to be distributed nearly equally between the UTR regions.
Figure 3.
Percentage distribution of Alu mediated events across the transcripts. In terms of base pair coverage, >88% of Alu exonization primarily occurs in the 3′UTR region with a small fraction in 5′UTR and a minimal fraction in the CDS region. Surprisingly, the 3′UTR is enriched in both antisense and A→I events. The coordinates for Alu antisense events were anchored on Alu-harbouring exons across the CDS and UTR regions. For a normalized count, percentage contribution to Alu antisense in each of the three regions is expressed as fraction of the Alu-harbouring exons that overlap with antisense coordinates. Percentage contribution to Alu editing in each of the UTR or CDS region is expressed as fraction of the Alu-harbouring exons that have occurrence of A→I editing events.
Table 1.
Number of Alu-exonized events in different regions of the genes
| Exonic regions | 5′UTR | CDS | 3′UTR |
|---|---|---|---|
| 5′UTR | 708T (522G) | ||
| CDS | 28T (32G) | 141T (102G) | |
| 3′UTR | 88T (110G) | 94T (99G) | 3610T (2328G) |
This table categorizes propensity of Alu-exonization events across different regions of the transcripts. Though there are exonization events observed in multiple region of the genes (G), the transcript isoform (T) involved in many cases is not the same. Hence, for cells denoting exonization in only one region, 5′UTR/CDS/3′UTR, number of transcripts are more than corresponding genes, whereas genes are higher in number when multiple regions are involved in exonization.
Alu as cis-antisense transcripts in the exonized genes
The virtual SAGE map data set (from NCBI ftp), which contains all possible SAGE tags from the 3′end generated by computational cleavage of all human transcripts using Nla III site was used for identification of antisense Alus (Supplementary methods). This data set comprises 9 million virtual SAGE tags screened from the complete human dbEST (hg18) comprising >7 million EST sequences, which ensures a comprehensive representation of transcripts not only from diverse tissues but also different conditions. Using the RepeatMasker (version 3.2.7) track, nearly 0.4 million EST sequences having Alu exonization were identified. When the aforementioned two data sets were overlaid, ∼89 900 ESTs with virtual SAGE tags overlapping to exonized Alus were obtained. From this data set, using strand information for the host gene and EST alignment 23 069 cis-antisense SAGE tags which had overlaps with Alu and mapped to 22917 ESTs from 5648 gene were identified. Sixty-seven percent of the antisense tags were either redundant or resembled Best Tag data for sense transcripts and hence were removed. Additionally, 5620 intronic/ambiguously located ESTs were also filtered out. Through this, we identified a total of 5602 antisense tags (ESTs) with Alus mapping to 3375 genes. Nearly 54% (3055) of the virtual antisense tags were observed to be actually present in >20 tissues across 124 samples in NGS SAGE data from CGAP. These antisense tags were mapped back to the genomic coordinates through ESTs. Subsequent mapping of the overlapped genomic coordinates to RefSeq mRNA resulted in 1162 antisense tags, which could be anchored to 607 RefSeq mRNA (Supplementary Table S3). We observe that nearly one-fourth of the Alu-containing exons (590 genes) in the 3′UTR have antisense tags. Antisense tags by themselves are predominant (96.8%) in the 3′UTR (Figure 3), whereas only 3.8 and 1.5% of the Alu-containing exons in the 5′UTR and CDS, respectively, have antisense tags. We observed seven genes (MDM4, PDLIM5, TCF7, MATR3, MRPL49, P2RX7 and CES2) to have antisense both in the 3′UTR and 5′UTR. Surprisingly, the sharing was at the gene level, and they mapped to distinct transcript isoforms (Table 2).
Table 2.
Number of Alu-antisense events in different regions of Alu-exonized genes
| Exonic regions | 5′UTR | CDS | 3′UTR |
|---|---|---|---|
| 5′UTR | 35T (14G) | ||
| CDS | 0T (0G) | 4T (3G) | |
| 3′UTR | 0T (7G) | 0T (1G) | 952T (582G) |
Each cell within the table represents Alu-exonized transcripts (T) and its corresponding genes (G) for incidence of antisense events across 5′UTR, CDS and 3′UTR regions. Even if there are antisense events observed in multiple regions of the genes, the transcript isoform involved is not the same. For example, seven genes have antisense event for both 5′UTR and 3′UTR, but not within the same transcript isoform.
Alu editing in exonized transcripts
Analysis of approximate genomic alignment blocks from 4 million exonized ESTs retrieved from dbEST was carried out to identify Alu editing within exonized transcripts (Supplementary methods). After filtering out 59 654 ESTs, which had ambiguous or non-unique genomic locations, 86% of the ESTs were retained. To consider an alignment block, a valid entry for A→I editing, we applied two threshold criteria of block size ≥41 bps, comprising Alu sequence of ≥25 bps and a non-Alu sequence of minimum 16 bps. From the filtered set of unambiguous ESTs, we could retain 77% (2 17 637) ESTs, which were used for identifying A→I editing events. Using a global alignment strategy implemented in Stretcher (EMBOSS suite), between the genomic alignment block and the corresponding EST stretch, a total of 1 22 047 A→G mismatch positions were identified (20% of all mismatches). Subsequent filtering through dbSNP database (release 129) led to removal of 6299 mismatch positions. Additionally, 20 721 positions were not within Alus, and hence these were also filtered out. Editing within Alu is characterized by presence of two Alus in head-to-tail orientation. Therefore, as an additional criterion, we also looked at these events in the context of presence of an opposite oriented Alu. This led to filtering out of 20 761 positions. After all, the aforementioned filtering criteria 74 266 possible A→I editing events were considered for further analysis. More than 95% of the Alus that are edited have an opposite oriented Alu within 5 kb with a median distribution of ∼660 bps. We observed 74 266 editing sites mapping to 26 537 ESTs. Querying these ESTs in the UniGene database resulted in 5988 Alu-edited ESTs with gene information. On further anchoring of these ESTs onto RefSeq exonic coordinates resulted a total of 1580 RefSeq transcripts belonging to 1003 genes. We observe that of the A→I editing events represented in the RefSeq exons, a predominant fraction (87.4%) targets the 3′UTR (Figure 3). In contrast to Alu antisense, 5′UTR contributes a substantial fraction (12.5%) of editing events. CDS exons, however, had negligible editing. We observed 22 genes to be target for A→I editing both in the 3′ and 5′UTR (Table 3). Of these, five genes (MDM4, PDLIM5, TCF7, MATR3 and CES2) had presence of antisense also, both in their 5′ and 3′UTR (Supplementary Table S4).
Table 3.
Number of Alu-editing events in different regions of Alu-exonized genes
| Exonic regions | 5′UTR | CDS | 3′UTR |
|---|---|---|---|
| 5′UTR | 148T (89G) | ||
| CDS | 0T (0G) | 17T (12G) | |
| 3′UTR | 9T (22G) | 0T(2G) | 1406T (878G) |
The table categorizes exonized transcripts (T) for occurrence of A→I editing events across the 5′UTR, CDS and 3′UTR regions. Here again, though there are events in multiple regions of the gene, the transcript isoforms involved in such cases are not the same.
Overlap of exonization, antisense and editing events in the different regions of the transcripts
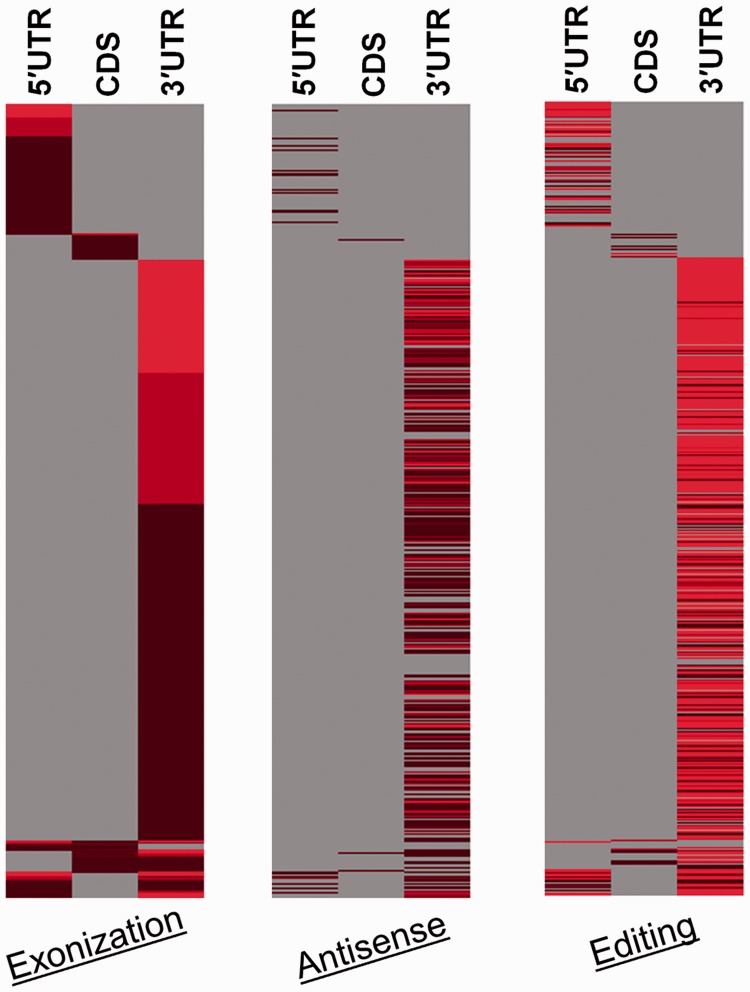
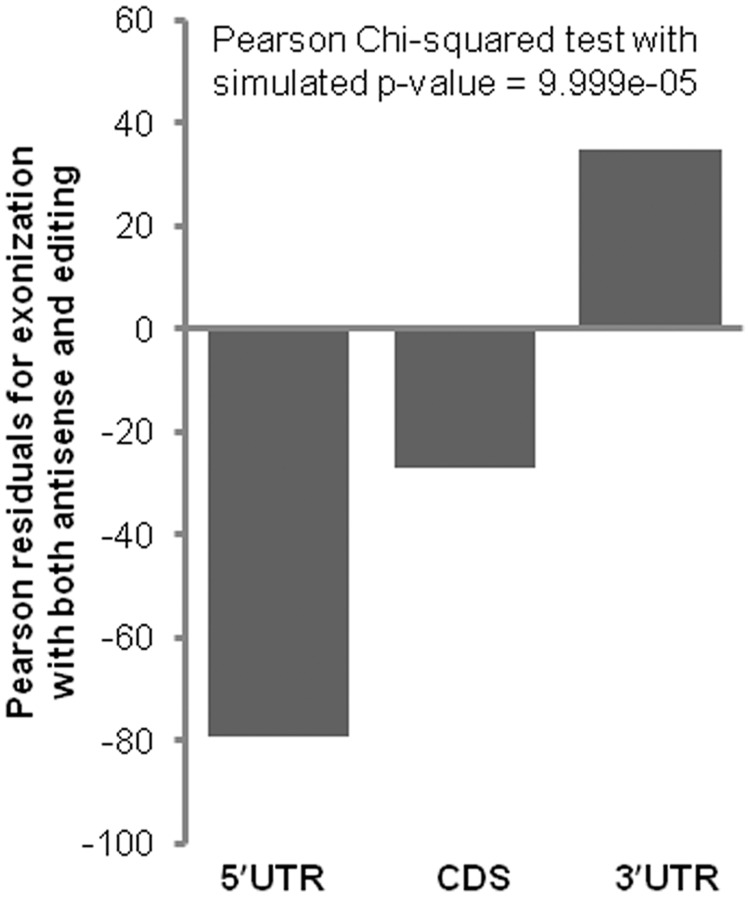
In nearly 40% of the exonized transcripts, we observed editing and antisense events. Of 3177 unique genes that were exonized, we detected the co-occurrence of both editing and antisense in 319 genes, whereas only antisense and editing were observed in 288 and 686 genes respectively (Supplementary Table S2). The overlap has been seen at the exon level, i.e. transcript isoform specific. Hence, though a gene might have both antisense and editing events, albeit in different isoforms, it has not been counted as co-occurring. In all the cases, the events were observed in overlapping regions with respect to 5′UTR, CDS and 3′UTR (a case e.g. is visualized in UCSC Genome Browser; Supplementary methods). Heatmap summarizes exonization, antisense and editing information and represents it in a gene-wise fashion (Figure 4). As is evident from the map, nearly 97% of these events were observed in the 3′UTR, which is statistically significant with P-value of <2 × 10−16 (Pearson chi-squared test) after category-wise analysis for Alu exons with incidence of both antisense and editing (Figure 5). The robustness of the test result was evaluated by re-computing the P-value after 10 000 Monte Carlo simulations, which was found to be ∼10−4. It is also discernible that these events are highly variable between the genes (Supplementary Table S5). Some of the genes had a large number of exonized isoforms that were extensively edited and also had a large number of antisense transcripts. Compared with editing, antisense events seem to be lower. This could be owing to the stringent methodology that has been adapted for defining antisense transcripts, and more importantly, all the antisense transcripts identified are in cis only.
Figure 4.
Occurrence of Alu-mediated transcriptional events in different regions of genes. The figure is a heatmap depicting density of occurrence for Alu exonization, antisense and editing across 5′UTR, CDS and 3′UTR regions in each of the 3177 genes. Each row of the heatmap is a gene with the number of exonization (first panel), antisense (second panel) and editing (third panel) events summarized. The colour gradient from light to dark represents the magnitude of the related event, i.e. number of exonized Alus, number of Alu antisense targets and number of A→I editing events, respectively, with lighter colour representing larger number of events. As is evident, majority of genes exhibit extensive editing and antisense events in the 3′UTR.
Figure 5.
Residuals plot for Pearson chi-squared test. The null hypothesis that there is no difference in co-occurrence of antisense and editing on Alu exons across the categories, is rejected at the P-value ∼10−4 threshold. The residuals give the direction of deviation between the observed and expected, in this case the co-occurrence events. Whereas 5′UTR and CDS show lower than expected incidence, 3′UTR is enriched for such events.
Though we observe an overwhelming majority of Alu-mediated events in the 3′UTR region compared with 5′UTR and CDS, this relative frequency of events could be possible owing to differential selection pressure on UTR versus CDS. This is in addition to the fact that the average 3′UTR exon size of 1 kb is much larger than 300 bp of an average exon. Hence, we set forth to test this hypothesis by analysing distribution of A→I editing within Alu in intronic region. We find that the A→I editing density of 3′UTR region introns is more than three times that of 5′UTR and CDS. This is especially significant, as the genomic space available to 3′UTR intron is 2–3 magnitudes lesser than that of 5′UTR and CDS (Table 4).
Table 4.
Comparison of A→I editing density in introns intervening CDS and UTR exonic regions
| Exonic region | Number of A→I editing events within introns | Intron length (bp) | A→I editing density (editing sites/ Mb of intron) |
|---|---|---|---|
| 5′UTR | 5885 | 2.9 × 108 | 20.21 |
| CDS | 34 983 | 1.8 × 109 | 19.43 |
| 3′UTR | 508 | 7.58 × 106 | 66.99 |
The A-to-I editing density observed for 3'UTR region introns (highlighted in bold) is much higher than expected from its genomic coverage.
Positional bias for Alu-mediated events varies across the transcript regions
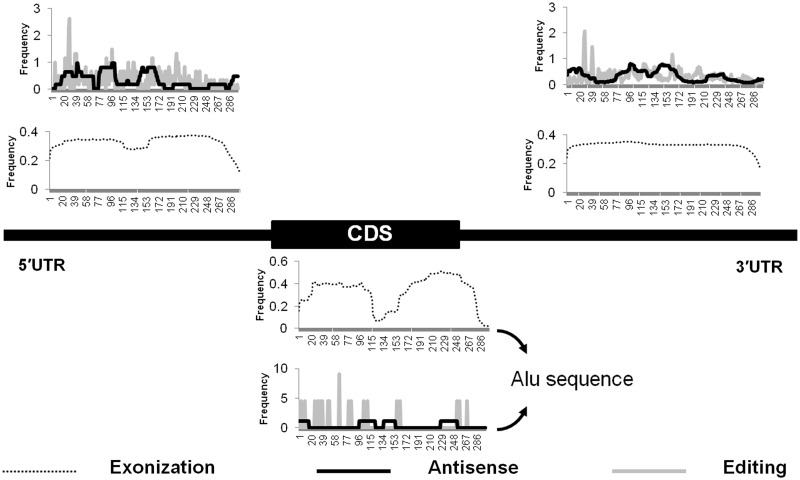
Aforementioned results indicated a possibility of crosstalk between the three events, as they were localized to the same regions. We therefore sought to determine whether there are preferred positional stretch/positions within the Alu sequences for occurrence of such events. For this, we mapped the genomic coordinates of all exonization, antisense or editing events onto the concerned Alu subfamily consensus sequence position using the RepeatMasker (version 3.2.7) alignment data from UCSC Table Browser (Supplementary Table S6). We observed that the preferred stretch for exonization, antisense and editing position varies depending on whether the Alu is present in the 5′UTR, CDS or 3′UTR exon (Figure 6).
Figure 6.
Positional preference of exonization, antisense or editing events within Alu repeats in different genic regions. Base positions of full-length Alu are represented in the 5′UTR, CDS and 3′UTR, and the graphs are plotted based on the frequency of exonization, editing and antisense at each of the positions in the three genic regions. Y-axis indicates the frequency of occurrence of the particular base position along the length of Alu. In the 5′UTR or the CDS, either of the monomer of the full-length Alu is exonized, whereas in the 3′UTR, the full-length Alu is mostly exonized. The antisense events peak around ∼80–100th and ∼140–170th base positions in both the 5′ and 3′UTR regions. A→I editing has a clear peak at ∼17–45th base position. The number of antisense and editing events identified in CDS is miniscule compared with that in UTR regions.
In the 3′UTR in nearly all the cases, the full-length Alus are exonized, whereas in the 5′UTR and CDS, there is a distinct preference for exonization of either the right arm monomer or the left arm monomer, and the regions from 113 to 145 seems to be excluded. Though the editing sites seem to be dispersed over the entire length of Alu, there are distinct peaks in the region from 17th to 33rd position in Alu. The antisense events are present as three distinct humps across the Alu length. The humps centred around 95th and 150th position are shared between the 5′ and 3′UTR Alus, whereas the hump at the start of Alu sequence shows variability. We find it significant to observe shared preference for A→I editing and antisense across the 5′ and 3′UTR from different genes, despite of the large difference in numbers. 3′UTR has 847 antisense and 18 642 editing events, whereas 5′UTR has 22 antisense and 2657 editing events only. We did not observe any discernible pattern in the CDS owing to the presence of low number of antisense and editing events (4 and 17, respectively).
Non-random distribution of Alu exonization in the transcriptome
Surprisingly when GO analysis was carried out on the genes that had Alu exonization at the 5′UTR, CDS or the 3′UTR region, we observed different processes to be enriched (Supplementary Table S5). The 3′UTR were enriched in processes related to cellular biosysnthesis, nucleotide metabolism, DNA integrity check point, negative regulation of homeostasis, metal ion binding and catalytic activity, whereas in the 5′UTR, processes related to positive regulation of fatty acid secretion and lipid transport as well as metal ion binding and poly-ubiquitin binding were observed. In the CDS, male gamete generation and cell cycle checkpoint genes were observed (Table 5). All the unique categories observed in the 5′UTR and CDS were lost when the genes were pooled from the three regions for GO analysis. The overlapping set of 319 genes (Supplementary Table S5), which had all the three events co-occurring at the exon level, was enriched in genes localizing to lysosomal and mitochondrial compartments and related to caspase regulatory activity (Table 6 and Supplementary Table S5). The exonized genes that showed editing had significant over-representation of genes with the KRAB domain.
Table 5.
GO analysis of Alu exonization with respect to position in the transcript
| Summarized GO categories (P ≤ 0.05) | Genes with Alu exonization in |
||
|---|---|---|---|
| 5′UTR | CDS | 3′UTR | |
| Biological process (BP) and molecular function (MF) | Positive regulation of Fatty acid secretion | Male gamete generation | Cellular biosynthesis |
| Positive regulation of Lipid transport | Cell cycle checkpoint | Nucleotide metabolism | |
| Metal ion binding | DNA integrity checkpoint | ||
| Poly-ubiquitin binding | Negative regulation of homeostasis | ||
| Metal ion binding | |||
| Catalytic activity | |||
Table 6.
Pathway analysis of genes with co-occurrence of exonization, antisense and editing at the exon level
| Enriched KEGG pathway | Genes |
|---|---|
| Lysosome | AP3S2, CTSB, CTSS, DNASE2, GGA1, GGA2, GM2A, LAMP3, SLC11A2, SLC17A5. |
| Apoptosis | APAF1, CASP8, CHP, CYCS, PIK3R2, TNFRSF10B, TNFSF10, TP53. |
Functional evidence for dynamic interaction between Alu-mediated events during viral recovery stage
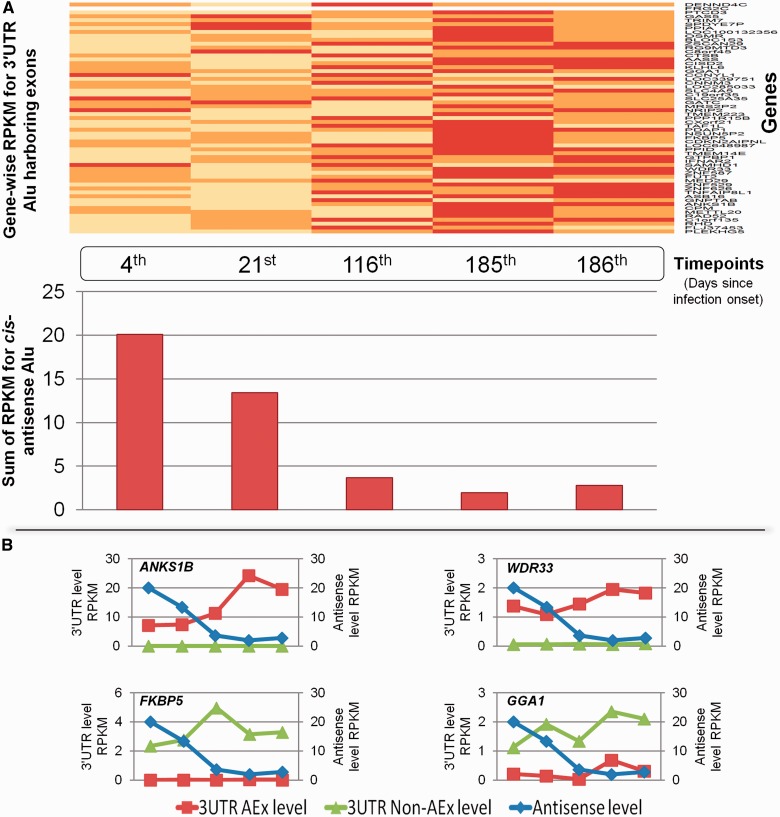
In view of previous knowledge of increased Alu transcription during stress, we chose the iPOP data set to investigate dynamic interaction between Alu exonization and Alu antisense. Comprising time course poly-A+ RNA-seq coupled to small RNA-seq of Peripheral Blood Mononuclear Cells (PBMCs) during recovery from viral load, iPOP data set allows unique opportunity to track dynamic interaction between exonized transcript isoform and their cis Alu antisense. We report 59 genes for which we detect cis-antisense Alu transcription against the exonized transcript isoform. Intriguingly, we find that the number of genes against whom antisense is detected decreases sharply as we move away from viral infection stage (Figure 7a). Based on the observation, we hypothesized that the level of exonized transcript isoforms should show an opposite trend if Alu-antisense interacts functionally with Alu-exonized transcripts. As predicted, we observed negative correlation (correlation score = −0.96, P-value = 0.007) between expression of Alu-exonized transcript isoforms and its antisense transcript. The dynamic and functional relevance of this result was strengthened by the observed differential expression pattern of Alu-exonized and non-Alu-exonized transcript isoforms in gene-specific manner. In genes like ANKS1B and WDR33, we find the exonized isoform to have elevated expression when antisense transcription is low, whereas in FKBP5 and GGA1, we find the non-exonized isoform to be elevated (Figure 7b).
Figure 7.
Dynamic levels of Alu-exonized and Alu-antisense transcripts using iPOP data set during viral recovery phase. (A) Upper panel represents analysis of time series data for poly-A+ RNA-seq from iPOP data set during viral recovery phase showing increase in expression levels along the time course for 59 Alu-exonized genes. Lower panel corresponds to time point matched analysis for small RNA-seq from the same data set, emphasizing that the combined expression level of antisense transcripts decreases as we move away from the viral infection stage. (B) The functional importance of different Alu-mediated events is evident from differential levels of Alu-exonized and non-Alu exonized transcripts during viral recovery stage. In upper panel, ANKS1B and WDR33 show that as we move away from viral infection stage, the levels of Alu antisense transcript decreases, whereas, its target, the Alu-exonized transcripts show elevated expression pattern. Interestingly, the levels of the non-Alu-exonized counterpart do not change during this phase. In lower panel, FKBP5 and GGA1 show that although non-Alu-exonized transcript levels show anti-correlation with Alu-antisense levels, the levels of Alu-exonized transcripts are unchanged.
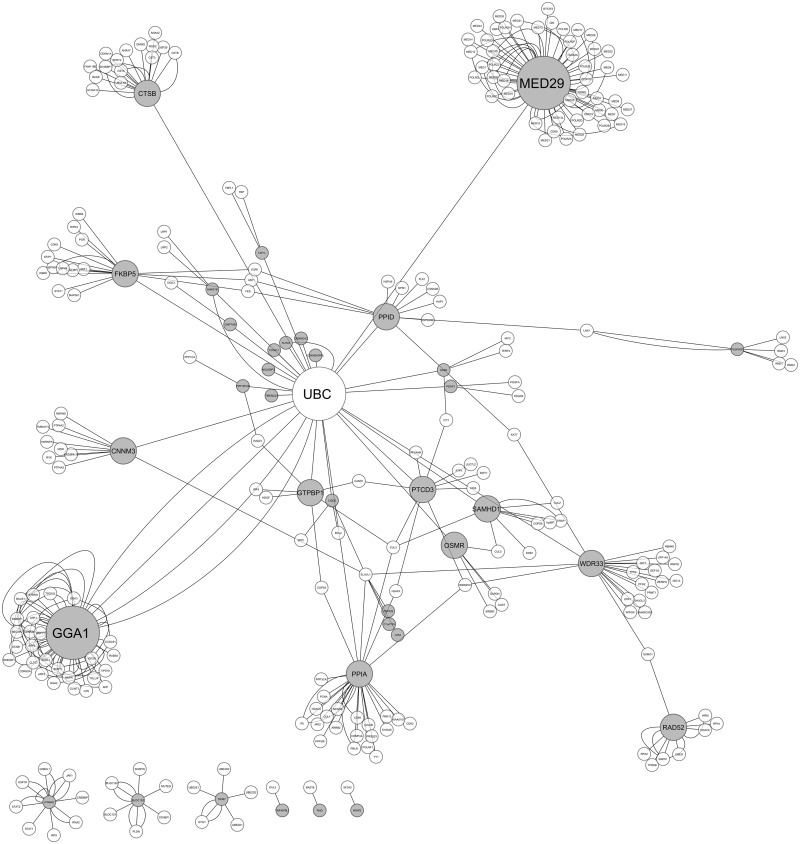
To understand the importance of the observed dynamic behaviour of Alu-containing exons, we used BioGRID to look for possible interaction between the genes (Figure 8). Interestingly, network analysis revealed that most of the genes are directly/indirectly interacting with the ubiquitin C gene, UBC (linked to ubiquitin-proteasome pathway). Some of the genes like ANKS1B have few, whereas WDR33 have multiple partners, which help to shape the ubiquitination pathway. Genes like IFNAR2 and TRIM7 are although not directly interacting with UBC, but they are important players in anti-viral response mechanism (65,66). It is already known that this pathway plays a major role in protein degradation in a complex, temporally controlled and tightly regulated manner as part of host defence against viral infection.
Figure 8.
Network analysis of Alu-containing genes responsive to viral recovery from iPOP experimental data set. Alu-containing genes with anti-correlated sense versus antisense levels (poly-A+ RNA-seq and small RNA-seq, respectively) during viral recovery were used as query in the BioGRID database for experimentally verified protein–protein interactions. The network analysis reveals that majority of such genes (shown in grey) interact either directly or via an intermediary (shown in white) to ubiquitin conjugating enzyme (UBC). Ubiquitin-proteasomal system is known to be an integral component of host defence against viral infections.
DISCUSSION
To our understanding, this study provides the first evidence of co-occurrence and functional interaction of Alu-mediated exonization, antisense and editing events in the transcriptome (Figure 9). Through an integrative analysis of RefSeq mRNAs, NCI-CGAP SAGE tags and dbEST, we could elucidate the extent and non-random distribution of these Alu-mediated events not only across genes but also at specific positions within transcripts and Alus. These events also show preferential enrichment in the 3′UTR of genes in specific biological processes. Moreover, the editing and antisense positions within Alu have an overlap, suggesting the likelihood of a functional crosstalk. Using an experimental data set, we also demonstrate the dynamic interaction between these events following recovery from viral infection.
Figure 9.
Schematic for identification, analysis and evidence for interaction between Alu-mediated transcriptional events. The figure summarizes the work carried out by representing it as schematic, subdivided into three parts. Transcriptome-wide Alu exonization events were mapped using RefSeq full-length mRNAs (Supplementary Table S2). Subsequently, Alu-antisense and A→I editing events were anchored onto Alu-exonized transcripts using NCI-CGAP SAGE tags (Supplementary Table S3) and dbEST (Supplementary Table S4), respectively. Preference for occurrence of these events was studied across the length of the transcript isoforms. Bias in the Alu positions involved was determined by comparing with the subfamily-specific consensus Alu sequence from RepBase. Finally, to find experimental evidence between these events, we used the time series RNA-seq coupled to small RNA-seq from the iPOP data set. Focussing our analysis on the Alu-harbouring 3′UTR region, we find significant anti-correlation between exonized transcript levels and cis-antisense Alu level from poly-A+ RNA-seq and small RNA-seq, respectively.
Alu elements can contribute substantially to transcriptome diversity through its inclusion in alternative exons, propensity for A→I editing and antisense transcripts. We hypothesized that a crosstalk between these Alu-mediated events could contribute to diverse transcript isoforms from the same gene with varying fates. Exonization of Alus has been implicated in diverse functions depending on their position in the genes. When present in the 5′UTR region of genes, Alu has been implicated in the translational efficiency of the exonized isoforms (67) and tissue-specific novel splice variant that can affect cell viability and induce apoptosis (68). In some cases, when present in CDS, the proteins encoded by these transcript isoforms have contrasting or specific functions. It has also been shown that Alu present in the 3′UTR region of human aspartyly tRNA synthetase can bind to a tRNA isodecoder and affect stability of mRNA (69). An extreme example of exaptation of Alu in the coding region is the case of neuronal thread protein AD7c-NTP, used as a biomarker in Alzheimer’s disease, where the entire coding region comprises only Alu repeats (70). We observed that top 10% of the genes with respect to Alu exonization counts were enriched for processes related to transcription and apoptosis. Interestingly, we also observe distinct enrichment of genes from specific biological processes for the 5′UTR, CDS and 3′UTR regions. Such diverse yet distinct enrichment of biological processes in genes harbouring Alu exonization in different locations can potentiate multiple modes of regulation, either through transcriptional or translational mechanisms. Our analysis also reveals that majority of these events are in the 3′UTR, and many genes harbour multiple exonization events. In contrast to our observation, Shen et al. recently reported 5′UTR enrichment of Alus. However, this was because they studied only those Alu-harbouring exons that are internal and flanked by constitutively spliced exons. In this scenario, with an average of 2.7 introns in 5′UTR to 1.8 in 3′UTR (Supplementary Table S7), the probability of observing an internal Alu-containing exon in the 3′UTR is further reduced. On the contrary, our study has included all Alu-harbouring exons without any pre-defined criteria.
Of all the editing events, A→I editing comprises the majority in the transcriptome (71–73). Global analysis of A→I editing reveals that these events are most frequent in 3′UTR or intronic regions, ∼90% of which are localized within Alu elements. Additionally, different positions have variable propensity to undergo editing (32,74). Predominance of this editing activity has been attributed to a human specific splice variant of ADAR2, created as a result of an Alu-derived exon, which accounts for 40% of all the ADAR2 transcripts (75). By virtue of recoding the information post-transcriptionally through editing, it can create scenarios such as evolution of novel exons, dsRNA stability, altered splice patterns in response to stress, nuclear retention of transcripts and tissue specificity (35,76–81). Recent studies have shown that editing is involved in regulation of processing and expression of mature miRNAs (82–85). Besides, it has also been shown that the RNAi and A→I editing pathways might compete for a common dsRNA (86–88). Editing in an exonized RNA may also regulate the expression of the transcripts by preventing sense-antisense pairing (89). In this context, it is noteworthy that ∼70% of the genome produces transcripts from both the strands, and many of such sense-antisense transcript pairs are co-ordinately regulated (90,91). Though a number of studies report the presence of Alu within antisense transcripts, a systematic genome-wide study of the phenomena has not been attempted. To explore this, we anchored the Alu editing and antisense events on to the exonized transcript isoforms and observed that 3′UTR is enriched for both the events. Furthermore, these two events also exhibit positional overlap within Alu elements. Difference in editing and antisense events in the same genes across tissues could create condition-specific regulatory switches. Moreover, as the 3′UTR are also the most preferred sites for miRNA binding, editing in Alu may be an additional mechanism for modulating these interactions (92). A functional crosstalk is plausible if Alu exonization, editing and antisense events map onto overlapping sites. More than 300 genes in our study showed all the three events in the same transcript isoform and also exhibited positional overlap within Alu sequences. It is worthwhile to note that co-occurrence of antisense and editing events on the template of exonized Alu is statistically over-represented in the 3′UTR. Being longer, it can be argued that multiple occurrences of Alu-mediated events in the 3′UTR are owing to lack of evolutionary constraint. Nevertheless, analysis of introns across 5′UTR, CDS and 3′UTR show that 3′UTR is enriched even in introns for A to I editing events. This implies that exonized Alu elements can add a novel dimension to the 3′UTR regulatory hotspot.
A variety of model systems have been investigated for elucidating the functional role of Alu-mediated events. We have earlier shown that the heat shock factor binding within Alu drive antisense transcription, which in turn leads to regulation of sense transcripts. This expands the role of elevated levels of Alu RNA during stress response, like viral infection, heat shock and cancer. To further probe the functional implication of our findings reported here, we selected an experimental data set corresponding to viral recovery. We observed a dynamic interaction between exonization and antisense, where the expression level of the Alu-harbouring transcript was negatively correlated with that of the cis-antisense through consecutive time points following recovery from viral infection. When segregated at the isoform level, we found instances, albeit for a couple of genes only, where either the Alu-exonized or the non-exonized-isoform to be expressing in response to the cellular stimulus. There are multiple other regulatory factors involved in host defence mechanisms, and primate-specific Alu could have evolved to complement the existing network of regulators. Hence, for genes like GGA1 and FKBP5, expression of non-exonized isoform points towards an alternative mode of regulation rather than cis-antisense Alu. This corroborates the earlier finding in our laboratory that cis-antisense Alu transcription can regulate the levels of the corresponding sense transcript. Our findings are further strengthened by the fact that these genes were found to be either involved directly in host response or as part of the ubiquitin-proteasomal pathway, integral to anti-viral response.
This work is important by virtue of its relevance in imparting a single transcript with multiple modular functions through Alu-mediated events. Thus, a gene with potential for Alu exonization can have at least four possible states: non-exonized, exonized, edited and different proportions of these isoforms through antisense. An Alu-exonized transcript has the potential to be regulated through cis-antisense Alus, which in turn is subject to variability if there are A→I editing events in the exonized transcripts. A genome-wide consequence of such a crosstalk could lead to altered dynamics and outcome of a regulatory network. For instance, in context of the total genes exonized in the transcriptome, we observed an abundance of these events in apoptosis (Figure 10) (Table 6), a pathway, which is not only important in cellular homeostasis but also assumes importance in various disease etiologies (93–95). Genes of apoptotic pathway are under accelerated evolution in humans (96,97). Our observations suggest that changes in the non-coding regions through insertion of Alu elements combined with functional and dynamic interplay of Alu-mediated events, during primate evolution, could further shape the transcriptome. For instance, of the genes transcript involved in apoptosis (98), we observe many genes in the DR3/DR4 receptor pathway as well as those involved in the extrinsic pathway and DNA damage to have these events (99,100). Especially in genes such as TRAIL, TRAIL-R, XIAP, CFLAR, CASP8, APAF, CYTC, MAVS, TP53 that are some key nodal points in apoptosis, variability in expression could affect the cell viability. Noteworthy, CFLAR or c-FLIP not only has multiple transcript isoforms but also cleaved protein isoforms, proportions of which determine whether the cells would undergo apoptosis or survive (101,102). We find that majority of the isoforms have four Alus in their 3′UTR, two of which are also edited. The larger isoform is pro-apoptotic, but its cleaved smaller product is anti-apoptotic. One of the small isoforms, which is anti-apoptotic does not have any of these Alus. The CFLAR protein is also responsive to dsRNA during viral infections. Alu element could be an important player in this dynamics. Examples like c-FLIP can be taken up as case study to understand the mechanism in detail. This will help us to highlight the role of Alu in multiple transcript isoforms of a single gene with respect to functional diversity/specificity in response to stimuli.
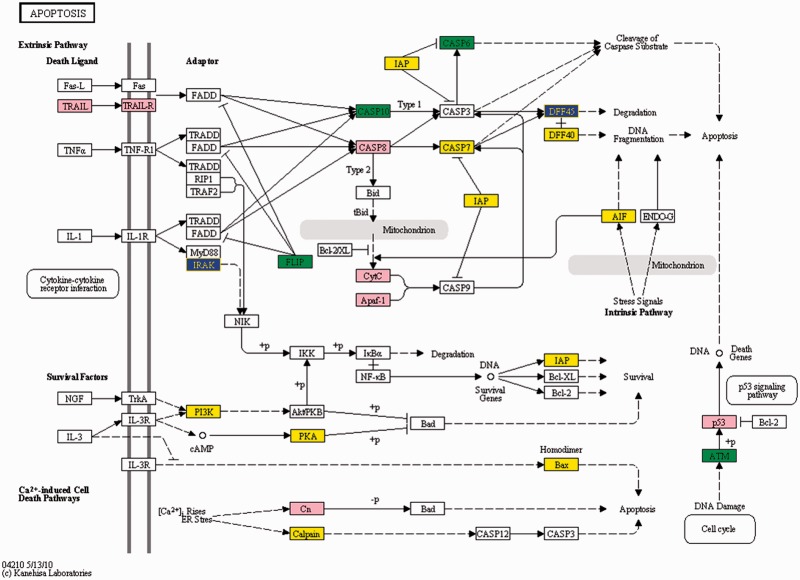
Figure 10.
Presence of a diversity of Alu-mediated events in genes involved in apoptosis. Genes identified for Alu-mediated events have been overlaid on the Apoptosis pathway (KEGG database). The colours represent different status of the genes; pink for presence of all the three events, yellow when Exonization only, green for exonization + antisense and blue for exonization + editing. The differential presence of these phenomena can provide an extra layer of regulation over cellular pathways.
Given the recent findings from the ENCODE project with respect to the non-coding regions of the genome (103), we believe that co-occurrence of Alu exonization, editing and antisense in the same transcript could provide additional insights into the novel primate specific regulatory switches. In conclusion, non-coding exonic Alu elements in the transcriptome can affect global responses through a unifying mechanism from specific biological processes. This concept would be important to explore during conditions of stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–10 and Supplementary Methods.
FUNDING
Funding for open access charge: Council of Scientific and Industrial Research (CSIR) (to M.M.) in the form of project (NWP-0036). Senior research fellowships from CSIR (to A.K.M. and R.P.); project assistantship from CSIR project [SIP-0006 to V.J.] is also duly acknowledged.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Walters RD, Kugel JF, Goodrich JA. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB Life. 2009;61:831–837. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasler J, Samuelsson T, Strub K. Useful ‘junk': Alu RNAs in the human transcriptome. Cell Mol. Life Sci. 2007;64:1793–1800. doi: 10.1007/s00018-007-7084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey R, Mukerji M. From ‘JUNK' to just unexplored noncoding knowledge: the case of transcribed Alus. Brief. Funct. Genomics. 2011;10:294–311. doi: 10.1093/bfgp/elr029. [DOI] [PubMed] [Google Scholar]
- 4.Shankar R, Grover D, Brahmachari SK, Mukerji M. Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol. Biol. 2004;4:37. doi: 10.1186/1471-2148-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez J, Vives L, Jorda M, Morales C, Munoz M, Vendrell E, Peinado MA. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De SF, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey R, Mandal AK, Jha V, Mukerji M. HSF binding in Alu repeats expands its involvement in stress through an antisense mechanism. Genome Biol. 2011;12:R117. doi: 10.1186/gb-2011-12-11-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de AA, Wang M, Bonaldo MF, Xie H, Soares MB. Genetic and epigenetic variations contributed by Alu retrotransposition. BMC Genomics. 2011;12:617. doi: 10.1186/1471-2164-12-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grover D, Kannan K, Brahmachari SK, Mukerji M. ALU-ring elements in the primate genomes. Genetica. 2005;124:273–289. doi: 10.1007/s10709-005-3086-8. [DOI] [PubMed] [Google Scholar]
- 11.Grover D, Majumder PP, Rao B, Brahmachari SK, Mukerji M. Nonrandom distribution of alu elements in genes of various functional categories: insight from analysis of human chromosomes 21 and 22. Mol. Biol. Evol. 2003;20:1420–1424. doi: 10.1093/molbev/msg153. [DOI] [PubMed] [Google Scholar]
- 12.Corvelo A, Eyras E. Exon creation and establishment in human genes. Genome Biol. 2008;9:R141. doi: 10.1186/gb-2008-9-9-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gal-Mark N, Schwartz S, Ram O, Eyras E, Ast G. The pivotal roles of TIA proteins in 5' splice-site selection of alu exons and across evolution. PLoS Genet. 2009;5:e1000717. doi: 10.1371/journal.pgen.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G. Intronic Alus influence alternative splicing. PLoS Genet. 2008;4:e1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 17.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS One. 2009;4:e5761. doi: 10.1371/journal.pone.0005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu M, Li L, Sun Z. Transposable element fragments in protein-coding regions and their contributions to human functional proteins. Gene. 2007;401:165–171. doi: 10.1016/j.gene.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Dagan T, Sorek R, Sharon E, Ast G, Graur D. AluGene: a database of Alu elements incorporated within protein-coding genes. Nucleic Acids Res. 2004;32:D489–D492. doi: 10.1093/nar/gkh132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: en route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XH, Chasin LA. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl Acad. Sci. USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y. Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl Acad. Sci. USA. 2011;108:2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panning B, Smiley JR. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology. 1994;202:408–417. doi: 10.1006/viro.1994.1357. [DOI] [PubMed] [Google Scholar]
- 25.Tang RB, Wang HY, Lu HY, Xiong J, Li HH, Qiu XH, Liu HQ. Increased level of polymerase III transcribed Alu RNA in hepatocellular carcinoma tissue. Mol. Carcinog. 2005;42:93–96. doi: 10.1002/mc.20057. [DOI] [PubMed] [Google Scholar]
- 26.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Conley AB, Miller WJ, Jordan IK. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet. 2008;24:53–56. doi: 10.1016/j.tig.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Kim DS, Hahn Y. Human-specific antisense transcripts induced by the insertion of transposable element. Int. J. Mol. Med. 2010;26:151–157. [PubMed] [Google Scholar]
- 29.Quere R, Manchon L, Lejeune M, Clement O, Pierrat F, Bonafoux B, Commes T, Piquemal D, Marti J. Mining SAGE data allows large-scale, sensitive screening of antisense transcript expression. Nucleic Acids Res. 2004;32:e163. doi: 10.1093/nar/gnh161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 31.Barak M, Levanon EY, Eisenberg E, Paz N, Rechavi G, Church GM, Mehr R. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–6915. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osenberg S, Paz YN, Safran M, Moshkovitz S, Shtrichman R, Sherf O, Jacob-Hirsch J, Keshet G, Amariglio N, Itskovitz-Eldor J, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS One. 2010;5:e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011;10:3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blankenberg D, Von KG, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996-2010. [Google Scholar]
- 42.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 43.Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riggins GJ, Strausberg RL. Genome and genetic resources from the Cancer Genome Anatomy Project. Hum. Mol. Genet. 2001;10:663–667. doi: 10.1093/hmg/10.7.663. [DOI] [PubMed] [Google Scholar]
- 46.Strausberg RL, Buetow KH, Emmert-Buck MR, Klausner RD. The cancer genome anatomy project: building an annotated gene index. Trends Genet. 2000;16:103–106. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- 47.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 50.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 52.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 53.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders Simon . HTSeq: Analysing High-Throughput Sequencing Data with Python. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hannon Gordon . FASTX-Toolkit: FASTQ/A Short-Reads Pre-processing Tools. 2009. [Google Scholar]
- 57.Andrews Simon . FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. [Google Scholar]
- 58.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 64.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 65.Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, Peng H, Rauscher FJ, III, Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 2011;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 67.Yi P, Zhang W, Zhai Z, Miao L, Wang Y, Wu M. Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett. 2003;534:61–68. doi: 10.1016/s0014-5793(02)03778-x. [DOI] [PubMed] [Google Scholar]
- 68.Mishra RR, Chaudhary JK, Bajaj GD, Rath PC. A novel human TPIP splice-variant (TPIP-C2) mRNA, expressed in human and mouse tissues, strongly inhibits cell growth in HeLa cells. PLoS One. 2011;6:e28433. doi: 10.1371/journal.pone.0028433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rudinger-Thirion J, Lescure A, Paulus C, Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc. Natl Acad. Sci. USA. 2011;108:E794–E802. doi: 10.1073/pnas.1103698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monte SM, Ghanbari K, Frey WH, Beheshti I, Averback P, Hauser SL, Ghanbari HA, Wands JR. Characterization of the AD7C-NTP cDNA expression in Alzheimer's disease and measurement of a 41-kD protein in cerebrospinal fluid. J. Clin. Invest. 1997;100:3093–3104. doi: 10.1172/JCI119864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc. Natl Acad. Sci. USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 73.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 74.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerber A, O'Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 76.Maas S. Posttranscriptional recoding by RNA editing. Adv. Protein Chem. Struct. Biol. 2012;86:193–224. doi: 10.1016/B978-0-12-386497-0.00006-2. [DOI] [PubMed] [Google Scholar]
- 77.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 78.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 79.Scadden AD, O'Connell MA. Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites. Nucleic Acids Res. 2005;33:5954–5964. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osenberg S, Dominissini D, Rechavi G, Eisenberg E. Widespread cleavage of A-to-I hyperediting substrates. RNA. 2009;15:1632–1639. doi: 10.1261/rna.1581809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Y, Lv J, Gui B, Yin H, Wu X, Zhang Y, Jin Y. A-to-I RNA editing alters less-conserved residues of highly conserved coding regions: implications for dual functions in evolution. RNA. 2008;14:1516–1525. doi: 10.1261/rna.1063708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 85.Dupuis DE, Maas S. MiRNA editing. Methods Mol. Biol. 2010;667:267–279. doi: 10.1007/978-1-60761-811-9_18. [DOI] [PubMed] [Google Scholar]
- 86.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 87.Wu D, Lamm AT, Fire AZ. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol. 2011;18:1094–1101. doi: 10.1038/nsmb.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawahara Y, Nishikura K. Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense-antisense duplex formation. FEBS Lett. 2006;580:2301–2305. doi: 10.1016/j.febslet.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 91.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daskalova E, Baev V, Rusinov V, Minkov I. 3'UTR-located ALU elements: donors of potential miRNA target sites and mediators of network miRNA-based regulatory interactions. Evol. Bioinform. Online. 2007;2:103–120. [PMC free article] [PubMed] [Google Scholar]
- 93.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 94.Mattson MP. Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- 95.DeLong MJ. Apoptosis: a modulator of cellular homeostasis and disease states. Ann. N. Y. Acad. Sci. 1998;842:82–90. doi: 10.1111/j.1749-6632.1998.tb09635.x. [DOI] [PubMed] [Google Scholar]
- 96.Vallender EJ, Lahn BT. A primate-specific acceleration in the evolution of the caspase-dependent apoptosis pathway. Hum. Mol. Genet. 2006;15:3034–3040. doi: 10.1093/hmg/ddl245. [DOI] [PubMed] [Google Scholar]
- 97.da Fonseca RR, Kosiol C, Vinar T, Siepel A, Nielsen R. Positive selection on apoptosis related genes. FEBS Lett. 2010;584:469–476. doi: 10.1016/j.febslet.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 98.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar R, Herbert PE, Warrens AN. An introduction to death receptors in apoptosis. Int. J. Surg. 2005;3:268–277. doi: 10.1016/j.ijsu.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 100.Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 101.Handa P, Tupper JC, Jordan KC, Harlan JM. FLIP (Flice-like inhibitory protein) suppresses cytoplasmic double-stranded-RNA-induced apoptosis and NF-kappaB and IRF3-mediated signaling. Cell Commun. Signal. 2011;9:16. doi: 10.1186/1478-811X-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lavrik IN, Krammer PH. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 2012;19:36–41. doi: 10.1038/cdd.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.