Abstract
Human mitochondrial RNA polymerase, POLRMT, is required for mitochondrial DNA (mtDNA) transcription and forms initiation complexes with human mitochondrial transcription factor B2 (h-mtTFB2). However, POLRMT also interacts with the paralogue of h-mtTFB2, h-mtTFB1, which is a 12S ribosomal RNA methyltransferase required for small (28S) mitochondrial ribosome subunit assembly. Herein, we show that POLRMT associates with h-mtTFB1 in 28S mitochondrial ribosome complexes that are stable in the absence of mitochondrial transcription and distinct from transcription complexes containing POLRMT and h-mtTFB2. Overexpression of POLRMT in HeLa cells increases 12S rRNA methylation by h-mtTFB1 and reduces the steady-state levels of mtDNA-encoded proteins and respiration, apparently because of a decrease in fully assembled 55S mitochondrial ribosomes. We propose that POLRMT interacts directly with h-mtTFB1 in 28S mitochondrial ribosomes to augment its 12S rRNA methyltransferase activity, and that together they provide a checkpoint for proper 28S and 55S mitochondrial ribosome assembly. Thus, POLRMT is multi-functional, forming distinct protein complexes that regulate different steps in mitochondrial gene expression, at least one of which does not involve transcription per se. The significance of these results is discussed with regard to the mechanism and regulation of human mitochondrial gene expression and the potential multi-functionality of RNA polymerases in general.
INTRODUCTION
Mitochondria are essential eukaryotic organelles that participate in a variety of cellular functions and play a central role in energy metabolism by producing adenosine triphosphate via the process of oxidative phosphorylation. Mitochondrial DNA (mtDNA) in mammals encodes thirteen critical oxidative phosphorylation components, as well as rRNAs and tRNAs required for mRNA translation by dedicated mitochondrial ribosomes in the matrix. Mitochondrial gene expression is a complex, multi-step process that requires a plethora of protein factors, all of which are encoded in the nucleus and imported into the organelle (1). Transcription of mtDNA-encoded genes is carried out by a single-subunit, bacteriophage-related RNA polymerase, POLRMT (2–4). Although the C-terminal catalytic domain of POLRMT shares significant sequence and structural homology with bacteriophage polymerases, the N-terminal extension is less conserved and, in mammalian mitochondrial RNA polymerases, contains two pentatricopeptide repeat (PPR) domains (3,5). Studies in yeast show that the N-terminal extension of mtRNA polymerase is a binding platform for factors involved in coupling transcription to RNA processing and translation (5–9). Although the function of the N-terminal extension in mammalian POLRMT remains to be determined and is not closely related in primary sequence to that of yeast, it seems reasonable to assume it too is involved in interactions that mediate additional functions of POLRMT beyond transcription.
Consistent with the structural differences between mitochondrial and bacteriophage RNA polymerases, the former requires additional transcription factors for initiation. In humans, two proteins, h-mtTFB1 and h-mtTFB2, were originally characterized as mitochondrial transcription initiation factors and both can support transcription initiation in vitro (10,11). Interestingly, both h-mtTFB1 and h-mtTFB2 are homologous to the bacterial KsgA enzyme, a site-specific rRNA adenine N6-dimethyltransferase that dimethylates the small subunit rRNA in bacteria at a conserved stem–loop (12–14). Although both h-mtTFB1 and h-mtTFB2 interact directly with POLRMT and cooperatively regulate mitochondrial gene expression and biogenesis (10,11), their functions have diverged, with h-mtTFB2 being the primary mitochondrial transcription initiation factor and h-mtTFB1 being the primary 12S rRNA methyltransferase required for the biogenesis of the mammalian 28S mitochondrial ribosomal subunit (15–19). Although the significance of the POLRMT–h-mtTFB2 interaction during transcription initiation is clear, the relevance of the POLRMT–h-mtTFB1 interaction in vivo (11) has not been established.
Mammalian mtDNA is transcribed as polycistronic transcripts that undergo extensive processing before the liberated mRNAs are translated by a dedicated set of 55S mitochondrial ribosomes (1). Although the two rRNAs (12S and 16S) are mtDNA- encoded, all mitochondrial ribosomal proteins (MRPs) are encoded in the nucleus and imported into mitochondria, where they assemble into the small (28S) and large (39S) mitochondrial ribosomal subunits. MRPs are designated as part of the large or small subunit by the letters ‘L’ or ‘S’ followed by a number (e.g. MRPL12 or MRPS10). The biogenesis and assembly pathways of mitochondrial ribosomes in mammalian mitochondria have not been characterized fully. Furthermore, steps involved in the final maturation of the small and large ribosomal subunits and checkpoint pathways that prevent association and translation initiation by immature ribosomes, which are operative in other systems (20,21), have not been characterized in detail for mitochondrial ribosomes. Here, we report that human POLRMT, along with h-mtTFB1, associates stably with 28S mitochondrial ribosomes independently of its role in transcription, implicating it as a new factor required for mitochondrial ribosome biogenesis and function.
MATERIALS AND METHODS
Cells and antibodies
HeLa cells expressing hemagglutinin-tagged POLRMT (HA-POLRMT), as well as MRPS10 and MRPL12 knockdown HeLa cells were generated as described previously (22). ND6, MRPS10, MRPL37 and MRPL45 antibodies were purchased from Sigma. MRPS17, MRPS18B, MRPS27, MRPL12, MRPL13 and MRPL48 antibodies were purchased from ProteinTech Group. The h-mtTFB1 and h-mtTFB2 rabbit polyclonal antibodies were provided by Dr Craig Cameron. POLRMT antibody was purchased from Abcam; NDUFA9 and COII antibodies were obtained from Mitosciences.
Cross-linking, whole-cell extract preparation, immunoprecipitation and western blotting
Cellular proteins were cross-linked with dithiobis succinimidyl propionate (DSP) (Thermo Scientific) for 30 min as described previously (22). Preparation of whole-cell extracts (WCE) and the immunoprecipitation (IP) conditions were also performed as described (22). Briefly, cells were lysed in WCE buffer [40 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM ethylenediaminetetraacetic acid, 1% NP40, 1× protease inhibitor cocktail (Roche)], and 1–2 mg of total protein was used for IP with antibodies and protein G agarose (Pierce). After overnight incubation at 4°C, beads were washed four times with WCE buffer and co-precipitated proteins analysed by western blotting. Western blotting was carried out as described (22). It is noted that ‘inputs’ shown in the figures are included only to demonstrate the relative loading and that the antibodies worked in the western blots (especially in control cases where no co-IP is observed). The input protein concentration does not necessarily correspond to the protein concentration of the co-IP experiment; hence, the western signals in the co-IP lanes in the figures often do not reflect the actual fold-enrichment. The co-IP experiments are, therefore, only interpreted as a qualitative measure of an interaction.
Sucrose density gradient fractionation
Linear sucrose gradients (10–30%) were prepared in 14 × 89 mm Beckman centrifuge tubes in the buffer described in (23). Protein (3–10 mg) from whole-cell extracts was used for density centrifugation for 17.5 h at 80 000g in a Beckman L8-M ultracentrifuge. Fractions (1 ml) were collected from top to bottom and analysed by western blotting or diluted 1:3 in WCE buffer and used for IP as described previously (22).
Quantitative reverse transcriptase-polymerase chain reaction and ethidium bromide treatment
To inhibit mitochondrial transcription, cells were treated with 0.04 µg of ethidium bromide (EtBr) per 1 ml of media for 3 h. Cells were then washed twice with phosphate-buffered saline and harvested for total cellular RNA extraction. High Capacity cDNA Reverse Transcription kit (Applied Biosystems) was used for reverse transcription of 1.5–2 µg of total cellular RNA. Steady-state levels of mitochondrial transcripts were measured as described previously (22). Quantitative polymerase chain reaction (PCR) results were analysed by unpaired two-tailed student’s t-test.
Mitochondrial 12S rRNA methylation and respiration assays
The 12S rRNA methylation-sensitive primer extension assay was performed as described previously (15). Mitochondrial oxygen consumption rates were measured using an YSI 5300A Biological Oxygen Monitor. Cells were resuspended in respiration buffer (25 mM glucose, 1 mM pyruvate in phosphate-buffered saline), and oxygen consumption was recorded for 8 min, normalized to total cell protein, and plotted as the percent O2 consumed/s/mg protein in Figure 5B.
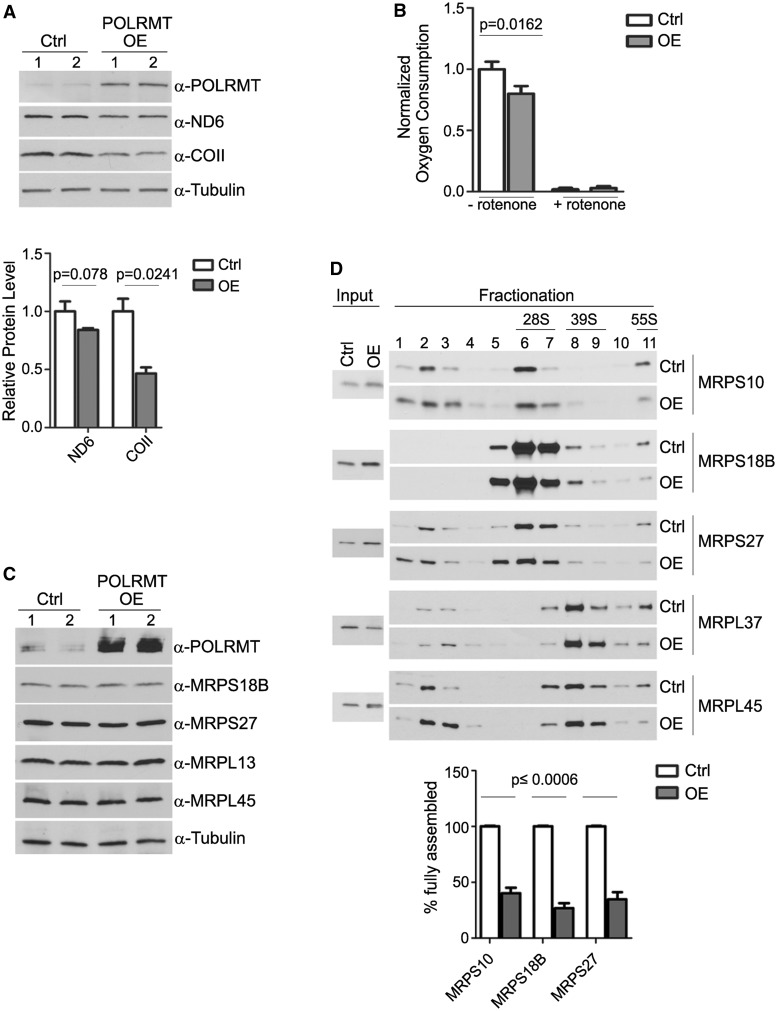
Figure 5.
POLRMT overexpression reduces the steady-state level of mtDNA-encoded proteins and mitochondrial respiration, and it perturbs 28S ribosome assembly dynamics. (A) Western blot analysis of indicated proteins (right) in cells expressing empty vector (Ctrl) or overexpressing POLRMT (POLRMT OE). Changes in ND6 and COII proteins were quantified and graphed with the ratio of each to tubulin in control cells set to a value of 1.0 (the mean ± standard deviation is shown with P-values indicated). Duplicate samples were analysed (indicated by 1 and 2). (B) Basal (-rotenone) and rotenone-inhibited (+rotenone) mitochondrial oxygen consumption rates normalized to total protein in control (Ctrl, white bars) and POLRMT overexpressing (OE, grey bars) cells. The mean ± standard deviation (n = 12) is plotted with P-values indicated. (C) Western blot analysis of indicated MRPs in control (Ctrl) and POLRMT overexpressing (OE) cells. Duplicate samples were analysed (indicated by 1 and 2). (D) Western blot analysis of small and large subunit MRPs in 10–30% sucrose density centrifugation fractions. Whole-cell extracts from empty-vector control HeLa cells (Ctrl) or HeLa cells overexpressing HA-POLRMT (OE) were used for fractionation. Fractions 6 and 7, which are enriched for small mitochondrial subunits are labelled as 28S. Antibodies used for immunoblotting are shown on the right. As an approximation of degree of 55S mitochondrial ribosome assembly, the ratio of signal of indicated small subunit MRPs in the higher-density gradient fraction 11 (which likely contains 55S ribosomes and polysomes) to the total MRP signal in all ribosome-containing fractions (fractions 5–11) is plotted (% fully assembled). The average of results from this and two other independent experiments presented in Supplementary Figure S3 is graphed below (the mean ± standard deviation and P-values are indicated). In each experiment, the ratio for each MRP in control cells was set to one.
RESULTS
Human mitochondrial RNA polymerase is associated with small mitochondrial ribosome subunit complexes containing the mitochondrial 12S rRNA methyltransferase h-mtTFB1
Human mitochondrial RNA polymerase, POLRMT, binds directly to both h-mtTFB1 and h-mtTFB2 (10). The POLRMT–h-mtTFB2 complex is generally accepted to be the primary complex involved in transcription initiation, but the nature and relevance of the POLRMT–h-mtTFB1 complex has not been established. To address this issue, we used a co-IP strategy with h-mtTFB1 and h-mtTFB2 to better define POLRMT complexes in HeLa cells. We found that POLRMT was able to efficiently co-IP with both h-mtTFB1 and h-mtTFB2 (Figure 1, lanes 3 and 4). However, h-mtTFB1 did not co-IP with h-mtTFB2 or vice versa, indicating that POLRMT forms separate complexes with each of these proteins. Although both h-mtTFB1 and h-mtTFB2 are related to rRNA methyltransferases (10–12), h-mtTFB1 is the primary mitochondrial 12S rRNA methyltransferase in mammalian cells and required for 28S mitochondrial ribosomal subunit biogenesis (13,15–17). We, therefore, considered that POLRMT might be associating with h-mtTFB1 in this context. Consistent with this hypothesis, we found that, in addition to POLRMT, proteins of the small mitochondrial ribosomal subunit were indeed co-precipitated with h-mtTFB1 (Figure 1A, lane 3). Large subunit mitochondrial ribosomal proteins did not co-IP with h-mtTFB1 (Figure 1A, lane 3), and no mitochondrial ribosomal proteins were co-immunoprecipitated with h-mtTFB2 (Figure 1A, lane 4). The specificity of these interactions was demonstrated by probing for two abundant mitochondrial proteins, heat shock protein 60 (HSP60) and NDUFA1, as negative controls, neither of which was co-immunoprecipitated with h-mtTFB1 or h-mtTFB2.
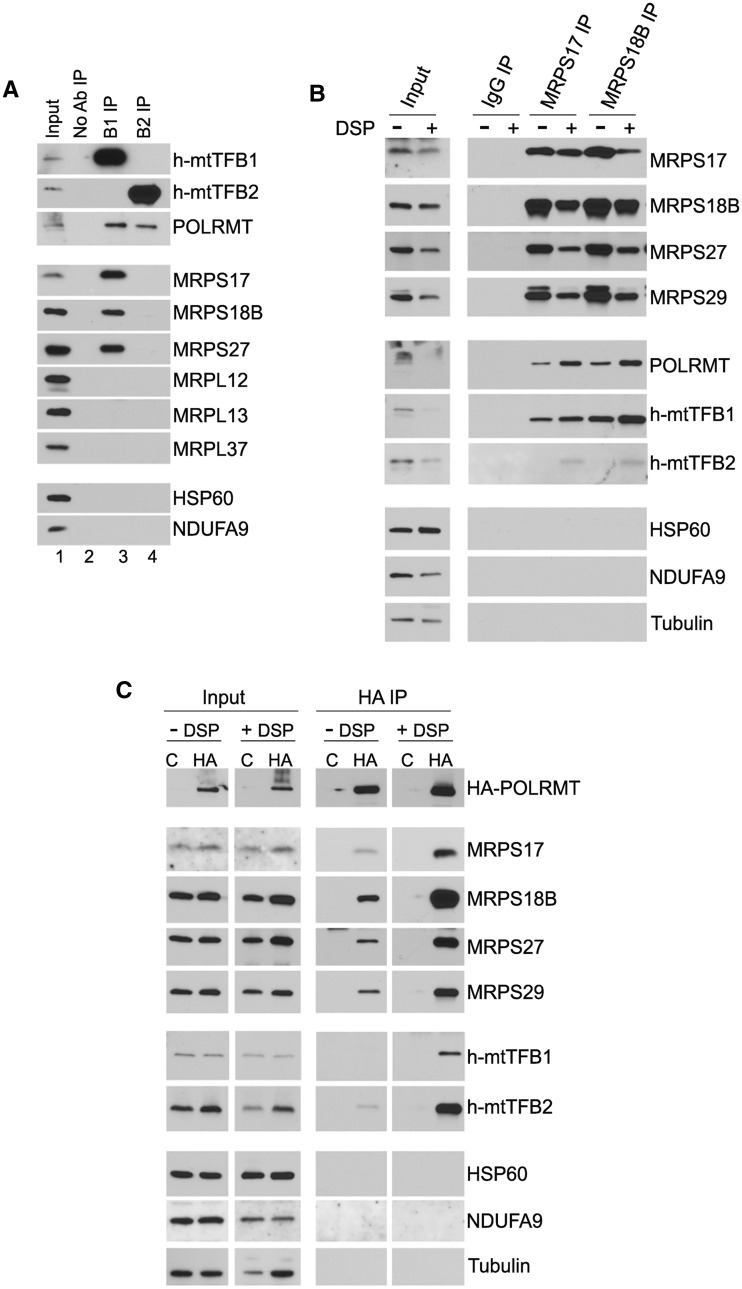
Figure 1.
POLRMT associates independently with both h-mtTFB2 and h-mtTFB1, as well as with small mitochondrial ribosomal subunit MRPs. (A) Western blot analysis of proteins co-precipitated with h-mtTFB1 (B1 IP) and h-mtTFB2 (B2 IP) from whole-cell HeLa extracts. (B) Immunoblotting of proteins co-immunoprecipitated with MRPS17 and MRPS18B from HeLa whole-cell extracts with and without DSP cross-linking (+/− DSP). IgG IP was used as a negative control. Antibodies used for analysis of input and immunoprecipitates are indicated. (C) Western blot analysis of proteins recovered in HA antibody IP from HeLa cell expressing HA-tagged POLRMT (HA) or empty vector as a negative control (C). Antibodies used for immunoblotting of input and pull-down fractions are shown.
To confirm association of POLRMT with small mitochondrial ribosomal subunits, we next performed co-IP experiments with the small mitochondrial proteins, MRPS17 and MRPS18B, in the presence or absence of DSP cross-linking to better trap transient interactions (Figure 1B). A significant amount of POLRMT was pulled down with MRPS17 and MRPS18B that also each co-IP other small ribosomal proteins (Figure 1B). Likewise, h-mtTFB1 was co-immunoprecipitated with both MRPS17 and MRPS18B, consistent with its specific interaction with 28S mitochondrial ribosomes (17). However, h-mtTFB2 did not efficiently co-IP with these small mitochondrial ribosomal proteins (i.e. only a small amount was detected in cross-linked extracts) and, therefore, was not considered significant compared with the strong interactions of POLRMT and h-mtTFB1 with 28S subunits observed in the absence of cross-linking. The specificity of these co-IP experiments was confirmed using an IgG as a negative control, which did not pull down any proteins, and by the fact that other abundant mitochondrial (HSP60 and NDUF9) and cytoplasmic (tubulin) proteins did not co-IP with any of these three MRPs even after cross-linking with DSP (Figure 1B).
We next performed the reciprocal experiment using POLRMT for IP and analysis of co-precipitated proteins. As none of the available antibodies against endogenous POLRMT that we tested were able to immunoprecipitate the protein, we used HeLa cells that overexpress hemagglutinin-tagged POLRMT (HA-POLRMT) described previously (22). When HA antibodies were used to immunoprecipitate HA-POLRMT (with or without DSP cross-linking), in addition to detecting the expected interaction with h-mtTFB2 (i.e. transcription complexes), multiple small subunit MRPs were co-immunoprecipitated from HA-POLRMT-expressing cells but not empty-vector control cells (Figure 1C), consistent with POLRMT interacting with small mitochondrial ribosomes. Importantly, similar results were obtained when sucrose gradients were performed on purified mitochondrial lysates from HA-POLRMT expressing HeLa cells (Supplementary Figure S1A). Thus, the interactions observed are not an artefact of using whole-cell lysates. Altogether, these data show that POLRMT interacts with small mitochondrial ribosomes along with its known direct-binding partner h-mtTFB1 (10,11,24).
Human mitochondrial RNA polymerase–ribosome complexes are distinct from those involved in transcription
If POLRMT, like h-mtTFB1, stably associates with small mitochondrial ribosome subunits, we predicted they would co-fractionate in sucrose gradients. To this end, we used sucrose gradient sedimentation of HA-POLRMT-expressing HeLa extracts (post–DSP-cross-linking), followed by co-IP analysis of small mitochondrial ribosome complex-containing fractions to test for interaction with POLRMT. Based on western blot analysis of sucrose gradient fractions with multiple small and large subunit mitochondrial ribosomal proteins (Figure 2A), we were routinely able to isolate fractions that contained only small ribosomal subunit complexes (e.g. fraction 7 in Figure 2A). That such fractions contained only small mitochondrial subunits was confirmed by co-IPs with MRPS18B, which pulled down other small subunit MRPs, but not any large subunit ribosomal proteins tested (Figure 2B, lane 3). Consistent with our initial, unfractionated co-IP experiments (Figure 1B), MRPS18B also co-immunoprecipitated POLRMT (Figure 2B, lane 3), clearly demonstrating that POLRMT interacts and co-fractionates with small mitochondrial ribosomal subunits. Similar results were obtained with endogenous POLRMT protein in HeLa cells that do not overexpress POLRMT (Supplementary Figure S2). Altogether, these data demonstrate that POLRMT physically associates with 28S mitochondrial ribosomes at some point during or after assembly in human mitochondria.
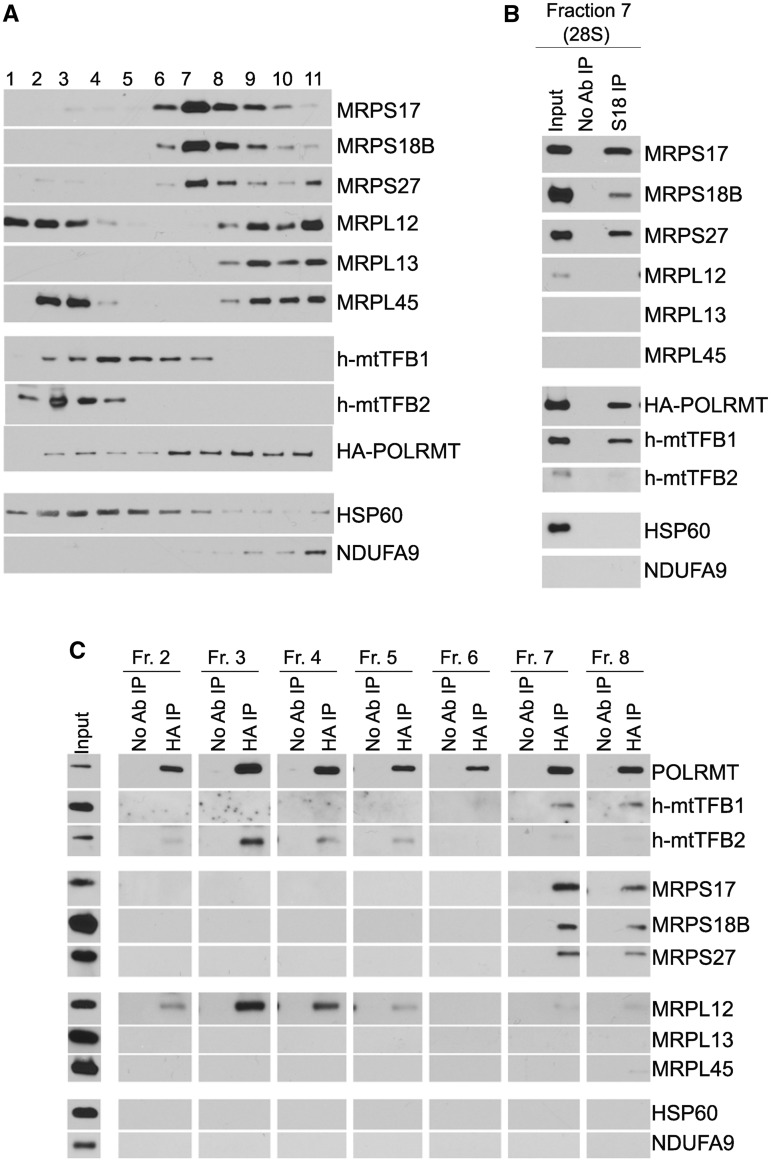
Figure 2.
POLRMT interacts with h-mtTFB1 in conjunction with small mitochondrial ribosome subunits. (A) Western blot analysis of 10–30% sucrose gradient sedimentation fractions used for immunoprecipitation in (B). HA-POLRMT HeLa cells were cross-linked with DSP before lysis and sucrose density sedimentation. Fractions numbers and antibodies used for analysis are shown. (B) Western blot analysis of proteins co-immunoprecipitated with MRPS18B (S18 IP) from sucrose gradient fraction number 7. (C) Immunoblotting of proteins recovered in IP with HA antibody (HA IP) from multiple sucrose fractions as indicated (Fr.2–Fr.8). Antibodies used for immunoblotting of co-precipitated proteins are shown. IP with no antibody added (No Ab IP) was used as a negative control. In (B and C), indicated fractions from (A) were used for immunoprecipitation.
We next performed co-IPs with POLRMT from multiple fractions of sucrose gradients to address the relationship between the POLRMT–ribosome–h-mtTFB1 complexes we identified herein to those involved in transcription. Confirming the results in Figure 2B, POLRMT co-precipitated h-mtTFB1 along with small subunit MRPs only in higher-density fractions containing small mitochondrial ribosomes (Figure 2C, fractions 7 and 8), consistent with a primary role of h-mtTFB1 in the biogenesis of the small ribosomal subunit and h-mtTFB1 and POLRMT interacting in this context. In contrast, h-mtTFB2 and free (non-ribosome associated) MRPL12, which both directly interact with POLRMT in transcription complexes (10,22,25), were associated with POLRMT only in lower-density fractions that do not contain mitochondrial ribosomes (Figure 2C, fractions 3–5). These data confirm that POLRMT forms ribosome-associated complexes with h-mtTFB1 that are distinct from transcription-associated protein complexes with h-mtTFB2 and ‘free’ MRPL12 (22).
Interaction of human RNA polymerase with small mitochondrial ribosomes occurs in the absence of active transcription
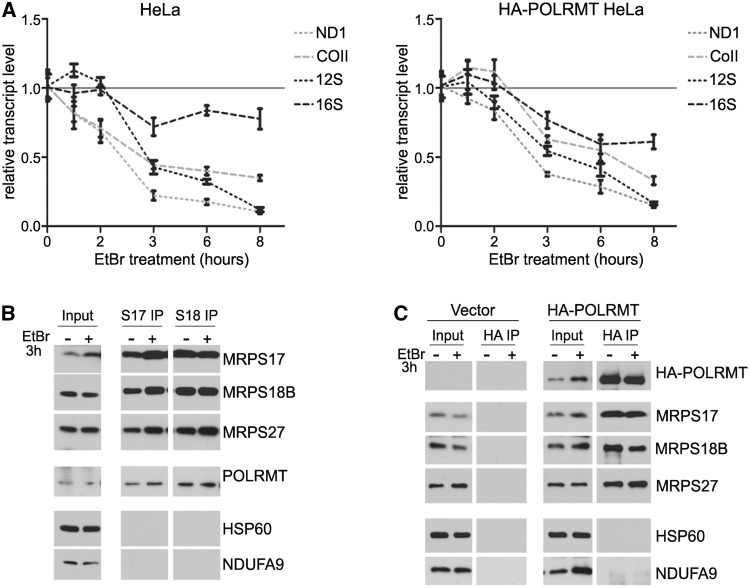
We next investigated whether the interactions between POLRMT and small mitochondrial ribosomes depends on active transcription by exposing cells to a low dosage of EtBr to inhibit mitochondrial transcription, without interfering with mitochondrial ribosome assembly or stability (26). As expected, steady-state levels of mitochondrial transcripts decreased in a time-dependent manner after drug treatment in HeLa cells expressing HA-tagged POLRMT or containing just an empty vector (Figure 3A). After 3 h of treatment, both mRNAs and rRNAs were significantly decreased, showing that transcription was successfully inhibited (Figure 3A). Therefore, at this time point, cells were lysed and used for co-IP experiments. All representative small subunit MRPs that we analysed were recovered in MRPS17 and MRPS18B co-IPs from EtBr-treated HeLa cells, confirming that 28S ribosomal subunits remained largely intact (Figure 3B). Similar amounts of endogenous POLRMT co-immunoprecipitated with MRPS17 or MRPS18B in the presence or absence of EtBr (Figure 3B). Likewise, in the reciprocal co-IP experiment, small mitochondrial ribosomes were similarly pulled down by HA antibodies in HA-POLRMT overexpressing cells in the presence and absence of EtBr (Figure 3C). These data show that the association of POLRMT with small mitochondrial ribosomes does not require active mitochondrial transcription.
Figure 3.
The interaction of POLRMT with small mitochondrial ribosomes occurs in the absence of active transcription. (A) Quantitative reverse transcriptase–PCR analysis of mitochondrial 12S and 16S rRNAs, and ND1 and COII mRNAs levels in EtBr-treated HeLa (left) and HA-POLRMT HeLa (right) cells. Time of EtBr treatment is indicated. For each transcript, the relative abundance at time zero is set to one. (B) Western blot analysis of MRPS17 (S17 IP) and MRPS18B (S18 IP) immunoprecipitates from HeLa cells with mitochondrial transcription inhibited by treatment with EtBr for 3 h. (C) Immunoblotting of proteins recovered in HA IP from empty vector control HeLa cells (Vector) or HA-POLRMT HeLa cells (HA-POLRMT) after 3 h EtBr treatment to inhibit mitochondrial transcription. In (B and C), antibodies used for detection are indicated on the right.
Human mitochondrial RNA polymerase facilitates 12S rRNA methylation by h-mtTFB1
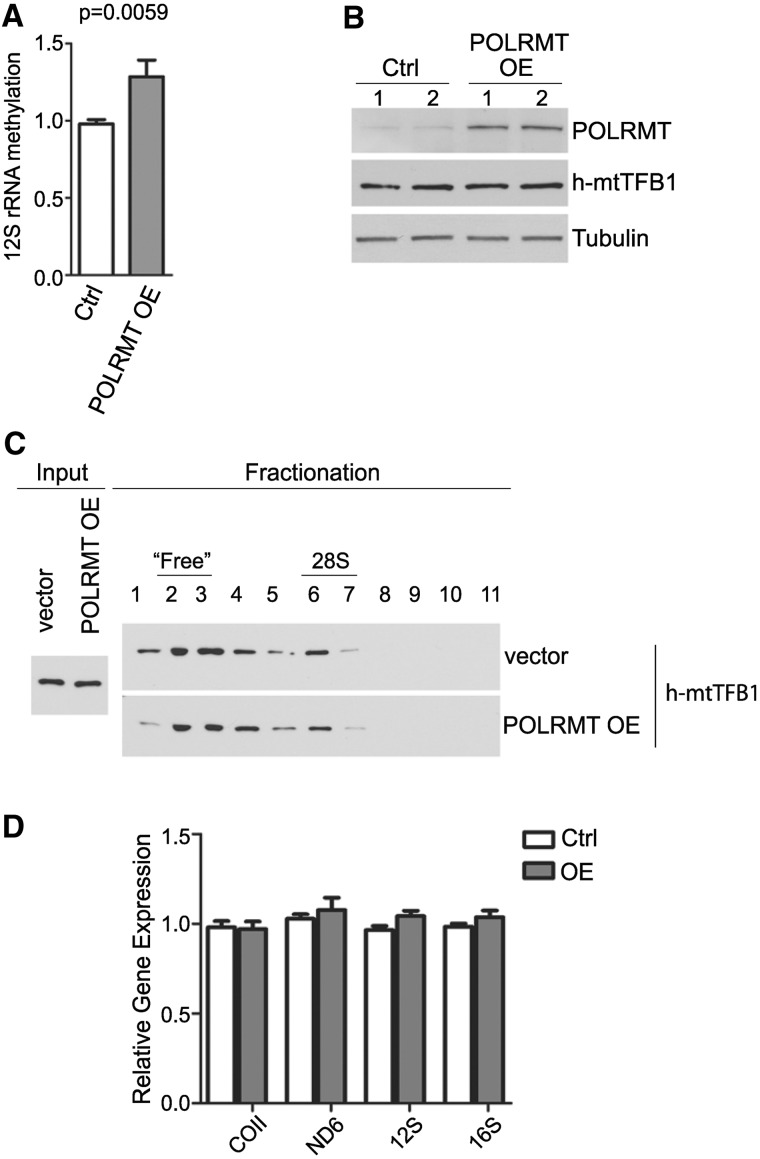
The transcription-independent co-association of POLRMT and h-mtTFB1 rRNA methyltransferase with small mitochondrial ribosomes led us to ascertain whether POLRMT can affect 12S rRNA methylation by h-mtTFB1. To do this, we again took advantage of HA-POLRMT overexpressing cells. In these cells, we found that the amount of 12S rRNA methylated at the specific stem–loop substrate by h-mtTFB1 was significantly increased (Figure 4A). This increase in methylation was not accompanied by an increase in the steady-state amount of h-mtTFB1 (Figure 4B) or an increase in its association with 28S mitochondrial ribosomes based on sucrose gradient fractionation (Figure 4C). There was also no change in the steady-state levels of mtDNA-encoded transcripts in response to POLRMT overexpression, including the 12S rRNA methylation substrate (Figure 4D). Altogether, these data indicate that POLRMT, through interaction with 28S mitochondrial ribosomes, enhances h-mtTFB1 enzymatic activity.
Figure 4.
Overexpression of POLRMT increases methylation of 12S rRNA by h-mtTFB1. (A) Quantification of methylation-specific primer extension analysis of mitochondrial 12S rRNA from control cells expressing empty vector (Ctrl) or cells overexpressing POLRMT (POLRMT OE). Shown is the methylated/unmethylated 12S rRNA ratio, which value in control cells was set to one. Mean ± standard deviation (n = 12) and t-test P-value are indicated. (B) Immunoblotting of indicated proteins in cells expressing empty vector (Ctrl) or overexpressing POLRMT (POLRMT OE). Duplicate samples were analysed (indicated by 1 and 2). (C) Western blot analysis of 10–30% sucrose density centrifugation fractions. Whole-cell extracts from empty vector control HeLa cells (vector) or HeLa cells overexpressing HA-POLRMT (POLRMT OE) were used for fractionation. Fractions numbers and fractions enriched for free MRPs and small (28S) mitochondrial ribosomes are indicated. h-mtTFB1 antibody was used for immunoblotting. (D) Steady-state levels of indicated mitochondrial transcripts in control (Ctrl) and POLRMT overexpression (OE) cells. Results from four biological replicates are shown (mean ± standard deviation). None of the transcripts analysed showed a statistically significant difference between wild-type cells and HA-POLRMT overexpressing cells.
Overexpression of human mitochondrial RNA polymerase perturbs mitochondrial ribosome assembly dynamics, and reduces mtDNA-encoded proteins and respiration
The functional consequences of overexpressing human mitochondrial RNA polymerase have not been reported to date. This could in principle affect transcription, ribosome biogenesis (based on the results detailed so far) or both. To address this, we analysed further HeLa cells that overexpress HA-tagged POLRMT. As already shown, this does not result in changes in the steady-state levels of mtDNA-encoded transcripts (Figure 4D). However, it did result in a decrease in two mtDNA-encoded proteins (ND6 and COXII; Figure 5A), and a small, but significant, reduction in basal mitochondrial oxygen consumption (Figure 5B). This suggested to us that it is the function of POLRMT in ribosomes, rather than in transcription, that was driving these latter phenotypes. To test this hypothesis, we investigated mitochondrial ribosome assembly status using sucrose density centrifugation in cell overexpressing POLRMT. As shown in Figure 5D, we were able to successfully separate the 28S mitochondrial ribosomal subunits from larger mitochondrial ribosomal assemblies (likely 55S ribosomes and/or polysomes) in these gradients. Analysis of these gradients revealed that, although the steady-state level MRPs were largely unchanged (Figure 5C), there was a disruption in mitochondrial ribosome assembly dynamics in cells overexpressing POLRMT compared with empty-vector negative control cells (Figure 5D). Quantification of this (Figure 5D, graph below) and two other independent experiments (Supplementary Figure S3) confirmed a significant decrease in the percentage of fully assembled mitochondrial ribosomes (i.e. less MRP signal in fraction 11, containing 55S and polysomes, versus lower-density fractions containing 28S and 39S subunits), suggesting that POLRMT negatively regulates 55S ribosome assembly (e.g. 28S/39S subunit joining) in human mitochondria.
DISCUSSION
The main conclusions we draw from the results of this study is that human mitochondrial RNA polymerase, POLRMT, has a transcription-independent role in the biogenesis and/or function of mitochondrial ribosomes that involves a physical and functional interaction with the h-mtTFB1 rRNA methyltransferase. Our interpretation of the specific results that lead to these conclusions is discussed later in the text. We also speculate on functions POLRMT may play in this previously unrecognized mitochondrial–ribosome context and the potential broader implications for RNA polymerase biology in terms of multi-functionality of these enzymes.
Our results provide multiple lines of evidence that POLRMT is stably associated with 28S mitochondrial ribosomes (Figures 1, 2B and 3 and Supplementary Figures S1 and S2). These complexes also contain the 12S rRNA methyltransferase h-mtTFB1, but not its transcription initiation factor paralogue h-mtTFB2 or MRPL12, a protein of the large mitochondrial ribosome we showed previously binds directly to POLRMT in its ‘free’ form to activate transcription (22,25). This separate pool of POLRMT being devoid of these transcription factors is also consistent with our results showing that its association with 28S mitochondrial ribosomes occurs independently of active transcription (Figure 3). We interpret these results to indicate that POLRMT forms multiple, distinct protein complexes that regulate different steps in mitochondrial gene expression, some of which are involved in transcription and some that are not. For example, h-mtTFB2 interacts with POLRMT in complexes that are devoid of mitochondrial ribosome components and h-mtTFB1 (Figures 1A and 2C), which almost certainly are those involved in transcription initiation, whereas POLRMT and h-mtTFB1 are involved in separate 28S mitochondrial ribosome complexes that are involved in ribosome biogenesis and function independently of transcription. The full spectrum of POLRMT complexes, which will likely involve other factors functioning in transcriptional, co-transcriptional and post-transcriptional processes will be important to define going forward.
Potential functions of POLRMT in association with 28S mitochondrial ribosomes are worthy of discussion, and our study provides some important initial insight. First, the fact that POLRMT binds h-mtTFB1 directly (10) and the POLRMT-28S mitochondrial ribosome complexes defined herein also contain h-mtTFB1, leads us to speculate that POLRMT and h-mtTFB1 are binding directly to each other in this context. That overexpression of POLRMT increases 12S rRNA methylation by h-mtTFB1 (Figure 4A) is consistent with these proteins interacting to modulate this methylation reaction. This seems to be a direct or indirect effect on enzymatic activity, as overexpression of POLRMT does not increase the steady-state amount or degree of small subunit association of h-mtTFB1 (Figure 4B–D). Although many possibilities exist to explain how POLRMT might modulate h-mtTFB1 methyltransferase activity, it is noteworthy that other site-specific RNA-modifying enzymes require protein co-factors or chaperones to target and modify their specific substrates. A salient example is the mitochondrial RNA methyltransferase NSUN4, which requires the mitochondrial transcription termination factor orthologue, MTERF4, to be targeted to its 16S rRNA substrate in the large mitochondrial ribosome subunit (27). By analogy, we speculate that POLRMT is serving a similar function in the small mitochondrial ribosome subunit with regard to the h-mtTFB1 rRNA methyltransferase. It is tempting to speculate further that h-mtTFB1 and POLRMT together are part of a checkpoint pathway for 28S subunit assembly that prevents its binding to the 39S subunit before being fully formed, which if allowed to occur could lead to formation of defective 55S ribosomes and aberrant translation. A similar checkpoint function has been proposed for the bacterial rRNA methyltransferase, KsgA (28), the homologue of h-mtTFB1, and for mouse mtTFB1 (17), and it is consistent with data from bacteria that this methylation event occurs during small ribosome subunit assembly (28). Therefore, we propose that 12S rRNA methylation occurs during the assembly of the human 28S mitochondrial ribosome, and that POLRMT both facilitates this reaction by h-mtTFB1 and then remains stably associated with the 28S ribosome to block its interaction with the 39S subunit until the 28S assembly is complete. Enhancing this ‘blocking’ function could explain the apparent disruption in 55S assembly we observe in cells overexpressing POLRMT (Figure 5D). It has been reported that a common polymorphism in the TFB1M gene (encoding h-mtTFB1) is associated with risk for type-2 diabetes (29). This certainly increases the stake in understanding how this methylation event in mitochondria is regulated and causally involved in disease pathogenesis, and whether its interaction with POLRMT is important in this regard.
It is clear at this point that various steps of mitochondrial gene expression might be functionally and physically linked and tightly coordinated, as there are several examples of multi-functional mitochondrial factors involved in transcription initiation and downstream gene expression events. For example, members of mTERF family of transcription termination factors have been implicated in regulation of transcription initiation (30–33), although their interaction with the core mitochondrial transcription machinery remains unclear. In addition, ‘free’ (non-ribosome associated) mitochondrial ribosomal protein MRPL12 selectively associates with POLRMT to modulate mitochondrial transcription, perhaps by facilitating the transition from initiation to elongation (22,25). Finally, LRP130/LRPPRC, a PPR domains containing protein implicated in post-transcriptional processes, such as mitochondrial transcript stability, polyadenylation and translation (34–39), also interacts with POLRMT and regulates mitochondrial transcription in vitro and in vivo (40,41). Our study demonstrates that POLRMT is also a multi-functional protein and central to complex interactions between the mitochondrial transcription machinery, post-transcriptional processing factors and mitochondrial ribosomes, which will certainly be important to consider going forward in attempts to fully understand how mitochondrial gene expression is regulated in mammals and is involved in human disease and aging.
Finally, the elucidation of a transcription-independent role for mitochondrial RNA polymerase may have implications beyond the mitochondrial system. This may hold significance in bacteriophage systems, whose RNA polymerases are homologous to mitochondrial RNA polymerases (42). Furthermore, the possibility that RNA polymerase may be involved in bacterial ribosome biogenesis via interaction with KsgA homologues seems likely based on the results of our study. Finally, ribosome biogenesis in eukaryotes occurs in the nucleolus and requires multiple RNA polymerases. We speculate that transcription-independent functions for these RNA polymerases may likewise exist to regulate the complex process of ribosome biogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
National Institutes of Health [R01 HL-059655 to G.S.S.] and [F32 GM-093590 to Y.V.S]. Funding for open access charge: Department of Pathology funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Craig Cameron for providing antibodies used in this study, Sophia Chen for experimental assistance, Dr Nuno Raimundo for helpful discussions and advice on the manuscript and Dr Jon Morrow for use of the ultracentrifuge.
REFERENCES
- 1.Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ. Mol. Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiranti V, Savoia A, Forti F, D’Apolito MF, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 3.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 4.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc. Natl Acad. Sci. USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–440. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 11.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 13.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 14.Shutt TE, Gray MW. Homologs of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethyladenosine methyltransferase of the mitochondrial endosymbiont. Mol. Biol. Evol. 2006;23:1169–1179. doi: 10.1093/molbev/msk001. [DOI] [PubMed] [Google Scholar]
- 15.Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Matsushima Y, Adan C, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J. Biol. Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 19.Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surovtseva YV, Shutt TE, Cotney J, Cimen H, Chen SY, Koc EC, Shadel GS. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc. Natl Acad. Sci. USA. 2011;108:17921–17926. doi: 10.1073/pnas.1108852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZM, Lightowlers RN. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 2010;430:551–558. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zylber E, Vesco C, Penman S. Selective inhibition of the synthesis of mitochondria-associated RNA by ethidium bromide. J. Mol. Biol. 1969;44:195–204. doi: 10.1016/0022-2836(69)90414-8. [DOI] [PubMed] [Google Scholar]
- 27.Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, O’Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 29.Koeck T, Olsson AH, Nitert MD, Sharoyko VV, Ladenvall C, Kotova O, Reiling E, Ronn T, Parikh H, Taneera J, et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 2011;13:80–91. doi: 10.1016/j.cmet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Wenz T, Luca C, Torraco A, Moraes CT. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 32.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 33.Roberti M, Bruni F, Polosa PL, Gadaleta MN, Cantatore P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res. 2006;34:2109–2116. doi: 10.1093/nar/gkl181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Camara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2011;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu F, Addis JB, Cameron JM, Robinson BH. LRPPRC mutation suppresses cytochrome oxidase activity by altering mitochondrial RNA transcript stability in a mouse model. Biochem. J. 2012;441:275–283. doi: 10.1042/BJ20110985. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Sanosaka M, Lei S, Bestwick ML, Frey JH, Jr, Surovtseva YV, Shadel GS, Cooper MP. LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J. Biol. Chem. 2011;286:41253–41264. doi: 10.1074/jbc.M111.276121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.