Abstract
Zinc-finger nucleases (ZFNs) and TAL effector nucleases (TALENs) have been shown to induce targeted mutations, but they have not been extensively tested in any animal model. Here, we describe a large-scale comparison of ZFN and TALEN mutagenicity in zebrafish. Using deep sequencing, we found that TALENs are significantly more likely to be mutagenic and induce an average of 10-fold more mutations than ZFNs. We observed a strong correlation between somatic and germ-line mutagenicity, and identified germ line mutations using ZFNs whose somatic mutations rates are well below the commonly used threshold of 1%. Guidelines that have previously been proposed to predict optimal ZFN and TALEN target sites did not predict mutagenicity in vivo. However, we observed a significant negative correlation between TALEN mutagenicity and the number of CpG repeats in TALEN target sites, suggesting that target site methylation may explain the poor mutagenicity of some TALENs in vivo. The higher mutation rates and ability to target essentially any sequence make TALENs the superior technology for targeted mutagenesis in zebrafish, and likely other animal models.
INTRODUCTION
Zinc-finger nucleases (ZFNs) and TAL effector nucleases (TALENs) have recently emerged as powerful tools for generating targeted genomic mutations. These proteins bind specific DNA sequences and induce double-strand DNA breaks that are repaired by non-homologous end joining (1), an error-prone process that often results in insertion or deletion (indel) mutations. ZFNs have been studied for several years, but their widespread use has been limited by the difficulty of targeting them to specific DNA sequences. Selection assays for identifying zinc fingers that bind specific targets are laborious and challenging for non-specialist laboratories (2,3). An alternative method, known as modular assembly, combines pre-selected zinc-finger modules into arrays (1). These ZFNs are relatively easy to generate but have low success rates (4), although significant progress has recently been made (5,6). Proprietary methods have also been used to generate ZFNs that are effective in zebrafish (7), but these nucleases must be purchased and are expensive. Another approach, context-dependent assembly (CoDA) (8), does not require selection assays and was claimed to have a success rate comparable with selection-based methods. However, the sequences that can be targeted using CoDA are limited (8) and, as for all ZFN technologies, there is no established code for specific zinc finger/DNA interactions. In contrast, TALENs contain a variable number of repeated modules that each preferentially binds a specific nucleotide. Therefore, TALENs can in principle be targeted to any DNA sequence without the need for selection assays. Furthermore, TALENs can be constructed using standard molecular biology techniques (9). Both ZFNs and TALENs can induce mutations in zebrafish (3,5–15), but these approaches have not been extensively tested and compared for mutagenicity in any animal model. Here, we describe a large-scale analysis and comparison of ZFN and TALEN mutagenicity in developing zebrafish embryos.
MATERIALS AND METHODS
Animal care
All experiments were performed by mating TL and AB wild-type zebrafish strains using standard protocols (16) in accordance with the California Institute of Technology Institutional Animal Care and Use Committee guidelines.
Construction of ZFNs and TALENs
ZFNs and TALENs were designed using the ZiFIT Targeter (http://zifit.partners.org/ZiFiT) (9,17). DNA fragments encoding zinc-finger arrays were synthesized (Epoch Life Science Inc.) and cloned into FokI EL/KK (18) heterodimeric expression vectors by BamHI/XbaI (pMLM290 and pMLM292) or NotI/XbaI (pMLM800 and pMLM802) digestion (19). Each ZFN contained three zinc fingers. TALE repeat arrays were constructed using the REAL Assembly TALEN Kit (9) and were cloned into the wild-type FokI expression vectors JDS70, JDS71, JDS74 and JDS78. All TALENs were sequence verified before mRNA synthesis. Plasmids were obtained from the non-profit plasmid repository Addgene.
RNA transcription and injection
ZFN and TALEN expression plasmids were linearized with PmeI and purified using the polymerase chain reaction (PCR) Purification Kit (Qiagen). mRNA was synthesized using 500 ng of purified linear DNA as template and the mMessage mMachine T7 Ultra kit (Ambion). The transcription reaction yielded ∼20 µg of polyA tailed mRNA, which was dissolved in 20 µl of nuclease-free water. mRNA synthesis and polyA tailing were verified by agarose gel electrophoresis. Final mRNA concentrations ranged from 0.8 to 1.2 µg/µl. Approximately 50–100 pg of each ZFN or TALEN mRNA was injected into the cell of zebrafish embryos at the one-cell stage. mRNA concentrations that were sufficient to cause developmental defects in 10–50% of injected embryos were used to assay for somatic mutations and to generate germ line mutants.
Analysis of somatic mutations
For each ZFN and TALEN, genomic DNA was prepared from 12 injected embryos at 72 h post-fertilization (h.p.f.) as previously described (19). Embryos were incubated in 500 µl of sodium dodecyl sulfate lysis buffer [10 mM of Tris pH 8.0, 200 mM of NaCl, 10 mM of ethylenediaminetetraacetic acid (EDTA), 0.5% of sodium dodecyl sulfate, 100 µg/ml of proteinase K] overnight at 50°C with occasional gentle mixing until no clumps were visible. Genomic DNA was then purified by phenol/chloroform extraction and dissolved in 40 µl of TE (10 mM of Tris pH 8.0, 0.5 mM of EDTA). Targeted genomic regions were amplified using Amplitaq (Invitrogen), with amplicons ranging in size from 175 to 350 bp. Because of the short Illumina sequence read length, for each PCR reaction, one primer was designed to anneal 6–12 bp away from the spacer. PCR products were purified using the PCR Purification Kit (Qiagen) and pooled at roughly equal molar ratios. Sequencing libraries were prepared by following the Illumina TruSeq Genomic DNA protocol without the DNA fragmentation step. The pooled PCR products were end repaired, A-tailed and ligated to TruSeq single-index adaptors. The adaptor-ligated DNA was gel purified and PCR amplified to produce finished libraries. The libraries were sequenced using an Illumina GAIIx machine in the single read 38-nt mode, producing 34.1 million reads, and using an Illumina HiSeq2000 machine in the single-read 50-nt mode, producing 144.1 million reads.
Detecting indels using short-read data is challenging because the commonly used alignment algorithms developed for high-throughput sequencing data, such as bowtie (20) and ELAND, cannot map discontinuous reads. Aligners that use the split-read method, which maps defined portions of the read separately before creating the final alignment, such as tophat (21) and ELAND2, suffer from poor sensitivity of indel detection because of the requirement for the indel site to fall between the mapped portions of the read. Custom implementations of the split-read method suffer from similar sensitivity problems. Also, many split-read aligners, including tophat, were designed for the alignment of RNA-Seq data and are much more sensitive in aligning discontinuous reads that span splice junctions, which is not the case for our data.
To detect indels rigorously, we used a combination of two methods. We first aligned reads to target genomic regions using the SHRiMP2 software package (22,23), which allowed us to identify small indels with high sensitivity. SHRiMP2 uses the vectorized Smith–Waterman algorithm for local alignment during the candidate mapping location identification phase, followed by the full Smith–Waterman alignment to detect single-nucleotide polymorphisms and indels. It has been shown to have the highest sensitivity of the currently available short-read aligners (22,23). Despite its high sensitivity for small indels, SHRiMP2 is unable to map reads across large insertions or deletions. To identify such events in our data, we mapped reads that failed to align with SHRiMP2 using BLAT, which is capable of finding regions of high similarity separated by large gaps (24). Mappings produced by SHRiMP2 were output in the SAM format, whereas the BLAT-native psl format was converted to SAM using the psl2sam.pl script provided by the samtools package (25). The SAM format-defined CIGAR representation of the alignment for each read was then used to identify insertions and deletions using a custom perl script (available on request). We then removed indels that were closer than 17 nt, the length of the shortest PCR primer, to the 5′-end of the read, or did not have at least a 5-nt continuous match on the 3′-end, which may correspond to incorrect alignments. We also filtered out 1-nt indels, which may result from PCR or sequencing errors. The filtered indels produced by SHRiMP and BLAT were merged to produce the final indel list.
Isolation of germ line mutants
Zebrafish embryos injected with a ZFN or TALEN pair were raised to adulthood and mated to other potential founders or wild-type fish. Depending on the somatic mutation rate, genomic DNA was isolated from a pool of 1–6 embryos at 72 h.p.f., with up to 96 embryos tested for each fish. Embryos were incubated in embryo lysis buffer (10 mM of Tris–HCl pH 7.5, 1 mM of EDTA, 50 mM of KCl, 0.3% of Tween 20, 0.3% of NP40) for 10 min at 98°C. Proteinase K was then added to a final concentration of 1 mg/ml, and the lysis reaction was incubated overnight at 55°C, followed by 10 min at 98°C. One microliter of this solution was used as template for PCR.
Targeted genomic regions were amplified using Amplitaq (Invitrogen) with amplicons ranging in size from 200 to 550 bp. In cases where an indel was expected to delete a restriction enzyme site, half of the PCR reaction was digested with the appropriate restriction enzyme and analysed by agarose gel electrophoresis. In cases where the targeted region did not overlap with a restriction enzyme site, PCR was performed using a standard forward primer and a reverse primer fluorescently labelled with 6-FAM, HEX (Integrated DNA Technologies) or NED (Applied Biosystems). For PCR using fluorescently labelled primers, we performed a final 1-h incubation at 60°C to ensure that an extra adenosine was added to the 3′-end of all PCR products. Fluorescent PCR products were run on an ABI 3730 DNA analyzer (Applied Biosystems), and PCR product sizes were analysed using Peak Scanner (Applied Biosystems). To confirm indel sequences, genomic DNA from single embryos was amplified and sub-cloned using the Strataclone PCR Cloning Kit (Stratagene), and DNA from several independent colonies was sequenced. DNA sequences were analysed using SeqBuilder and SeqMan (DNAStar Lasergene).
Statistical methods
Mutation rate and size data do not follow a normal distribution; therefore, we used non-parametric methods to analyse these data. The non-parametric Wilcoxon rank-sum test (also known as Mann–Whitney U-test or Mann–Whitney–Wilcoxon test) was used to examine statistically significant differences in measurement variables (mutation rate or size) between two nominal variables (ZFN versus TALEN). Correlations were used to test whether pairs of variables co-vary. Pearson’s linear correlation was used to test interval data, whereas Spearman’s rank correlation was used to test ordinal data. Results of the correlations are reported as the r2-value (coefficient of determination) and P-value. Data are displayed as scatter or box plots, and significant linear correlations were additionally displayed with a regression line. All tests were two-tailed, and alpha level was P < 0.05.
RESULTS
Few CoDA ZFNs induce somatic mutations in vivo
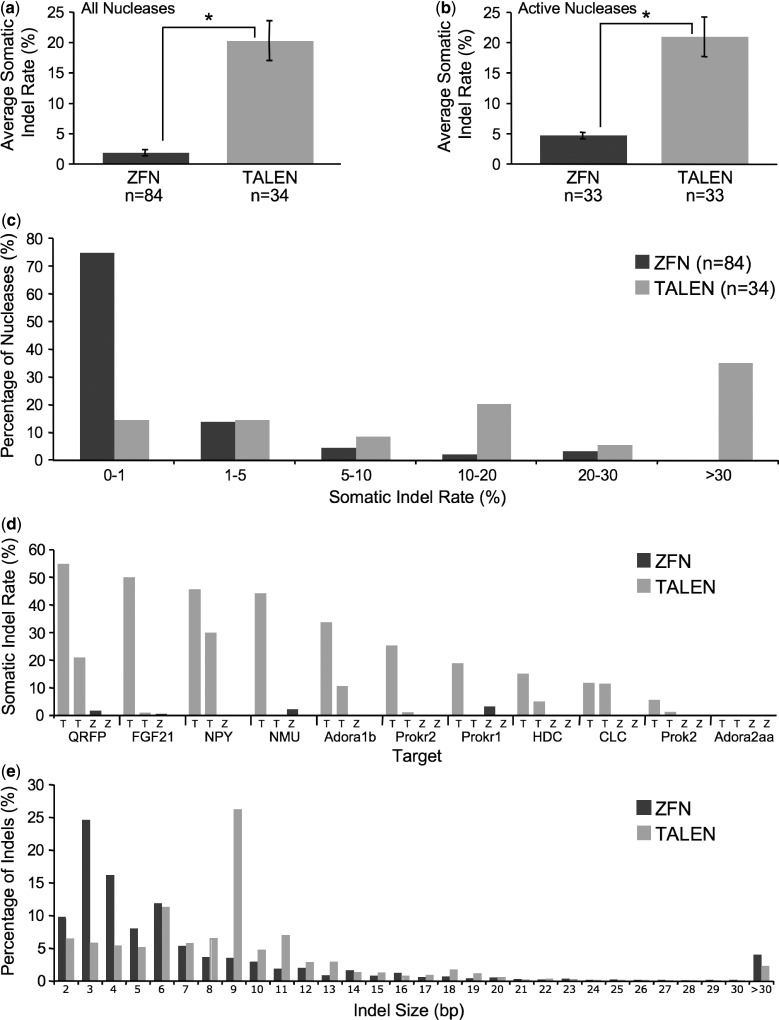
In a previous study, 12 of 24 (50%) CoDA ZFNs induced somatic mutations in zebrafish, with indel rates between 1 and 17% (8). However, these ZFNs were pre-screened for activity using a bacterial reporter assay. Because only ∼75% of CoDA zinc fingers were found to be active in the reporter assay (8), the success rate of CoDA ZFN pairs that are not pre-screened was estimated at ∼28% (50% × 75% left ZFN × 75% right ZFN). Another study found that 3 of 17 (18%) CoDA ZFNs that were not pre-screened induced somatic mutations in zebrafish, with indel rates of 1–3% (11). To more comprehensively evaluate CoDA ZFN mutagenicity in vivo, we generated 84 ZFN pairs targeting 66 zebrafish genes. We screened for indels using deep sequencing, generating an average of 1 200 000 reads per target. In contrast, most studies have analysed ≤96 sequence reads; hence, ZFNs that induced indels at rates <1% would likely not have been identified as mutagenic.
Of the 84 ZFN pairs tested, 21 (25%) induced indels in >1% of sequence reads, in close agreement with the predicted rate of 28% (8,11), and only 5 (6%) produced indel rates >10% (Figure 1c, Supplementary Figure S1 and Supplementary Table S1). Thus, CoDA ZFNs have relatively low success rates and few CoDA ZFNs induce mutations at high rates. A genomic region that was not targeted by a ZFN gave an indel rate of 0.009% (Supplementary Table S1), indicating a false-positive rate of ∼0.01%. Surprisingly, many ZFNs induced indels at low frequencies (Figure 1c, Supplementary Figure S1 and Supplementary Table S1). Fifty-four ZFNs (64%) induced indels at rates between the negative control value of 0.01% and 1%, and 18 (21%) induced indels at rates between 0.1% and 1%. Only nine (11%) produced indels at frequencies below the negative control. Thus, most CoDA ZFNs are mutagenic in vivo but induce mutations at frequencies below the commonly used threshold of 1%. A caveat to using Illumina-based deep sequencing for indel detection is that one primer must be located close to the targeted region because of the short-sequence read length. Therefore, deletions removing more than ∼9 bp beyond the spacer on one side of the targeted region will not be detected. Indeed, of the 20 ZFN-induced germ line mutations in 8 genes that we confirmed using Sanger sequencing (see below), 2 would not have been detected using deep sequencing, suggesting that our methodology may underestimate indel rates by ∼10%, although the sample size is modest. An additional caveat is that our requirement that both the 5′-and 3′-ends of a sequence read align to its reference sequence (see ‘Materials and Methods’ section) will result in a failure to detect large insertions. However, none of the 91 germ line mutations that we identified (see below) contain insertions that are too large to align to their reference sequence using our approach, suggesting that this is unlikely to significantly affect quantification of somatic mutation rates.
Figure 1.
TALENs induce somatic indels at higher rates than ZFNs in zebrafish embryos. Average somatic indel rates are shown for all 84 ZFN and 34 TALEN pairs tested (a) and for the 33 ZFN and 33 TALEN active pairs (b). Nucleases that induce somatic indels at rates >0.27% are defined as active, because this rate is sufficient to generate germ line mutations. The difference in mutation rates for ZFNs and TALENs is statistically significant, indicated by asterisks, with P = 5.1 × 10−12 for all nucleases (a) and P = 6.0 × 10−5 for active nucleases (b) using the Wilcoxon rank-sum test. (c) Distribution of somatic indel rates for ZFNs and TALENs. Most ZFNs induced somatic indels at frequencies <1%, whereas most TALENs induced indels at significantly higher rates. (d) Somatic indel rates for 11 genes that were targeted with one or two pairs of ZFNs and TALENs. For 10/11 genes, a TALEN pair induced higher indel rates than a ZFN pair. Z and T indicate ZFN and TALEN data. See Supplementary Table S4 for indel rate values. (e) Distribution of somatic indel sizes for ZFNs and TALENs. TALEN-induced indels were significantly larger than ZFN-induced indels, with P = 2.2 × 10−16 using the Wilcoxon rank-sum test. Median indel sizes were 4nt and 9nt for ZFNs and TALENs, respectively. Data are based on 5 527 940 and 2 893 241 sequence reads containing indels induced by TALENs and ZFNs, respectively.
Germ line mutations can be generated using ZFNs that have low somatic indel rates
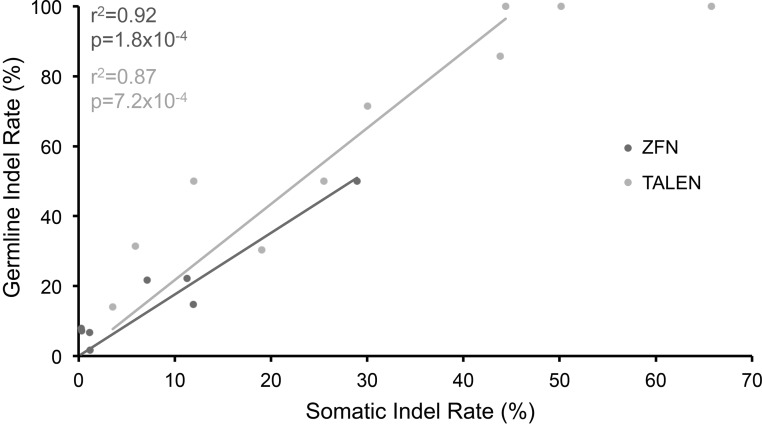
To determine the relationship between somatic and germ line mutation rates, we isolated germ line mutants using ZFNs that induced somatic mutations at a range of frequencies. We observed a strong correlation between somatic and germ line mutation rates (Figure 2, r2 = 0.92, P = 1.8 × 10−4 for eight ZFN pairs). Surprisingly, ZFNs with somatic indel rates as low as 0.27% and 0.33% produced germ line mutations in 8% (3/38) and 7% (3/42) of fish, respectively. Thus, ZFNs whose somatic indel rates are well below the commonly used threshold of 1% can be used to isolate germ line mutants at reasonable frequencies. As a result, it should be possible to isolate germ line mutations using at least 39% (33/84) of the ZFNs that we tested (Supplementary Table S1); we refer to these ZFNs as active. Because our false-positive rate is 0.01%, ZFNs with somatic indel rates <0.27% might be useful for generating germ line mutations. For all mutations analysed, the number of mutant embryos produced by each founder was <50%, indicating that mutations were present in a subset of founder germ cells (Supplementary Table S2a). Germ line indels ranged in size from a 5-bp insertion to a 61-bp deletion (Figure 3). ZFNs induced similar somatic and germ line indel sizes regardless of mutation rate (Figure 3 and Supplementary Figure S2a).
Figure 2.
Relationship between rates of somatic and germ line mutations. There is a significant correlation between rates of somatic and germ line indels for ZFNs (r2 = 0.92, P = 1.8 × 10−4, n = 8 ZFNs using Pearson’s correlation) and TALENs (r2=0.87, P = 7.2 × 10−4, n = 8 TALENs using Pearson’s correlation). The linear regression lines are shown. Three TALENs induced germ line mutations in 100% of injected fish. Among these three TALENs, only the TALEN with the lowest somatic mutation rate was used in the Pearson’s correlation calculation and to generate the linear regression line to avoid a ceiling effect.
Figure 3.
Sequences of ZFN-induced germ line mutations. ZFN target sequences and spacer sequence are highlighted in yellow and grey, respectively. Deletions are indicated by red dashes and insertions are highlighted in blue. Only mutations that were analysed using Sanger sequencing are shown.
Most TALENs induce somatic mutations in vivo
TALEN technology has recently been applied to zebrafish (9,11–15). We sought to comprehensively assess the use of TALENs for zebrafish mutagenesis and compare them to ZFNs. We used the REAL method (9) to generate 34 TALEN pairs that target 18 genes. Twenty-one (62%) of the TALENs induced somatic indels at rates >10%, 29 (85%) at rates >1% and all at rates >0.1% (Figure 1c, Supplementary Figure S1 and Supplementary Table S3). 33/34 (97%) induced somatic indels at rates >0.27%, and should therefore generate germ line mutations at reasonable frequencies. The single TALEN pair below this threshold had an indel rate of 0.16%, which is still above the false-positive rate of 0.01%. We conclude that most, if not all, TALENs can induce somatic mutations, and in this respect TALENs are superior to CoDA ZFNs. A potential caveat to this conclusion is that we compared ZFNs containing FokI EL/KK heterodimers with TALENs containing FokI homodimers. However, this difference is unlikely to underlie the higher mutagenicity observed for TALENs, as TALENs containing FokI heterodimers exhibit similar or higher mutation rates than those containing homodimers (13), and the EL/KK heterodimers used in our study and the ELD/KKR FokI heterodimers primarily used by Cade et al. (13) induce mutations at similar rates in zebrafish (11). Nevertheless, ELD/KKR heterodimers have been shown to be more active than EL/KK heterodimers in some contexts (26), and they might partially account for the difference in ZFN and TALEN mutation rates in our study.
Somatic and germ line mutation rates are similarly correlated for ZFNs and TALENs
We isolated germ line mutations using several TALEN pairs and observed a strong correlation between somatic and germ line mutation rates (Figure 2, r2 = 0.87, P = 7.2 × 10−4 for eight TALEN pairs). Notably, the slopes of the linear regression lines and correlation coefficients are similar for TALENs and ZFNs, indicating that the relationship between somatic and germ line mutation rates is similar for ZFNs and TALENs. TALEN-induced germ line indels ranged in size from a 23-bp insertion to a 203-bp deletion (Supplementary Figure S3 and Supplementary Table S2b). TALENs induced similar somatic and germ line indel sizes regardless of mutation rate (Supplementary Figures S2b and S3).
Mutagenic TALENs exhibit higher indel rates than mutagenic CoDA ZFNs
Having found that TALENs are much more likely than CoDA ZFNs to be active (i.e. somatic indel rate >0.27%), we next compared their somatic mutagenicity. The average somatic indel rates for the 84 ZFNs and 34 TALENs were 2% and 20%, respectively (Figure 1a and Supplementary Figure S1). If we only compare active nucleases, the indel rates were 5% and 21%, respectively (Figure 1b). These differences are statistically significant (P = 5.1 × 10−12 for all nucleases, P = 6.0 × 10−5 for active nucleases) and result from a shift in the distribution of indel rates for TALENs compared with ZFNs and higher rates for the most mutagenic TALENs (Figure 1c and Supplementary Figure S1). For 10/11 genes that were targeted using both ZFNs and TALENs, at least one TALEN pair was more active than a ZFN pair, in most cases by a large margin (Figure 1d and Supplementary Table S4). Therefore, not only are TALENs more likely than CoDA ZFNs to be active, they also induce mutations at higher rates. The distribution of indel sizes is also significantly different for ZFN- and TALEN-induced mutations (P = 2.2 × 10−16), with median indel sizes of 4nt and 9nt, respectively (Figure 1e).
Published guidelines do not predict ZFN or TALEN mutagenicity in vivo
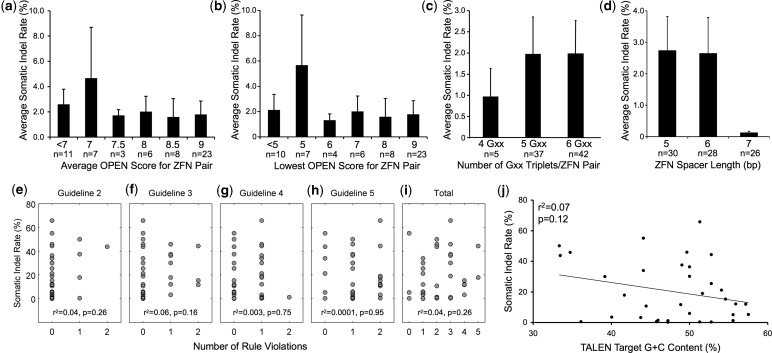
Several guidelines have been proposed to select optimal ZFN and TALEN target sites but have not been extensively tested in an animal. We used our data set to evaluate how well these guidelines predict success in generating mutations in vivo. First, we used the confidence score provided by the ZiFiT Targeter for ZFNs that are generated using the OPEN method (27). This score is based on analysis of zinc fingers that activate transcription of a reporter gene in a bacterial assay, which is correlated with ZFN activity (2,10,28–30). Confidence scores range from 0 to 9, with 9 indicating the greatest likelihood of mutagenicity. We compared scores for the 58 CoDA ZFN pairs that we tested that could have been generated using the OPEN method, but found no correlation between confidence score and indel rate (Figure 4a and b, P = 0.69 and P = 0.65 for average and lowest score of ZFN pairs). Second, it has been suggested that ZFNs are more likely to be mutagenic if most or all nucleotide triplets in the target sequence start with a guanine (3,4,31–36). We found no correlation between mutation rate and target sites containing four, five or six Gxx triplets (Figure 4c, P = 0.30). ZFN targets containing only four Gxx triplets might have lower mutation rates, but our data set contains few of these cases, and the difference that we observed is not statistically significant (Figure 4c). However, we did observe a significant negative correlation between the target sequence spacer length and mutagenicity (Figure 4d, P = 0.005). ZFN targets with a 7-bp spacer had much lower mutation rates than those with 5- or 6-bp spacers.
Figure 4.
ZFN and TALEN targeting guidelines do not predict mutagenicity in zebrafish embryos. There is no correlation between somatic indel rate and the average (a) or lowest (b) OPEN score for each ZFN pair [r2 = 0.003, P = 0.69 for (a) and r2 = 0.004, P = 0.65 for (b) using Spearman’s rank correlation]. Data for 58 ZFN pairs, for which ZiFiT OPEN scores are available, are shown. (c) There is no correlation between somatic indel rate and the presence of four, five or six Gxx triplets in the ZFN target sequence (r2 = 0.01, P = 0.30 using Spearman’s rank correlation). ZFN targets containing four Gxx triplets may have lower mutation rates but do not reach a significant threshold. (d) There is a correlation between mutation rate and ZFN spacer length (r2 = 0.09, P = 0.005 using Spearman’s rank correlation). ZFN targets containing 7-bp spacers have lower mutation rates than those containing 5- or 6-bp spacers. There is no significant difference in mutation rates for ZFN targets containing 5- or 6-bp spacers (P = 0.42 using the Wilcoxon rank-sum test). For (a–d), n indicates number of ZFN pairs. Panels (e–i) show somatic indel rates for each TALEN pair for which neither, one or both target half-sites violate one of four design guidelines: (e) no T at position 1, (f) no A at position 2, (g) T at last position and (h) nucleotide composition within 2 standard deviations of natural TALE targets (A = 0 to 63%, C = 11 to 63%, G = 0 to 25%, T = 2 to 42%) (37). Panel (i) shows the total number of guideline violations for each target site (maximum number of violations = 8). There is no correlation between somatic mutation rate and violation of any of these guidelines, as determined using Spearman’s rank correlation (see r2 and P-values on graphs e–i). (j) There is no correlation between TALEN-induced mutation rate and target sequence G + C content (r2 = 0.07, P = 0.12 using Pearson’s correlation). The linear regression line is shown. Analysis in (e–j) was performed using 34 TALEN pairs.
We also evaluated guidelines proposed for designing mutagenic TALENs. First, Cermak et al. (37) proposed guidelines based on TAL effectors found in nature. In addition to the well-established requirement that TALEN binding sites should be preceded by a T (Guideline 1) (37–39), which we followed for all TALENs tested, they suggest that TALEN binding sites should not have a T at position 1 (guideline 2) or an A at position 2 (guideline 3) and should have a T at the last position (guideline 4). They also suggest that TALEN targets should have a nucleotide composition within 2 standard deviations of the average nucleotide composition of natural TALE targets (guideline 5). We failed to detect correlations between TALEN mutagenicity and any of these guidelines (Figure 4e–i and Supplementary Table S5, P > 0.16 for all guidelines). Second, Streubel et al. (40) generated artificial TALE arrays and tested their ability to activate a reporter gene in plant cells. Based on their results, they proposed that TALENs should contain at least three to four repeats that bind C or G, whereas stretches of repeats that interact with A or T should be avoided, especially at the ends of target sites. We found no correlation between indel rate and target G + C content (Figure 4j, r2 = 0.07, P = 0.12), although none of our targets contained fewer than four G + C nucleotides. We also did not observe a relationship between indel rates and A/T repeats. Two TALEN target sites contained stretches of six or seven A/T nucleotides, yet the TALENs targeting these sites exhibited high-somatic mutation rates (Supplementary Table S5). We note, however, that the guidelines suggested by Streubel et al. are based on monomeric TALE proteins and may have less impact in the context of TALEN dimers. Finally, we found that diverse sequences can serve as nuclease targets (Supplementary Figure S4), similar to observations in human cells (41). We conclude that these guidelines have little or no predictive power for TALEN mutagenicity in zebrafish.
TALEN mutagenicity is negatively correlated with the number of CpG repeats in the target site
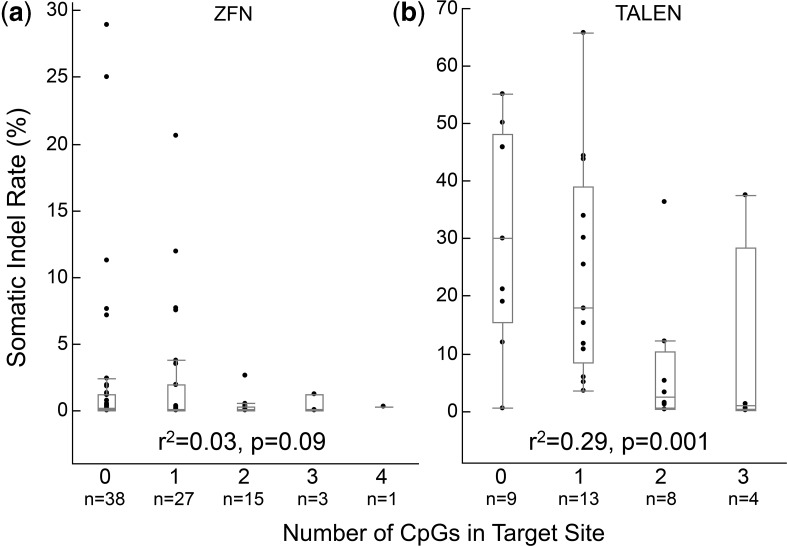
It has been shown that binding of TALE domains to their targets is inhibited by 5-methylated cytosine (5mC) and that demethylation of CpG repeats can improve TALE activity in human and rodent cells (42,43). Consistent with these reports, we observed a significant negative correlation between TALEN-induced somatic mutation rates and the number of CpG repeats in target sites (Figure 5b and Supplementary Table S6, r2 = 0.29, P = 0.001). TALEN targets containing zero or one CpG repeat exhibited significantly higher mutation rates than those containing two or three CpG repeats (Figure 5b and Supplementary Table S6). In support of this observation, we found that three smaller zebrafish studies that used the same TALEN architecture as our study (9,11,13) showed a similar effect (Supplementary Table S7). In particular, in one study that targeted 10 zebrafish genomic sites, the three targets containing no CpG repeats had the highest mutation rates, whereas the two targets containing five CpG repeats were not mutagenic [Supplementary Table S7 (13)]. We also analysed data from a large-scale study of TALEN mutagenicity in human cells (41) but found no correlation between mutation rates and the number of CpG repeats in the target (Supplementary Figure S5b and Supplementary Table S8, r2 = 0.01, P=0.25), although targets with no CpG repeats had higher average and median mutation rates than those containing 1—4 CpGs. The basis for the discrepancy between zebrafish and human cells is unknown, but may result from different CpG methylation patterns. In contrast to TALENs, we did not observe a significant correlation between ZFN mutagenicity and the number of CpG repeats in target sites (Figure 5a, Supplementary Figure S5a and Supplementary Table S9, r2 = 0.03, P = 0.09). We conclude that, at least for zebrafish studies, TALEN target sites should contain no more than one CpG repeat.
Figure 5.
Somatic mutation rate of TALENs but not ZFNs is negatively correlated with the number of CpG repeats in the target site. There is no significant correlation between somatic mutation rate and the number of CpG repeats in ZFN target sequences (a, r2 = 0.03, P = 0.09 using Spearman’s rank correlation), but there is a significant negative correlation for TALEN target sequences (b, r2 = 0.29, P = 0.001 using Spearman’s rank correlation). Data points represent somatic indel rates for each of the 84 ZFN (a) and 34 TALEN (b) target sites that we tested. The box within each plot indicates the middle half of the data, and the line in each box indicates the median. The lines extending from the box indicate the farthest data point that is within 1.5 interquartile ranges from the first and third quartiles. Individual data points outside these lines are possible outliers. N indicates number of target sites in a category.
DISCUSSION
ZFNs and TALENs have been used to generate somatic and germ line mutations in zebrafish (3,5–13,15) and relatively small-scale studies have suggested that TALENs may be superior mutagens (9,11–15). However, large-scale comparisons of ZFN- and TALEN-induced mutations have not been reported in any animal. Here, we describe a large-scale, deep sequencing-based comparison of ZFN and TALEN mutagenicity in zebrafish that provides several insights.
First, as has been suggested by other studies (9,11–13), we found that TALENs are significantly more likely to be mutagenic than ZFNs generated using CoDA. Eighty-five percent of the 34 TALEN pairs that we tested induced somatic indels at rates >1%, compared with only 25% of the 84 ZFNs that we tested. Furthermore, all TALENs induced mutations at rates significantly greater than our false-positive rate of 0.01%. We also found that TALENs generated significantly more mutations than ZFNs. The average somatic mutation rates were 20% and 2% for TALENs and ZFNs, respectively, indicating that TALENs are on average 10-fold more mutagenic than ZFNs. Second, by using deep sequencing to screen for somatic mutations, we found that germ line mutants can readily be isolated using ZFNs whose somatic mutation rates are at least as low as 0.27%, which is well below the standard cut-off of 1%, in accordance with observations using a different ZFN methodology (5). If we use a somatic indel rate of 0.27% as a threshold to define nucleases that can generate germ line mutants at reasonable frequencies, 97% of the TALENs and 39% of the ZFNs that we tested are active in zebrafish, with average mutation rates of 21% and 5%, respectively. This analysis also revealed that 89% of ZFNs induced somatic indels at rates above our false-positive rate of 0.01%, suggesting that most ZFNs generated using CoDA are mutagenic, although it may be difficult to isolate germ line mutations using ZFNs that have somatic indel rates <0.27%. Third, we used our relatively large data set to test whether several guidelines that have been proposed to select optimal TALEN and ZFN target sites are useful predictors of mutagenicity in vivo (27,37,40). Our results indicate that none of these guidelines have strong predictive power in zebrafish, and we conclude that they should not be a factor in choosing ZFN and TALEN target sites.
Although published guidelines did not predict ZFN or TALEN mutagenicity in vivo, we did find that TALEN mutagenicity is negatively correlated with the number of CpG repeats in the target sequence. TALEN targets containing zero or one CpG repeat exhibited significantly higher mutation rates than those containing two or three CpG repeats. Although not noted in their analysis, three published zebrafish studies that used the same TALEN architecture as our study showed a similar effect (Supplementary Table S7) (9,11,13). These results suggest that target CpG methylation may inhibit TALEN mutagenicity. We note that two TALENs with two or three CpG repeats exhibited high mutation rates in our study (Supplementary Table S6), but it is possible that these sites are not methylated in vivo. Consistent with these observations, the presence of 5mC in target DNA can inhibit TALE activity, and demethylation can improve TALE activity, in human and rodent cells (42,43). It has been noted (42) that CpG methylation may be associated with some of the non-mutagenic TALENs described in a large-scale analysis of TALENs in human cells (41), although we found no significant correlation between mutagenicity and the number of target site CpGs in the overall data set (Supplementary Table S8). The cause of the discrepancy between the zebrafish and human cell culture results is unclear, but may result from different CpG methylation patterns in the human cell line that was used compared with developing zebrafish embryos. It has been shown that use of the TALE repeat N* rather than HD at 5mC residues can increase TALEN activity, likely because of reduced steric hindrance of N* compared with HD with the 5mC methyl moiety (42). Taken together, these observations suggest that CpG methylation could be a significant factor in the low mutagenicity of some TALENS, and that targeting CpG residues using N* rather than HD could significantly improve mutagenicity at these targets. Alternatively, we suggest that TALEN targets should not contain more than one CpG repeat, at least for zebrafish studies. In contrast to TALENs, we did not observe a significant correlation between ZFN mutagenicity and the number of target site CpG repeats (Supplementary Table S9).
Studies using plasmid-based and artificial genomic reporters in human cell lines and Xenopus oocytes found that ZFNs whose targets contain 5- or 7-bp spacers were less mutagenic than those containing 6-bp spacers (44–46). In contrast, we found that endogenous ZFN targets containing 5- or 6-bp spacers exhibited similar mutation rates, consistent with several studies using animal models (5,8,11). We also found that ZFN targets containing 7-bp spacers were 4- to 5-fold less likely to be mutagenic and had significantly lower mutation rates than targets containing 5- or 6-bp spacers, even though we used ZFN architectures that were shown to be optimal for 5-, 6- and 7-bp spacers in human cells (44), suggesting that targets containing 7-bp spacers should be avoided.
Our results are consistent with the largest test of TALEN mutagenicity to date, which targeted 96 human genes in cell culture (41). In this study, 88% of TALENs produced mutation rates of 2–56%, with an average of 22%. In comparison, 85% of the TALENs that we tested produced mutation rates of 1–66%, with an average of 20%. Similar to our observations, this study failed to detect correlations between target selection guidelines (37) and mutation rates (41). These results indicate that TALENs mutate endogenous genes at similar rates in human cells and developing zebrafish embryos.
Our results demonstrate that TALENs are highly effective in generating mutations in zebrafish, and that essentially all TALENs are capable of inducing mutations, although mutation rates vary considerably. This variation is likely not completely due to the number of CpG repeats in target sequences, because targets containing the same number of CpG repeats exhibit a wide range of mutation rates (Supplementary Tables S6 and S7), although this might result from different methylation patterns in vivo. Further work is therefore needed to understand the basis of this variability, which should be facilitated by high-throughput methods for TALEN construction (41,47,48).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–9 and Supplementary Figures 1–5.
FUNDING
National Institutes of Health (NIH) [R00 NS060996, R01 NS070911 and R01DA031367 to D.A.P., F31NS077842 to S.C., F31NS077844 to J.L.]; Edward Mallinckrodt Jr., Rita Allen and Brain and Behavior Research Foundations (to D.A.P.); Della Martin Fund (to D.A.P. and G.O.); Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jason Stein for statistical assistance, Scot Wolfe for sharing sequence alignment algorithms and Catherine Oikonomou for comments on the manuscript.
REFERENCES
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 13.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug Ii RG, Tan W, Penheiter SG, Ma AC, Leung AY, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th edn. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- 17.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 19.Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat. Protoc. 2009;4:1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput. Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics. 2011;27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 24.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 27.Sander JD, Reyon D, Maeder ML, Foley JE, Thibodeau-Beganny S, Li X, Regan MR, Dahlborg EJ, Goodwin MJ, Fu F, et al. Predicting success of oligomerized pool engineering (OPEN) for zinc finger target site sequences. BMC Bioinformatics. 2010;11:543. doi: 10.1186/1471-2105-11-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 32.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 33.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Stormo GD. Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics. 2008;24:1850–1857. doi: 10.1093/bioinformatics/btn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 39.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 40.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 41.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valton J, Dupuy A, Daboussi F, Thomas S, Marechal A, Macmaster R, Melliand K, Juillerat A, Duchateau P. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J. Biol. Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu Y, Bhakta MS, Segal DJ. Restricted spacer tolerance of a zinc finger nuclease with a six amino acid linker. Bioorg. Med. Chem. Lett. 2009;19:3970–3972. doi: 10.1016/j.bmcl.2009.02.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Li J, Huang H, Wang G, Jiang M, Yin S, Sun C, Zhang H, Zhuang F, Xi JJ. An integrated chip for the high-throughput synthesis of transcription activator-like effectors. Angew. Chem. Int. Ed. Engl. 2012;51:8505–8508. doi: 10.1002/anie.201203597. [DOI] [PubMed] [Google Scholar]
- 48.Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.