Abstract
The Saccharomyces cerevisiae 2 micron plasmid exemplifies a benign but selfish genome, whose stability approaches that of the chromosomes of its host. The plasmid partitioning locus STB (stability locus) displays certain functional analogies with centromeres along with critical distinctions, a significant one being the absence of the kinetochore complex at STB. The remodels the structure of chromatin (RSC) chromatin remodeling complex, the nuclear motor Kip1, the histone H3 variant Cse4 and the cohesin complex associate with both loci. These factors appear to contribute to plasmid segregation either directly or indirectly through their roles in chromosome segregation. Assembly and disassembly of the plasmid-coded partitioning proteins Rep1 and Rep2 and host factors at STB follow a temporal hierarchy during the cell cycle. Assembly is initiated by STB association of [Rsc8-Rsc58], followed by [Rep1-Rep2-Kip1] and [Cse4-Rsc2-Sth1] recruitment, and culminates in cohesin assembly. Disassembly starts with dissociation of RSC components, is followed by cohesin disassembly and Cse4 exit during anaphase and late telophase, respectively. [Rep1-Rep2-Kip1] persists through G1 of the ensuing cell cycle. The de novo assembly of the ‘partitioning complex’ is cued by the innate cell cycle clock and is dependent on DNA replication. Shared functional attributes of STB and centromere (CEN) are consistent with a potential evolutionary link between them.
INTRODUCTION
The high copy selfish plasmid 2 micron circle of Saccharomyces cerevisiae propagates itself stably with the help of a partitioning system and an amplification system (1,2). The partitioning system is composed of the plasmid-coded proteins Rep1 and Rep2 and a cis-acting locus STB (stability locus). Despite its apparent simplicity of organization, the partitioning system is able to confer nearly chromosome-like stability on the plasmid (a loss rate of 10−4 to 10−5 per cell division). The Rep proteins appear to orchestrate the channeling of chromosome segregation factors to STB, presumably as a means to coupling plasmid segregation and chromosome segregation (3–10). Although current evidence is consistent with plasmids hitchhiking on chromosomes, alternative chromosome independent mechanisms of plasmid segregation cannot be ruled out.
The Flp site-specific recombination system harbored by the plasmid rectifies copy number decrease resulting from rare missegregation events by a DNA amplification mechanism (11,12). Amplification is thought to be triggered by a carefully timed recombination event that converts bidirectional replication into unidirectional rolling circle replication. The tandem plasmid copies comprising the amplified DNA may be resolved into individual monomers by Flp-mediated recombination or by the host’s homologous recombination machinery. Transcriptional regulatory mechanisms mediated by Rep1, Rep2 and an additional plasmid-coded protein Raf1 provide for a prompt amplification response when required without the danger of a runaway increase in plasmid copy number (13–15).
Host contributions towards equal segregation of the 2 micron plasmid include the Rsc2 subunit of the remodels the structure of chromatin (RSC) chromatin remodeling complex, the mitotic spindle, the spindle-associated motor protein Kip1, the histone H3 variant Cse4 and the yeast cohesin complex (3–8,10,16,17). These host factors play important functional roles at centromeres as well. The RSC complex appears to regulate chromatin organization at centromere (CEN) and CEN-proximal regions to promote normal chromosome segregation (18–20). Kip1 contributes to the typical bi-lobed organization of centromere clusters during metaphase, presumably by bundling kinetochore microtubules (21). Together with the other kinesin motors (Cin8, Kip3 and Kar3), Kip1 promotes the assembly and dynamics of the mitotic spindle (22). Cse4 is the hallmark of a specialized nucleosome assembled at each point centromere (23–25) and is essential for kinetochore assembly. Recent estimates suggest that additional Cse4-containing nucleosomes may occupy regions neighboring the centromere (26,27). Although the mitotic spindle directly facilitates sister chromatid segregation, the role of the spindle in STB plasmid segregation is likely indirect. The spindle, together with the Kip1 motor, may help transport the plasmid to its specific nuclear address (3). Segregation of single copy STB plasmid reporters suggest that the binary counting mechanism by which cohesin ensures equal chromosome segregation likely operates on STB-mediated plasmid segregation as well (4,5).
Distinctions between CEN and STB in their DNA organizations and functions are also significant. The ∼125-bp CEN comprises three characteristic DNA elements—centromere DNA element (CDE) I, CDE II and CDE III (28,29). STB, which is much larger, can be divided into two sub-regions, proximal and distal with respect to the plasmid replication origin (30). STB-proximal, composed of five tandem copies of a 60-bp consensus element, is where Cse4 interaction occurs (16). Interestingly, two directly repeated regions with 97% identity between them, each 124-bp long and thus equal in size to CEN, are contained within STB-proximal. The Rep proteins have no role to play in CEN-mediated segregation, and kinetochore proteins are apparently unassociated with STB. Furthermore, only a small fraction of the plasmid molecules appears to be associated with Cse4 or cohesin (5,16). Yet, CEN and STB serve as the platforms for assembling high-order DNA–protein complexes dedicated towards the equal segregation of chromosomes and of plasmids, respectively. The conserved traits between CEN and STB, contrasted by their present functional distinctions, may suggest their potential divergence from a common evolutionary ancestor (16,31).
We have identified, using tandem affinity purification and mass spectrometry, the interaction of the Rep proteins with subunits of the RSC chromatin remodeling complex and subsequently verified the functional relevance of this interaction to plasmid segregation. Furthermore, we have delineated temporal hierarchies in the assembly and disassembly of the Rep proteins and host factors at STB during the cell cycle. The sequence of protein associations and dissociations at STB, conjoined with observations from previous studies, suggests a coordinated and regulated program for the plasmid partitioning pathway. This program is initiated by innate cell cycle cues and is dependent on DNA replication. The partitioning clock is reset at the G1-S transition stage of each cell cycle, but it can be started without requiring any input from a previous partitioning cycle.
MATERIALS AND METHODS
Yeast strains
The strains used in this study and their relevant genotypes are assembled in Supplementary Table S1 (Supplementary Material).
Tandem affinity purification-tagging REP1 or REP2 locus in the 2 micron circle genome
Two strategies were used to engineer the 2 micron circle genome by fusing the tandem affinity purification (TAP) tag of Protein A and calmodulin-binding peptide at the N-terminus of Rep1 or Rep2. The first targeted the native plasmid; the second used a 2 micron circle derivative that also harbors the ADE2 marker (32), but no other extraneous sequences. The latter plasmid, which we have designated as p2µ-ADE2, is one of the most stable among 2 micron circle-based artificial plasmids. Under non-selective growth, its loss rate was found to be within an order of magnitude or two of that of the 2 micron plasmid (unpublished data). In the plasmid constructions described later in the text, the template for obtaining the TAP sequences was the plasmid pSB1761 described previously (33), designed for tagging target proteins at their amino termini.
Strategy 1
The DNA cassettes containing the dual tag (33) for fusion to the N-terminus of Rep1 or Rep2 were amplified in two separate polymerase chain reaction (PCR) reactions. The ‘forward’ and reverse primers used for amplification were designed to contain terminal 50-bp homologies to requisite regions of the 2 micron plasmid. The two linear DNA fragments were introduced individually into a [cir+] recipient yeast strain (containing endogenous 2 micron plasmid) in separate transformation steps. The homologous ends would promote double strand break repair using 2 micron circles as the template. The end result would be the generation of modified 2 micron circle genomes carrying the TRP1 marker along with the tagged REP1 locus in one case and the tagged REP2 locus in the other. The separation of the tagged plasmid from the untagged ones was accomplished in a second transformation step using a [cir0] recipient strain (lacking native 2 micron circle) and total DNA isolated from a first-step transformant. Colonies that harbored the desired Trp+ plasmid alone (and not the native 2 micron circle) were identified by PCR-based screens using isolated total DNA and diagnostic primer sets. The authenticity of the plasmid constructs was ascertained by DNA sequencing.
The tagged Rep1 or Rep2 was functional in plasmid partitioning. Under non-selective growth in glucose (Rep1 or Rep2 not induced), there was high instability of the TRP1 marker harbored by the plasmid, whereas growth in galactose (Rep1 or Rep2 induced) suppressed this instability (unpublished data).
The tagged plasmids were further modified by excising the TRP1 marker and the GAL promoter via Cre-mediated site-specific recombination between two directly oriented LoxP target sites. In the resulting plasmids, expression of the tagged REP1 or REP2 was under the control of the respective native promoter.
TAP analyses were performed using strains that expressed the tagged REP loci from their native promoters, or those that expressed these loci from the GAL promoter, after growing them in glucose and galactose, respectively.
Strategy 2
For construction of a Rep-tagged derivative of p2µ-ADE2, two overlapping DNA fragments were first prepared by PCR amplification of total DNA isolated from the strains harboring p2µ-ADE2 in one case and 2 micron circle containing the amino-terminal TAP tag on REP1 or REP2 (see strategy1 earlier in the text) in the other. One of the fragments so generated contained sequences from p2µ-ADE2 including the ADE2 marker. Its partner fragment contained the tagged REP1 or REP2 under control of the corresponding native promoter. A roughly equimolar mixture of the two fragments was used to transform an ade2 [cir0] strain to reconstitute the TAP-tagged versions of p2µ-ADE2 by homologous recombination/repair in vivo. DNA from a subset of the Ade+ transformants was analysed by PCR and DNA sequencing to identify those that had correctly reconstructed the requisite plasmids, p2µ-ADE2(TAP-REP1) and p2µ-ADE2(TAP-REP2), harboring the engineered REP1 or REP2 locus, respectively.
The stability of p2µ-ADE2(TAP-REP2) was comparable with that of p2µ-ADE2; that of p2µ-ADE2(TAP-REP1) was 2–3-fold lower, but still considerably higher than that of most standard 2 micron circle-derived plasmids (unpublished data). For TAP analysis, strains were grown in media lacking adenine.
Enrichment of Rep1 and Rep2 partners by TAP
Extracts for affinity purification were prepared essentially according to published procedures (33). In a standard run, a 4 l culture grown to an OD600 of ∼1.0 was processed. Harvested cells were washed twice with sterile water, once with A10 buffer (10 mM HEPES, pH 8.0; 10 mM KCl; 300 mM NaCl; 1.5 mM MgCl2; 0.2 mM ethylenediaminetetraacetic acid; 10% v/v glycerol) plus protease inhibitors (complete mini ethylenediaminetetraacetic acid-free protease inhibitor mix from Roche) and resuspended in equal w/v of A10 buffer. The cell suspension was quickly frozen by dropping it into liquid nitrogen using a pipette to form frozen cell ‘popcorns’. They were either stored at −80°C for future use, or immediately carried through the TAP protocol (33). Cells within the frozen popcorns were disrupted by hand grinding (∼100 strokes) using a mortar and pestle. Disruption was performed in a cold room with the mortar placed on ice. Cleared lysates obtained by centrifugation of cell extracts at 100 000× g for 1 h were subjected to the following steps: chromatography on IgG beads, tobacco etch virus protease digestion and chromatography of eluted proteins on calmodulin beads.
The TAP enrichment data provided in Figure 1 comprised expression of the Rep1 and Rep2 baits from the native promoters as well as the GAL promoter.
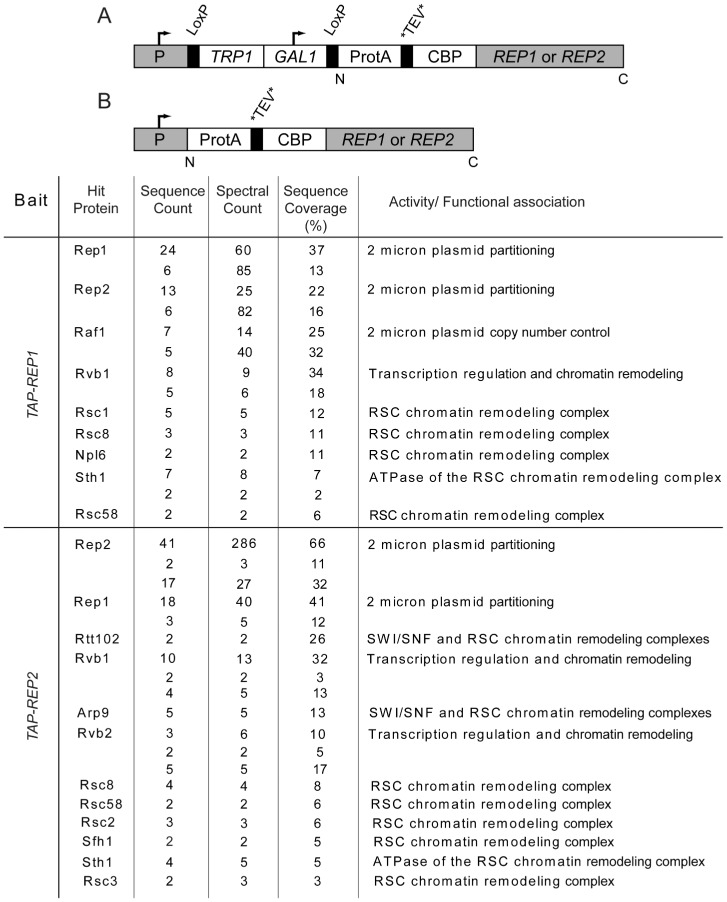
Figure 1.
Interacting partners of the 2 micron circle partitioning proteins Rep1 and Rep2. The two types of constructs used for TAP tagging, with the tagged REP locus under control of the GAL promoter (A) or the native promoter (B), are schematically diagrammed. Identification of tryptic peptides by mass spectrometry was performed on the unfractionated ensemble of proteins enriched by a two-step affinity purification of tagged Rep1 or Rep2 (see Materials and Methods). The list, except for Rep1, Rep2 and Raf1, comprises a subset of interacting partners that are constituents of, or are associated with, chromatin remodeling complexes. Sequence count refers to the number of distinct peptides, spectral count to the number of total peptides and sequence coverage to the percentage of amino acids of the full-length protein represented by the sequenced peptides. Those cases where more than one value is shown for each of these parameters refer to separate experiments. This analysis used strains MJY4013 to MJY4025 listed in Supplementary Table S1.
Mass spectrometry analysis of proteins
Eluant fractions from the ethylene glycol tetraceticacid-elution buffer washing of the calmodulin beads (see earlier in the text) were concentrated by trichloroacetic acid precipitation, processed further for tryptic digestion, and the digest was subjected to mass spectrometry analysis (34) using mutli-dimensional protein identification technology. The spectra, refined using the PARC filter (35), was searched against S. cerevisiae proteins from Saccharomyces Genome Database using the SEQUESTTM algorithm (36–38). Further analytical refinements, including imposition of the trypsin specificity constraint, were performed as described previously (34,39). Data collected from individual runs are deposited at http://depts.washington.edu/yeastrc/.
Plasmid stability assays
The stability assays were carried out in rsc8 and rsc58 Ts strains. The STB reporter plasmid was p2µ-ADE2, on which the TAP-tagging (strategy 2) was based (see earlier in the text). The autonomously replicating sequence (ARS) reporter plasmid (p2µ-ADE2-ARS) was derived from p2µ-ADE2 by deleting STB-proximal (bordered by HpaI and AvaI) from it. The CEN reporter plasmid (p2µ-ADE2-CEN) was obtained by replacing STB-proximal in p2µ-ADE2 by the centromere sequence derived from chromosome VI (CEN6). As described for the plasmids containing the affinity-tagged REP genes, these plasmids were also constructed from two PCR-amplified DNA fragments, one of which contained the requisite modification at the STB locus, by recombining them in vivo. Except for the deletion of STB-proximal or its replacement with CEN6, all three reporter plasmids were identical in organization and sequence. Their replication was promoted by the 2 micron circle replication origin.
For a quantitative estimate of plasmid loss rates, the following protocol was used. From an overnight culture of an Ade plus transformant containing a reporter plasmid grown under selection at 26°C, ∼7 × 104 cells were inoculated into 10 ml of the non-selective medium (generation = 0) and grown for eight generations at 26°C or 35°C. Cells were seeded on non-selective medium (supplemented with adenine; ∼200 cells per plate), incubated for 72 h at 26°C and transferred to 4°C for 40 h. The fraction of fully red colonies, those that arose from progenitor cells that had suffered plasmid loss, at generations 0 and 8 was estimated on each plate. The numbers were verified by replicating the master plates on medium lacking adenine. The loss rate per generation (or the instability index) was estimated as I = (1/n) × [ln (f0/fn)], where f0 and fn denote the fractions of plasmid bearing cells at generation ‘0’ and after ‘n’ generations of non-selective growth (30).
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (6). Anti-HA and Anti-Myc antibodies purchased from Covance (Princeton, NJ) were used after 1:100 dilution of the stock titer. For cell cycle ChIP, cells grown to OD600 = ∼0.4 were arrested in G1 using α factor (10 µg/ml), washed four times with sterile water, released into fresh medium and incubated in a shaking water bath maintained at 26°C. In all, 40 OD600 units of cells were harvested for performing ChIP at each time point. Initial standardizations were done to keep the amount of immunoprecipitated DNA used as the template in PCR reactions to be within the linear range of amplification. For the ChIP assays, the native 2 micron circle served as the reporter plasmid.
Strains for bromo-deoxyuridine (BrdU)-directed ChIP were engineered to harbor three copies of the herpes simplex virus thymidine kinase gene, each expressed from the GPD1 promoter (40). In addition, they contained the human equilibrative nucleoside transporter 1 gene expressed from the ADH1 promoter. In this ChIP analysis, the amounts of immunoprecipitated DNA used as templates for PCR amplification corresponded to the same number of cells for the individual samples compared. Furthermore, a standardization assay was carried out to ensure that the aliquot of each sample used for PCR was within the linear range of amplification.
Cell cycle arrest in G1 in a Cdc6 depleted state
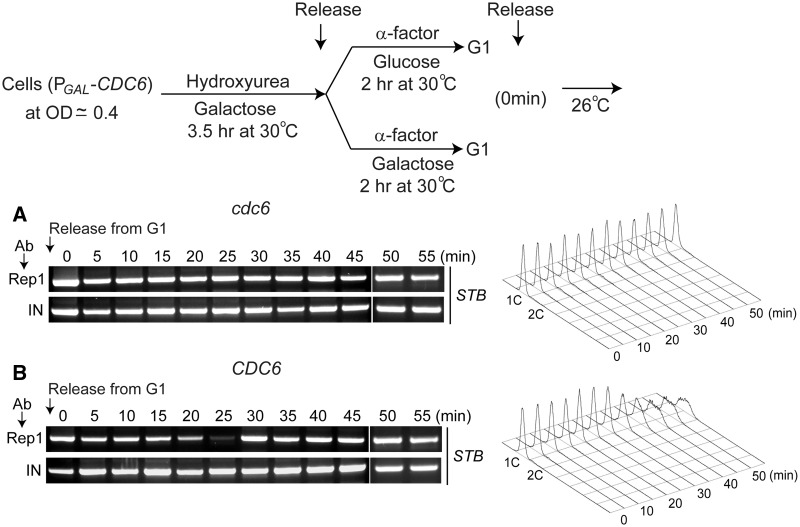
The procedure used for arresting cells in G1 in a Cdc6 depleted state was similar to that described by Severin et al. (41). The strain harboring the CDC6 gene under the control of the GAL promoter was grown in galactose at 30°C to an OD600 of ∼0.4, and hydroxyurea was added in powder form, with shaking, to a final concentration of 0.2 M. Cells were incubated for 3.5 h at 30°C, when the OD600 reached 0.8–1.0. Cells were harvested, washed four times with sterile water, resuspended in glucose medium in presence of α factor (10 µg/ml), and incubated at 30°C to arrest them in G1.
Other miscellaneous procedures
Cell cycle arrest in G1 (except under Cdc6 depletion) using α factor and in G2/M using nocodazole were performed according to previously published protocols (9). Bacterial and yeast transformations, yeast genomic DNA and plasmid DNA preparations, curing yeast strains of endogenous 2 micron circle, fluorescence-activated cell sorting analysis and culturing of yeast and bacterial strains were carried out according to the standard procedures used by the Jayaram laboratory (http://www.sbs.utexas.edu/jayaram/jayaramlab.htm).
RESULTS
TAP reveals components of the RSC chromatin remodeling complex as interaction partners of Rep1 and Rep2
In a search for host factors potentially involved in 2 micron plasmid segregation, we carried out enrichment of protein complexes using Rep1 and Rep2 baits fused to Protein A and calmodulin-binding peptide as tandem affinity tags (Figure 1; Materials and Methods). The putative protein partners identified by mass spectrometry, following a two-step enrichment of Rep1 or Rep2 expressed from the GAL promoter, included components of the RSC chromatin remodeling complex (Figure 1). Also detected, under conditions of native or galactose induced expression of the bait, were Rvb1 and Rvb2, AAA(+) DNA helicases associated with transcriptional regulation and chromatin remodeling. Furthermore, Rvb1 has been shown to interact with Rsc2 (42). None of the RSC components were identified in a mock run using untagged Rep1 or Rep2 as the bait. In one of the control runs, peptide signals for Rep2 were detected. However, the peptide and spectral counts of 2 each and the sequence coverage of 4.2% were much lower than those from experimental samples. Additionally, confidence in the observed associations was bolstered by the baiting of Rep2 and Raf1 by the tagged Rep1 and of Rep1 by the tagged Rep2. Dihybrid and affinity pull-down assays have documented bipartite cross-interactions among Rep1, Rep2 and Raf1 (43–46) (also unpublished data). Even though Rep1 and Rep2 were overexpressed in the assays that revealed their associations with RSC complex subunits, these associations are functionally relevant, as demonstrated by the analyses described later in the text. The potential effects of Rvb1 and/or Rvb2 on 2 micron plasmid stability and/or copy number control remain to be investigated.
The Rep1- and Rep2-interacting proteins revealed in this study (Figure 1), together with the related observations noted earlier in the text, argue in favor of the RSC2 complex being an authentic constituent of the 2 micron plasmid partitioning complex. The presence of Rsc1 among the Rep1-associated proteins (Figure 1) could potentially denote an incidental interaction mediated through a shared component (or components) of the RSC1 and RSC2 complexes. Alternatively, this may be an authentic interaction that is indirectly related to plasmid stability. In addition to their role in segregation, the Rep1 and Rep2 proteins also act cooperatively to regulate 2 micron circle gene expression (13–15). Thus, Rsc1 might potentially influence plasmid copy number control by affecting FLP gene transcription. There is considerable evidence for the association of the RSC complex with activated and repressed promoters and for its role in positive and negative regulation of gene expression (47). Assuming that Rsc1 does play a role in plasmid physiology, it is certainly subservient to Rsc2, as indicated by the apparently normal plasmid stability in the rsc1Δ strain, at least over a limited number of generations (17).
In our analyses presented later in the text addressing the role of host factors in 2 micron plasmid partitioning, Rsc2, Rsc8, Rsc58 and Sth1 were chosen as representative subunits signifying the function of the RSC2 complex as a whole.
Inactivation of RSC2 complex subunits Rsc8 or Rsc58 causes missegregation of an STB-reporter plasmid
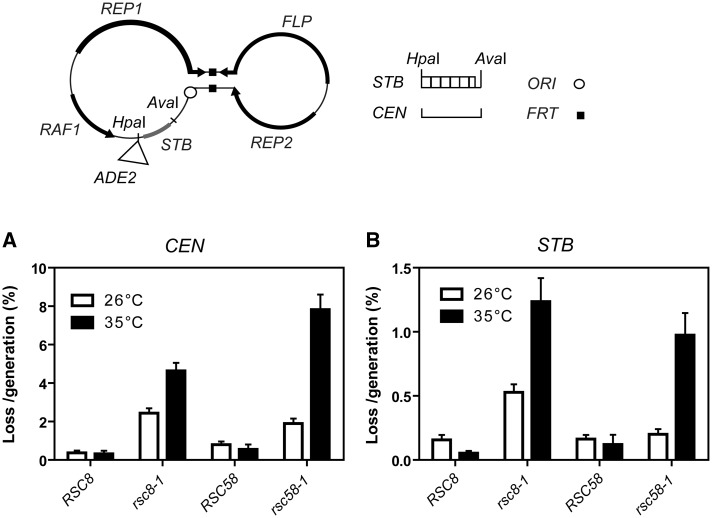
To verify the possible role for chromatin remodeling by the RSC2 complex in 2 micron circle segregation suggested by mutational and interaction analyses (17,48) (Figure 1), and to examine potential distinctions between STB and CEN with respect to RSC function, we followed the segregation of STB and CEN reporter plasmids in rsc8 and rsc58 Ts mutant strains at the permissive (26°C) and semi-permissive (35°C) temperatures.
The STB reporter plasmid used, p2µ-ADE2 (see ‘Materials and Methods’ section), contained the ADE2 marker, inserted at the HpaI site in the 2 micron circle genome, as the only locus non-native to the plasmid. The CEN reporter plasmid differed from the STB reporter in lacking STB-proximal containing the repeat elements but harboring CEN6 in its place. The loss rate per generation was increased for both the CEN and STB reporter plasmids in the rsc8 and rsc58 mutant backgrounds at 35°C (semi-permissive) compared with 26°C (Figure 2). The ARS reporter plasmid, in which STB-proximal was deleted, was highly unstable at both temperatures (data not shown). The stability of the CEN reporter plasmid was lower than that of the STB plasmid even at the permissive temperature. This may be owing to the contextual effects from replacing STB-proximal by CEN6. The relative increases in the loss rates owing to the rsc mutations were comparable between the CEN and STB reporter plasmids.
Figure 2.
Effects of the rsc8 and rsc58 mutations on plasmid stability. The genetic organizations of the reporter plasmids used in stability assays are schematically diagrammed. Plasmid p2µ-ADE2 contains the ADE2 gene inserted at the HpaI site of the native 2 micron circle. In p2µ-ADE2-CEN, the HpaI-AvaI region, comprising the repeat elements of STB, is replaced by CEN6. These reporter plasmids are capable of undergoing Flp-mediated recombination at the FRT sites. Plasmid loss rates were estimated by counting the fraction of cells harboring the reporter plasmid at time zero and following eight generations of growth under non-selection (30). The time zero values were provided by overnight cultures of purified transformants grown in liquid media under selection at 26°C. The mutant strains used in these assays were MJY5056 to MJY3024 (Supplementary Table S1). The control wild-type strains were constructed by replacement of the mutant rsc8 and rsc58 genes by the corresponding wild type genes, RSC8 and RSC58, respectively.
The replication of the reporter plasmids was driven by the 2 micron plasmid origin in its native location. We have verified that plasmid replication was not affected by the rsc mutations by assaying the copy number of the CEN reporter plasmid relative to a chromosomal locus in G1 and metaphase cells by real-time PCR (Supplementary Figure S1; Supplementary Material). The rsc mutations also showed no obvious effect on the performance of a chromosomal replication origin, as ascertained from the mean intensities of a fluorescence-tagged CEN plasmid harboring ARS1 (derived from chromosome IV) in G1 and metaphase cells (Supplementary Figure S2; Supplementary Material).
Previous work demonstrated that the absence of Rsc2 or conditional inactivation of the Sth1 (ATPase) subunit of the RSC complex by a mutation in its bromodomain (L1346A) results in poor maintenance of the 2 micron plasmid (17,48). These findings, augmented by the now-revealed plasmid instability in the rsc8 and rsc58 mutant backgrounds (Figure 2) and the corroborating interactions of Rep1 and Rep2 (Figure 1), authenticate the functional contribution of the RSC2 complex towards plasmid partitioning.
The plasmid loss rate reported for rsc2Δ (15–26% per cell per generation) (17) is considerably higher than that we have seen for rsc8 and rsc58 at 35°C. The difference may, in principle, be accounted for by the complete versus partial inactivation of the RSC2 complex. Alternatively, Rsc2 may play a specific role in 2 micron plasmid partitioning that is independent of its function as a component of the RSC2 complex. In our strain backgrounds, the magnitude of the rsc2Δ effect on 2 micron plasmid stability is smaller than the reported values (unpublished data).
In principle, the STB chromatin structure promoted by the RSC2 complex could be important in regulating 2 micron plasmid gene expression. A possible role for such a transcriptional control in plasmid stability has not yet been explored. As 2 micron plasmid segregation is tightly coupled to that of chromosomes, the potential effects of the rsc mutations on CEN cannot be dissociated from their direct effects on STB. Additionally, changes in the levels of host factors stemming from global transcriptional changes induced by these mutations could indirectly affect plasmid stability.
Temporal sequence in the maturation of the plasmid partitioning complex
The cell cycle dependent assembly of the plasmid partitioning complex is initiated at the G1-S window. Components left over from the previous partitioning cycle, Rep1, Rep2 and Kip1, are extruded from STB, presumably as part of setting the new assembly program in motion (3,10). Initially, we attempted to discern the potential functional hierarchy in the association of partitioning factors (Rep1, Rep2, Kip1, RSC components, Cse4 and the cohesin subunit Mcd1) at STB by ChIP when each was inactivated, one at a time, by deletion or by a temperature sensitive mutation (data not shown). In a number of previously published studies, these factors have been shown to interact specifically with STB, and not with other regions of the plasmid genome, in wild-type genetic backgrounds. The results shown in Supplementary Figure S3 (Supplementary Material) further verify the authenticity of these interactions. However, the ChIP results from individual mutant strains were too complex to order proteins into a simple assembly scheme in series. One interpretation of these results is that the analysed protein associations at STB are not functionally interrelated. However, more likely, the disruption of a particular interaction in the multi-protein assembly at STB may not only eliminate a relevant subsequent interaction but may also unmask out of sequence, and potentially non-productive, interactions. We then wished to query the temporal sequence of protein-STB interactions in cells going through a synchronous cell cycle.
We performed short 5 min interval ChIP using cells released from G1 arrest to map the times of recruitment of a given protein at STB relative to the time of Rep1 recruitment. Previous analyses showed that Rep1 and Rep2 mirror each other in their associations with STB (10). The assays were performed in cells grown at 26°C, rather than at the 30°C norm, to improve time resolution by slowing down the cell cycle and expanding the G1-S window. As the partitioning proteins are all wild-type in this analysis, it is free of the potential indirect effects of mutations that could have vitiated our functional hierarchy assays.
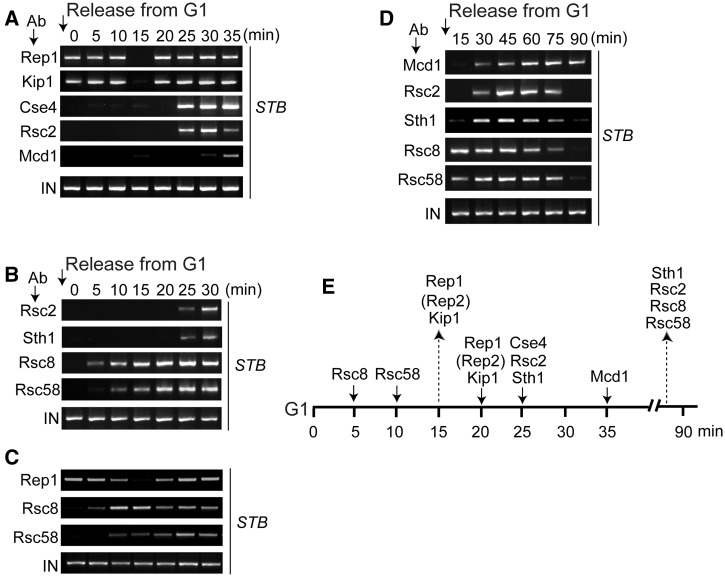
The data assembled in Figure 3A were obtained from pairwise ChIP assays, simultaneously targeting Rep1 and a particular test protein of interest within a given cell population. By including a common reference protein in all of the assays, any potential variability in the cell cycle timing from assay to assay could be corrected. The results are presented with 20 min after release from G1 as the normalized time point for de novo Rep1–STB association. For the data shown in Figure 3B, the reference protein was Rsc2. The timing of its association with STB was clocked at 25 min after release from G1, as judged by the Rep1 reference frame (Figure 3A). The temporal order of protein association at STB, within the resolution permitted by the ChIP assays, was as follows: Rsc8-Rsc58, followed by [Rep1-Rep2-Kip1], [Cse4-Rsc2-Sth1] and finally Mcd1 (or cohesin). This scheme is consistent with a previous study indicating that the recruitment of Sth1 at STB can be temporally resolved from the subsequent assembly of cohesin at STB (48). The association of Rsc8 and Rsc58 with STB earlier than Rsc2 and Sth1 or before transient Rep1 dissociation was unexpected. However, similar results were obtained when the ChIP assays were repeated with Rep1 (rather than Rsc2) as the reference protein (Figure 3C).
Figure 3.
Temporal sequence of events at STB during the assembly and disassembly of the plasmid partitioning complex. ChIP assays were performed in cells arrested in G1 (time zero), and at every 5 min time point after their release into the cell cycle. In each panel, the top row indicates the reference protein that was paired with each of the others in individual assays. (A) Rep1 was not epitope-tagged; Mcd1 and Cse4 harbored HA3- and Myc12-epitopes, respectively, and the others Myc13-epitope. (B) Rsc2 harbored Myc13-epitope; the others HA3-epitope. (C) Rep1 was not epitope-tagged; Rsc8 and Rsc58 were tagged with Myc-13 epitope. (D) Mcd1 harbored HA3-epitope; the others Myc13-epitope. In each individual assay, after splitting samples into equal portions, the reference protein was immunoprecipitated in one set and the test protein in the other. Rep1 was immunoprecipitated using an antibody to the native protein. Each of the other proteins was immunoprecipitated using an antibody to the epitope tag harbored by it. (E) The cumulative results from A–D are placed along the time line for plasmid segregation. Rep2, placed in parentheses, was not analysed in the present assays. Based on previous analyses, it is expected to follow the Rep1 pattern. The strains for these assays were MJY3162 to MJY5042 (Supplementary Table S1).
The sequence of host factor interactions with STB suggests that the RSC2 complex is not recruited in a fully pre-assembled form. However, it is possible that the early association of Rsc8 and Rsc58 with STB signify incidental interactions that precede the functionally relevant interaction of the RSC2 complex as a whole. Alternatively, the association of Rsc8 and Rsc58 with STB may be important for triggering the dissociation of Rep1, Rep2 and Kip1 from STB (15 min time point in Figure 3A). Cohesin is the last host factor, among those analysed, to be recruited at STB.
Sequence of disassembly of the plasmid partitioning complex
The disassembly of the plasmid partitioning complex is also temporally regulated. Results from previous studies indicate that the Sth1 subunit of RSC2 dissociates from STB before, and non-RSC2 components of the partitioning complex do so subsequent to, cohesin disassembly (3,6,7,10,48). Cse4 exits STB in late telophase before cytokinesis, whereas Rep1, Rep2 and Kip1 persist at STB beyond cytokinesis and all through G1. It is not known whether RSC2 subunits leave STB in unison, or some of these are retained at STB until after cohesin disassembly. To resolve this issue, we have now determined the lengths of association of Rsc2, Rsc8, Rsc58 and Sth1 with STB during a cell cycle relative to Mcd1 (cohesin) as the reference. As disassembly of the partitioning complex spans a large stretch of the cell cycle, in contrast to the relatively narrow window of assembly, the relevant ChIP assays were performed at 15 min intervals.
As shown in Figure 3D, Rsc2, Rsc8, Rsc58 and Sth1 dissociated from STB more or less simultaneously (within the time resolution of the ChIP assay), well before cohesin disassembly. This timing is consistent with that reported for Sth1 in a previous study (48). The current results, together with those from earlier work (3,6,7,10,48), suggest the following order for disassembly of the plasmid partitioning complex: RSC2 complex first, followed sequentially by cohesin, Cse4 and [Rep1-Rep2-Kip1]. The data in Figure 3D are consistent with the RSC2 complex being released from STB as a single entity. Alternatively, the time intervals separating the release of individual subunits are shorter than the resolution of the assay.
The temporal demarcations during the assembly phase of the plasmid partitioning complex and the onset of the disassembly phase by release of RSC components, as inferred from Figure 3A–D, are depicted in Figure 3E.
Delaying DNA replication delays the assembly of the partitioning complex
The timing of the transient dissociation of Rep1/Rep2/Kip1 from STB in cells released from G1 arrest suggests that this event may be linked to, or triggered by, plasmid replication in early S phase (49). We have therefore investigated the effects of altering the time of DNA replication on the dissociation of Rep1 from STB during G1 exit and entry into S.
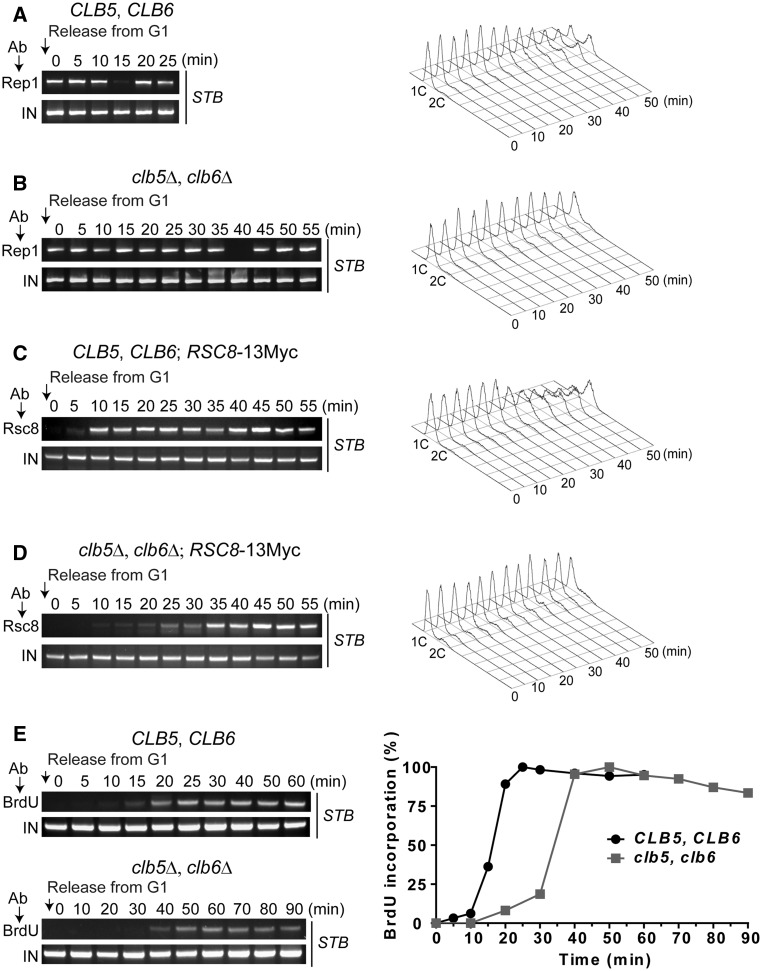
For delaying DNA replication, we used deletions of the B-type cyclin genes CLB5 and CLB6. In the clb5Δ, clb6Δ background, bud emergence is not delayed, but the interlude before the onset of replication is lengthened (50) (see also Supplementary Figure S4; Supplementary Material). ChIP analysis revealed ∼25 min delays in the dissociation and de novo reassociation of Rep1 at STB in the clb5Δ, clb6Δ strain (Figure 4A and B), with a similar delay in the association of Rsc8 with STB (Figure 4C and D). Rsc8–STB association preceded Rep1 exit from STB in the replication delayed cell cycle, as in the normal one. The replication time of STB was also pushed back by ∼25 min in the deletion strain, as verified by BrdU incorporation (Figure 4E).
Figure 4.
Delay in the de novo assembly of the plasmid partitioning complex in the clb5Δ, clb6Δ host strain. (A, B) The transient dissociation and reassociation of Rep1 at STB in cells released from G1 arrest was assayed by ChIP in wild-type and clb5Δ, clb6Δ strains. Gross DNA replication was followed by fluorescence-activated cell sorting analysis. (C, D) Association times of Rsc8 with STB after release from G1 were monitored by ChIP in wild-type and clb5Δ, clb6Δ strains. (E) Replication times of STB were followed by ChIP using an antibody against BrdU. In the plot for BrdU incorporation, the band intensities of the PCR amplified DNA from an assay were normalized to the highest band intensity from that assay, which was assigned a value of 100%. The experiments were conducted using strains K699 to MJY5048 listed in Supplementary Table S1.
Thus, the timing of DNA replication and the timing of the assembly of the plasmid partitioning complex are well correlated.
Blocking of DNA replication blocks the assembly of the plasmid partitioning complex
To further verify the relationship between DNA replication and the assembly of the plasmid partitioning complex, we programmed a cell cycle in the absence of replication. As in previous experiments, the transient dissociation of Rep1 from STB provided the marker for the initiation of the assembly pathway.
We blocked DNA replication by depriving cells of Cdc6, which associates with replication origins at the initiation step of DNA replication (51,52), before releasing them from G1. In the experimental strain, the only functional CDC6 gene was placed under the control of the GAL promoter, and cells were maintained viable by growing them in galactose containing medium. Cdc6 depletion was accomplished by hydroxyurea treatment of cells in presence of galactose, followed by their release in presence of glucose and α factor to arrest them in G1, deprived of Cdc6 (Materials and Methods). After release from G1 in glucose, dissociation of Rep1 from STB was not observed for up to 55 min (Figure 5A). In control cells (expressing Cdc6) released from G1 in galactose, this event occurred at 25 min (Figure 5B).
Figure 5.
Prevention of de novo assembly of the plasmid partitioning complex in the absence of Cdc6. To initiate a cell cycle in the absence of Cdc6, cells arrested in G1, with Cdc6 depleted, were released in glucose. In the control assay, galactose grown cells (expressing Cdc6) were arrested in G1, and released from arrest in galactose medium. The dissociation of Rep1 from STB was monitored by ChIP as the indicator of a critical event in the de novo assembly of the partitioning complex. The strain for conditional depletion of Cdc6 was K4675 (Supplementary Table S1).
Thus, the disassembly of the partitioning complex from a previous cell cycle does not occur when DNA replication is blocked.
A prior round of partitioning is not required for the de novo assembly of the partitioning complex at a naive STB
As pointed out, a subset of the components of the plasmid partitioning complex remains at STB after plasmid apportioning has been accomplished and cytokinesis completed. Their exit from STB is correlated with the initiation of a new round of assembly of the partitioning complex. A relevant question is whether a previous round of partitioning, and thus an experienced STB, is a pre-requisite for a new round of successful partitioning. We addressed this question by following the segregation of an STB reporter plasmid excised from the chromosome in host strains expressing the Rep proteins either constitutively or inducibly during a particular cell cycle.
The reporter plasmid pSTB[LacO]256 was present as an integrant at the chromosomal HIS3 locus in two host strains harboring a REP1-REP2 expression cassette, controlled by the constitutive ADH promoter in one case and the inducible bidirectional GAL promoter in the other (Figure 6). These otherwise isogenic [cir0] strains were also engineered to express GFP-LacI from the HIS3 promoter and the R recombinase from the GAL promoter. In the integrated state, with cells grown in glucose or raffinose, the plasmid would segregate as the chromosome that houses it, indifferent to the continuous expression of Rep1 and Rep2 in one strain and the absence of their expression in the other. The Rep proteins, when present, orchestrate the assembly of the plasmid partitioning factors at the integrated STB locus. In previous experiments, we observed the Rep1- and Rep2-dependent association of Cse4 and the cohesin subunit Mcd1 at a copy of STB inserted at the HIS3 locus (16).
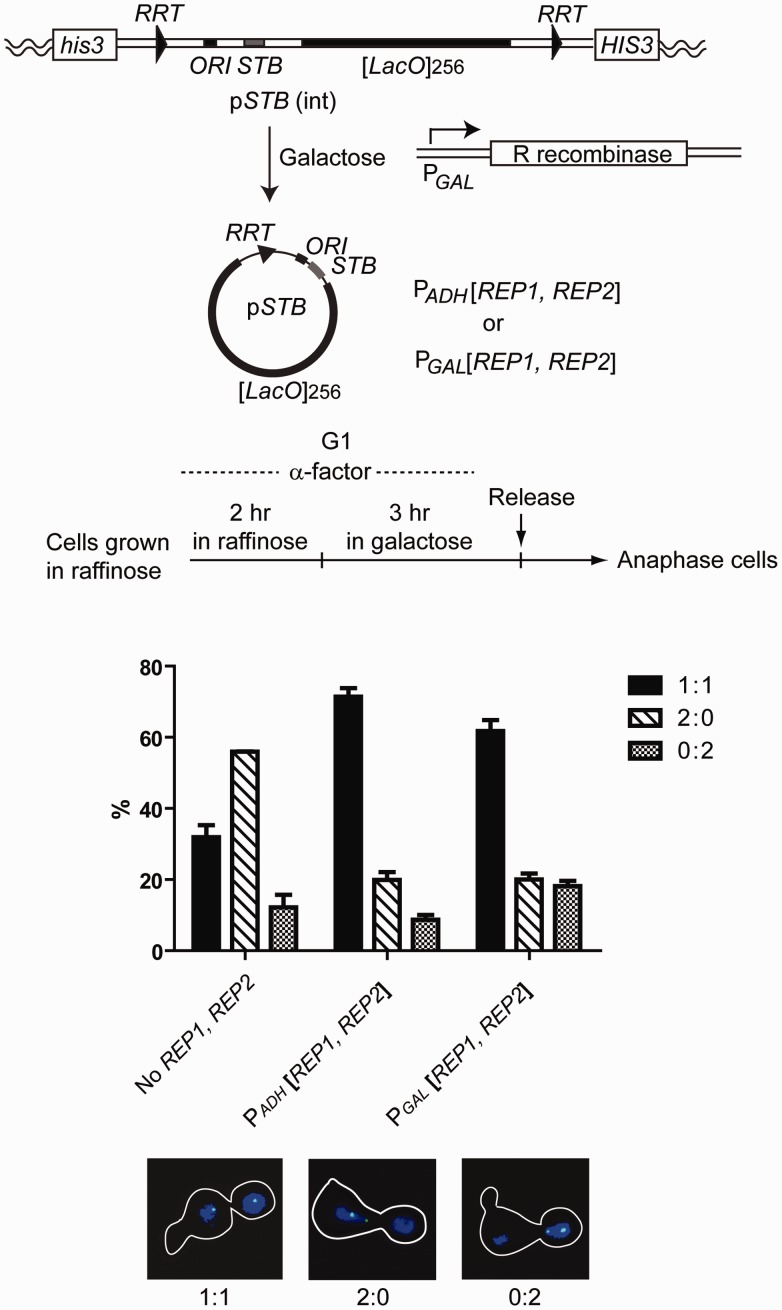
Figure 6.
Segregation of an STB reporter plasmid during a cell cycle immediately following its excision from a chromosomally integrated state during G1. The organization of the DNA cassette harboring STB, 2 micron plasmid replication origin (ORI) and [LacO]256 flanked by the target sites for the R recombinase present at the HIS3 locus of a [cir0] host strain expressing GFP-LacI is schematically diagrammed. The strain also contained the R recombinase under the GAL promoter and GFP-LacI under the HIS3 promoter. In two individual derivatives of this strain, the REP1 and REP2 genes were placed under the GAL promoter or under the ADH promoter. The experimental scheme for arresting raffinose grown cells in G1, inducing the R recombinase and assaying plasmid segregation in released cells at anaphase, is outlined. The frequencies of equal (1:1) and unequal (2:0 or 0:2) segregations scored in anaphase cells are plotted. 2:0 and 2:0 types of missegregation denote the bias towards the mother and daughter, respectively. The strains for this set of assays were MJY5060 to MJY5062 (Supplementary Table S1).
Cells arrested in G1 were shifted to galactose, to induce R recombinase in both strains and the Rep proteins in one, for a time sufficient to excise the plasmid in >95% of the cells. After release from arrest, the percentage of 1:1 plasmid segregation scored in anaphase cells was nearly the same whether Rep proteins were present during the previous cell cycle (Figure 6). By contrast, in cells lacking the REP1-REP2 expression cassette, the frequency of 1:1 plasmid segregation was markedly lower. When the assay was performed in glucose (i.e. without recombinase induction), the segregation pattern was 1:1 in every anaphase cell, as expected for the high fidelity of chromosome segregation (data not shown).
Thus, the plasmid segregation machinery functions competently even when all preceding cell cycles had been completed in the absence of the Rep proteins, and the reporter plasmid was never before partitioned by the Rep-STB system. Neither the memory of a previous partitioning event nor the association with the Rep proteins during the preceding cell cycle is important for STB function.
DISCUSSION
In this study, we have revealed the interactions between the 2 micron plasmid partitioning system and the RSC2 chromatin remodeling complex and established their functional relevance to plasmid segregation. The recruitment of RSC2 subunits as well as additional host factors at STB and their subsequent release from STB during the cell cycle follow a temporal sequence. Although the assembly of the partitioning complex is completed within a rather narrow window of the cell cycle, the disassembly phase covers a much wider time span. Initiating the partitioning pathway at a cell cycle requires DNA replication, but not a previous round of plasmid partitioning.
As previously noted, as host factors that interact with STB also function at CEN and contribute to chromosome segregation, the caveat that they affect 2 micron plasmid segregation indirectly through their effects on chromosome segregation cannot be discounted. Except for inactivating the Rep-STB partitioning system, very few conditions have been successful in uncoupling plasmid segregation from chromosome segregation. These include running the cell cycle under spindle disassembly from G1 through G2/M followed by spindle restoration (8) or under inactivation of the Kip1 motor (3) or the Rsc2 protein (17). All three conditions increase plasmid missegregation with no effect or only minor effects on chromosome segregation. Delayed spindle assembly does not block cohesin-mediated pairing of sister chromatids or their bi-orientation on the spindle, whereas the absence of the Kip1 motor and the RSC2 complex is largely compensated for by the redundant Cin8 motor and RSC1 complex, respectively. The recruitment of cohesin at STB is dependent on Kip1 and Rsc2 (3,10), whereas it is delayed, and rendered non-effectual, by delayed spindle assembly (8). These results suggest that at least a subset of the host factors that associate with STB in a cell cycle dependent-manner are germane to plasmid partitioning. However, the suggested potential kinship between CEN and STB (31) raises the possibility that some of the host factor–STB interactions might be no more than evolutionary relics without functional significance in plasmid partitioning.
The role of the RSC2 complex in plasmid partitioning
Two forms of the SWI/SNF-related RSC chromatin remodeling complex have been identified in S. cerevisiae, RSC1 and RSC2, with multiple shared components including the ATPase Sth1 and at least two unique components Rsc1 and Rsc2, as indicated by their respective designations (53,54). Although deletion of either RSC1 or RSC2 is tolerated by the cell, their simultaneous deletion causes lethality. Previous work demonstrated that rsc2Δ, but not rsc1Δ, point mutations in Rsc2 and conditional inactivation of Sth1 lead to increased loss of the 2 micron plasmid (17,48). These findings are consistent with the lack of cohesin assembly at STB in the rsc2Δ background or when Sth1 is inactivated (10,48). The association of Sth1 with the 2 micron plasmid has been shown to be restricted to the STB region (48). The recruitment of Rsc2 or Rsc8 at STB is blocked in the presence of a non-functional form of Cse4 (6). Furthermore, in the absence of Rsc2, Rep1 fails to associate with STB, but Rep2 association is not abrogated (10). The micrococcal nuclease digestion pattern at STB-proximal, spanning the repeat elements of STB, is altered in an rsc2Δ strain (17). Cleavages at two strong sensitive sites present at its borders and a weaker site present internally are conspicuously diminished in the absence of Rsc2, whereas such changes are not observed in other regions of the 2 micron plasmid genome. These observations, as well as the interactions of RSC subunits with the Rep proteins and the increased plasmid loss in rsc8 and rsc58 mutants revealed in the present study, argue for the RSC2 complex being an authentic 2 micron plasmid partitioning factor.
The Rsc1 and Rsc2 proteins share 45% identity, and each harbors two bromodomains, an AT-hook motif, a bromodomain-adjacent homology region and a C-terminal domain (54). The specific role for Rsc2, despite the organizational similarities between Rsc1 and Rsc2, in 2 micron plasmid partitioning is rather surprising. However, there is precedent for the use of only one of two redundant or nearly redundant host factors by the plasmid, e.g. the Kip 1 but not Cin8 nuclear motor (3). This selectivity in selfishness is perhaps a safeguard against potentially disrupting the host’s normal physiology.
A potential alternative role for Rsc2 in plasmid partitioning
If the 2 micron plasmid segregates by hitchhiking on chromosomes, as would be consistent with currently available evidence, the possibility that Rsc2 might potentially mediate plasmid attachment to chromosomes cannot be ruled out. By analogy to the episomes of Epstein–Barr virus, bovine papilloma virus and Kaposi’s sarcoma-associated herpes virus, which also use the hitchhiking mode of segregation (55–59), the AT-hook motif and the bromodomains harbored by Rsc2 would be consistent with such a role. The mechanisms used by the partitioning proteins of these viruses to tether viral episomes to host chromosomes include binding to AT-rich chromosomal regions through the AT-hook activity as well as interactions with the chromatin-associated bromodomain containing Brd4 protein (58–60). Rsc2-mediated tethering of the 2 micron plasmid to chromosomes, if true, cannot last through the entire segregation period, as the protein dissociates from STB before anaphase. If the hitchhiking model holds for the plasmid, there must be some additional mechanism for maintaining plasmid-chromosome association, at least after Rsc2 leaves STB. The higher plasmid instability caused by rsc2Δ reported previously (17) compared with rsc8 or rsc58 (this study) could be owing to the dual role of Rsc2, in tethering plasmids to chromosomes and as a component of the RSC2 complex.
Cell cycle cued assembly of the plasmid partitioning complex at STB: parallels to kinetochore assembly at CEN
Dissociation of components from the spent partitioning complex to set a new plasmid partitioning cycle in motion during G1-S is reminiscent of analogous events that occur at centromeres. Outer kinetochore proteins such as Ctf19 and Mtw1 are disassembled during replication of centromeres, causing them to detach from microtubules (61). Subsequent kinetochore reassembly permits recapture of centromeres by the spindle. Furthermore, the turnover of Cse4 at CEN during early S phase suggests renewal of inner kinetochore proteins as well (62,63). Although kinetochore turnover appears to be restricted to a short early period of the cell cycle, the disassembly of the plasmid partitioning complex is spread over a larger time span: from S phase though the following G1-S transition period. Dissociation of RSC2 components from STB precedes plasmid segregation, disassembly of cohesin triggers (and coincides with) this event, and dissociation of Cse4, Kip1 and the Rep proteins occurs following segregation.
Based on several tantalizing pieces of circumstantial evidence, including the unique presence of 2 micron related plasmids in Saccharomycetaceae, it has been speculated that the point centromere of budding yeasts and STB might share a common evolutionary history (31). The association of common protein factors with CEN and STB, despite their obvious functional differences at present, would be consistent with their divergence from a common ancestor.
Replication dependent sequential association of partitioning factors at STB
The final steps in the disassembly of the preceding partitioning complex and the initial steps in the assembly of a new partitioning complex are cued by DNA replication itself or by cell cycle signals that set the time of replication. It is possible that proteins present at STB could be mechanically dislodged by the advancing replication fork. Alternatively, the activation of the 2 micron circle replication origin, without requiring the act of replication, may be sufficient to communicate the signal for protein reorganization at STB, located less than a kbp away.
Within the resolution permitted by the short interval ChIP assays, Rsc8 and Rsc58 associate with STB before Cse4, whereas Rsc2 and Sth1 do so concurrently with Cse4. As Sth1 is the ATPase component of the RSC2 complex, this sequence of events is consistent with the RSC2 complex remodeling STB chromatin following its interaction with Cse4. There is evidence to suggest that RSC promotes remodeling of centromeric and flanking nucleosomes subsequent to the incorporation of Cse4 into the CEN nucleosome (19).
The temporal separation in the association of RSC components with STB is unexpected. However, the association of Sth1 with STB in the absence of Rsc2 has been described in a previous study (48). According to this study, Rsc8 associates with STB at the same time as Sth1, whereas in our analysis, Rsc8 precedes Sth1 in its occupancy at STB. This discrepancy may be owing to the lower time resolution (15 min rather than 5 min intervals) used in the earlier study.
2 micron plasmid segregation: current status
Although several attributes of the 2 micron plasmid partitioning system have been characterized, we are still ignorant of the precise molecular mechanisms of plasmid partitioning. The association of Cse4 and cohesin with STB (5–7,16,48) and the one-to-one segregation of single copy reporter plasmids (4) would be consistent with a model in which sister copies of the plasmid paired by cohesin associate with sister chromatids that are also bridged by cohesin. The organization of a fluorescence tagged multi-copy STB reporter plasmid suggests that the native plasmid molecules form a limited number of individual groups within the nucleus (9). How the sister chromatid association model would operate in this context is unclear. Furthermore, the highly substoichiometric nature of Cse4 and cohesin association with STB (5,16) raises uncertainties regarding the functional relevance of these associations. Perhaps they are relics of STB and CEN evolution from a common ancestral partitioning locus and are unrelated to present day plasmid partitioning. However, it is possible that the assembly of multiple plasmid molecules into functional groups, thus effectively reducing copy number, might help overcome the deficit in stoichiometry.
Single copy (or close to single copy) STB plasmids harboring a conditional CEN that we have previously used are valuable for probing partitioning mechanisms, as they report unambiguously equal segregation (1:1) and unequal segregation (2:0) in anaphase cells (3,4). However, there are potential drawbacks to introducing a CEN sequence within an STB plasmid to obtain this low copy number. The CEN-inactive plasmid may not be strictly under Rep-STB control during a cell cycle that immediately follows CEN annulment. For example, the carryover effects of CEN-mediated plasmid localization, host factor associations, kinetochore assembly and segregation from previous cell cycles may not be inconsequential.
Our current thinking on possible models of 2 micron plasmid segregation has been strongly influenced by the premise that every segregation event is equal or almost equal. If unequal segregations are frequent, there should be continuous operation of the plasmid amplification system. However, biochemical and physical analyses indicate that nearly all plasmid molecules (≥90%) in a cell population replicate only once per cell cycle (49). The quantitative meaning of ‘equal’ segregation will be determined by how tight or broad the copy number distribution is around the mean (∼40–60 molecules per cell), what threshold drop in copy number is required for triggering amplification and what the rate of amplification per generation is? Depending on these parameters, it is possible that the Rep-STB system can confer chromosome-like stability on the plasmid even without chromosome-like fidelity in segregation and without rapid corrections of copy number decline by amplification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–4, Supplementary References [64–66].
FUNDING
The National Institutes of Health (NIH) [GM064363 to M.J., P41 RR0111823 to J.Y.]; Robert F Welch Foundation [F-1274 to M.J. (in part)]; National Science Foundation [MCB1049925 to M.J. (in part)]. Funding for open access charge: NIH [GM064363 to M.J., P41 RR0111823 to J.Y.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Biggins, M. Carlson, A. Kikuchi, B. Laurent, K. Nasmyth, K. Shirahige and T. U. Tanaka and T. Taneda for gifts of yeast strains and plasmids used in this study. We are grateful to S. Stevens for providing advice on the tandem affinity purification strategies and protocols.
REFERENCES
- 1.Ghosh SK, Hajra S, Paek A, Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 2.Jayaram M, Yang X-M, Mehta S, Voziyanov Y, Velmurugan S. The 2 micron plasmid of Saccharomyces cerevisiae. In: Funnell BE, Phillips G, editors. Plasmid Biology. Washington DC: ASM Press; 2004. pp. 303–324. [Google Scholar]
- 3.Cui H, Ghosh SK, Jayaram M. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J. Cell Biol. 2009;185:251–264. doi: 10.1083/jcb.200810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh SK, Hajra S, Jayaram M. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc. Natl Acad. Sci. USA. 2007;104:13034–13039. doi: 10.1073/pnas.0702996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh SK, Huang CC, Hajra S, Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 2010;38:570–584. doi: 10.1093/nar/gkp993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajra S, Ghosh SK, Jayaram M. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-micron circle partitioning locus and promotes equal plasmid segregation. J. Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta S, Yang XM, Chan CS, Dobson MJ, Jayaram M, Velmurugan S. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta S, Yang XM, Jayaram M, Velmurugan S. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2 micron plasmid-cohesin association. Mol. Cell. Biol. 2005;25:4283–4298. doi: 10.1128/MCB.25.10.4283-4298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velmurugan S, Yang XM, Chan CS, Dobson M, Jayaram M. Partitioning of the 2-micron circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang XM, Mehta S, Uzri D, Jayaram M, Velmurugan S. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol. Cell. Biol. 2004;24:5290–5303. doi: 10.1128/MCB.24.12.5290-5303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futcher AB. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 12.Volkert FC, Broach JR. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 13.Murray JA, Scarpa M, Rossi N, Cesareni G. Antagonistic controls regulate copy number of the yeast 2 micron plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds AE, Murray AW, Szostak JW. Roles of the 2 micron gene products in stable maintenance of the 2 micron plasmid of Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Som T, Armstrong KAF, Volkert C, Broach JR. Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 16.Huang CC, Hajra S, Ghosh SK, Jayaram M. Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: implications for centromere evolution. Mol. Cell. Biol. 2011:1030–1040. doi: 10.1128/MCB.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong MC, Scott-Drew SR, Hayes MJ, Howard PJ, Murray JA. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 micron plasmid maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:4218–4229. doi: 10.1128/MCB.22.12.4218-4229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- 23.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 26.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbon J, Clarke L. Centromere structure and function in budding and fission yeasts. New Biol. 1990;2:10–19. [PubMed] [Google Scholar]
- 29.Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray JA, Cesareni G. Functional analysis of the yeast plasmid partition locus STB. EMBO J. 1986;5:3391–3399. doi: 10.1002/j.1460-2075.1986.tb04655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat. Struct. Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 32.Tsalik EL, Gartenberg MR. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast. 1998;14:847–852. doi: 10.1002/(SICI)1097-0061(19980630)14:9<847::AID-YEA285>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 34.Diop SB, Bertaux K, Vasanthi D, Sarkeshik A, Goirand B, Aragnol D, Tolwinski NS, Cole MD, Pradel J, Yates JR, 3rd, et al. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 2008;9:260–266. doi: 10.1038/embor.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bern M, Goldberg D, McDonald WH, Yates JR., 3rd Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20(Suppl. 1):i49–54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 36.Eng J, McCormack A, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein data base. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 37.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome. Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 38.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR., 3rd Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome. Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 39.Cociorva D, Tabb DL, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics. 2007 doi: 10.1002/0471250953.bi1304s16. Chapter 13, Unit 13.4. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Severin F, Hyman AA, Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn YT, Wu XL, Biswal S, Velmurugan S, Volkert FC, Jayaram M. The 2 micron-plasmid-encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J. Bacteriol. 1997;179:7497–7506. doi: 10.1128/jb.179.23.7497-7506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott-Drew S, Murray JA. Localisation and interaction of the protein components of the yeast 2 micron circle plasmid partitioning system suggest a mechanism for plasmid inheritance. J. Cell Sci. 1998;111:1779–1789. doi: 10.1242/jcs.111.13.1779. [DOI] [PubMed] [Google Scholar]
- 45.Huang CC, Chang KM, Cui H, Jayaram M. Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc. Natl Acad. Sci. USA. 2011;108:13671–13676. doi: 10.1073/pnas.1101944108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velmurugan S, Ahn YT, Yang XM, Wu XL, Jayaram M. The 2 micron plasmid stability system: analyses of the interactions among plasmid- and host-encoded components. Mol. Cell. Biol. 1998;18:7466–7477. doi: 10.1128/mcb.18.12.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 48.Huang J, Hsu JM, Laurent BC. The RSC nucleosome-remodeling complex is required for Cohesin’s association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 49.Zakian VA, Brewer BJ, Fangman WL. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979;17:923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]
- 50.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 53.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 54.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 55.Botchan M. Hitchhiking without covalent integration. Cell. 2004;117:280–281. doi: 10.1016/s0092-8674(04)00410-6. [DOI] [PubMed] [Google Scholar]
- 56.Frappier L. Viral plasmids in mammalian cells. In: Funnell BE, Phillips G, editors. Plasmid Biology. Washington DC: ASM Press; 2004. pp. 325–340. [Google Scholar]
- 57.McBride AA. Replication and partitioning of papillomavirus genomes. Adv. Virus Res. 2008;72:155–205. doi: 10.1016/S0065-3527(08)00404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 59.You J, Srinivasan V, Denis GV, Harrington WJ, Jr, Ballestas ME, Kaye KM, Howley PM. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 63.Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell. 43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buvelot S, Tatsutani SY, Vermaak D, Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piatti S, Bohm T, Cocker JH, Diffley JF, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 66.Taneda T, Kikuchi A. Genetic analysis of RSC58, which encodes a component of a yeast chromatin remodeling complex, and interacts with the transcription factor Swi6. Mol. Genet. Genomics. 2004;271:479–489. doi: 10.1007/s00438-004-0999-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.