Abstract
Cytochrome c oxidase (COX) is the terminal enzyme of the electron transport chain, made up of 13 subunits encoded by both mitochondrial and nuclear DNA. Subunit 4 (COX4), a key regulatory subunit, exists as two isoforms, the ubiquitous isoform 1 and the tissue-specific (predominantly lung) isoform 2 (COX4I2). COX4I2 renders lung COX about 2-fold more active compared with liver COX, which lacks COX4I2. We previously identified a highly conserved 13-bp sequence in the proximal promoter of COX4I2 that functions as an oxygen responsive element (ORE), maximally active at a 4% oxygen concentration. Here, we have identified three transcription factors that bind this conserved ORE, namely recombination signal sequence–binding protein Jκ (RBPJ), coiled-coil-helix-coiled-coil-helix domain 2 (CHCHD2) and CXXC finger protein 5 (CXXC5). We demonstrate that RBPJ and CHCHD2 function towards activating the ORE at 4% oxygen, whereas CXXC5 functions as an inhibitor. To validate results derived from cultured cells, we show using RNA interference a similar effect of these transcription factors in the gene regulation of COX4I2 in primary pulmonary arterial smooth muscle cells. Depending on the oxygen tension, a concerted action of the three transcription factors regulates the expression of COX4I2 that, as we discuss, could augment both COX activity and its ability to cope with altered cellular energy requirements.
INTRODUCTION
Cytochrome c oxidase (COX, complex IV) is the terminal enzyme of the electron transport chain (ETC) and couples electron transfer, from cytochrome c to oxygen, to the generation of a proton gradient across the inner mitochondrial membrane during oxidative phosphorylation (OxPhos). COX is composed of 13 subunits, three encoded in the mitochondria and the remaining 10 in the nucleus. The largest of the nuclear encoded subunits, COX4, is known to function as an allosteric subunit, inhibiting the enzyme at high adenosine triphosphate concentrations (1). This property of COX4 places it in a pivotal physiological position to adjust cellular energy production to demand.
COX4 genes exist as isoform 1 (COX4I1), whose precursor protein is 169 amino acids long, and isoform 2 (COX4I2), which has 171 amino acids. COX4I1 is ubiquitous in its expression, whereas COX4I2 is tissue-specific and highly expressed in lung, trachea and placenta, but can also be detected at low levels in heart and brain (2).
We recently performed a comprehensive investigation on the role of COX4I2, and analysed the effect of Cox4i2 gene knockout in mice (3). In lung, Cox4i2 knockout reduces COX activity to 50%, despite the presence of the ubiquitously expressed subunit isoform, Cox4i1. Knockout mice show a considerable (∼30%) reduction in lung energy levels. As a consequence, and because airway constriction (as occurs, for instance, during asthma) requires energy, knockout mice showed significantly reduced airway responsiveness. Furthermore, the knockout mice developed severe lung pathology and presented with Charcot–Leyden crystals (3), which are also seen in patients with chronic lung diseases. Apparently, increased COX activity mediated through COX4I2 is essential for normal function of the lung, a tissue that has the lowest mitochondrial content compared with other tissues, such as heart, liver and brain (4).
At the gene level, a distinct feature of COX4I2 is its responsiveness to cellular oxygen tension. Its expression is stimulated under low oxygen conditions. Our labs have previously attributed this to the presence of a 24 base pair conserved sequence in the proximal promoter of COX4I2 that contains an Sp1 element plus the 13-bp sequence analyzed here and termed the oxygen responsive element (ORE) (5). This ORE is highly conserved in COX4I2 promoters among rat, mouse, cow and human. To further investigate promoter regulation of COX4I2 in response to varying oxygen concentrations, we carried out a one-hybrid screen with the ORE as bait. We identified three transcription factors, two of which had not previously been associated with the ETC. These are recombination signal sequence–binding protein Jκ (RBPJ), CXXC5 and coiled-coil-helix-coiled-coil-helix domain 2 (CHCHD2).
RBPJ is a key player in the evolutionarily conserved NOTCH signaling pathway and was first isolated as a DNA binding protein from murine B-cells (6). CXXC5 is a nuclear zinc-finger protein that is not well characterized. CHCHD2 was identified as one of the genes that were co-expressed with other genes of the OxPhos pathway using a computational expression screening technique (7). In this study, we show their binding to the ORE of the COX4I2 promoter and their interaction with each other in regulating the COX4I2 promoter under the influence of the oxygen concentration. Finally, we discuss these findings with respect to the role of COX4I2 in the functioning and regulation of OxPhos related to clinical syndromes resulting from hypoxic [We use the term hypoxia, without pathological implication, to indicate a decrease in the availability of oxygen and normoxia, a return to ambient levels. Specifically, we refer to 4% as hypoxia and atmospheric 20–21% as normoxia. In general, 4% oxygen in cells is within normal levels or can be compensated by physiological adaptations (8)] stress.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney 293 cells (293 cells) were cultured in Dulbecco’s modified Eagle's medium with 10% fetal bovine serum and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA); 293-RBPJ-KD and 293-CHCHD2-KD are 293 cells with a stable knockdown of RBPJ and CHCHD2, respectively.
Effector and reporter plasmids
The minimal 118-bp COX4I2 promoter luciferase reporter plasmid has been described previously (5) and harbors the conserved ORE (Figure 2A). A mutant reporter plasmid used in this study has a 3-bp mutation in the conserved ORE of the COX4I2 promoter (Figure 4A). The proteins RBPJ and CXXC5 were expressed from the pCI-neo vector (Promega, Madison, WI, USA). These expression plasmids were constructed to express a (HIS)6-tag at the C-terminus of the expressed protein. The CHCHD2 expression plasmid was cloned in the pCI-Neo vector with a C-terminal (FLAG)3 epitope. Hypoxia inducible factor (HIF)-1α was purchased from DNASU Plasmid Repository, Tempe, AZ, USA, clone ID: HsCD00074691. pCMX-RBPJ-VP16, from Dr T. Honjo (Kyoto University, Japan), expresses a transcriptionally active form of RBPJ (9,10). All expression plasmids were purified using the EndoFree plasmid purification kit (Qiagen, Valencia, CA, USA).
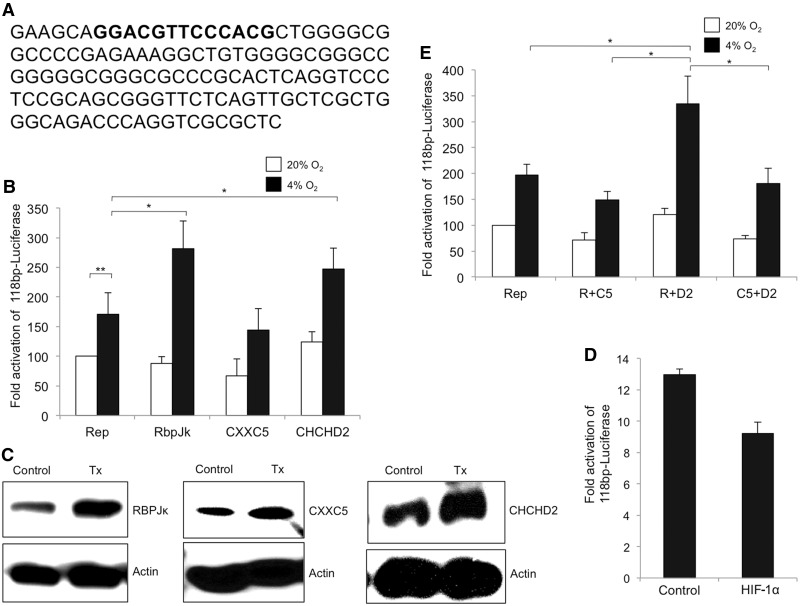
Figure 2.
Transactivation of the 118 bp minimal luciferase reporter. (A) Sequence of the 118-bp minimal ORE used in the study with the conserved ORE depicted in boldface. (B) Effect of overexpression of each of the three proteins on the ORE reporter was analysed in transient transfection assays in 293 cells. Reporter constructs and the effector plasmids were co-transfected (1 µg each). Reporter activity (Rep) observed over control at normoxia was set to 100% in this and subsequent figures. Open bars show activity at 20% oxygen and filled bars show activity at 4% oxygen in all figures. The data represents an average of at least three experiments. (C) Representative blots for overexpression of RBPJ, CXXC5 and CHCHD2 as used in reporter assays. (D) Overexpression of HIF-1α does not affect the activity of the 118 bp-conserved luciferase reporter. The 293 cells were co-transfected with the reporter and HIF-1α and maintained at 4% O2. Cells were analysed for reporter activity as described in the ‘Materials and Methods’ section. Data represents an average of three experiments. (E) Effect of co-overexpressing the three proteins on the ORE luciferase reporter. Cells were co-transfected with 1 µg each of RBPJ and CXXC5 (R+C5), RBPJ and CHCHD2 (R+D2) and CXXC5 and CHCHD2 (C5+D2) along with the 118-bp reporter. The data represent an average of at least three experiments (*P < 0.05; **P < 0.01).
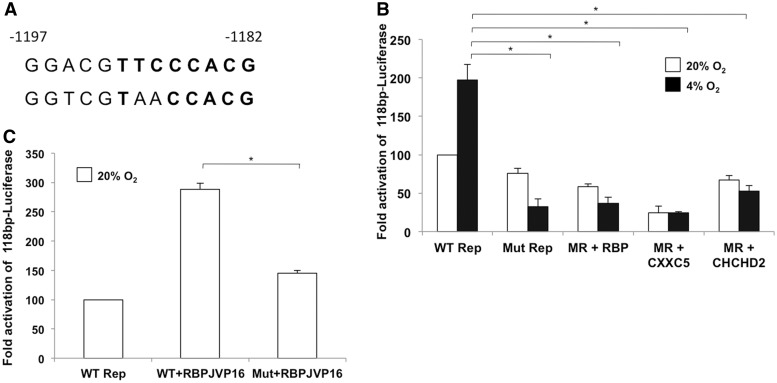
Figure 4.
A mutant ORE abolishes the hypoxic response. (A) The wild-type (top) and mutant (bottom) ORE used in this study. Two of the three mutations are in the consensus RBPJ binding site. (B) Activity of the mutant reporter in cells overexpressing the individual proteins that bind the ORE. The hypoxic effect is completely abolished in cells transfected with 1 µg each of the reporter alone or with overexpressed individual proteins. (C) Effect of RBPJ-VP16 on the mutant reporter. RBPJ-VP16 was tested for its ability to activate the mutant ORE under normoxia, a condition where this chimera activates the reporter (*P < 0.05).
Transient transfection of 293 cells
The 293 cells were transfected with the indicated plasmids using TransFast transfection reagent (Promega) following the manufacturer's protocol. Unless indicated otherwise, cells were plated onto 12-well culture plates at a concentration of 105 cells per well a day before transfection. On the day of transfection, the TransFast reagent was mixed with the indicated concentrations (µg) of DNA at a ratio of 3:1 in serum- and antibiotic-free medium. Following incubation at room temperature for 15 min, the mixture was overlaid onto the cells. The plates were incubated for 1 h at 37°C, and the cells were transferred to complete medium for further incubation.
Luciferase reporter assays
Luciferase assays were performed using the dual-luciferase reporter assay kit (Promega) per the manufacturer's instructions. In brief, cells were lysed using 250 µl of 1× Passive Lysis Buffer per well and incubated on a shaker for 15 min. The lysate was then transferred to a tube and spun down at 5000 rpm for 5 min. The supernatant was analysed for luciferase levels using an Optocomp 1 luminometer (MGM instruments, Sparks, NV, USA). Transfection efficiency was normalized to Renilla luciferase activity (pRL-SV40; a Renilla luciferase expression plasmid was co-transfected with the reporter construct).
Normoxia and hypoxia experiments
For these experiments, cells were incubated for the indicated times under 20% oxygen (traditionally referred to as normoxia) or 4% oxygen (representing hypoxia relative to 20%). To achieve hypoxic conditions, the incubation chamber was placed at 37°C and infused with CO2 and N2. The gas flow was controlled with a proOX110 gas controller (BioSpherix, Redfield, NY, USA) to achieve 4% oxygen and 5% CO2. Oxygen equilibration time (3–4 h) was accounted for in the hypoxia experiments.
RNAi assays
The 293-RBPJ-KD and 293-CHCHD2-KD were generated by transfecting 293 cells with an shRNA plasmid (Santa Cruz Biotechnology, Santa Cruz, CA, USA) followed by selection of antibiotic resistant clones. Luciferase levels are presented as the ratio observed in the knockdown cells compared with wild-type.
Real-time polymerase chain reaction
Total cellular RNA was extracted using Trizol (Invitrogen) per the manufacturer’s instructions. Complementary DNA (cDNA) was generated by reverse transcriptase polymerase chain reaction (PCR) using the ProtoScript Mu-MLV first strand cDNA synthesis kit (NEB, Ipswich, MA, USA). Transcript levels were measured by real time PCR using SYBR-green on an ABI7500 system. Real time analysis was performed by the ΔΔCt method. The primer sequences (5′→3′) used were as follows (F, forward; R, reverse). RBPJ (F-TGCATTGCCTCAGGAACAAAGGTG, R-CCACTGCTGTGAACTGGCATGAAA), CXXC5 (F-AACCCAGGCCTCTTCATTATGACC, R-TTCCTACAACTGCTGCACTGCT), CHCHD2 (F-CACACATTGGGTCACGCCATTACT, R-TTCTGGGCACACTCCAGAAACTGT) and 18SrRNA (F-AGTCCCTGCCCTTTGTACACA, R-GATCCGAGGGCCTCACTAAAC).
Immunoblotting and co-immunoprecipitation
Immunoblotting using equal protein concentrations was performed as described previously (11). Unless specified otherwise, the indicated primary antibodies were diluted 1:1000, whereas the secondary antibodies were at 1:5000. For co-immunoprecipitation, the immunoprecipitating antibody was complexed to the immunoprecipitating matrix (Santa Cruz Biotechnology) in a micro-centrifuge tube by incubating the mixture overnight at 4°C on a shaker. For immunoprecipitation of the FLAG-tagged proteins, ANTI-FLAG M2–Agarose (Sigma Aldrich, St. Louis, MO, USA) was used. The cell lysate was incubated with the matrix–antibody complex overnight at 4°C on a shaking platform. The matrix was pelleted and washed using phosphate buffered saline. Bound proteins were eluted by resuspending the matrix in 2× electrophoresis sample buffer followed by boiling for 2–3 min. After a brief spin, the supernatants were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and immunoblotted using the indicated antibody. The immunoprecipitated protein was also probed as a loading control.
DNA binding assay
DNA binding assays were performed as described (12), with a few modifications. The 293 cells were co-transfected in six-well plates with 100 ng of the 2.8-kb reporter gene construct (5), a non-specific control plasmid and 1 µg of RBPJ, CXXC5 or CHCHD2 expression plasmid. After incubation, 1% formaldehyde (v/v) was added for 10 min to induce cross-linking. The reaction was halted with 125 mM glycine for 5 min. Cells were collected and washed in ice-cold phosphate buffered saline supplemented with a protease inhibitor cocktail. The cell pellet was then resuspended in ice-cold swelling buffer and kept on ice for 10 min. Plasmid DNA was fragmented by incubation with 500 U each of Mbo I and Dpn I for 16 h at 37°C. Nuclei were lysed in lysis buffer on ice for 20 min. For immunoprecipitation, the antibody–matrix complex was prepared as described under co-immunoprecipitation. Tag-specific antibodies were used for immunoprecipitation. The nuclear lysate was diluted 1:10 with dilution buffer, and an aliquot of the antibody–matrix complex was added. The mixture was shaken overnight at 4°C. A small fraction of the nuclear lysate was saved as the input fraction. The immunoprecipitates were collected and washed sequentially at 4°C on a shaker for 5 min, once each using low-salt wash buffer, high-salt wash buffer and LiCl wash buffer, and then twice in Tris-ethylenediaminetetraacetic acid. The immunoprecipitated protein was eluted twice with elution buffer for 15 min each on a shaker at 65°C. Cross-linking was reversed by incubating the elution fraction with 200 mM NaCl at 65°C for 5 h. Proteins were digested using Proteinase K. The precipitated DNA was purified using the QIAquick purification kit (Qiagen) and amplified with primers encompassing the segment harboring the ORE. The control primers amplified a segment of the co-transfected non-specific plasmid. The input fraction was also purified and amplified using the ORE and control primers.
Reagents
The anti-CHCHD2, DRBP76 and the anti-FLAG antibodies were purchased from Proteintech Group Inc (Chicago, IL, USA) and Sigma Aldrich, respectively. COX4I2 (monoclonal) and β-tubulin antibodies were purchased from Abnova (Taipei City, Taiwan) and Cell Signaling (Danvers, MA, USA), respectively. The COX1 antibody was purchased from Abcam (Cambridge, MA, USA). All other antibodies were purchased from Santa Cruz Biotechnology.
Yeast one-hybrid screen
The bait sequence with a 3xORE [ATAAGAATGCGGCCGC 3X(GCAGGACGTTCCCACGCT) ACTAGTCCA] was cloned into pHIS3NB, constructed as described (13), driving the HIS3 selection gene. A HeLa cell cDNA library (14) in the pB42A vector was used to analyse the interactions. Positive clones were selected for growth on medium lacking histidine.
Isolation of rat pulmonary smooth muscle cells and siRNA treatment
Smooth muscle cells were isolated from pulmonary precapillary arteries and cultured as described previously (15). Briefly, rats were anesthetized intraperitoneally using ketamine (50 mg/kg) and medetomidin (100 µg/kg, Pfizer, Karlsruhe, Germany). Lungs were filled with a mixture of 5 mg/ml glow melting point agarose and 5 mg/ml Fe3O4 in medium M199 (Invitrogen) through the pulmonary artery. A magnetic holder collected pulmonary arteries containing the iron particles and agarose. Fibroblasts were disrupted by incubation at 37°C for 60 min with 80 U/ml collagenase (Sigma-Aldrich). The medial layer of the pulmonary artery attached to iron particles was suspended in M199 medium with 10% fetal calf serum (FCS, PromoCell, Heidelberg, Germany), transferred to T75 culture flasks, and incubated at 37°C with 5% CO2 in air for 4–5 days. On reaching 80% confluence, cells were trypsinized and divided into fresh T75 culture flasks. PASMC were verified by co-staining with alpha-smooth muscle actin and von Willebrand factor. Rat PASMC were used to conduct the experiments after 1–2 passages.
Selective targeting of CXXC5, RBPJ and CHCHD2 was performed using a pool of specific siRNAs. As a control, scrambled siRNA was used that do not target any gene in the rat and mouse genome. The siRNAs were commercially purchased from Invitrogen and Thermo Scientific. Transfection of siRNAs was performed in low-serum and antibiotic-free medium [1% (v/v) FCS in M199]. siRNAs (100 nM CHCHD2, CXXC5 and 200 nM RBPJ) were transfected using 1 µl X-tremeGENE siRNA Transfection Reagent (Roche, Mannheim, Germany) per cm2 of the well. Both siRNAs and the transfection reagent were diluted in OPTI-MEM medium (Gibco, Karlsruhe, Germany). After 6 h of transfection, the medium was changed to 10% FCS M199 containing antibiotics [1% (v/v) penicillin and streptomycin]. After 36 h of incubation, PASMC were transferred to a 4% O2 cell incubator (Thermo Scientific) for 36 h. A normoxic control was maintained at 21% O2. COX4I2 and β-tubulin levels were analysed by immunoblotting and evaluating band density with BIO-RAD Quantity One analysis software (BIO-RAD, Hercules, CA, USA).
Statistical analysis
All statistical analyses were performed using MSTAT version 5.4 (N. Drinkwater, University of Wisconsin). The Wilcoxon rank-sum test was applied to calculate the P-value for statistical significance; ** indicates P < 0.01 and * indicates P < 0.05.
RESULTS
RBPJ, CXXC5 and CHCHD2 bind the ORE in the COX4I2 promoter
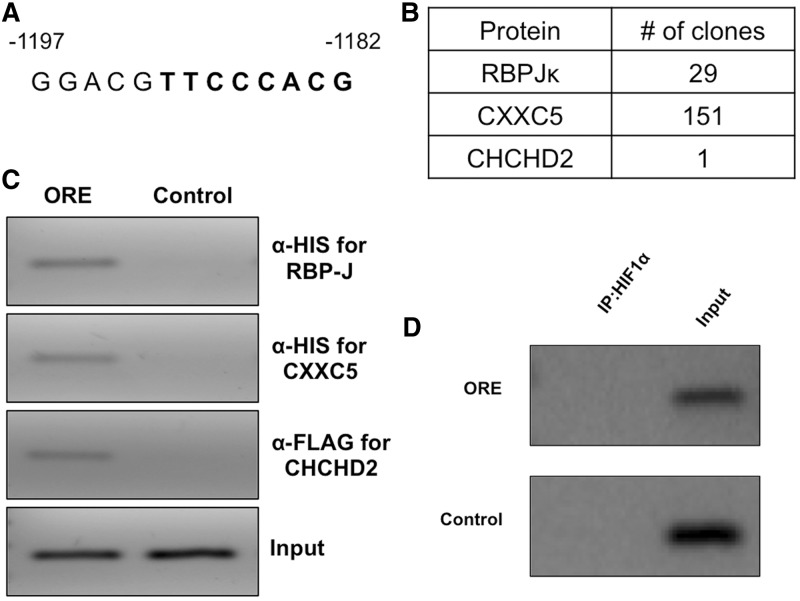
We previously identified a novel, highly conserved ORE in the COX4I2 promoter (Figure 1A). We here applied a yeast one-hybrid screen to detect transcription factors that bind this conserved ORE in the human promoter. Three binding proteins were identified from a HeLa cell yeast one-hybrid library (Figure 1B). Of the 200 clones analysed, 151 were for CXXC5, 29 for RBPJ and 1 for CHCHD2. The proteins were confirmed by a DNA-binding assay (see ‘Materials and Methods’ section) in which positive amplification was observed for immunoprecipitated DNA harboring the conserved ORE for each of the three factors from the one-hybrid screen (Figure 1C). Furthermore, HIF-1α failed to bind the conserved ORE (Figure 1D).
Figure 1.
The COX4I2 promoter ORE binds three proteins, RBPJ, CXXC5 and CHCHD2. (A) The conserved 13-bp ORE in human COX4I2 promoter upstream of exon I is shown, with coordinates from the translation start site located in exon II. Bases shown in boldface form the consensus RBPJ binding site. (B) HeLa cell yeast one-hybrid analysis was performed to identify binding proteins of the ORE. Three proteins were identified that specifically bound the bait sequence. (C) DNA binding assay was performed in 293 cells to confirm binding to the ORE of the proteins identified in the one-hybrid screen. (D) HIF-1α fails to bind the ORE in the DNA binding assay described in the ‘Materials and Methods’ section.
RBPJ and CHCHD2 function as activators, whereas CXXC5 functions as a repressor at the COX4I2 ORE
To determine the role of each of the individual proteins, 293 cells were cotransfected with the 118-bp luciferase reporter (Figure 2A) and an expression plasmid for RBPJ, CXXC5 or CHCHD2, along with pRL-SV40 Renilla luciferase for normalization of transfection efficiency. Cells were maintained at normoxia (20% oxygen), or hypoxia (4% oxygen) for 48 h, followed by a dual luciferase assay to measure reporter activity. The activity of the 118-bp reporter under hypoxia was ∼2-fold higher than that observed under 20% oxygen (Figure 2B). In cells overexpressing RBPJ under normoxia, the activity of the reporter was not significantly different than that observed in its absence. However, at 4% oxygen, the hypoxic response of the minimal ORE reporter was enhanced 3-fold in the presence of RBPJ (Figure 2B). When CHCHD2 was overexpressed, the reporter activity was enhanced both at normoxia and hypoxia compared with baseline levels. By contrast, CXXC5 had a suppressive effect on the promoter. There was a reduction in reporter activity under both normoxic (∼40% decrease) and hypoxic conditions (∼16% decrease) (Figure 2B). Increased protein levels deriving from exogenously expressed RBPJ, CXXC5 and CHCHD2 are shown (Figure 2C). We also examined the effect of HIF-1α on events at the ORE. Overexpression of HIF-1α does not enhance the minimal ORE reporter activity (Figure 2D).
To further explore these findings, the individual proteins were overexpressed in combination with each other. Co-expression of CXXC5 with RBPJ or CHCHD2 caused a reduction in the luciferase activation levels under both normoxia and hypoxia (Figure 2E). The effect was more pronounced under hypoxia, where CXXC5 caused an ∼50% reduction in the level of activation observed with RBPJ alone (Figure 2B and E). Similarly, an ∼30% reduction was observed in the hypoxic response of CHCHD2 at the ORE (Figure 2B and E). However, the activation observed when both CHCHD2 and RBPJ were co-transfected was not significantly higher than that seen after overexpression of each individual protein, suggesting the absence of an additive effect.
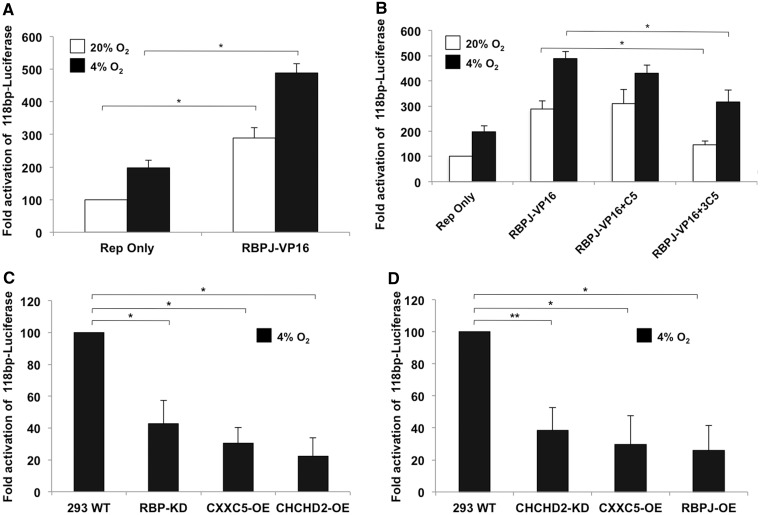
Role of RBPJ in transcriptional activation of the COX4I2 ORE
RBPJ was found to augment the hypoxic response of the COX4I2 promoter (Figure 2B and E). On binding the intracellular domain of NOTCH (NOTCH ICD), RBPJ recruits co-activators to cause transcriptional activation at promoters. Hence, a constitutively active version of RBPJ, RBPJ-VP16 was used. The chimeric protein enhanced the level of activation under both normoxic and hypoxic conditions to about 3- and 5-fold, respectively (Figure 3A). We tested whether the activation of RBPJ-VP16 was sensitive to CXXC5 under hypoxia as is RBPJ (Figure 2C) and observed reduction in the activation levels of RBPJ-VP16 in a concentration-dependent manner (Figure 3B), suggesting the mechanism of RBPJ-VP16 activation at the ORE is similar to that of RBPJ, albeit at a stronger level. A major portion of the ORE is the consensus RBPJ binding sequence. To further test the role of RBPJ on the COX4I2 ORE, we generated a cell line that stably knocks down RBPJ. In these cells under hypoxia, the minimal reporter shows an ∼50% reduction in luciferase activity compared with wild-type cells. Overexpressing CHCHD2 in these cells fails to overcome the repressive effect on the promoter. Further, overexpressing CXXC5 in these cells causes a reduction in transcriptional activation of the ORE (Figure 3C). A stable knockdown of CHCHD2 also reduces the reporter activity under hypoxia (Figure 3D). In these cells, overexpression of RBPJ failed to reverse transcriptional suppression of the reporter. Taken together, the results suggest RBPJ and CHCHD2 function as transcriptional activators, and CXXC5 functions as a repressor. Both RNA and protein levels of RBPJ and CHCHD2 are knocked down in shRNA transfected versus control cells, indicating a robust effect at both the transcript and protein level (Supplementary Figure S1).
Figure 3.
Role of RBPJ and CHCHD2 in the regulation of the COX4I2 ORE. (A) The effect of a constitutively active version of RBPJ (RBPJ-VP16) was used to dissect the role of this protein in the hypoxic activation of COX4I2. RBPJ-VP16 shows the activation of RBPJ-VP16 under 20% O2 (open bars) and 4% O2 (filled bars) conditions. (B) Effect of the repressor CXXC5 on RBPJ-VP16. CXXC5 overexpression repressed the activator effect of RBPJ-VP16 on the COX4I2 promoter. A higher concentration (3 µg of reporter construct; 3C5) was required for the inhibitory effect. (C) Reporter assay in 293 RBPJ-KD cells under hypoxic conditions. Reporter plasmid (100 ng) was transiently transfected and analysed for reporter levels. CXXC5 and CHCHD2 (100 ng) were co-transfected along with the reporter (CXXC5-OE and CHCHD2-OE). (D) Reporter assay in 293 CHCHD2-KD cells under 4% O2. Reporter plasmid (100 ng) was used in the assay along with 100 ng of CXXC5 and RBPJ (CXXC5-OE and RBPJ-OE) (*P < 0.05; **P < 0.01).
A mutation in the conserved ORE abrogates the hypoxic response of RBPJ, CXXC5 and CHCHD2
A reporter construct with three point mutations in the conserved ORE of the COX4I2 promoter (Figure 4A) was used in transient transfection assays to further examine the role of RBPJ, CXXC5 and CHCHD2 in the hypoxic response of COX4I2. The mutant construct failed to demonstrate a hypoxic response at 4% oxygen (Figure 4B). In addition, there was a ∼20% and 50% reduction in the activity of the reporter in cells overexpressing RBPJ under normoxia and hypoxia, respectively. A reduction was also observed in the level of reporter activity at 20% and 4% oxygen in cells overexpressing CXXC5 (Figure 4B). The hypoxic response was abrogated in cells transfected with CHCHD2 such that the level of reporter activity was lower under hypoxia than normoxia (Figures 2B and 4B). Transient mutant construct reporter assays were also performed on cells overexpressing RBPJ-VP16 under normoxia, where this chimeric protein functions as an activator of the wild-type ORE (Figure 3A). RBPJ-VP16 failed to activate the mutant reporter (Figure 4C), confirming that the activation function of RBPJ-VP16 parallels that of RBPJ.
Novel conserved ORE in the COX4I2 promoter is maximally active at 4% oxygen
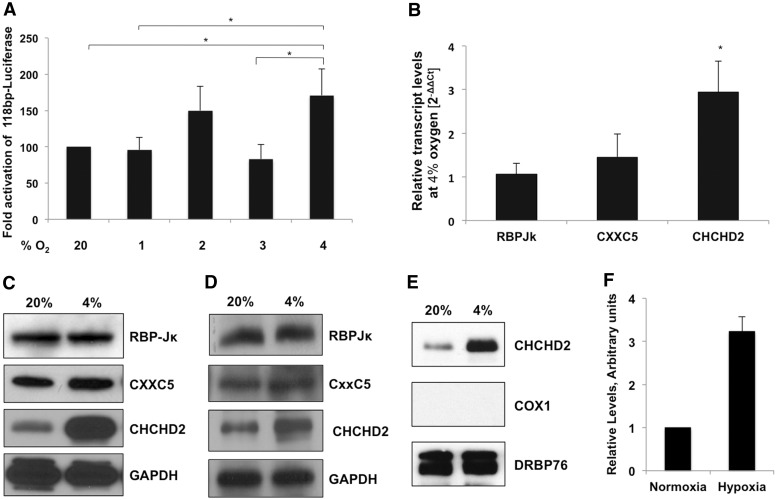
HIFs are among the best-studied transcription factors, adjusting expression for a still growing number of genes during episodes of hypoxia. To test whether the hypoxic responsiveness of the minimal ORE in the COX4I2 promoter was dependent on HIFs that generally affect gene regulation at lower oxygen tensions (e.g. ≤2%), we tested the reporter activity at 1, 2, 3 and 4% oxygen. In each case, the 118-bp reporter activity was lower than that observed at 4% oxygen (Figure 5A).
Figure 5.
The minimal ORE of the COX4I2 promoter is maximally active at 4% oxygen. (A) ORE reporter activity at varying oxygen tensions. The 293 cells were transfected with 1 µg of the reporter plasmid and maintained at the indicated oxygen tensions for 48 h, followed by analysis of reporter activity relative to normoxia. (B) Real-time PCR analysis of COX4I2 transcript levels of RBPJ, CXXC5 and CHCHD2 at 4% oxygen. (C) Endogenous protein levels of RBPJ, CXXC5 and CHCHD2 analysed by immunoblotting of whole cell lysates from cells maintained at 20% and 4% oxygen for 48 h. (D) Expression levels of the individual proteins in transfected cells maintained under normoxic and hypoxic conditions for 48 h. GAPDH was used as a loading control (*P <0 .05). (E) CHCHD2 is localized in the nucleus. Nuclear fraction of cells maintained at 20% and 4% oxygen were immunoblotted for the levels of CHCHD2. DRBP76 is the nuclear marker and COX1 is the mitochondrial marker. A representative blot has been shown. (F) Quantitation of the level of nuclear CHCHD2 at normoxia (20% O2) and hypoxia (4% O2).
To gain further insight into the observed effects, we examined the transcript and protein levels of each of the transcription factors at 20% and 4% oxygen. Only CHCHD2 showed a substantial change in the transcript levels when oxygen concentration was decreased from 20 to 4% (Figure 5B). A similar increase was seen in the protein levels of endogenous (Figure 5C) as in overexpressed CHCHD2 (Figure 5D). The levels of CHCHD2 specifically in the nuclear fraction is also increased at 4% O2 (Figure 5E and F).
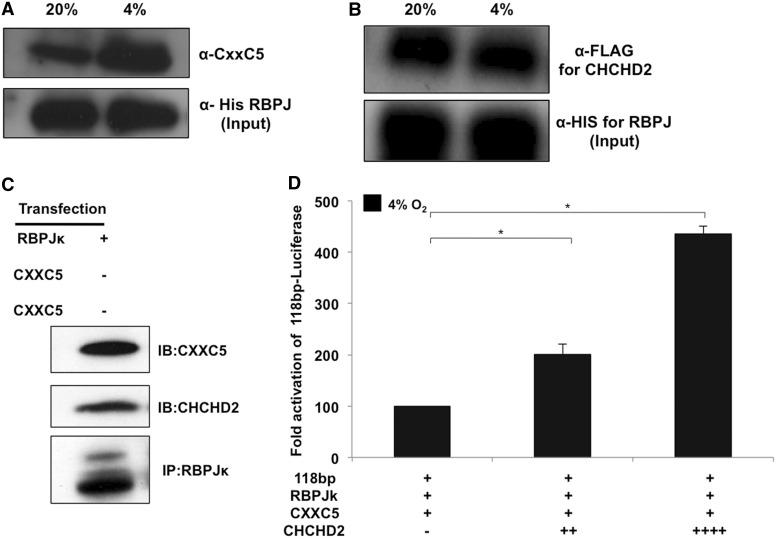
RBPJ interacts with CXXC5 and CHCHD2
To examine whether RBPJ and the other two proteins interact, co-immunoprecipitation was performed on 293 cell lysates co-transfected with an expression plasmid for RBPJ and either CXXC5 or CHCHD2. RBPJ was immunoprecipitated from cell lysates maintained at 20% and 4% oxygen for 48 h. The bound proteins were analysed by immunoblotting using the anti-CXXC5 or the anti-FLAG (for CHCHD2) antibodies. Interestingly, RBPJ interacted with both CXXC5 (Figure 6A) and CHCHD2 (Figure 6B), suggesting they act in concert in regulating the COX4I2 ORE. To further confirm the aforementioned co-immunoprecipitation findings, RBPJ was pulled down from cells transfected with an expression plasmid for RBPJ and probed for binding of endogenous CXXC5 and CHCHD2 (Figure 6C). To explore their individual roles, we next examined whether CHCHD2 reduces the inhibitory effect of CXXC5 on the ORE. We found that overexpression of CHCHD2 negates the inhibitory effect of CXXC5 under hypoxic conditions in a concentration-dependent manner (Figure 6D).
Figure 6.
CXXC5 and CHCHD2 interact with RBPJ in vivo. (A) The 293 cells were transfected with C-terminal His-tag expression plasmids for RBPJ and either CXXC5 or CHCHD2. After 48 h at 20% or 4% oxygen, RBPJ was immunoprecipitated using the anti-His-tag antibody. The eluted protein was immunoblotted and probed using anti-CXXC5 antibody. (B) Immunoprecipitation was performed as described above for cell extracts at 20% and 4% oxygen and the eluted protein was probed for CHCHD2 using the anti-FLAG antibody. The blot was reprobed using the anti-His antibody to show the input fraction. (C) RBPJ interacts with endogenous CXXC5 and CHCHD2. The 293 cells were transfected with an expression plasmid for RBPJ followed by an immunoprecipitation of the nuclear extracts. The elution fraction was probed for the presence of endogenous CXXC5 and CHCHD2. (D) CHCHD2 overrides CXXC5 mediated inhibition under hypoxia. The 293 cells were transfected with 200 ng of the CXXC5, RBPJ and the reporter plasmids along with increasing concentrations of the CHCHD2 expression plasmid. Cells were analysed for reporter activity 48 h after incubation under hypoxic conditions (*P < 0.05).
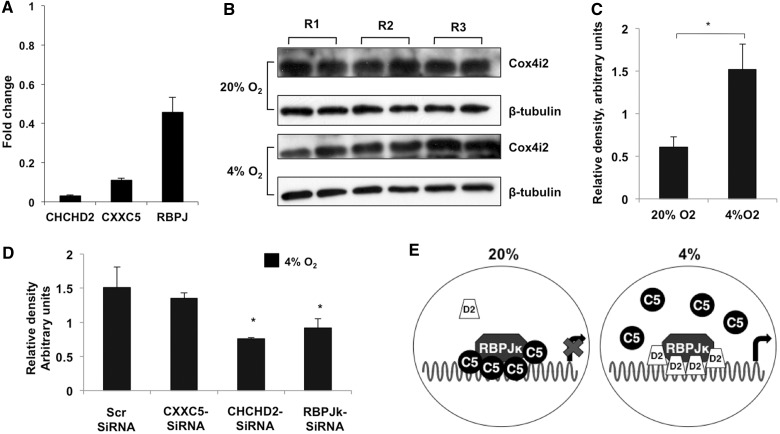
RBPJ, CXXC5 and CHCHD2 affect the levels of COX4I2 in rat primary lung cells
To extend the cell culture observations to primary rat pulmonary smooth muscle cells (PASMCs), the effect of a knockdown of the individual factors—assessed by real-time PCR (Figure 7A)—on COX4I2 expression was analysed. COX4I2 expression in cells treated with scrambled siRNA was ∼2-fold higher at 4% oxygen compared with normoxia (Figure 7B and C). To analyse the effect of the transcription factors, cells were transfected with the siRNAs, and endogenous COX4I2 expression levels were analysed under hypoxic conditions. The levels of COX4I2 were found to decrease in cells harboring siRNA for RBPJ (∼40%) and CHCHD2 (∼50%) (Figure 7D). These results obtained with PASMCs are consistent with those obtained in transient reporter assays in cultured cells (Figure 3C and D).
Figure 7.
Knockdown of RBPJ and CHCHD2 affects the hypoxic induction of COX4I2 in rat primary lung cells. (A) Real-time analysis of the level of knockdown of the individual proteins in PASMCs transfected with siRNA. Analysis was performed using the ΔΔCt method. (B) Immunoblot analysis of COX4I2 levels in primary cells isolated from three rats (R1, R2 and R3). (C) Density of the COX4I2 bands relative to β-tubulin. (D) Quantitative analysis of the COX4I2-β-tubulin ratio in cells transfected with the indicated siRNAs as detailed in the ‘Materials and Methods’ section (*P < 0.05). (E) Model for the regulation of the COX4I2 ORE. CXXC5 (C5)-bound RBPJ (left) is not able to activate transcription, whereas CHCHD2 (D2) binding to RBPJ (right) displaces C5 and activates transcription.
DISCUSSION
Most cellular oxygen is used by the mitochondrial ETC for the generation of adenosine triphosphate. Regulatory systems that can sense and respond to varying oxygen levels are of great interest, as oxygen availability, delivery and utilization are among the most basic functions in which multicellular animals engage and depend on. Much attention has focused on HIF and its network but less is known about other systems. Lung is an organ in which the need for oxygen regulation is distinct compared with other organs, as the pulmonary tissues are exposed to a significantly higher oxygen tension than others such as heart, liver, spleen, brain and the lymphatic system. The oxygen concentration of air that reaches the upper respiratory tract is ∼20 kPa (∼20% oxygen), and oxygen levels at the alveolar surface are ∼13 kPa. In contrast, tissues such as spleen, brain and the lymph nodes are exposed to an oxygen tension of ∼≤2% (16). However, certain pathophysiological conditions exists in which lungs experience oxygen tensions that significantly deviate from the normal values. For example, under physiological conditions in early development, lungs are exposed to 3–5% oxygen during embryogenesis, called physiologic hypoxia, which is necessary for normal development of mammalian embryos (17,18). Furthermore, lungs have been shown to experience oxygen levels as low as 3–5% at high altitudes (19). Pathological conditions often also cause alveolar hypoxia as seen, for example, in chronic obstructive pulmonary disease (20).
We previously identified an isoform of COX subunit 4, COX4I2, with highest expression in the bronchial and arterial smooth muscle of the lung, whose promoter is transcriptionally responsive to oxygen and shows maximum response at 4% oxygen (2,5). We have also shown that lung COX exhibits a 2–2.5 times higher activity compared with its liver counterpart that does not express COX4I2 (3). This allowed us to attribute the increase in enzyme activity to the presence of COX4I2 protein. Here, we identified three proteins that bind the conserved ORE in the COX4I2 promoter upstream of exon I, namely RBPJ, CXXC5 and CHCHD2. Each of the proteins binds the COX4I2 promoter at or in the vicinity of the ORE to regulate the response of the promoter to changes in oxygen tension.
Relatively little is currently known about the three transcription factors we identified. Only RBPJ, which plays a dual role as a transcription factor (activator and repressor) in the NOTCH signaling pathway, has previously received much attention (21–24). Hypoxia is a crucial factor in the regulation of the NOTCH signaling pathway, including stabilizing and enhancing the activity of the NOTCH ICD (25). CXXC5 was initially identified on genomic screens of modulators of Wnt (26), NFκB (27) and retinoic-acid receptor signaling pathways (28). Andersson et al. have shown CXXC5 to be a downstream effector of bone morphogenic protein 4 that belongs to the transforming growth factor beta family of signaling proteins. For CHCHD2, its silencing in a human fibroblast cell line decreased the oxygen consumption rate by ∼50% and significantly reduced the activity of COX, indicating an important role of CHCHD2 in the regulation of OxPhos (7). A homolog of CHCHD2, called MRP10, is present in yeast and serves the function of translating mitochondrial genes (29,30). It is also up-regulated in breast cancer cells exposed to estrogen and hypoxic conditions (31,32).
Our data suggest that RBPJ and CHCHD2 act in concert at the ORE to enhance the hypoxic activation of COX4I2. CXXC5, in contrast, could act by itself as an ORE repressor, an effect that was enhanced under normoxic conditions (Figure 2B). The knockdown of RBPJ attenuated the activation by CHCHD2 under hypoxic conditions and vice versa, suggesting that both RBPJ and CHCHD2 are necessary for optimal hypoxic regulation of COX4I2. However, knockdown of neither RBPJ nor CHCHD2 affect the inhibitory function of CXXC5.
Protein levels for CHCHD2 were only higher at 4% oxygen compared with 20% (Figure 5C). Under normoxic conditions, only overexpression of CXXC5 results in a significant change (∼40% reduction) in reporter activity, showing the ORE is negatively regulated by the action of CXXC5. Under reduced oxygen tension, however, there is an enhanced expression of the activator CHCHD2 that appears to overcome the inhibitory effect of CXXC5 on the ORE. Furthermore, masking of the inhibitory effect of CXXC5 is also seen by the additional activation when CHCHD2 is overexpressed (Figure 6D).
To validate our findings from cell culture experiments, we used rat lung primary cells in which COX4I2 is endogenously expressed. We found that primary cells incubated under low oxygen tension (4%) showed enhanced expression of COX4I2 (Figure 7). Furthermore, on knocking down CHCHD2 or RBPJ, reduced COX4I2 expression was observed at 4% oxygen. Figure 7E depicts a model for the regulation of the COX4I2 ORE. At 20% oxygen, CXXC5 is bound to the promoter to cause a repressive effect that is reversed at 4% by an increased expression of CHCHD2 that displaces CXXC5.
Under conditions of low oxygen, the transcriptional regulator HIF plays a critical role in the regulation of hypoxic genes. The genes regulated by HIFs include GLUT1, iNOS, VEGF, CXCR4 and ETS1 (33), which are up-regulated at oxygen tensions lower than 1% where HIFs are stable and bind the hypoxia responsive elements in their promoters (34). However, a study by Elvidge et al. (35) points out the possibility of hypoxic gene regulation independent of HIFs. Furthermore, one of the oxygen sensors for the regulation of ATF4 was found to be PHD3 (36). Similarly, the hypoxic induction of ATF3 is independent of HIF-1 (37). The hypoxic regulation of NFκB also occurs via reduced hydroxylation of the IκB Kinases (IKKs) (38), suggesting the presence of an alternative regulatory systems. Fukuda et al. (39) have shown that hypoxia, in addition to enhancing the expression of COX4I2, also increases the expression of the mitochondrial protease LON, which causes the degradation of COX4I1. They identified two sequences in the COX4I2 gene, present in the human promoter but not conserved in other mammalian species, that bind HIFs to regulate COX4I2 activity under oxygen tensions as low as 1%. However, the role of HIFs in COX4I2 regulation under physiological conditions is not fully clear because 1% oxygen is not typical in normal lung. As an extreme example, the partial pressure of oxygen in the alveoli of subjects at the summit of Mt. Everest was found to be ∼35 torr (∼4–7% oxygen) (19), a level of oxygen used in the current study. Results from a study using a HIF-luciferase reporter construct harboring three HIF binding sites indicated that HIF is not significantly induced at 4% oxygen (5). It should be noted that the minimal 118 bp-ORE-luciferase promoter construct used in this study does not contain any HIF binding sites. Nevertheless, a role for HIFs in the regulation of COX4I2 could be possible at lower oxygen tension (≤1%) found under pathological conditions.
Expression of COX4I2 is essential for normal lung function because knockout of the gene causes severe lung pathology (3). Our previous finding that COX4I2-containing cytochrome oxidase is at least twice as active as the more common COX4I1-containing holoenzyme (5) suggests that the COX4I2 subunit might be expressed in environments where increased activity is needed to satisfy other constraints. Lung, a fibrous tissue with relatively few mitochondria, may need the ability to prevent physiological hypoxia from descending into pathological hypoxia, as occurs, for example, in chronic obstructive pulmonary disease. Brain, also reported to express some COX4I2 (2,40), may need a more active enzyme under reduced blood oxygen supply and/or increased workload to supplement the physiological responses of increased blood flow and increased cerebral blood volume (37,38).
In summary, we have characterized a novel and conserved HIF-1-independent ORE in the promoter of COX4I2 that responds to physiological hypoxia via the action of at least three cellular proteins—RBPJ, CXXC5 and CHCHD2—to regulate the expression of COX4I2 and thereby the activity of COX to adapt to cellular energy requirements under varying oxygen concentrations. Other transcription factors are likely to be involved in COX4I2 regulation because tissue specificity is not present in the ones so far identified.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Funding for open access charge: Henry L. Brasza endowment (in part).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Human embryonic kidney 293 cells were a kind gift from Dr. Assia Shisheva, Wayne State University. The authors thank Dr. Yuling Meng for conducting yeast one-hybrid screens and constructing the expression plasmids for RBPJ and CXXC5.
REFERENCES
- 1.Napiwotzki J, Kadenbach B. Extramitochondrial ATP/ADP-ratios regulate cytochrome c oxidase activity via binding to the cytosolic domain of subunit IV. Biol. Chem. 1998;379:335–339. doi: 10.1515/bchm.1998.379.3.335. [DOI] [PubMed] [Google Scholar]
- 2.Hüttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–123. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- 3.Hüttemann M, Lee I, Gao X, Pecina P, Pecinova A, Liu J, Aras S, Sommer N, Sanderson TH, Tost M, et al. Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J. 2012;26:3916–3930. doi: 10.1096/fj.11-203273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooperstein SJ, Lazarow A. A microspectrophotometric method for the determination of cytochrome oxidase. J. Biol. Chem. 1951;189:665–670. [PubMed] [Google Scholar]
- 5.Huttemann M, Lee I, Liu J, Grossman LI. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 2007;274:5737–5748. doi: 10.1111/j.1742-4658.2007.06093.x. [DOI] [PubMed] [Google Scholar]
- 6.Hamaguchi Y, Matsunami N, Yamamoto Y, Honjo T. Purification and characterization of a protein that binds to the recombination signal sequence of the immunoglobulin J kappa segment. Nucleic Acids Res. 1989;17:9015–9026. doi: 10.1093/nar/17.22.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK. A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet. 2009;5:e1000590. doi: 10.1371/journal.pgen.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 9.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 10.Carroll KD, Bu W, Palmeri D, Spadavecchia S, Lynch SJ, Marras SA, Tyagi S, Lukac DM. Kaposi's Sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J. Virol. 2006;80:9697–9709. doi: 10.1128/JVI.00746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aras S, Singh G, Johnston K, Foster T, Aiyar A. Zinc coordination is required for and regulates transcription activation by Epstein-Barr nuclear antigen 1. PLoS Pathog. 2009;5:e1000469. doi: 10.1371/journal.ppat.1000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang SH, Vieira K, Bungert J. Combining chromatin immunoprecipitation and DNA footprinting: a novel method to analyze protein-DNA interactions in vivo. Nucleic Acids Res. 2002;30:e44. doi: 10.1093/nar/30.10.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijer AH, Ouwerkerk PB, Hoge JH. Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast. 1998;14:1407–1415. doi: 10.1002/(SICI)1097-0061(199811)14:15<1407::AID-YEA325>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 15.Weissmann N, Dietrich A, Fuchs B, Kalwa H, Ay M, Dumitrascu R, Olschewski A, Storch U, Mederos y Schnitzler M, Ghofrani HA, et al. Classical transient receptor potential channel 6 (TRPC6) is essential for hypoxic pulmonary vasoconstriction and alveolar gas exchange. Proc. Natl. Acad. Sci. USA. 2006;103:19093–19098. doi: 10.1073/pnas.0606728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinnula VL, Crapo JD. Superoxide dismutases in the lung and human lung diseases. Am. J. Respir. Crit. Care Med. 2003;167:1600–1619. doi: 10.1164/rccm.200212-1479SO. [DOI] [PubMed] [Google Scholar]
- 17.Stenmark KR, Gebb SA. Lung vascular development: breathing new life into an old problem. Am. J. Respir. Cell Mol. Biol. 2003;28:133–137. doi: 10.1165/rcmb.F259. [DOI] [PubMed] [Google Scholar]
- 18.Gebb SA, Jones PL. Hypoxia and lung branching morphogenesis. Adv. Exp. Med. Biol. 2003;543:117–125. doi: 10.1007/978-1-4419-8997-0_8. [DOI] [PubMed] [Google Scholar]
- 19.West JB, Hackett PH, Maret KH, Milledge JS, Peters RM, Jr, Pizzo CJ, Winslow RM. Pulmonary gas exchange on the summit of Mount Everest. J. Appl. Physiol. 1983;55:678–687. doi: 10.1152/jappl.1983.55.3.678. [DOI] [PubMed] [Google Scholar]
- 20.Tuder RM, Yun JH, Bhunia A, Fijalkowska I. Hypoxia and chronic lung disease. J. Mol. Med. (Berl) 2007;85:1317–1324. doi: 10.1007/s00109-007-0280-4. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 22.Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 25.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Andersson T, Sodersten E, Duckworth JK, Cascante A, Fritz N, Sacchetti P, Cervenka I, Bryja V, Hermanson O. CXXC5 is a novel BMP4-regulated modulator of Wnt signaling in neural stem cells. J. Biol. Chem. 2009;284:3672–3681. doi: 10.1074/jbc.M808119200. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 28.Pendino F, Nguyen E, Jonassen I, Dysvik B, Azouz A, Lanotte M, Segal-Bendirdjian E, Lillehaug JR. Functional involvement of RINF, retinoid-inducible nuclear factor (CXXC5), in normal and tumoral human myelopoiesis. Blood. 2009;113:3172–3181. doi: 10.1182/blood-2008-07-170035. [DOI] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin C, Myers AM, Tzagoloff A. Cloning and characterization of MRP10, a yeast gene coding for a mitochondrial ribosomal protein. Curr. Genet. 1997;31:228–234. doi: 10.1007/s002940050199. [DOI] [PubMed] [Google Scholar]
- 31.Seifeddine R, Dreiem A, Tomkiewicz C, Fulchignoni-Lataud MC, Brito I, Danan JL, Favaudon V, Barouki R, Massaad-Massade L. Hypoxia and estrogen co-operate to regulate gene expression in T-47D human breast cancer cells. J. Steroid Biochem. Mol. Biol. 2007;104:169–179. doi: 10.1016/j.jsbmb.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 32.Ghazoui Z, Buffa FM, Dunbier AK, Anderson H, Dexter T, Detre S, Salter J, Smith IE, Harris AL, Dowsett M. Close and stable relationship between proliferation and a hypoxia metagene in aromatase inhibitor-treated ER-positive breast cancer. Clin. Cancer Res. 2011;17:3005–3012. doi: 10.1158/1078-0432.CCR-10-1704. [DOI] [PubMed] [Google Scholar]
- 33.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF-1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J. Biol. Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 36.Koditz J, Nesper J, Wottawa M, Stiehl DP, Camenisch G, Franke C, Myllyharju J, Wenger RH, Katschinski DM. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110:3610–3617. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- 37.Ameri K, Hammond EM, Culmsee C, Raida M, Katschinski DM, Wenger RH, Wagner E, Davis RJ, Hai T, Denko N, et al. Induction of activating transcription factor 3 by anoxia is independent of p53 and the hypoxic HIF signalling pathway. Oncogene. 2007;26:284–289. doi: 10.1038/sj.onc.1209781. [DOI] [PubMed] [Google Scholar]
- 38.Cummins EP, Comerford KM, Scholz C, Bruning U, Taylor CT. Hypoxic regulation of NF-kappaB signaling. Methods Enzymol. 2007;435:479–492. doi: 10.1016/S0076-6879(07)35025-8. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 40.Singh S, Misiak M, Beyer C, Arnold S. Brain region specificity of 3-nitropropionic acid-induced vulnerability of neurons involves cytochrome c oxidase. Neurochem. Int. 2010;57:297–305. doi: 10.1016/j.neuint.2010.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.