Abstract
Single-stranded DNA (ssDNA) binding proteins are important in basal metabolic pathways for gene transcription, recombination, DNA repair and replication in all domains of life. Their main cellular role is to stabilize melted duplex DNA and protect genomic DNA from degradation. We have uncovered the molecular function of protein domain family domain of unknown function DUF2128 (PF09901) as a novel ssDNA binding domain. This bacterial domain strongly associates into a dimer and presents a highly positively charged surface that is consistent with its function in non-specific ssDNA binding. Lactococcus lactis YdbC is a representative of DUF2128. The solution NMR structures of the 20 kDa apo-YdbC dimer and YdbC:dT19G1 complex were determined. The ssDNA-binding energetics to YdbC were characterized by isothermal titration calorimetry. YdbC shows comparable nanomolar affinities for pyrimidine and mixed oligonucleotides, and the affinity is sufficiently strong to disrupt duplex DNA. In addition, YdbC binds with lower affinity to ssRNA, making it a versatile nucleic acid-binding domain. The DUF2128 family is related to the eukaryotic nuclear protein positive cofactor 4 (PC4) family and to the PUR family both by fold similarity and molecular function.
INTRODUCTION
Single-stranded DNA (ssDNA) binding proteins, termed SSBs, are ubiquitous in nature and are essential in transcription, repair and recombination metabolism (1). SSBs interact strongly and non-specifically with unwound DNA, thereby preventing the formation of secondary structure elements and its degradation by nucleases. In Escherichia coli, SSBs play an integral role as genome maintenance agents that initiate and stimulate the DNA repair machinery. The oligosaccharide/oligonucleotide-binding domain (OB) fold is the recognized structural signature of SSBs in eubacteria.
Single-stranded-binding domains that deviate from the canonical OB fold were identified more recently. Among these domains are the positive cofactor 4 (PC4)/Sub1 (2), the PUR-α (3) and Deinococcus radiodurans DdrB (4). The PC4 domain binds non-specifically ssDNA as dimers, whereas PUR (purine-rich binding) domains preferentially bind purine-rich (NGG)n ssDNA and RNA repeats (5). DdrB is an SSB with a novel fold and is key to D. radiodurans resistance to ionizing radiation damage (6). The PC4 domain was thought to be unique in the eukaryotic domain (7), whereas the PUR superfamily was shown to have representatives in both the eukaryotic and prokaryotic kingdoms (8). These multifunctional domains play a number of distinct roles as transcription co-regulators by interacting with basal factors, in mRNA transport and in DNA repair pathways (3). PC4 has shown disparate functions, acting as both a co-activator of transcription factor-mediated RNA Pol-II transcription (9) and as a repressor of Pol-II-mediated transcription by preventing its phosphorylation (10). Although their affinity to double stranded DNA (dsDNA) may only be sufficient to weaken the helix (11), the domains have the ability to sequester ssDNA while sliding or translocating freely along the chain (12).
Before this work, protein domain family domain of unknown function DUF2128 (PF09901) was a family of functionally uncharacterized proteins found exclusively in prokaryotes (13). The domain family was targeted for structural studies by the Protein Structure Initiative (14) as part of a broad effort in structural coverage of proteins identified in the human gut metagenomic sequencing projects (15). The sequence homology of this domain family was too low to be matched with sufficient accuracy to any other known superfamily, but clues to its biochemical function could be gleaned from the knowledge of its structure. The 72-residue (8.40 kDa) YdbC protein from Lactococcus lactis is a representative member of this protein domain family. Because of its use in dairy fermentations and its GRAS (generally regarded as safe) status, L. lactis is an important industrial microorganism. Its uses are increasingly expanding to applications in medicine, including the delivery of recombinant proteins to humans (16). Many features in the proteome of this important microorganism remain to be uncovered. Here, we present solution nuclear magnetic resonance (NMR) structural and ssDNA binding studies of L. lactis YdbC. The protein exhibits unexpectedly high-structural similarity to the symmetric homodimer structures of PC4 and PUR-α eukaryotic ssDNA-binding domains, suggesting a potential ssDNA binding function for this protein. We demonstrate that L. lactis YdbC forms a tight complex with ssDNA, adopting a structure that closely resembles that of PC4 and characterize the binding energetics by microcalorimetry. Moreover, we show that YdbC can partially disrupt a 26-base DNA duplex sequestering the resulting single strands and is capable to bind weakly to ssRNA. Using structure-based sequence and phylogenetic analyses, we place the DUF2128 protein domain family in its proper evolutionary context and merge the DUF2128 and the PC4 domain into the same superfamily.
MATERIALS AND METHODS
Sample preparation
The full-length YdbC protein from L. lactis, including a C-terminal His6 tag (LEHHHHHH), was cloned, expressed and purified following standard protocols in the literature to prepare [U-13C,15N]- and [U-5%-13C,100%-15N]-YdbC samples for NMR spectroscopy (17). Detailed descriptions of sample preparation and results of biophysical characterization, including analytical gel filtration, analytical ultracentrifugation, isothermal titration calorimetry (ITC) and NMR T1/T2 measurements can be found in Supplementary Methods and Supplementary Figure S1–S4. Protocols for the preparation of YdbC:ssDNA, YdbC:dsDNA and ssRNA samples are also detailed in the Supplementary Methods. This expression vector is available as KR150.21.1 from the Protein Structure Initiative Materials Repository (http://psimr.asu.edu/).
Structure determination and analysis
The solution NMR structures of apo-YdbC and YdbC:dT19G1 complex were calculated using NOESY data collected under identical conditions and parameters. NMR protocols are detailed in the Supplementary Methods section. Initial apo-YdbC structures were calculated with CYANA 3.0 (18) using resonance assignments, NOESY peak lists from 3D 13C-edited, 15N-NOESY and F1-13C/15N-filtered, F3-13C-edited NOESY spectra, dihedral restraints derived from TALOS+ (19) and two sets of 1H-15N residual dipolar couplings (RDCs). Symmetry identity dihedral and distance restraints were imposed between the two protomers to calculate 100 initial structures within CYANA 3.0. The final 20 structures with the lowest target functions were, subsequently, refined by restrained molecular dynamics (rMD) in explicit water, non-crystallographic symmetry and the PARAM19 parameters using CNS 1.3 (20,21). Identical protocol was followed for initial YdbC:dT19G1 structures calculations. The structure was computed with the knowledge that a single species in solution must include symmetric protein dimer and symmetric ssDNA units bound to each YdbC protomer. Symmetry was enforced both during initial CYANA calculations and later during energy refinement in explicit water bath. The program was supplied with the new chemical shifts (CS) resonance list, including ambiguous resonance assignments for thymidine, NOESY peak lists 13C/15N-edited 3D NOESY, 2D 1H-1H NOESY and 3D F1-13C/15N-filtered, F3-13C/15N-edited NOESY spectra and the revised TALOS+ dihedral restraints set for the complex. Symmetry identity dihedral and distance restraints were imposed between the two protomers and between the two dT chains. The ‘KEEP’ sub-routine was used in CYANA 3.0 to enforce the manually assigned protein:dT X-filtered peaks. The best 20 structures from the final cycle were then refined by rMD in a water bath, non-crystallographic symmetry and C2 symmetry and OPLSX parameters using the HADDOCK web server (22). For both the apo- and ssDNA-bound YdbC structure refinements, experimental restraints (nuclear Overhauser effect (NOE)-derived distance, dihedral and empirical hydrogen bond) were used in the final rMD calculations. Structural statistics and global structure quality scores for apo-YdbC and YdbC:dT19G1 were computed using the PSVS 1.4 software package (23). The global RDC statistics for apo-YdbC were computed using PALES (24). Single-stranded DNA geometry was analysed with the program 3DNA (25). The final coordinates (excluding the C-terminal hexa-His polypeptide segment) for the ensemble of 20 structures and NMR-derived restraints for both apo- and holo-YdbC were deposited to the Protein Data Bank (PDB) with IDs 2ltd and 2ltt, respectively. The CS assignments were deposited to the Biological Magnetic Resonance Data Bank with entries 18 469 and 18 496, respectively. Pairwise structure-based sequence alignments and coordinate superimpositions were obtained from the jCE server (26,27). 3D protein structure comparison of the apo-YdbC structure with structures in the Protein Data Bank was conducted using the DaliLite server (28). Conserved residue analysis was performed using the ConSurf server (29,30) using full-length sequences from the entire PF09901 (DUF2128) protein domain family (Pfam 26.0; 414 sequences) re-aligned with the ClustalW 2.0 server (31). Electrostatic surface potentials were computed for the first (lowest energy) model of the apo-YdbC ensemble using the APBS version 1.2.1 software package (32) and PDB2PQR version 1.6 server (33). Structure figures were made using PyMOL version 1.4 (www.pymol.org).
Isothermal titration calorimetry
ITC measurements were conducted at 25°C on an iTC200 microcalorimeter (MicroCal Inc., Northampton, MA, USA). All ITC measurements were performed in 10 mM of Tris buffer at pH 7.5 containing either 0, 50, 150 or 300 mM of NaCl. In each experiment, aliquots of a 220-µM solution of YdbC were sequentially injected from a 40-µl rotating syringe (1000 r.p.m.) into an isothermal sample chamber containing 210 µl of 8 µM of an ssDNA oligonucleotide either dT19G1, dC20, dA20 (The Midland Certified Reagent Company) or d(A–C)10 (Integrated DNA Technologies). In each experiment, the initial injection was 0.4 µl and 0.8 s in duration, whereas the remaining 19 injections were 2 µl and 4 s in duration with a 180 s delay between each injection. Each titration experiment was accompanied by the corresponding control experiment, in which YdbC was injected into a solution of buffer alone. Each injection generated a heat burst curve (µcal/s versus s), the area under which was determined by integration [using Origin version 7.0 software (MicroCal Inc., Northampton, MA, USA)], to obtain a measure of the heat associated with that injection. The measure of the heat associated with each YdbC-buffer injection, as estimated using a linear regression analysis of the integrated data, was subtracted from that of the corresponding heat associated with each YdbC–ssDNA injection to yield the heat of ssDNA binding for that injection. After removal of the point corresponding to the first low volume injection, the buffer-corrected ITC profiles for the binding of each YdbC–ssDNA experiment were fit models for either one set or two sets of binding sites.
Sequence analysis
Representative homologues of the L. lactis subspecies lactis sequence YdbC (ID 15672295), of the Homo sapiens PC4 (ID 62088150), and of the Borrelia burgdorferi PUR-α (ID 308198561) were selected in diverse taxonomic groups. BLASTP (34) was used to identify and retrieve these sequence homologues in genome and protein databases at NCBI (35). Furthermore, bacterial homologues of PC4 were identified with Protein Structure Initiative (PSI)-Basic Local Alignment Search Tool (BLAST). Sequences within each family were first aligned using Clustal W (36). Because of the low sequence similarity between the three families, these three alignments were manually aligned with BioEdit version 7.1 (Ibis Biosciences) on the basis of their structural similarity derived from jCE server (26,27). Sequence analysis was based on partial protein sequences encompassing the full-length DUF2128 domain and corresponding regions in PC4 and PUR-α sequences. Sixty-two positions were included in the analysis. Programs of the PHYLIP package (37) were used for tree construction. The final alignment was re-sampled 100 times with Seqboot (37). A matrix of distances was obtained with Protdist (37), and used for tree construction with the neighbour-joining program Neighbor (37), and a consensus tree was derived using the program Consense (37).
RESULTS
Apo-YdbC
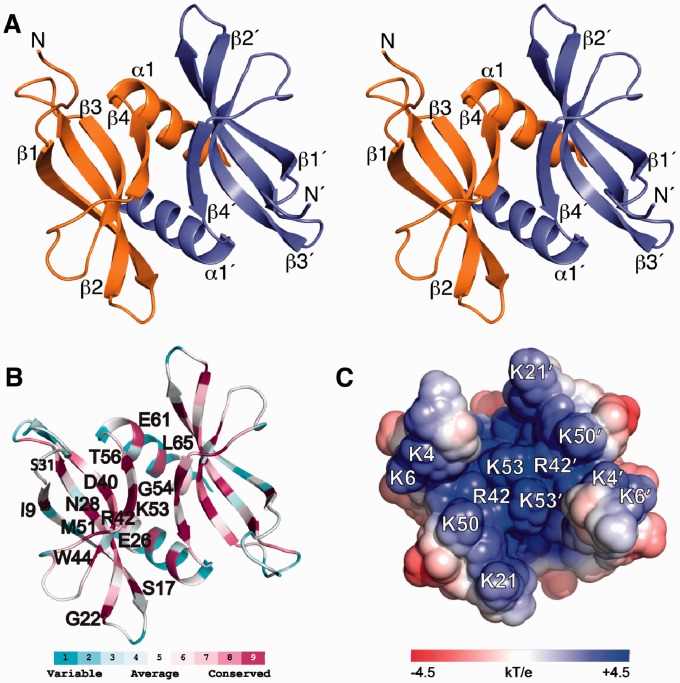
The structure of L. lactis YdbC adopts the dimeric PC4 fold as presented in the stereoview in Figure 1A. Secondary structure elements are as follows: 7–19 (β1, β1′), 22–32 (β2, β2′), 37–44 (β3, β3′), 51–57 (β4, β4′), 59–72 (α1, α1′). Each 72-residue protomer has a concave four-stranded antiparallel sheet followed by a C-terminal helix. Helices (α1, α1′) and strands (β4, β4′) from each subunit form the main dimer interface, which has a buried surface area of ∼2000 Å2. Structure statistics for apo-YdbC are listed in Table 1; the assignment and NOE maps are shown in Supplementary Figure S5; and the structure ensemble is shown in Supplementary Figure S6.
Figure 1.
Solution NMR structure of L. lactis apo-YdbC shown in the identical top-view orientation. (A) Stereoview of dimeric YdbC with labelled secondary structure elements and amino termini. (B) ConSurf (29,30) amino acid conservation mapped onto the lowest energy NMR structure. Highly conserved residues are labelled on the protein backbone of a single protomer. (C) Solvent exposed electrostatic potential (32) mapped onto the surface of apo-YdbC. Only the ssDNA-binding epitope is shown for clarity.
Table 1.
Summary of NMR Structural Statistics for apo-YdbC and YdbC:dT19G1 ensemblesa
| Data Type | apo | holo |
|---|---|---|
| Completeness of resonance assignmentsb | ||
| Backbone (%) | 97 | 91 |
| Side-chain (%) | 93 | 93 |
| Aromatic (%) | 100 | 100 |
| Stereospecific methyl (%) | 100 | 100 |
| Conformationally restricting restraintsc | ||
| NOE restraints | ||
| Total | 3142 | 2258 |
| Intra-residue (i = j) | 673 | 362 |
| Sequential (|i − j| = 1) | 819 | 562 |
| Medium range (1 < |i − j| < 5) | 480 | 288 |
| Long range (|i − j| ≥ 5) | 1170 | 1046 |
| NOE restraints/residue | 42 | 14 |
| Interchain protein/protein NOEs | 244 | 184 |
| Interchain protein/ssDNA NOEs | 254 | |
| Dihedral angle restraints | 330 | 446 |
| Hydrogen bond restraints | 128 | 120 |
| NH RDC restraints (polyethylene glycol (PEG)+phage) | 222 | |
| Number of restraints/residue (total/long range) | 48/16.6 | 17.4/6.5 |
| Residual constraint violationsc | ||
| Average distance restraint violations/structure | ||
| 0.1–0.2 Å | 19.8 | 30.9 |
| 0.2–0.5 Å | 3.6 | 11.7 |
| >0.5 Å | 0.0 | 0.3 |
| Average RMS of distance violation/restraint (Å) | 0.02 | 0.03 |
| Maximum distance violation (Å) | 0.45 | 0.61 |
| Average RMS dihedral angle violations/structure | ||
| >1°–10° | 18.1 | 40.1 |
| >10° | 0.4 | 1.35 |
| Average RMS dihedral angle violation/restraint | 1.0 | 1.1 |
| Maximum dihedral angle violation (°) | 11.0 | 20.5 |
| Model qualityc | ||
| RMSD from average coordinates (Å) | ||
| All backbone atoms (ordered/all) | 0.6/1.7 | 1.0/1.4 |
| All heavy atoms (ordered/all) | 0.9/2.3 | 0.4/0.4 |
| RMSD bond lengths (Å) | 0.018 | 0.004 |
| RMSD bond angles (°) | 1.3 | 0.7 |
| Molprobity Ramachandran plotd | ||
| Most favoured regions (%) | 95.7 | 90.9 |
| Additionally allowed regions (%) | 4.2 | 9.1 |
| Disallowed regions (%) | 0.1 | 0.0 |
| Global quality scores (Raw/Z-score)c | ||
| Procheck G-factor (ϕ,ψ)d | −0.47/−1.53 | −0.55/−1.85 |
| Procheck G-factor (all dihedrals)d | −0.18/−1.06 | −0.43/−2.54 |
| Verify3D | 0.38/−1.28 | 0.39/−1.12 |
| ProsaII | 0.40/−1.03 | 0.53/−0.50 |
| MolProbity clashscore | 14.52/−0.97 | 21.6/−2.18 |
| RPF scorese | ||
| Recall/Precison | 86.8/89.1 | |
| F measure/DP score | 87.9/71.8 | |
| Residual Dipolar Couplings (RDC) Scoresf | ||
| Q-factor (PEG/phage) | 0.20/0.18 | |
| R (PEG/phage) | 0.97/0.98 |
aStructural statistics were computed for the ensembles of 20 deposited structures (PDB ID: 2ltd and 2ltt) using PSVS (23).
bComputed for residues 1–74. Resonances that were not included were exchangeable protons (N-terminal NH3+, Lys NH3+, Arg NH2, Cys SH, Ser/Thr/Tyr OH) and Pro N, C-terminal carbonyl, side-chain carbonyl and non-protonated aromatic carbons.
cAverage distance constraints were calculated using the sum of r−6.
dOrdered residue ranges [S(ϕ) + S(ψ) > 1.8]:3–74 (chain A), 3–74 (chain B). Secondary structure elements APO: 7–19 (β1, β1′), 22–32 (β2, β2′), 37–44 (β3, β3′), 51–57 (β4, β4′), 59–72 (α1, α1′). Secondary structure elements HOLO: 7–17 (β1, β1′), 24–32 (β2, β2′), 36–44 (β3, β3′), 55–57 (β4, β4′), 59–72 (α1, α1′).
eRPF scores (38) reflecting the goodness-of-fit of the final ensemble of structures (including disordered residues) to the NOESY data and resonance assignments.
fResidual dipolar coupling quality scores (24).
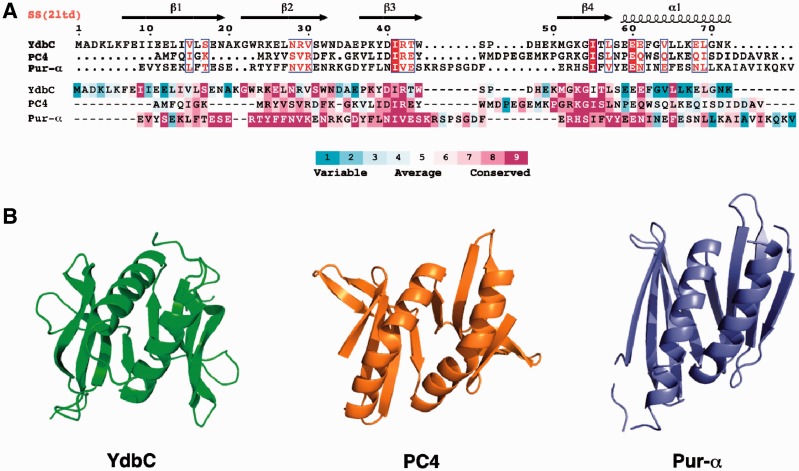
ConSurf (29,30) analysis of the DUF2128 sequences for the entire protein domain family is mapped onto the structure (Figure 1B) and YdbC sequence of L. lactis YdbC (Figure 2A). Conserved residues occur both in the centre of the concave β-sheet scaffold with side-chains extending into the concave side and in the β-strand that is part of the dimer interface (Figure 1B). Conservation within the DUF2128 is especially strong in the β3 (Asp40, Arg42 and Trp44) and β4 (Met51, Lys53, Gly54 and Thr56) strands. Within helix α1, conservation is limited to Glu61 and Leu65, which maybe key to fold stability. Several conserved positively charged residues are involved in ssDNA binding as discussed below. Clustering of basic residues Lys4, 6, 21, 50 and 53 and Arg42 bias the electrostatic distribution and produce strong, uniform positive charge on one face of the molecule (Figure 1C and Supplementary Figure S7). PC4-like fold and charge characteristics provide the first evidence for the function of YdbC as a nucleic acid-binding protein. The sequence identity determined by structure-based alignment (DALI or jCE) to the PC4 and PUR-α domains was found to be 15.3 and 11.8%, respectively (Figure 2B), and the corresponding Cα root-mean-square-deviation (RMSD) was found to be 2.6 and 4.0 Å. Significant residue conservation in the ssDNA-binding site, particularly on β3, and β4 was also found between YdbC and PC4, whereas between YdbC and PUR-α conservation is remote.
Figure 2.
(A) Structure-based sequence alignment (26,27) of L. lactis YdbC (DUF2128; PF09901), H. sapiens PC4 (PF02229) and B. burgdorferi PUR-α (DUF3276; PF11680). (Top) Sequence alignment rendered by ESPript (42) using default parameters for residue similarity calculations, where boxed residues represent identical (red box, white character) and similar (red character) amino acid conservation. (Bottom) Sequence alignment rendered using ConSurf (29,30) where residue conservation across individual protein domain families range from highly conserved (magenta) to variable (cyan). (B) Comparison of the solution NMR structure of L. lactis YdbC with crystal structures structurally similar apo-forms of dimeric ssDNA-binding proteins, H. sapiens PC4 (PDB ID: 1pcf) (43) and B. burgdorferi PUR-α (PDB ID: 3nm7) (8).
YdbC:dT19G1 complex
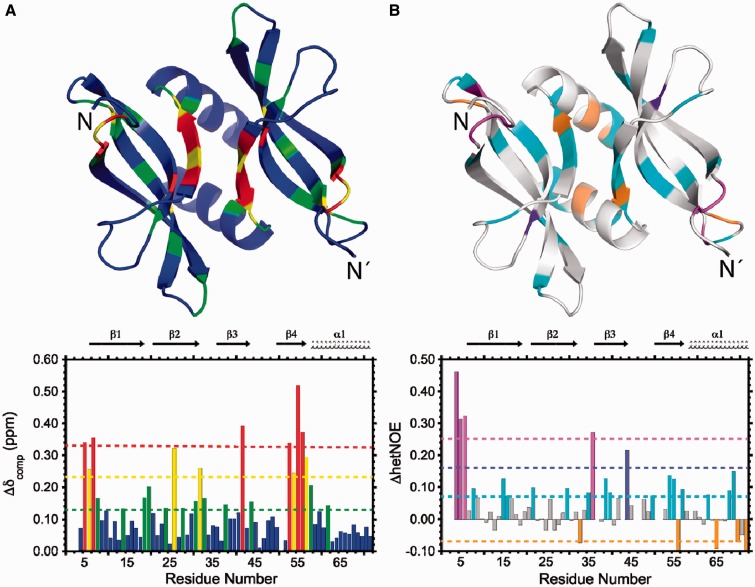
Strong backbone and side-chain chemical shift perturbations (CSPs) are observed on YdbC as a result of ssDNA binding. A 1H-15N heteronuclear single quantum coherence (HSQC) comparison of apo versus complex YdbC (Supplementary Figure S8) shows large variations in amide chemical shifts on binding, typical of slow exchange on the chemical-shift timescale and consistent with the nanomolar affinity of YdbC for poly-dT at low-salt NMR buffer conditions. Similar strong perturbations are visible in the 1H-13C HSQC for the YdbC residues both at the protein:protein and protein:DNA interface (i.e. Leu5 and others; data not shown). Full backbone CS perturbations for apo versus complex YdbC were computed (41) and mapped onto the apo-YdbC structure (Figure 3A). The strongest backbone CS differences are localized in the N-terminal region (residues 5–7) and at the dimer interface in β4 (residues 53–58). In addition, {1H}-15N heteronuclear NOEs (hetNOE) were measured for both apo- and dT19G1-bound YdbC (Supplementary Figure S9) and their difference (ΔhetNOE) is mapped onto the apo-YdbC structure (Figure 3B). To first approximation, the average increase in {1H}-15N hetNOE ratio (average ∼0.07) effect of complex versus apo indicates an overall increase in structural ordering on poly-dT binding. Ordering on poly-dT binding is predominant in the N-terminal region (residues 4–6) and, in addition, in the β2–β3 loop (residues 35–36) as discussed later in the text. We predict that these findings would be general for a variety of ssDNA sequences that bind with affinity similar to that of poly-dT as measured by ITC. CS assignment strategy and findings of bound-dT19G1 are described in Supplementary Figure S10 and S11.
Figure 3.
NMR characterization of poly-dT binding to L. lactis YdbC. (A) CSPs (Δδcomp) histogram. The bottom panel shows colour-coded residues defined according to the magnitude of the deviation from the mean CSP (green dotted line); yellow dotted line: mean + 1σ; red dotted line: mean + 2σ. The CSPs are mapped onto the apo-YdbC structure in tube representation. (B) {1H}-15N heteronuclear NOE difference (ΔhetNOE) between ssDNA-bound and apo-YdbC. The histogram (bottom panel) shows colour-coded residues defined according to magnitude of the deviation from the mean ΔhetNOE (cyan dotted line); purple dotted line: mean + 1σ; magenta dotted line: mean + 2σ. The ΔhetNOEs are mapped onto the apo-YdbC structure in tube representation with the same colouring scheme.
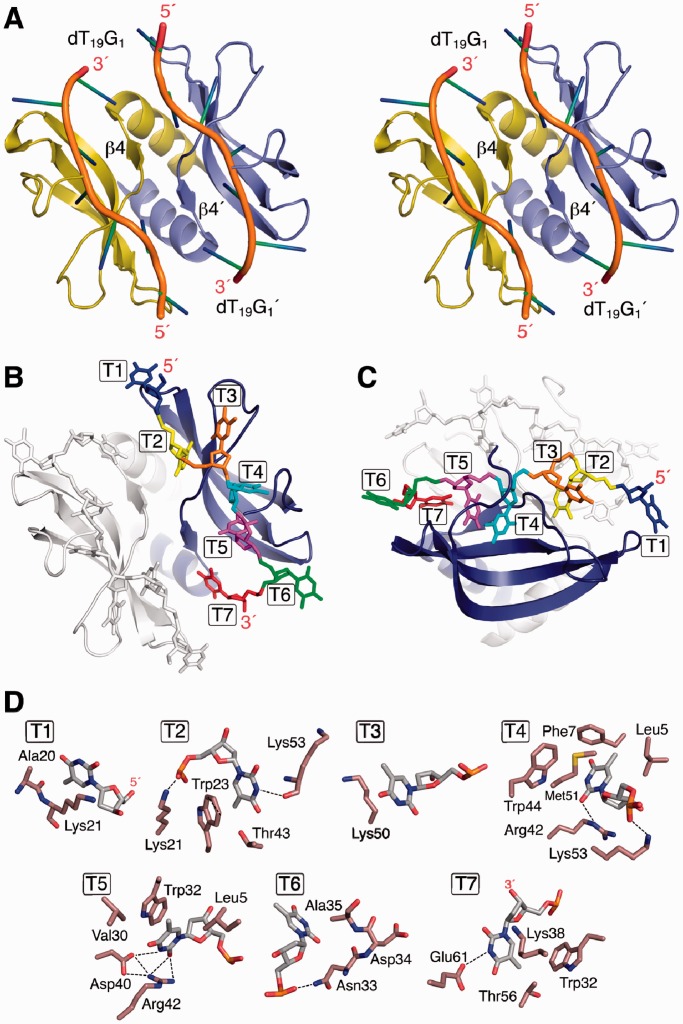
The complex structure is shown in Figure 4A, a top and side view of the complex assembly, Figure 4B and C show the numbering of the two symmetric poly-dT segments. Structural statistics for the protein–ssDNA complex are reported in Table 1, and a view of the final ensemble is shown in Supplementary Figure S12. CS averaging and degeneracy impede the structural characterization of the ssDNA loop and terminal regions and the identification of position-specific protein:ssDNA contacts. Site-specific protein to ssDNA contacts are shown in Figure 4D. YdbC to poly-dT hydrogen bond interactions, that were identified in the NOE assignment protocol, are indicated with dashed lines. Seven YdbC:dT interaction sites were identified. The protein:ssDNA interactions that are fully supported by NMR data include (i) strong aromatic stacking interactions between Trp23:T2 and Trp32:T5; and (ii) hydrophobic contacts Leu5(Hδ1,2):T4-T5 Phe7(Hδ,ε):T4, Ala20(Hβ):T1, Ala35(Hβ):T6, Thr43(Hγ2):T2, Met51(Hε):T4 and Thr56(Hγ2):T7. Strongly conserved Asp40(Oγ):T5 and Arg42(Hε, Hη):T4,T5 contacts form key side-chain to base hydrogen bond interactions in the core site of the complex. Lys21, Asn33, Lys50, Lys53 and Glu61 are active participants in complex formation via hydrogen bonding and/or hydrophobic side-chain stacking to dT. Cross-peaks between HN and Hβ, γ, δ, ε of these residues and the dT H1′, H7 and H6 are identified in the X-filtered NOESY spectrum. The protein to ssDNA surface contact area is ∼4200 Å2. Single-strand DNA (dT19G1) dihedral angles and sugar angles and puckering conformations are listed with the usual numbering convention (T1–T6 and T1′–T6′) in Supplementary Table S1 and S2, respectively. The bases were found to be in the ‘anti’ conformation for the χ torsion angle with the exception of T6 (T6′) and ‘endo’ sugar ring puckering except for T3 (T3′). The base-to-protein contacts are mapped as schematic view in Supplementary Figure S13.
Figure 4.
Solution NMR structure of YdbC:dT19G1 complex. (A) Cartoon stereoview with labelled β4 dimer interface element and structured ssDNA segments and their termini. (B and C) Top and side view of complex with labelled and coloured dT bases (T1–T7). For visual clarity, one side has been greyed out. (D) Detailed view of each dT:protein interaction sites for dT1–dT7. Residues showing hydrophobic interactions <5 Å have been included. Dashed lines represent H-bond interactions within typical range (2.7–3.1 Å). Base–base stacking between dT4 and dT5 was found; protein aromatic to base stacking was present between Trp23 and dT2 and Trp32 and dT5.
The structures of YdbC apo and complex were superposed using the combinatorial extension (CE) algorithm (27) in PyMol as shown in Supplementary Figure S14A. Changes in the β4 secondary structure length are apparent together with difference in the β3–β4 loop orientation and the β1 positioning. Overall, the β structure, more concave in the apo form becomes slightly more open in the complex, and similarly to PC4 (42), the N-terminus becomes highly ordered in the complex. YdbC retains structural similarity to human PC4 [PDB ID: 1pcf (apo) or 2c62 (complex)] (7) as clearly seen in Supplementary Figure S14B, but with a higher root-mean-square deviation because of differences in the secondary and tertiary structures of the termini.
EF_3132 of Enterococcus faecalis from the same DUF2128 family exhibits an even more dramatic relaxation behaviour, as the 1H-15N HSQC spectrum is broadened beyond detection and becomes observable only in the presence of dT19G1 (Supplementary Figure S15), indicating binding causes a change in the conformational exchange properties.
ssDNA binding properties of YdbC
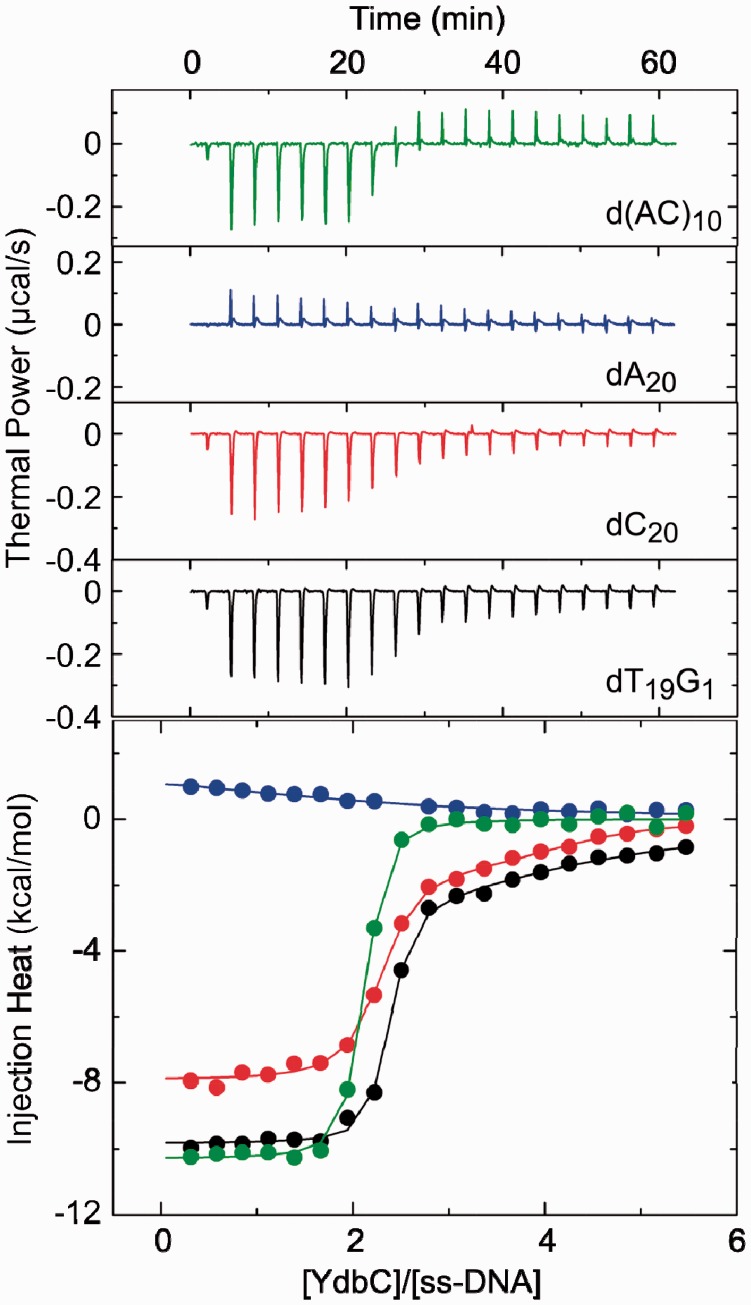
To assess the affinity and sequence specificity of YdbC for ssDNA, the energetics of the DNA-binding interaction between YdbC and selected 20mer single-stranded oligonucleotides were determined using ITC (Figure 5 and Table 2). The primary binding event for each interaction studied has a stoichiometry (N) of two YdbC to one oligonucleotide, indicating that YdbC binds to ssDNA as a dimer, as expected from the high-association affinity of YdbC subunits. Additional low-affinity interactions occur when dT19G1 and dC20 are used (KD > 1 µM). The presence of secondary interactions is evident in the integrated plots for dT19G1 and dC20 as non-linear portions in the [YdbC]/[ssDNA] >2 region of the curve. The secondary interactions between YdbC and both dT19G1 and dC20 show a high degree of uncertainty and salt concentration dependence. The interactions are eliminated by increasing the NaCl concentration to 300 mM (Supplementary Figure S16), indicating that these weak interactions are non-specific and electrostatically driven and might not be physiologically relevant for the function of YdbC. The primary interactions between YdbC and dT19G1, dC20 and d(A-C)10 oligonucleotides each have dissociation constants (KD) within a ∼4-fold range, from 11 to 39 nM, under physiologically relevant conditions (pH 7.5, 150 mM of NaCl). In contrast, the affinity of YdbC for dA20 (KD = 11 µM) is markedly less than that observed for the other oligonucleotides. Although indicative of reduced specificity for polypurine sequences, low affinities and unfavourable enthalpic contributions to binding for poly-A sequences are common features of non-specific ssDNA-binding proteins because of the coupled energetic cost of de-stacking adjacent adenine residues on protein binding (43,44). The similar affinity of YdbC for the alternating purine–pyrimidine sequence d(A–C)10 to the pyrimidine rich sequences, dT19G1 and dC20, provides further evidence that the lack of affinity of YdbC for dA20 is mechanistic in nature and does not reflect the presence of sequence-specific contacts in the YdbC:ssDNA complex.
Figure 5.
ssDNA-binding profiles for YdbC at 25° C and 150 mM of NaCl. (Top) Thermal power versus time with legend added for clarity. ITC thermograms for the injection of 220 µM YdbC into 8-µM solutions of d(AC)10 (green), dA20 (blue), dT19G1 (black) and dC20 (red). Each heat burst curve corresponds to the injection of 2 µl of a solution of YdbC into a solution of the ssDNA oligo. (Bottom) Injection heat versus YdbC/ssDNA ratio. The thermograms in the top panel were integrated to create the binding isotherms with the same colour-coding as in the top panel. The binding isotherms were fit (solid lines) with models for one [d(AC)10 and dA20] or two (dT19G1 and dC20) sets of binding sites. Top and bottom panels use identical colour-coding.
Table 2.
ITC-derived parameters for the binding of YdbC to selected 20mer oligonucleotides
| Oligonucleotide | Binding site | KD (M) | ΔG (kcal/mol) | ΔH (kcal/mol) | ΔS (cal/mol•K) | n |
|---|---|---|---|---|---|---|
| dT19G1 | 1 | (1.6 ± 0.6) × 10−8 | −10.6 ± 0.3 | −10.1 ± 0.1 | 1.8 ± 1.3 | 2.2 ± 0.1 |
| 2 | (7.7 ± 3.0) × 10−6 | −7.0 ± 0.3 | −4.3 ± 0.6 | 9.0 ± 3.0 | 2 fixed | |
| dC20 | 1 | (1.1 ± 0.6) × 10−8 | −10.8 ± 0.4 | −7.8 ± 0.1 | 10.2 ± 1.7 | 2.2 ± 0.1 |
| 2 | (1.7 ± 0.9) × 10−6 | −7.9 ± 0.4 | −1.6 ± 0.2 | −21.0 ± 2.0 | 2 fixed | |
| dA20 | 1 | (1.1 ± 0.6) × 10−5 | −6.8 ± 0.5 | −1.8 ± 0.6 | 16.6 ± 3.6 | 2.1 ± 0.5 |
| d(A–C)10 | 1 | (3.9 ± 0.5) × 10−8 | −10.1 ± 0.1 | −10.3 ± 0.1 | −0.6 ± 0.6 | 2.0 ± 0.1 |
The ITC profiles shown in Figure 5 were fit with models for either one [dA20 and d(A−C)10] or two (dT19G1 and dC20) independent sets of binding sites. All parameters were allowed to float during the fitting routines except for values of n for site 2 in dT19G1 and dC20, which were manually varied to yield the best fit (as reflected by minimization of χ2). The indicated uncertainties in the fitted values reflect the standard deviation of the experimental data from the fitted curves. Values for ΔG and ΔS were calculated using the standard formalisms containing the maximum errors as carried through the equations.
Binding of YdbC to dsDNA and ssRNA
PC4 has the capacity to disrupt duplex DNA at low ionic strength and micromolar protein concentrations (11). Analogously, we found that YdbC can disrupt a 26-base DNA duplex with 5′-GGATTTGGTTTCAAAAAGAAAAAAGG-3′sequence (and complementary) and bind to the resulting ssDNA while retaining the same overall structure to that of the YdbC:dT19G1 complex (Supplementary Figure S17). At 0.3 mM of YdbC and 100 mM ionic strength a 35 kDa YdbC:dsDNA complex consistent with the combined masses is formed that shows nearly identical HSQC amide chemical shifts compared with the YdbC:dT19G1 complex. In addition, despite the different DNA sequence, the key Trp–base stacking interactions seem to be re-capitulated based on the position of the Trp23, Trp32 and Trp44 side-chain ε1 amides. These are markedly distinct from the positions in the apo-YdbC spectrum (Supplementary Figure S17). These spectral features are consistent with a model in which the dsDNA structure has been disrupted to form a YdbC:ssDNA-type complex.
Given the overall fold similarity of YdbC to PUR-α (Figure 2) and to establish their function relationships more clearly, we examined the binding of YdbC to ssRNA. YdbC binding to an ssRNA, with sequence AGACAGCAUUAUGGUGUCUUU, was studied by analytical gel filtration and titrations monitored by 1H-15N HSQC (Supplementary Figure S18). Interestingly, we found that YdbC binds ssRNA with low to moderate affinity. The complex can be isolated by gel filtration chromatography at ∼0.3 mM of YdbC:ssRNA concentration. The CS perturbations mapped onto the structure point to a similar binding region for both ssRNA and ssDNA. The linear trajectory change in 1H-15N chemical shifts versus ssRNA:protein ratio indicates a two-state fast exchange binding model (45). A two parameters equation was used to fit the data and derive a value of KD ∼70 µM. The authors thank anonymous reviewers for suggesting detailed characterization of dsDNA and ssRNA binding to YdbC.
Taxonomic distribution and sequence analysis
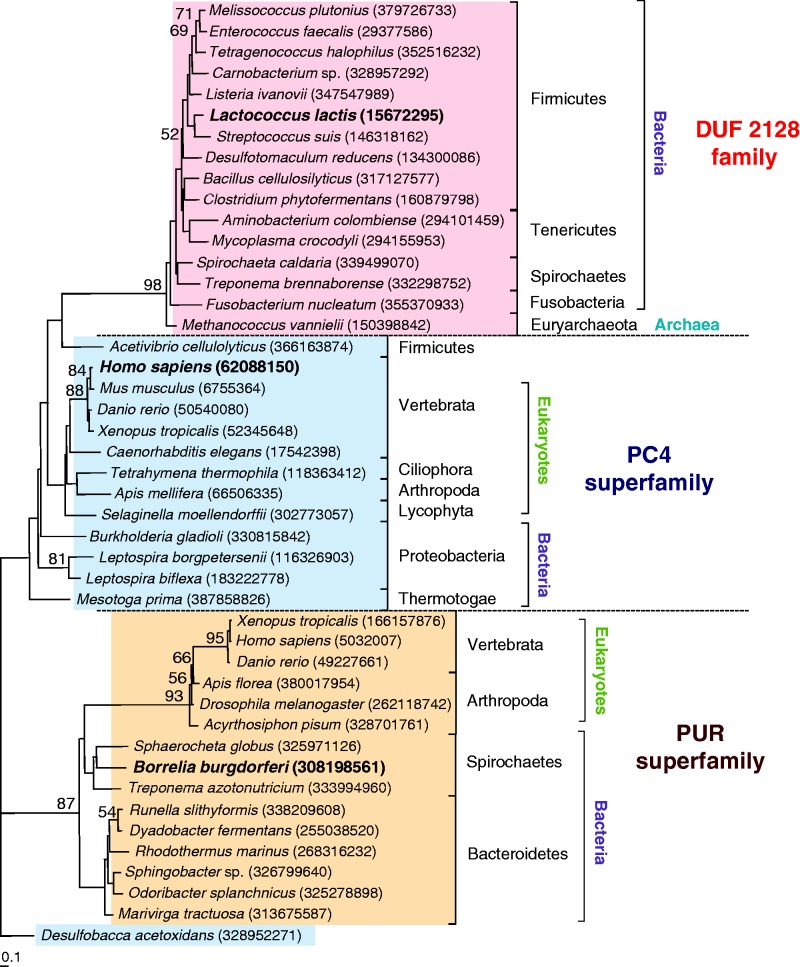
A search of sequenced genomes was conducted with the current (May 2012) NCBI database (35), to assess the extent of the taxonomic distribution of homologues of L. lactis YdbC, within the DUF2128 (PF09901) family. The genomes of 1831 bacterial, 101 archaeal and 181 eukaryotic species were searched using YdbC. Homologues were found in prokaryotic strains of the phyla Firmicutes (226 among bacilli, clostridia and others), spirochaetes (8 strains), Tenericutes (7 strains) and fusobacteria (5 strains). Four members of the archaeal genus Methanococcus also possess a homologue of YdbC. No related sequences were found in other prokaryotic phyla or in the eukaryotic genomes searched. Details of the search results are provided in Supplementary Table S3. Interestingly, the prokaryotic species encoding YdbC homologues also possess the homologue of SSB (GenBank 37999773), suggesting that YdbC plays a complementary role to that of SSB in these species. In addition, PC4 and PUR-α, two proteins known to bind ssDNA, are structurally similar to YdbC. Both PC4 and PUR-α are found in eukaryotes and in bacteria, but absent in archaea. Initial BLASTP searches of PC4 homologues in bacteria returned no significant results; therefore, we conducted BLAST-PSI and protein domain searches using the conserved Domain Architecture Retrieval Tool at NCBI (46) and Pfam 26.0. Twenty-four PC4 sequences were found in bacteria, mostly in proteobacteria (10 sequences) and spirochaetes (10 sequences). In addition, the PC4 sequence of the Firmicute Acetivibrio cellulolyticus was only found in Pfam. The PC4 domain occurs as a single unit or as part of multidomain proteins, where it can be present in tandem repeats. All the bacterial sequences are single-domain proteins containing only the PC4 domain. The distribution of putative PUR-α homologues in bacteria is also limited to few phyla, namely, in Bacteroidetes and spirochaetes. To better understand the relationships between the DUF2128, PC4 and PUR-α proteins families, putative YdbC homologues from representative strains were analysed together with sequences from PC4 and PUR-α families of ssDNA-binding proteins. Although these three families are structurally similar, they differ at the level of amino acid sequence, and accordingly they form three distinct clusters (Figure 6). However, the DUF2128 and PC4 clusters seem to be more closely related to each other than to the PUR-α clade. Within the PUR-α and the PC4 clusters, eukaryotic and bacterial sequences branch separately. Furthermore, the bacterial PC4 homologues constitute a loose group, with the A. cellulolyticus PC4 sequence forming a deep branch with sequences of the DUF2128 family.
Figure 6.
Neighbour-joining tree of YdbC homologues compared with sequences within the PC4 and Pur-α families. Sequence accession (GenBank ID) numbers are in parenthesis. Sequences and DUF2128 in bold to highlight significance to this study. The PC4 homologue of Desulfobacca acetoxidans was used as the outgroup. Bootstrap values >50 are shown. Bar indicates 0.1 substitutions per amino acid position.
To further clarify the function of YdbC, the genomic context of YdbC homologues was examined in the microbial chromosomes. This analysis was carried out with the YdbC amino acid sequence to search the database of Protein Clusters at NCBI, followed by retrieval of genomic neighbourhoods using the ProtMap function. The results show that the genome context of all the YdbC homologues differs, suggesting that YdbC is encoded by a monocistronic transcript. This observation is also consistent with the presence of the putative ribosomal binding site AGAAAGGA (47) located six nucleotides upstream from the start codon of the ydbC gene, and the fact that the gene downstream is transcribed in the opposite direction with respect to ydbC. A similar analysis of the genome context was also performed using PC4, SSB and PUR-α protein sequences. Similar to what is observed for YdbC, the genome context of PUR-α homologues differs among strains, suggesting that the bacterial PUR-α is not part of an operon. In the genomes of all Firmicutes, SSB is consistently encoded between two ribosomal proteins, but this arrangement is not maintained in other phyla and might not have functional meaning. The genome context for PC4 also varies within strains. One interesting observation is that in some Burkholderia and Leptospira strains, the sequences immediately upstream from the PC4 gene are phage-related integrases or transposases, raising the question whether these sequences might have been acquired by lateral gene transfer.
DISCUSSION
L. lactis YdbC representative of the DUF2128 family is a remarkably versatile nucleic acid-binding domain that binds ssDNA with sufficient strength to disrupt DNA duplex and also ssRNA, albeit more weakly. Remarkable structure–function similarity was found between L. lactis YdbC, the H. sapiens PC4 and the of B. burgdorferi PUR-α domains at low sequence similarity. PC4 is a well-characterized ssDNA-binding domain, whereas PUR-α is known to bind both ssDNA and RNA.
Short amino acid stretches (see Asp40–Ile41–Arg42 and Lys53–Gly54–Ile55–Thr56 in the sequence alignment) of YdbC and PC4 are identical (Figure 2A) and highly conserved within the DUF2128 and PC4 family, indicating a possible evolutionary link (see later in the text). The YdbC/PUR-α relationship is much more remote, although Ile41, Ile55 and Glu60 are strictly conserved among all three proteins, and Ile41 is also strongly conserved within each individual family, which may be incidental or may point to a fold stability role of Ile41. The conserved residue locations along key elements of the secondary structure involved in nucleotide binding underscores the importance of the residue type at these specific locations for proper functioning of the domain. Particularly, residues Lys38, Lys50 and Lys53 have critical functions to create the positively charged solvent-exposed surface required for interactions with ssDNA and ssRNA.
The L. lactis YdbC dimer binds ssDNA with nanomolar affinity at physiological conditions and non-specifically with no measurable bias for pyrimidine and mixed purine/pyrimidine oligonucleotides by ITC (48) (Figure 5 and Table 2). Although complete temperature-dependent characterizations were not performed, the binding energetics for the YdbC interactions with pyrimidine and mixed purine/pyrimidine oligonucleotides seem to be consistent with those obtained for other non-specific ssDNA-binding proteins (43,44). These protein–ssDNA interactions are largely enthalpically driven and have large negative-binding heat capacities (ΔCp) likely because of induced conformational changes in the bound oligonucleotides and unrelated to binding specificity. In ssDNA binding proteins, the lack of base preference for particular sites on the protein can produce chain translocation and weakening of the ssDNA electron density in diffraction data (7,12). The dT19G1 terminal guanine is known to promote uniform crystallization by slowing/preventing chain sliding and was originally sourced for use in crystallization trials in this study (7). Here, the strategy fails to provide adequate YdbC:dT19G1 crystals for X-ray diffraction.
Topologically, the binding mode of dT19G1 to YdbC is similar to that reported for PC4 (7) and covers the entire positively charged (top) face of the protein (Figure 1C and 4A). As no attempt was made at enforcing similar dihedral angle, slight differences were found in the ssDNA backbone, sugar and exocyclic angle in the YdbC and PC4 complexes. In either case, the conformation is dominated by the common anti base orientation and C2′,C3′-endo puckering (Supplementary Table S1 and S2). The C1′-exo conformation for the T3 nucleotide indicates dynamics of the sugar ring at that site. Strong symmetric protein:ssDNA contacts extend along the top centre β-ridge (positively charged surface) from the β1–β2 loop to the β3–β4 loop a total of seven bases on each side of the dT hairpin contact the symmetric YdbC protomer (Figure 4B and C and Supplementary Figure S14). The N-terminal Lys4–Leu5–Lys6 participates in complex formation and become ordered on binding. Four of seven nucleotides form base-aromatic stacking interactions with the protein. Bases at T4 and T5 positions are stacked and buried in the centre of the protein concave β face. The Asp40–Ile41–Arg42 site of conservation between YdbC and PC4 forms key hydrogen-bond interactions to the T5 pyrimidine ring. The T3 position is the most solvent exposed showing only interactions with Lys50 (Figure 4D). There is no evidence that higher order oligomers are formed in the presence of ssDNA. Although binding ssDNA in a manner analogous to the PC4 structure (7), YdbC forms more extensive contacts with ssDNA, and its interactions are dominated by aromatic stacking. Analogous to PC4, YdbC is capable of disrupting duplex DNA and binding to the resulting open strands (Supplementary Figure S17) (11). Here, we provide NMR evidence that the overall fold of YdbC in the YdbC:dsDNA versus YdbC:ssDNA complex is preserved while the protein sequesters the open strands.
The binding of YdbC to ssRNA is weaker (in the 100 μM range) for a mixed purine/pyrimidine 21-nt oligonucleotide. Similar YdbC binding epitopes for ssRNA versus ssDNA were deduced by CS perturbation mapping (Supplementary Figure S18). Although the PUR-α interaction with nucleic acids has not been structurally characterized, its similarity to the well-studied Whirly proteins in plants suggests completely different binding modes (49) to those of YdbC/PC4.
The findings reported herein for YdbC are likely to characterize the entire DUF2128 domain family. Analysis shows that ssDNA binding is occurring for Enterococcus faecalis EF_3132, another member of the DUF2128 protein family (Supplementary Figure S15). An important question arises with domains that are structurally and functionally similar, but whose sequence identity is <15%: do they/should they be grouped under the same superfamily, or differences are sufficient to claim the discovery of a novel ssDNA binding domain? Here, we show that YdbC and PC4 share strongly conserved short-sequence motifs that are clearly poised to impact the function. Structure-based sequence alignment is proven a useful starting point for bioinformatics characterization with sequence similarity that would normally be too low for meaningful examination. The sequence analysis built around structurally aligned sequences, shows that YdbC (DUF2128), PC4 and PUR families cluster in distinct regions of the sequence space (Figure 6). However, both DUF2128 and PC4 seem closer to each other than the PUR domain. The phylogenetic distribution of PC4 and PUR domains extends to both the prokaryotic and eukaryotic domains, although it seems to be restricted to only few well-defined prokaryotic phyla in both cases, whereas DUF2128 has so far only been identified in prokaryotes, primarily in Firmicutes. In addition, the PC4 sequence of A. cellulolyticus that form a branch with the DUF2128 cluster suggests that DUF2128 and PC4 are distant members of the same superfamily. Our findings were communicated to the Pfam group that independently validated our results. In the upcoming database release (Pfam 27.0), the DUF2128 (PF09901) will be merged with the PC4 (PF02229) family. The genome context of the genes encoding YbdC, PC4 and Pur-α is consistent with these genes being expressed as monocistronic transcription units. For YbdC, the finding is also supported by the presence of a ribosomal-binding site upstream of the translation start site, and a gene encoded in opposite orientation downstream of YdbC.
E. coli transformed to contain the human PC4 gene have shown enhanced protection from oxidative damage (50). It is conceivable that YdbC could have similar or general DNA repair functions in L. lactis and other prokaryotic members of the DUF2128 family. The biological implications of the newly uncovered YdbC ability to bind to ssRNA require further study but may be unique to the prokaryotic branch in the context of this new PC4 superfamily.
In summary, the structural, thermodynamic and bioinformatics analyses presented here demonstrate that YdbC, and indeed most members of the prokaryotic DUF2128 domain family, is a multifunctional nucleic acid-binding domain with high affinity for ssDNA. Given the industrial and biomedical applications of this microorganism, further functional characterization of YdbC should be of general interest.
ACCESSION NUMBERS
PDB ids, 2ltd, 2ltt.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–18 Supplementary Methods, Supplementary Results and Supplementary References [51–69].
FUNDING
National Institute of General Medical Sciences Protein Structure Initiative [U54-GM094597, to G.T.M.]; National Science Foundation [MCB0843678 in part to E.B.]; Hatch Project [NJ01136 to E.B.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Alexander M. J. J. Bonvin, for insights on conducting HADDOCK refinement, Marco Punta for helpful correspondence and Pfam reclassification of DUF2128 domain, Roberto Tejero for CNS refinement support, Xiang-Jun Lu for help with 3DNA analysis, Li-Chung Ma for kindly providing the ssRNA oligonucleotide, Phil Kostenbader for cluster computing support and Huang Wang and Eitan Kohan for contributions in sample production.
REFERENCES
- 1.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conesa C, Acker J. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol. 2010;7:287–290. doi: 10.4161/rna.7.3.11491. [DOI] [PubMed] [Google Scholar]
- 3.White MK, Johnson EM, Khalili K. Multiple roles for Pur-alpha in cellular and viral regulation. Cell Cycle. 2009;8:1–7. doi: 10.4161/cc.8.3.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugiman-Marangos S, Junop MS. The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins. Nucleic Acids Res. 2010;38:3432–3440. doi: 10.1093/nar/gkq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graebsch A, Roche S, Niessing D. X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc. Natl Acad. Sci. USA. 2009;106:18521–18526. doi: 10.1073/pnas.0907990106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norais CA, Chitteni-Pattu S, Wood EA, Inman RB, Cox MM. DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J. Biol. Chem. 2009;284:21402–21411. doi: 10.1074/jbc.M109.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werten S, Moras D. A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat. Struct. Mol. Biol. 2006;13:181–182. doi: 10.1038/nsmb1044. [DOI] [PubMed] [Google Scholar]
- 8.Graebsch A, Roche S, Kostrewa D, Soding J, Niessing D. Of bits and bugs–on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure. PloS One. 2010;5:e13402. doi: 10.1371/journal.pone.0013402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 10.Schang LM, Hwang GJ, Dynlacht BD, Speicher DW, Bantly A, Schaffer PA, Shilatifard A, Ge H, Shiekhattar R. Human PC4 is a substrate-specific inhibitor of RNA polymerase II phosphorylation. J. Biol. Chem. 2000;275:6071–6074. doi: 10.1074/jbc.275.9.6071. [DOI] [PubMed] [Google Scholar]
- 11.Werten S, Langen FW, van Schaik R, Timmers HT, Meisterernst M, van der Vliet PC. High-affinity DNA binding by the C-terminal domain of the transcriptional coactivator PC4 requires simultaneous interaction with two opposing unpaired strands and results in helix destabilization. J. Mol. Biol. 1998;276:367–377. doi: 10.1006/jmbi.1997.1534. [DOI] [PubMed] [Google Scholar]
- 12.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 13.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol. Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells JM, Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 2008;6:349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Cryst. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 22.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 24.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 25.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prlić A, Bliven S, Rose PW, Bluhm WF, Bizon C, Godzik A, Bourne PE. Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics. 2010;26:2983–2985. doi: 10.1093/bioinformatics/btq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 38.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 39.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 40.Brandsen J, Werten S, van der Vliet PC, Meisterernst M, Kroon J, Gros P. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat. Struct. Biol. 1997;4:900–903. doi: 10.1038/nsb1197-900. [DOI] [PubMed] [Google Scholar]
- 41.Evenäs J, Tugarinov V, Skrynnikov NR, Goto NK, Muhandiram R, Kay LE. Ligand-induced structural changes to maltodextrin-binding protein as studied by solution NMR spectroscopy. J. Mol. Biol. 2001;309:961–974. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]
- 42.Werten S, Wechselberger R, Boelens R, van der Vliet PC, Kaptein R. Identification of the single-stranded DNA binding surface of the transcriptional coactivator PC4 by NMR. J. Biol. Chem. 1999;274:3693–3699. doi: 10.1074/jbc.274.6.3693. [DOI] [PubMed] [Google Scholar]
- 43.Kozlov AG, Lohman TM. Adenine base unstacking dominates the observed enthalpy and heat capacity changes for the Escherichia coli SSB tetramer binding to single-stranded oligoadenylates. Biochemistry. 1999;38:7388–7397. doi: 10.1021/bi990309z. [DOI] [PubMed] [Google Scholar]
- 44.Ferrari ME, Lohman TM. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–12910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi I, Kuroiwa S, Lindfors HE, Ndamba LA, Hiruma Y, Yajima T, Okishio N, Ubbink M, Hirota S. Modulation of protein-ligand interactions by photocleavage of a cyclic peptide using phosphatidylinositol 3-kinase SH3 domain as model system. J. Pept. Sci. 2009;15:411–416. doi: 10.1002/psc.1132. [DOI] [PubMed] [Google Scholar]
- 46.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikkonen M, Vuoristo J, Alatossava T. Ribosome binding site consensus sequence of Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H. FEMS Microbiol. Lett. 1994;116:315–320. doi: 10.1111/j.1574-6968.1994.tb06721.x. [DOI] [PubMed] [Google Scholar]
- 48.Ballard DW, Philbrick WM, Bothwell AL. Identification of a novel 9-kDa polypeptide from nuclear extracts. DNA binding properties, primary structure, and in vitro expression. J. Biol. Chem. 1988;263:8450–8457. [PubMed] [Google Scholar]
- 49.Cappadocia L, Marechal A, Parent JS, Lepage E, Sygusch J, Brisson N. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell. 2010;22:1849–1867. doi: 10.1105/tpc.109.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang JY, Sarker AH, Cooper PK, Volkert MR. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol. Cell. Biol. 2004;24:6084–6093. doi: 10.1128/MCB.24.13.6084-6093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J. Biomol. NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 52.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry. Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 53.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 55.Johnson ML, Straume M. Comments on the analysis of sedimentation equilibrium experiments. In: Schuster TM, Laue TM, editors. Modern Analytical Ultracentrifugation. Boston, MA: Birkhäuser; 1994. pp. 37–65. [Google Scholar]
- 56.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Goddard TD, Kneller DG. Sparky 3. San Francisco, CA: University of California; 2006. [Google Scholar]
- 58.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 60.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 61.Stuart AC, Borzilleri KA, Withka JM, Palmer AG., III Compensating for variations in 1H-13C scalar coupling constants in isotope-filtered NMR experiments. J. Am. Chem. Soc. 1999;121:5346–5347. [Google Scholar]
- 62.Zwahlen C, Legault P, Vincent SJ, Greenblatt J, Konrat R, Kay LE. Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage λ N-Peptide/boxB RNA Complex. J. Am. Chem. Soc. 1997;119:6711–6721. [Google Scholar]
- 63.Tjandra N, Grzesiek S, Bax A. Magnetic field dependence of nitrogen-proton J splittings in N15-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 1996;118:6264–6272. [Google Scholar]
- 64.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 65.Rückert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- 66.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 67.Fushman D, Ohlenschlager O, Ruterjans H. Determination of the backbone mobility of ribonuclease T1 and its 2'GMP complex using molecular dynamics simulations and NMR relaxation data. J. Biomol. Struct. Dyn. 1994;11:1377–1402. doi: 10.1080/07391102.1994.10508074. [DOI] [PubMed] [Google Scholar]
- 68.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 69.Saenger W. Principles of Nucleic Acids Structure. 1984. Springer-Verlag New York, Inc, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.