Abstract
Zinc-finger nucleases (ZFNs) have been used for genome engineering in a wide variety of organisms; however, it remains challenging to design effective ZFNs for many genomic sequences using publicly available zinc-finger modules. This limitation is in part because of potential finger–finger incompatibility generated on assembly of modules into zinc-finger arrays (ZFAs). Herein, we describe the validation of a new set of two-finger modules that can be used for building ZFAs via conventional assembly methods or a new strategy—finger stitching—that increases the diversity of genomic sequences targetable by ZFNs. Instead of assembling ZFAs based on units of the zinc-finger structural domain, our finger stitching method uses units that span the finger–finger interface to ensure compatibility of neighbouring recognition helices. We tested this approach by generating and characterizing eight ZFAs, and we found their DNA-binding specificities reflected the specificities of the component modules used in their construction. Four pairs of ZFNs incorporating these ZFAs generated targeted lesions in vivo, demonstrating that stitching yields ZFAs with robust recognition properties.
INTRODUCTION
Zinc-finger nucleases (ZFNs) are chimeric fusions between a programmable zinc-finger array (ZFA) and the nuclease domain of FokI (1). These artificial nucleases are powerful tools for genome modification, as they can generate a site-specific double-strand break (DSB) within the genome to promote a number of different types of genome editing (2,3). ZFNs can disrupt the function of a protein-coding gene when an imprecisely repaired DSB creates a frameshift in the coding sequence (2,3). These DSBs can also be used for the introduction of tailor-made changes to the genome by dramatically stimulating the rate of homologous recombination at a locus with an exogenously supplied donor DNA (2,3). ZFNs have been used in a variety of model and non-model organisms to facilitate reverse genetic approaches to study gene function or construct disease models for analysis (3–8). Engineered nucleases also have potential application as gene therapy-based therapeutics (9–15), where the first of these reagents are now in advanced clinical trials for treatment of AIDS (16).
The use of ZFNs has primarily been limited by the ease with which ZFAs can be created to selectively target a desired genomic region. Excluding purchase from commercial sources, selection-based approaches provide the most reliable method for creating ZFAs with novel DNA-binding specificity (17–23). Bacterial-based selection systems have somewhat simplified the process of creating ZFAs with novel specificity (24–26), but these systems still require effort on the part of end-users to generate functional constructs. Many zinc-finger proteins bind to DNA in an apparently modular fashion (27–32). Based on this supposition, a comprehensive archive of single-finger modules should enable the ready assembly of any multi-finger ZFA, where the resultant recognition site is a composite of the specificities of the incorporated finger modules (31,33–35). Using this approach, many laboratories have generated ZFAs for incorporation into ZFNs (36–43). However, success rates with such modularly assembled ZFNs have typically been modest (<30%) (39–41). The inconsistency in these systems could reflect insufficient specificity or affinity of the modules within the published archives (44), or incompatibility between the assembled fingers, which has been dubbed ‘context-dependent effects’ (17,28,31,45). The primary source of this incompatibility likely resides with mismatched residues at the finger–finger interface that degrade or alter specificity (41) in some cases because of recognition overlap between neighbouring fingers (30,46–48).
The impact of interface incompatibility can be reduced by limiting the number of unproven finger–finger interfaces that are generated during assembly (2). The use of two-finger modules for ZFA assembly reduces, but does not eliminate, the number of unproven interfaces (49–51). Unproven interfaces can be eliminated entirely through the context-dependent assembly (CoDA) of two-finger modules, where three-finger ZFAs are constructed from two-finger units with a common overlapping central finger (52). CoDA-generated ZFNs have favourable success rates (∼50%) in vivo; however, the vast majority of the modules that have been tested as ZFNs recognize NG-type (gnNGnn) dinucleotide junctions at the finger–finger interface, which are well understood and, therefore, are not the limiting interface for the expansion of ZFN targeting density. Moreover, some of the CoDA modules assigned to target non-NG junctions actually prefer NG junction sequences (50).
Recently, we described the selection of two-finger modules recognizing GRNNYG dinucleotide junctions using our bacterial one-hybrid (B1H) system (50). This approach focused on randomizing the recognition residues at the finger–finger interface in a library of two-finger modules to select optimal modules for each target sequence. The specificity of the selected modules was, subsequently, validated to identify modules with the most favourable recognition properties. Additional mutagenesis expanded the breadth of this archive to contain modules spanning 162 six base pair target sites that can be used together or in conjunction with other single-finger archives (41) for the assembly of ZFAs. This archive expands the current collection of publicly available two-finger modules, in particular those that recognize non-NG interfaces. However, their assembly into larger ZFAs remains predicated on the concatenation of modules at dinucleotide junctions that have well-defined sequence preferences (e.g. GK or AN) (50). Appropriate interface residues for many of the other dinucleotide junctions remain poorly defined. Moreover, based on our analysis of zinc-finger specificity, it seems that the appropriate choice of these recognition residues can be impacted by the identity of the residues at position +3 in each finger (Gupta and Wolfe, unpublished results), confirming the complexity in zinc-finger–DNA recognition that has been observed in other mutagenesis studies (45,53,54).
Most ZFA-assembly methods use finger archives composed of single- or multi-finger modules, where these units are delimited by the structural motif of the zinc finger. However, if finger–finger interfaces represent critical grammar for the successful assembly of functional ZFAs, then construction units wherein this most dynamic feature of recognition is fixed may yield higher success rates. This type of assembly approach has been used in the construction of three-finger ZFAs from 1.5 finger modules by Choo and colleagues (19), where an intervening phage display selection step could be used to optimize the recognition properties of the assembled proteins. We extended this approach by choosing units of assembly with fixed elements of overlap, such that an additional selection is not required, and ZFAs containing any number of fingers could be assembled if complementary monomeric units were present in the archive. Moreover, this method allows the construction of hybrid fingers with novel specificity that is not present within the archive. To serve as an initial archive for this assembly approach, we selected a set of GANNAG two-finger modules that can be assembled into ZFAs either through the standard modular-assembly or via finger ‘stitching’. We demonstrated that ZFAs assembled through either method function robustly as ZFNs when assembled into nucleases. These results highlight the advantage of using defined finger–finger interfaces for the construction of artificial ZFAs.
MATERIALS AND METHODS
Animal husbandry
Zebrafish were handled according to established protocols (55) and in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines of the University of Massachusetts Medical School.
2F-library construction
The two-finger (2F) library was designed with scheme RSDNLXX XXXNLTR, using codons VNS(+5)VNS(+6) NNW(−1)NNW(+1)NNW(+2) (V: A/C/G, N:A/C/G/T, S:G/C and W:A/T) for the five randomized residues. 2F-libraries were constructed as previously described (50). Briefly, individual F1 and F2 libraries were independently constructed via cassette mutagenesis of annealed randomized oligonucleotides into pBluescript vector containing the appropriate zinc finger backbone derived from Zif268. The 2F-library was constructed from these single-finger libraries by overlapping polymerase chain reaction (PCR) assembly. This 2F-library was then ligated into the B1H expression vector 1352-omega-UV2 between unique BssHII and Acc65I restriction enzyme sites, such that the ω-subunit of the RNA polymerase is fused at the N-terminus of the two zinc fingers, and the Engrailed homeodomain follows the fingers at the C-terminus (Figure 1). After electroporation into bacterial cells, 1 × 108 cells (five times the theoretical size of the library) were plated on ten 2xYT–carbenicillin plates (150 × 15 mm) and grown at 37°C for 14 h. 1352-omega-UV2 plasmids containing the 2F-library were isolated from pooled surviving colonies and used for selections.
Figure 1.
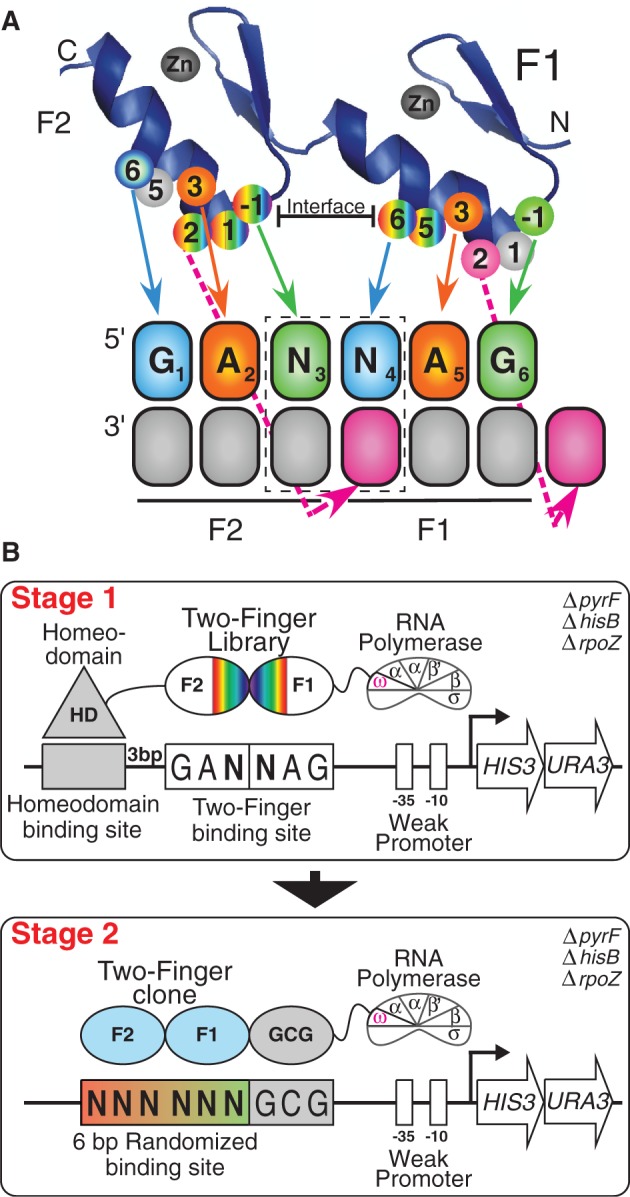
Selection of GANNAG finger sets. (A) Schematic of two-finger ZFP library used in these selections with the specificity determinants mapped to their recognition positions in their binding site. The dashed box indicates the position of the dinucleotide junction. This library contains randomized amino acids at the finger–finger interface at positions +5 and +6 of finger 1 (randomized with VNS codons) and positions −1, +1 and +2 of finger 2 (randomized with NNW codons), where the numbering scheme refers to the position of the residue relative to the start of the recognition helix. The finger 1 residues at positions −1, 1 and 2 (R, S and D) represent the N-terminal cap, and the finger 2 residues at positions 5 and 6 (T and R) represent the C-terminal cap. (B) Schematic representation of the two-stage process used to identify two-finger modules with the desired sequence preference. In Stage 1, the B1H system is used to select two-finger modules complementary to each target site. The randomized two-finger module library is fused between the DNA-binding domain of the Engrailed homeodomain and the ω-subunit of the RNA polymerase. The fixed 6-bp GANNAG target site is present on the His3/Ura3 reporter plasmid between the homeodomain binding site and the −35 box. In Stage 2, the DNA binding specificity of candidate two-finger modules obtained from the first stage of the selection are interrogated. Each two-finger module is fused to an N-terminal finger (RSDTLAR) that binds to the ‘GCG’ triplet adjacent to the 6 bp randomized zinc-finger binding region on the reporter plasmid. The recovered binding sites are determined by Illumina sequencing, and then a binding site motif is calculated from these sequences (56).
Zinc-finger binding site cloning and 2F-module B1H selections
The 16 GANNAG zinc-finger binding sites (ggccTAATTACCTGANNAGGacg) were cloned between the EcoRI and NotI sites in the pH3U3-mcs reporter vector (57). The homeodomain (Engrailed) binding site TAATTA (underlined) is present 3 bp away and on the strand opposite to the zinc-finger binding site to minimize any interference between the homeodomain and the zinc fingers. Selections for 2F-modules were performed as described previously (50). The zinc-finger library (20 ng) and the reporter vector (1 µg) containing the zinc-finger target site were co-transformed via electroporation in the selection strain that lacks endogenous expression of the ω-subunit of RNA polymerase (US0ΔhisBΔpyrFΔrpoZ). The 2 × 107 co-transformed cells were plated on selective NM minimal medium plates containing different concentrations of competitive inhibitor (3-aminotriazole; 3-AT) and inducer (isopropyl-β-d-thiogalactoside; IPTG) and grown at 37°C until a moderate number of colonies were visible. Selections were performed under different stringencies for each target site by varying 3-AT or IPTG concentration to achieve about surviving 1000 colonies per plate. In some instances, it was necessary to further increase stringency by adjusting the strength of the binding site. This was accomplished by altering either the engrailed binding site from canonical sequence TAATTA to mutant sequence TAATGC or the 6 bp 2-finger module target site from GANNAG to TANNAG. Post-selection, 2F-modules from eight surviving colonies of various sizes (typically four large, two medium and two small) were sequenced to identify functional amino acid sequences for further evaluation. The success of the selection was judged by the diversity of sequences obtained from these selections, with the expectation that successful selections will converge on a small number of functional residues at the critical recognition positions.
Cloning B1H-selected 2F modules into 3F F1-GCG constructs
To determine the binding specificities of 2F-modules, a ‘GCG’ binding anchor zinc finger (recognition helix: RSDTLAR) was fused at the N-terminus of the 2F-module via overlapping PCR. After overlapping PCR, the 3F-ZFA was cloned into 1352-omega-UV2 vector between the Acc65I and BamHI sites for expression as an omega fusion.
Constrained variation-B1H method
To determine binding site specificities of 2F-modules, the constrained variation-B1H assay was performed as described previously (56). After transformation into the selection strain, 1 × 106 cells containing the zinc-finger plasmid (1352-omega-UV2-ZFP) and the 6 bp randomized binding site library plasmid (pH3U3) were plated on selective NM minimal medium plates (100 × 15 mm) containing 50 µM of IPTG and 1 or 2 mM of 3-AT and grown at 37°C for 22–30 h. The surviving colonies were pooled, and the binding site plasmid was isolated for identification of the functional DNA sequences. The binding site region was PCR amplified, barcoded and sequenced via Illumina sequencing, and then binding specificities were determined from these data using the log-odds method (50,56,58).
Creating multi-finger ZFPs
All stitching finger ZFPs and six-finger traditional-modular-assembly ZFPs were created by gene synthesis through Genscript USA (Piscatacwa, NJ, USA) or Invitrogen (Calsbad, CA, USA). In some cases, the specificity of the two-finger module was adjusted from GDNNMG to GDNNMA by altering the residues at positions −1, 1 and 2 in the N-terminal cap from RSD to QRG (50). The traditional-modular-assembly four- or five-finger ZFPs were created based on existing six-finger ZFPs by PCR amplification of the desired finger subsets. These ZFPs were flanked by Acc65I/BamHI sites to facilitate the cloning of these ZFPs into 1352-omega-UV2 vector for B1H-binding site selection or pCS2-DD or RR vectors for creating ZFNs for activity assay.
B1H-binding site selections using the 28 bp library
The selections for 3F and 4F ZFAs were performed as previously described (41,58). The 1–5 × 107 US0 selection strain cells co-transformed with the 1352-omega-UV2 ZFA expression plasmid and the 28 bp pH3U3 library plasmid were plated on NM minimal medium selective plates lacking uracil and containing 3-AT (2.5, 5 or 10 mM) as the competitor and grown at 37°C for 36–72 h. The number of surviving bacterial colonies on each plate was estimated, and then these colonies were pooled, and the population of recovered DNA sequences was determined via Illumina sequencing. Unique sequences were ranked based on the number of recovered reads. From this list, an overrepresented sequence motif was determined with MEME (59,60) using as input the number of unique sequences from the top of the list that correspond to the estimated number of colonies on the selection plate (typically >1000).
ZFN injections and lesion analysis
For gene targeting in zebrafish, ZFAs were cloned in pCS2 vectors containing the DD/RR obligate heterodimer version of the FokI nuclease domain (61,62). pCS2–ZFN constructs were linearized with NotI, and mRNA was transcribed using the Message Machine SP6 kit from Ambion. ZFN mRNAs were injected into the blastomere of one-cell-stage zebrafish embryos as previously described (24). ZFN-injected embryos (8–30) with normal appearance and uninjected embryos were collected 24 h post-fertilization (h.p.f.) and incubated in 50 mM of NaOH (15 µl/embryo) for 15 min at 95°C to isolate genomic DNA and then neutralized with 0.5 M of Tris–HCl (4 µl/embryo). The DNA solution was centrifuged for 1 min at 13 000 r.p.m., and supernatant was taken for lesion analysis. For initial validation of ZFN activity, the region flanking the ZFN target site was amplified using the Phire Hot Start DNA Polymerase (New England Biolabs), and restriction fragment length polymorphism (RFLP) analysis or T7 Endonuclease I (or T7E1, New England Biolabs) assay was performed (39,63). For T7E1 assay, The PCR products were denatured and re-annealed using the following program: 95°C for 180 s, 85°C for 20 s, 75°C for 20 s, 65°C for 20 s, 55°C for 20 s, 45°C for 20 s, 35°C for 20 s and 25°C for 20 s with a 0.1°C/s decrease rate in between steps. This allows the hybridization of mutant and wild-type DNA strands to form duplex DNA, which then can be detected by T7E1 nuclease assay. After re-annealing, 10 μl of the products were then treated with 2U of T7E1 for 1 h in the presence of NEB buffer 2. The T7E1 treated and untreated DNA was then subjected to electrophoresis on 3% agarose gel (Ultra pure 1000, Invitrogen). The gel images were analysed using ImageJ, and lesion rate was calculated as described previously (64).
Transfection of ZFNs in HEK 293 T cells
Low passage HEK 293T cells were plated in 12-well plates overnight, such that they were 75% confluent the next day. Transfection was performed using TransIt-LT1 (Mirus Bio) according to the manufacturer’s protocol with 1 μg of total DNA made up of 40% for each ZFN and 20% of a GFP transfection-efficiency reporter. Cells were passaged once to a larger plate and harvested ∼3 days post-transfection by lysis in QuickExtract (EpiCentre) solution using 150 μl per million cells before PCR amplification of the ZFN cut sites.
LacZα blue–white assay
To further determine the types of the mutations induced by the ZFNs in the zebrafish or human cell genome, we also cloned the target fragments, such that they generate a sequence lacking stop codons in-frame with LacZα gene on pBluescript-KS(-) vector between XbaI and KpnI sites. The lengths of the target fragments cloned were between 60 and 90 bp, such that it would have minimal impact on the function of the translated LacZ peptide in the α-complementation assay using XL1-Blue Escherichia coli cell (the amplicons were chosen, such that there are no stop codons in the reading frame). The small indels induced by the ZFN through non-homologous end joining (NHEJ) pathway would disrupt the reading frame of the LacZα on the pBluescript-KS (-) vector, so that there is no functional LacZα peptide produced and consequently no active β-galactosidase. E. coli colonies harbouring such fragments seem to be white on plates containing X-gal and IPTG. On the other hand, colonies containing wild-type sequence without indels would produce active LacZα peptide and appear to be blue colonies on these plates. The identities of the sequence (or the types of the lesions) are then identified through sequencing the inserts from white colonies.
RESULTS
Selection of two-finger modules recognizing GANNAG target sites
Leveraging our previous success in selecting functional two-finger modules recognizing GRNNYG sites using the B1H system (50), we used a similar two-stage approach for the identification of modules targeting GANNAG sites (Figure 1). In the first stage, modules complementary to each of the 16 GANNAG interfaces were selected from a two-finger library that consists of ∼2 × 107 variants, where positions +5 and +6 of the N-terminal finger and positions −1, +1 and +2 of the C-terminal finger were randomized. The other recognition positions within the fingers were fixed, where Asn is present at position +3 of each finger to mediate Adenine recognition. The stringency of the selection conditions for each target site was optimized to obtain a few hundred surviving colonies on each selection plate to restrict survival to the most favourable fingers for target recognition.
The resulting pools of selected modules for the sixteen gANNAg target sites trended towards a consensus sequence (Figure 2, Supplementary Table S1). Comparison of these recovered sequences with clones recovered from the corresponding gANNCg selections (50) reveals the complexity inherent in recognition at the finger–finger interface. Fundamentally, the two-finger libraries used in these selections differed at only a single position (either Thr or Asn present at position +3 of Finger 1), yet in some cases, this led to a dramatic difference in the recovered residues at the interface positions. Some of the dinucleotide junctions, in particular those for the AN-type junctions, result in the recovery of similar finger interface residues in both the ANNA and ANNC selections. However, others, in particular those for the CN-type junctions, result in the recovery of different residue sets at the finger–finger interface from each library. This disparity alludes to the presence of context-dependent effects at the finger-finger interface.
Figure 2.

Comparison of the preferred residues recovered for the dinucleotide junctions in gANNCg (50) and gANNAg selections. In both selections, residues were selected at positions +5 and +6 of finger 1 and positions −1, +1 and +2 of finger 2. Amino acids at positions +3 of the two fingers were fixed in the library. The recovered sequences are displayed as frequency logos. The only fixed difference between the two-finger libraries used in these selections occurs at position +3 of the finger 1, which is threonine (T) in the gANNCg selections and asparagine (N) in gANNAg selections.
Identification of two-finger modules preferring each dinucleotide junction
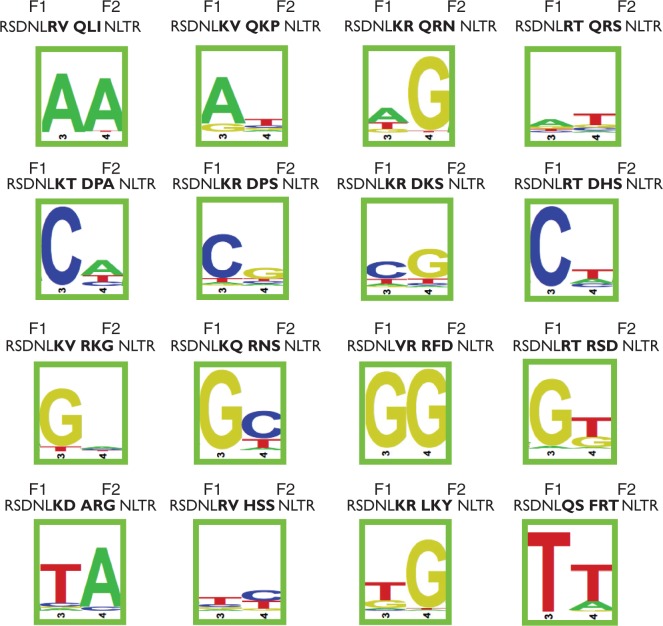
To identify two-finger modules with the most favourable recognition properties for each GANNAG dinucleotide junction, we analysed their DNA-binding specificity using the B1H system (41,57). We objectively selected a small number of clones from each recovered pool for analysis. The DNA-binding specificity of each module was characterized as a three-finger ZFA by appending a single finger recognizing a ‘GCG’ triplet to the N-terminus. In this context, each two-finger module was characterized using a reporter system containing a six base pair randomized binding site library flanking the finger 1 recognition sequence (50,56) (Figure 1B). Recovered binding sites from the randomized library for each two-finger module were pooled and then characterized by Illumina sequencing to determine the preferred recognition motif. Using this approach, we characterized 77 candidate two-finger modules to identify those with the strongest preference for each of the 16 GANNAG dinucleotide junctions (Supplementary Figure S1). Based on this analysis, we have identified modules that are compatible with each of the 16 dinucleotide junctions (Figure 3; Supplementary Table S2). For 14 of these modules, the desired dinucleotide junction is the most prevalent sequence recovered in the binding site selections, although in some cases, the preference for this sequence is only modest. For the two remaining junctions (gaACag & gaCCag), we identified modules that recognize the intended target site as the second and third most preferred site, respectively. These modules constitute our validated GANNAG archive for ZFA assembly.
Figure 3.
DNA binding specificities of the two-finger GANNAG modules with the most favourable specificity for each of the 16 different dinucleotide junctions. The DNA binding specificities were determined using B1H system (Supplementary Figure S1), and the sequence logo at the 2-bp interface is shown. Amino acid residues at positions −1 to +6 of the recognition helix of each finger are shown. The five amino acid residues recovered in the ANNA interface selection are indicated in bold font.
Demonstrating the functionality of the GANNAG two-finger modules
The validated GANNAG two-finger modules expand our existing archives (41,50) for generating ZFAs via modular assembly. To demonstrate the functionality of these new modules, we generated ZFNs via a traditional modular assembly approach for three targets (IRS2, met and sim1a) in the zebrafish genome, where each ZFA incorporates at least one new GANNAG module (Figure 4). In some cases, the specificity of the two-finger module was adjusted from GANNAG to GANNAA by altering the residues (from RSD to QRG) in the N-terminal cap (50). These targets were chosen, such that six-finger ZFAs can be constructed for each half site providing the opportunity to compare the activity of four-, five- and six-finger proteins (Supplementary Table S3). In two of these ZFAs, we used a recently described THPRAPIPKP linker between fingers from Sangamo BioSciences that allows a single base pair to be skipped between intervening modules (65).
Figure 4.
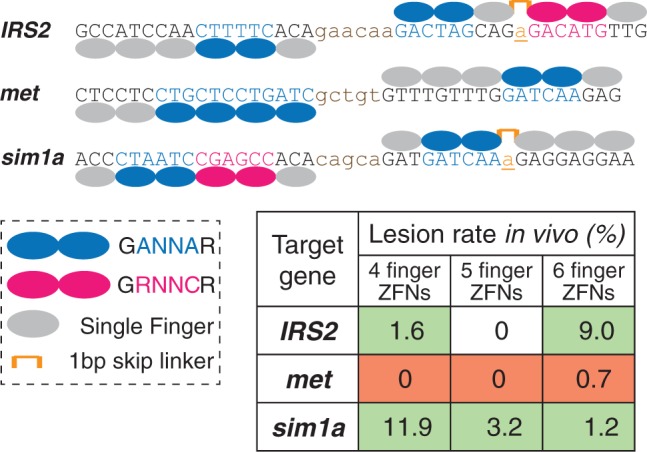
Schematic representation of the three pairs of ZFNs targeting IRS2, met and sim1a. Only the six-finger constructs are shown. Finger number was reduced by progressively removing fingers from the N-terminus of these constructs. The positions of two-finger modules (GANNAG) described herein or fingers from other archives [GRNNCG (50) and single fingers (41)] within these ZFAs are indicated, as is the position of the THPRAPIPKP linker that allows a single base pair (underlined lowercase base) to be skipped between intervening modules (65). The efficiency of lesion generation in zebrafish embryos by these ZFAs as a function of the number of fingers is indicated in the chart, where red highlights indicate the high toxicity of the met ZFNs.
ZFNs containing these ZFAs were tested as pairs of four-, five- or six-finger proteins for each target site in zebrafish embryos. Dose response curves were used to identify an optimal concentration of ZFN mRNA for each target. This initial survey revealed that all of the met ZFNs were highly toxic to embryos. Based on this analysis, each ZFN dose was calibrated to a level where ∼50% of the embryos developed normally at 24 h.p.f.. ZFN-injected normal embryos were subsequently analysed for lesions by T7 Endonuclease I (T7EI) analysis of PCR products spanning the target site (39,63) (Supplementary Figure S2). This revealed that the IRS2 and sim1a ZFNs were active, whereas the met1 ZFNs showed minimal activity (Figure 4). Interestingly the IRS2 and sim1a ZFNs displayed opposite trends with regards to activity and finger number. The IRS2 ZFNs had the greatest activity with the longer ZFAs, whereas the sim1a ZFNs displayed the opposite trend.
Stitching together ZFAs with novel DNA binding specificity
These newly characterized two-finger modules expand the archive of modules for generating ZFNs via traditional modular assembly (50). In addition these newly identified two-finger modules can serve as building blocks for a novel ZFA assembly method: finger stitching (Figure 5A). This new strategy takes advantage of a common feature of the two-finger modules targeting GANNAG interfaces. Each finger contains an asparagine (Asn) at position +3 of the recognition helix that recognizes A2 and A5 of the ‘G1A2N3N4A5G6’ 6-bp target site (Figure 1A). We envisioned that the two Asn residues might serve as bookends for the selected interface residues recognizing the dinucleotide junction N3N4 and thereby preserve their DNA binding specificity on incorporation into multi-finger proteins. Thus, in this approach, ZFAs are assembled by joining interfaces from compatible two-finger modules that share identical residues at the +3 position to create large arrays. For example, a three-finger protein recognizing a sequence GAN3N4AN6N7AG would be constructed from two two-finger modules recognizing GAN3N4AG and GAN6N7AG, where the bold A indicates the position of recognition overlap. Because our previously selected GANNCG modules also contain Asn at position +3 within the C-terminal finger of the two-finger module, these can be incorporated to generate ZFAs recognizing GAN3N4AN6N7CG target sites. In addition, we have developed an alternate N-terminal cap that is specific for adenine (50); consequently, the 3′-most base recognized by a stitched array can be G or A depending on the N-cap that is used. Similarly, the finger stitching can be used to create ZFPs of any desired length by extending the array through the overlap of additional modules off either terminus.
Figure 5.
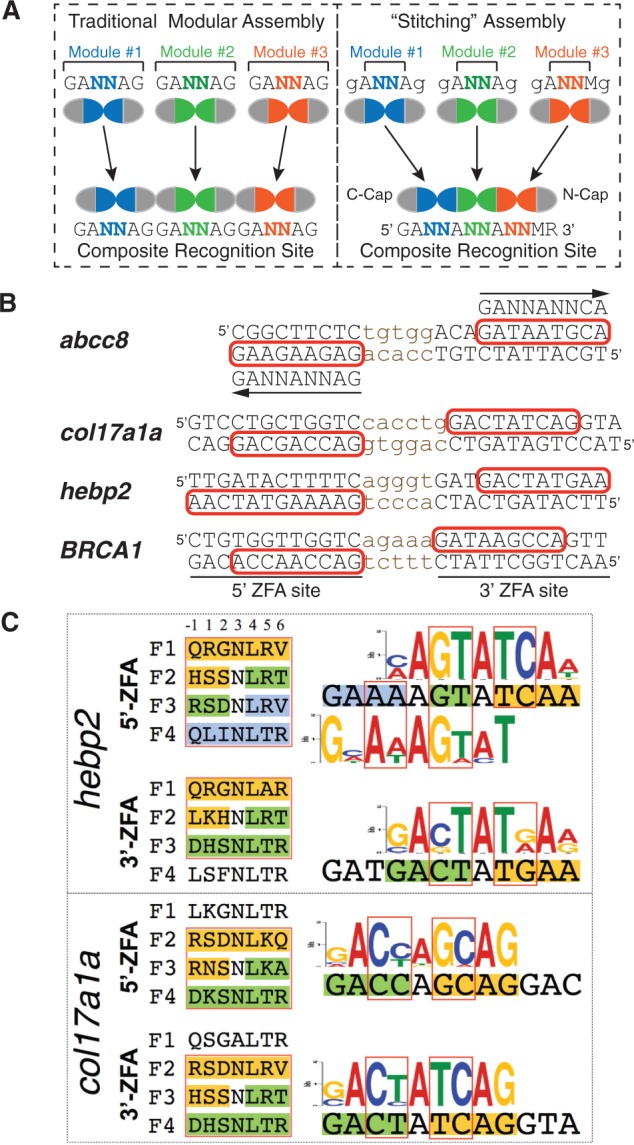
‘Finger stitching’ for ZFA assembly using GANNAG modules. (A) Schematic comparison of the traditional modular assembly approach (left) with our stitching approach (right). Instead of assembling whole finger units, stitching assembles segments between the +3 positions of the neighbouring recognition helices and then caps these at the N- and C- terminus (N-cap and C-cap). Two additional components extend the targetable sequences: the ability to incorporate GANNCG modules as the last unit in a stitched array, and the use of an alternate N-cap specific for a 3′ adenine (50), which allows the final specified base to be either G or A depending on the choice of the N-cap. (B) Target sites for the four pairs of ‘stitched’ ZFNs, where the binding site for each monomer is indicated in capital letters and the recognition element of the three or four finger stitched ZFA is boxed in red on the primary recognition strand. In some cases, a ZFA contains three stitched fingers and one additional single-finger module. For the abcc8 target site, the composite recognition site for the stitched portion of the array is indicated above or below the primary recognition sequence, where the arrow denotes the 5′–3′ orientation. (C) To assess the quality of ZFAs generated through this approach, we assembled three-finger stitched ZFAs spanning portions of the target site and determined their DNA binding specificities using the B1H system. The recognition helices for these fingers are indicated to the left of the target sites, where the positions of the stitched fingers are boxed in red. The segments of the stitched fingers that arise from a common 2F-module share a common colour. Likewise, the positions of the dinucleotide junctions between fingers in the recognition motifs for these fingers are boxed in red, and the subsites recognized by the stitched finger segments are differentially coloured.
To demonstrate that this novel assembly approach can create functional ZFAs and ZFNs, we chose three target genes (abcc8, col17a1a and hebp2) in the zebrafish genome containing ZFN sites that could be targeted using ‘stitched’ ZFAs, and where these target sites contain non-NG interfaces within the stitched fingers (Figure 5B). We also designed a pair of ZFNs for a human target gene, BRCA1, applying the same criteria. Genes encoding these ZFAs were generated by gene synthesis, where canonical TG(E/Q)KP linkers were used to connect all fingers in these arrays. As a first step in validating this approach, we determined the DNA binding specificities of these ZFAs in the B1H system using a randomized 28 bp library (41,50). Because our 28 bp library (∼108 unique members) can more effectively sample all possible recognition sequences for a three-finger than four-finger ZFA, we determined the DNA binding specificities of three-finger subsets for many of these ZFAs to provide a clearer assessment of their specificity. The determined DNA-binding specificities demonstrate in many cases that ZFAs assembled using the stitching method recognize their intended target sites with reasonable fidelity both at the dinucleotide junction sequences and the neighbouring adenines (Figure 5C and Supplementary Figure S3). However, there are instances where the dinucleotide preference is more degenerate in the stitched fingers than observed in the parent modules (e.g. abcc8-3p, Supplementary Figure S3), suggesting that in some cases, these assemblies are influenced by context-dependent effects.
Stitched ZFAs yield functional ZFNs
Given the favourable specificity of the stitched ZFAs, we evaluated their functionality as ZFNs. For each of the ZFNs targeting the zebrafish genomic sites, the optimal mRNA concentration was determined via a dose response curve as previously described, and ZFN activity was assessed at the optimal dose in healthy embryos at 24 h.p.f. The activity of the BRCA1 ZFNs was assessed by transfection of expression plasmids encoding these ZFNs into HEK 293T cells. Genomic DNA was harvested 64 h after transfection for lesion analysis. For all samples, lesion rates were determined by enzymatic digestion of PCR products spanning the target region (either T7EI or site-specific restriction enzyme) relative to untreated control samples. All four ZFN pairs induced lesions at frequencies between 1 and 11.4% (Table 1 and Supplementary Figures S4–S7), where the lesion sequences were consistent with the types of mutations expected for ZFN activity (Supplementary Figure S8).
Table 1.
Lesion rates for ‘stitched’ ZFNs
| Target gene | Lesion rate in vivo (%) |
|---|---|
| abcc8 | 1.1 |
| col17a1a | 11.4 |
| hebp2 | 2.8 |
| brca1* | 2.7 |
*In 293T cells.
DISCUSSION
Although ZFN technology has been successfully used in a multitude of systems for genomic modification (2,3), one of the major barriers in adoption is the need for a simple approach to generate functional ZFNs for nearly any target site. Traditional-modular-assembly of ZFAs, although becoming more facile as the quality of the finger archives improves, still suffers from either moderate success rates (39–41,51) or moderate targeting density (50). The functional assembly of these units is complicated by the influence of context-dependent effects at the finger–finger interface (17,28,31,41,45). The CoDA method described by the Zinc Finger Consortium bypasses this problem by using two separate archives of two-finger modules that share common N-terminal or C-terminal fingers, which permits the assembly of three-finger proteins through overlap at these common units (52). Although straightforward, this system is inherently limited to the creation of three-finger ZFAs, which can restrict the precision of these ZFNs in complex metazoan genomes, and assessment of its modules has focused on ZFAs recognizing NG-type junctions (Supplementary Table S4). We have sought to bypass this limitation through the development of a new assembly method wherein finger–finger interface units provide the grammar for assembly, which ensures that the finger–finger interface is always compatible. We assembled four ZFN pairs using this stitching approach focused on ZFAs that contain non-NG junctions at the finger recognition sequences. Remarkably, the specificity of the modules comprising these ZFAs on assembly was generally preserved when compared with the determined specificities of the primary two-finger modules that compose the archive. Moreover, all of these ZFNs were functional when tested in zebrafish or in human cells, demonstrating that this approach can produce ZFAs that can function in the context of a complex genome.
We believe that the success of this stitching approach stems primarily from using the +3 positions to demarcate the units of assembly. The recognition preference of residues is probably best understood for the +3 position in canonically binding fingers (54). Obviously, our stitching approach ignores potential context-dependent effects along each recognition helix that is splinted together from two different modules. For example, if one considers position +3 as a pivot point for the docking of the zinc finger within the major groove, the length and bulk of the residues at the flanking recognition positions (−1 and +6) may influence the geometry of finger binding, which could lead to sub-optimal recognition in some instances for stitched fingers. Thus, we do not expect that this new methodology will be completely free of complications, but we anticipate that it will perform favourably when compared with the traditional modular assembly approach by minimizing the incompatibility of fingers that are joined together.
Our strategy is adapted from the 1.5-finger assembly method described by Isalan et al. (19); however, their strategy focused solely on the assembly of three-finger ZFAs and used selection in many cases to generate a functional three-finger protein. Our approach does not require selection and can be adapted for the creation of ZFAs of any desired length. One limitation of our approach is the requirement for a suitable archive of two-finger modules that can be used for targeting the desired DNA sequence. Currently, our archive is limited to the GANNAG and GANNCG two-finger modules, but this set can be readily expanded through the selection of additional archives of modules or by co-opting two-finger modules with good specificity from archives that have been generated for other systems (39,52). Although the available archive for this assembly is currently somewhat sparse, subsets of stitched modules can be combined through standard modular assembly with one- and two-finger units from existing archives to broaden the sequences that can be readily targeted.
To facilitate the discovery of ZFN target sites that are accessible using this approach, we have modified our existing website for the identification of ZFN target sites within a user-input sequence element (http://pgfe.umassmed.edu/ZFPmodularsearchV2.html) to include the incorporation of stitched finger sets or subsets within a ZFA. The Web interface ranks a set of target sites based on the quality of the ZFA that can be assembled and outputs the amino acid and DNA sequence for the ZFAs to facilitate their creation through gene synthesis. This new assembly method coupled with the standard modular assembly approach increases the density of ZFN target sites in the zebrafish genome to approximately one every 110 bp, where 98% of the protein coding genes have a ZFN target site (Supplementary Table S5). The number of target sites that are accessible could be greatly expanded through the creation of additional two-finger module archives, where it should be readily feasible to generate a validated set of all 256 possible GNNNNG units allowing virtually any site to be targeted by varying the number of fingers and the spacer between ZFN binding sites.
Although our new archive of modules and our new assembly method increases the density of ZFN target sites, our zinc finger-based systems do not have the flexibility in targeting that has recently been demonstrated with the Transcription Activator-Like Effector Nuclease (TALEN)-based platform (63,66–69). However, ZFNs remain an important platform for targeted genomic editing that may have advantages over TALENs for certain applications, in particular therapeutics. Because each zinc finger recognizes three base pairs as opposed to one base pair for each TALE module (70–73), ZFNs are inherently more compact than TALENs. Thus, for nuclease-based gene therapy applications using viral delivery systems (74), ZFNs constitute a more compact cargo than TALENs, and as such, they may prove to be more amenable to use in certain settings.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–8.
FUNDING
U.S. National Institutes of Health (NIH) [GM068110 to S.A.W., HL093766 to N. Lawson and S.A.W.], [HG000249 to G.D.S.]; Breast Cancer Research Foundation and Silvian Foundation (to C.K.). Funding for open access charge: NIH [GM068110].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the other members of the Wolfe laboratory for insightful comments and discussions. They thank N. Lawson and his laboratory for their insightful advice and zebrafish husbandry training. They thank M. Noyes for his advice for improving the stringency of the B1H selections. They thank J. Zhu for her assistance with the website construction.
REFERENCES
- 1.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 3.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochiai H, Sakamoto N, Fujita K, Nishikawa M, Suzuki K, Matsuura S, Miyamoto T, Sakuma T, Shibata T, Yamamoto T. Zinc-finger nuclease-mediated targeted insertion of reporter genes for quantitative imaging of gene expression in sea urchin embryos. Proc. Natl. Acad. Sci. USA. 2012;109:10915–10920. doi: 10.1073/pnas.1202768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, et al. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat. commun. 2012;3:1017. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, et al. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 2011;108:7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merlin C, Beaver LE, Taylor OR, Wolfe SA, Reppert SM. Efficient targeted mutagenesis in the monarch butterfly using zinc finger nucleases. Genome Res. 2013;23:159–168. doi: 10.1101/gr.145599.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straimer J, Lee MC, Lee AH, Zeitler B, Williams AE, Pearl JR, Zhang L, Rebar EJ, Gregory PD, Llinas M, et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat. Methods. 2012;9:993–998. doi: 10.1038/nmeth.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NR, Rouhani FJ, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 14.Handel EM, Gellhaus K, Khan K, Bednarski C, Cornu TI, Muller–Lerch F, Kotin RM, Heilbronn R, Cathomen T. Versatile and efficient genome editing in human cells by combining zinc-finger nucleases with adeno-associated viral vectors. Hum. Gene Ther. 2012;23:321–329. doi: 10.1089/hum.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connelly JP, Barker JC, Pruett-Miller S, Porteus MH. Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol. Ther. 2010;18:1103–1110. doi: 10.1038/mt.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon P, June C. Chemokine receptor 5 knockout strategies. Current Opin. HIV AIDS. 2011;6:74–79. doi: 10.1097/COH.0b013e32834122d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 18.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 19.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nature Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal DJ, Dreier B, Beerli RR, Barbas CF. Toward controlling gene expression at will: Selection and design of zinc finger domains recognizing each of the 5 '-GNN-3 ' DNA target sequences. Proc. Natl. Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sepp A, Choo Y. Cell-free Selection of Zinc Finger DNA-binding proteins using in vitro compartmentalization. J. Mol. Biol. 2005;354:212–219. doi: 10.1016/j.jmb.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann F, Garriga-Canut M, Baumstark R, Fajardo-Sanchez E, Cotterell J, Minoche A, Himmelbauer H, Isalan M. p53 Gene repair with zinc finger nucleases optimised by yeast 1-hybrid and validated by Solexa sequencing. PloS One. 2011;6:e20913. doi: 10.1371/journal.pone.0020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamieson AC, Kim SH, Wells JA. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 24.Meng XD, Noyes MB, Zhu LHJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid Open-source engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durai S, Bosley A, Abulencia AB, Chandrasegaran S, Ostermeier M. A bacterial one-hybrid selection system for interrogating zinc finger-DNA interactions. Comb. Chem. High Throughput Screen. 2006;9:301–311. doi: 10.2174/138620706776843147. [DOI] [PubMed] [Google Scholar]
- 27.Pavletich NP, Pabo CO. Zinc Finger DNA Recognition—Crystal-Structure of a Zif268-DNA Complex at 2.1-A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 28.Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc. Natl. Acad. Sci. USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 30.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein–DNA complex refined at 1.6å: a model system for understanding zinc finger–DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 31.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, Koksch B, Lund CV, Magnenat L, Valente D, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 32.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protocols. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 33.Liu Q, Xia Z, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J. Biol. Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 34.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 35.Blancafort P, Segal DJ, Barbas CF. Designing transcription factor architectures for drug discovery. Mol. Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 36.Bibikova M, Beumer K, Trautman JK, Carroll D. enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 37.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim Y-G, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 39.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu Y, Sollu C, Meckler JF, Adriaenssens A, Zykovich A, Cathomen T, Segal DJ. Adding fingers to an engineered zinc finger nuclease can reduce activity. Biochemistry. 2011;50:5033–5041. doi: 10.1021/bi200393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandell JG, Barbas CF., 3rd Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 46.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc. Natl. Acad. Sci. USA. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5'-ANN-3' family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 49.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S, Lee MJ, Kim H, Kang M, Kim JS. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat. Methods. 2011;8:7. doi: 10.1038/nmeth0111-7a. [DOI] [PubMed] [Google Scholar]
- 52.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng C, Young ET. A single amino acid substitution in zinc finger 2 of Adr1p changes its binding specificity at two positions in UAS1. J. Mol. Biol. 1995;251:1–8. doi: 10.1006/jmbi.1995.0410. [DOI] [PubMed] [Google Scholar]
- 54.Miller JC, Pabo CO. Rearrangement of side-chains in a zif268 mutant highlights the complexities of zinc finger-DNA recognition. J. Mol. Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 55.Westerfield M. The Zebrafish Book. Eugene: University of Oregon Press; 1993. OR. [Google Scholar]
- 56.Christensen RG, Gupta A, Zuo Z, Schriefer LA, Wolfe SA, Stormo GD. A modified bacterial one-hybrid system yields improved quantitative models of transcription factor specificity. Nucleic Acids Res. 2011;39:e83. doi: 10.1093/nar/gkr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller JC. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 62.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nature Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 63.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. Engineered Zinc Finger Proteins. 2010. 649, pp. 247–256. [Google Scholar]
- 65.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 68.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug Ii RG, Tan W, Penheiter SG, Ma AC, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 71.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 72.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Atkinson H, Chalmers R. Delivering the goods: viral and non-viral gene therapy systems and the inherent limits on cargo DNA and internal sequences. Genetica. 2010;138:485–498. doi: 10.1007/s10709-009-9434-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.