Abstract
Riboswitches are regulatory RNA elements typically located in the 5′-untranslated region of certain mRNAs and control gene expression at the level of transcription or translation. These elements consist of a sensor and an adjacent actuator domain. The sensor usually is an aptamer that specifically interacts with a ligand. The actuator contains an intrinsic terminator or a ribosomal binding site for transcriptional or translational regulation, respectively. Ligand binding leads to structural rearrangements of the riboswitch and to presentation or masking of these regulatory elements. Based on this modular organization, riboswitches are an ideal target for constructing synthetic regulatory systems for gene expression. Although riboswitches for translational control have been designed successfully, attempts to construct synthetic elements regulating transcription have failed so far. Here, we present an in silico pipeline for the rational design of synthetic riboswitches that regulate gene expression at the transcriptional level. Using the well-characterized theophylline aptamer as sensor, we designed the actuator part as RNA sequences that can fold into functional intrinsic terminator structures. In the biochemical characterization, several of the designed constructs show ligand-dependent control of gene expression in Escherichia coli, demonstrating that it is possible to engineer riboswitches not only for translational but also for transcriptional regulation.
INTRODUCTION
Synthetic biology has become a rapidly developing branch of life sciences. At its heart is the rational design and construction of biosynthetic and regulatory networks, pathways and their constituent components. As RNA plays a central role in a diverse set of regulatory pathways, efforts in synthetic biology have produced novel RNA components capable of regulating gene expression in vivo (1). Riboswitches are a particularly attractive type of gadget for synthetic biology applications (2,3). These naturally occurring RNA elements determine the expression state of a gene in response to an external signal (in most cases, a small molecule that is specifically recognized) through a conformational change, regulating either transcription or translation. The fundamental architecture of a riboswitch combines a sensor (aptamer domain) for the external stimulus and an actuator that influences gene expression. Both elements can in principle be freely combined. Depending on the response to the trigger molecule, gene expression is either enhanced (ON switch) or repressed (OFF switch). In transcription-regulating riboswitches, the actuator element contains a possible intrinsic transcription terminator structure consisting of a stem-loop element followed by a U-rich region. Depending on the bound or unbound state of the sensor, this terminator structure is either masked or formed, leading to activation or inhibition of gene expression, respectively.
Although structural features of the conserved aptamer domains are well described, the organization of the regulatory platforms is in many cases unclear. In particular, transcription terminator elements can vary substantially, not only as isolated elements but also in the context of a specific riboswitch (4–6). Hence, the design of transcription regulating riboswitches is a difficult task. This was demonstrated by Fowler et al. (7), who intended to design transcriptional ON switches by fusing a theophylline aptamer to a Bacillus subtilis intrinsic terminator. Detailed analysis revealed, however, that the regulatory mechanism of the selected riboswitches was independent of the terminator structure and was probably targeting translation. The sole artificial combination of aptamer and terminator elements that showed transcriptional regulation consists of two naturally occurring components, where the aptamer region of the Streptococcus mutans tetrahydrofolate riboswitch was fused to the intrinsic terminator region of the B. subtilis metE riboswitch. This resulted in an artificial ON switch that controls transcription termination on folinic acid binding (8). Accordingly, currently available riboswitches consisting of synthetic components generated by rational and/or evolutionary approaches exclusively regulate translation (9–14).
We used the well-characterized theophylline aptamer as a sensor element to construct synthetic riboswitches that regulate gene expression at the transcriptional level. This aptamer has been successfully incorporated in different regulatory mechanisms for gene expression in vivo, demonstrating its functionality within the cell. Besides representing a sensor platform in translational riboswitches (15–17), it was used to modulate mRNA cleavage catalysed by yeast RNase III (18) as well as RNA splicing (19,20) and to control non-coding RNA activity (21). Fusing this aptamer to a computer-designed spacer element followed by an aptamer-complementary sequence, which together form a transcriptional terminator structure, several synthetic riboswitches were generated. Biochemical analysis revealed that these elements are able to efficiently regulate transcription of a reporter gene. Hence, although several in vivo screening approaches have failed so far, our data show that a strategy based on in silico design is successful in constructing functional transcriptional riboswitches in E. coli.
MATERIALS AND METHODS
Riboswitch design
The rational design algorithm starts from the known sequence and secondary structure of the aptamer and constructs the sequence of the remaining part of the riboswitch, i.e. the spacer and the terminator. It proceeds by proposing a large set of candidates that are then evaluated with respect to their secondary structure and properties of their folding paths, which are estimated using the RNAfold program of the Vienna RNA package (22). At the proposal stage, it incorporates some knowledge of sequence and structure of terminator elements for the given biological system in which the construct is supposed to function, here E. coli. In particular, we used the fact that the terminator hairpin must have a minimal size and that it is followed by a U stretch. First, a spacer database containing randomly generated sequences with lengths between 6 and 20 nucleotides (nt) is created. The size of this database is user defined. For each position k of the 3′-part of the aptamer, we form the sequence 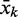 that is complementary to the subsequence from k to the end of the aptamer. For the theophylline aptamer, values of k ranging from position 21 to 32 were used. The sequences of the aptamer, a spacer from the database, a complementary sequence
that is complementary to the subsequence from k to the end of the aptamer. For the theophylline aptamer, values of k ranging from position 21 to 32 were used. The sequences of the aptamer, a spacer from the database, a complementary sequence 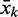 and the U stretch are concatenated to form a complete riboswitch candidate.
and the U stretch are concatenated to form a complete riboswitch candidate.
Each of these constructs is then evaluated by folding simulations. In the current implementation, folding paths are represented as a sequence of secondary structures computed for sub-sequences starting at the 5′-end. We used individual transcription steps of 5–10 nt to simulate co-transcriptional folding with varying elongation speed. Secondary structures are computed by RNAfold, a component of the Vienna RNA Package (22), with parameter settings -d2 -noPS -noLP. If one of the transcription intermediates forms base pairs between aptamer and spacer, it is likely that this will interfere with the ligand-binding properties; hence, such a candidate is rejected. On the other hand, if the full-length transcript does not contain a single hairpin structure composed of the aptamer 3′-part, the spacer and the sequence 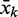 complementary to the 3′-part of the aptamer, the construct cannot form a functional terminator and is also rejected. For each construct that passes these filters, additional features are calculated that are used as further selection criteria. The energy difference between the minimum free energy (MFE) fold of the full-length construct and a structure where the aptamer region is constrained to form the ligand-binding competent fold is calculated. This parameter provides a rough measure for the stabilizing effect required to prevent terminator formation when the ligand is bound. Furthermore, a z-score of the structure downstream of the constrained aptamer fold is estimated as follows: The stability (MFE) of the candidate sequence is compared with the mean μ and standard deviation σ of the folding energies computed for a set of 1000 di-nucleotide shuffled sequences. A positive value of the z-score z = (MFE − μ)/σ indicates a structure less stable than expected by chance. This is used to estimate whether stable local structures downstream of the constrained aptamer fold might interfere with transcription.
complementary to the 3′-part of the aptamer, the construct cannot form a functional terminator and is also rejected. For each construct that passes these filters, additional features are calculated that are used as further selection criteria. The energy difference between the minimum free energy (MFE) fold of the full-length construct and a structure where the aptamer region is constrained to form the ligand-binding competent fold is calculated. This parameter provides a rough measure for the stabilizing effect required to prevent terminator formation when the ligand is bound. Furthermore, a z-score of the structure downstream of the constrained aptamer fold is estimated as follows: The stability (MFE) of the candidate sequence is compared with the mean μ and standard deviation σ of the folding energies computed for a set of 1000 di-nucleotide shuffled sequences. A positive value of the z-score z = (MFE − μ)/σ indicates a structure less stable than expected by chance. This is used to estimate whether stable local structures downstream of the constrained aptamer fold might interfere with transcription.
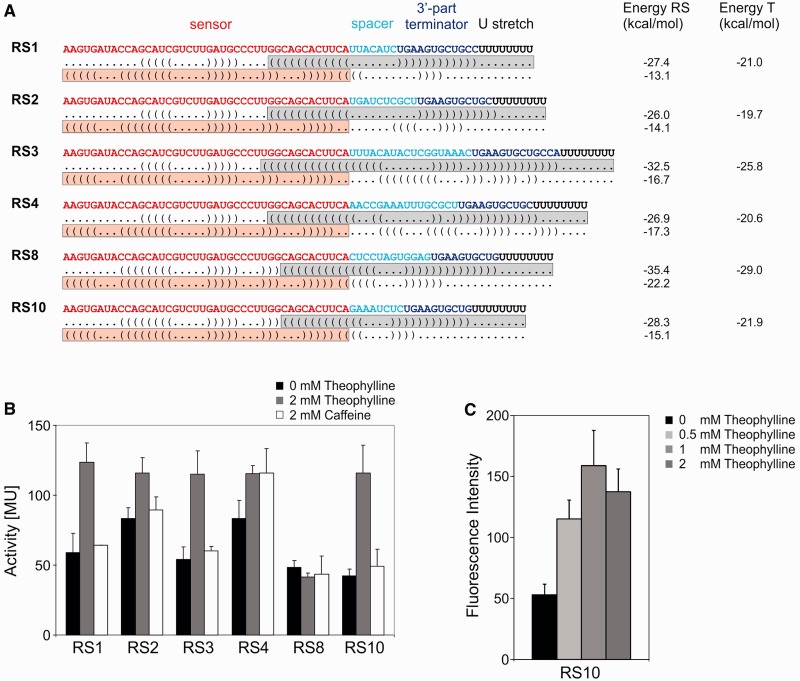
The six experimentally tested synthetic riboswitch constructs are summarized in Figure 2A. Secondary structures and free energy values of individual sequences were calculated using RNAfold 2.0.7.
Figure 2.
Riboswitch constructs and activity tests. (A) The theophylline aptamer represents the sensor (red), followed by a variable spacer sequence (cyan) and the 3′-part of the terminator (blue), forming a hairpin structure with a subsequence of the sensor domain. The terminator element is completed by a stretch of eight U residues (black). Below each construct, dot bracket notations of the secondary structures with terminator or antiterminator formation are indicated. The overlapping situation of sensor (theophylline aptamer, red box) and actuator element (terminator element, grey box) is required for a mutual exclusion of the two alternative structures. For each construct, calculated energy values of complete riboswitch elements (RS) and isolated terminators (T) are indicated. (B) Activity tests of the reporter enzyme β-galactosidase. Activities are indicated in Miller units (MU). All synthetic riboswitches were tested in the absence (black bars) and presence (gray bars) of 2 mM theophylline. As a control, the structurally related caffeine was offered (white bars). (C) Activity test of RS10 under the control of the Prrn promoter and GFP as reporter gene. Relative fluorescence signals are indicated as a function of theophylline concentration. The riboswitch shows a similar regulation profile as in (B), indicating the general functionality of the construct.
Given the dissociation constant Kd, the binding energy is given by the following: ΔG = RTln Kd. With T = 298 K, R = 1.98717 cal/K and the experimentally determined value Kd = 0.32 µM (23) for the theophylline aptamer, we obtain ΔG = −8.86 kcal/mol for the stabilization of the aptamer structure.
Chemicals
Oligonucleotides were purchased from biomers.net, and dNTP were obtained from Jena Biosciences. LB medium was purchased from Applichem; theophylline and caffeine were obtained from Sigma-Aldrich. Ampicillin and Arabinose were purchased from Carl Roth.
Plasmid construction
Riboswitch constructs were generated by overlap extension polymerase chain reaction and fused to bgaB reporter in pBAD2_bgaB (24) by QuickChange site-directed mutagenesis (Stratagene). To analyse riboswitch performance in a second promoter/reporter gene system, riboswitch RS10 was subcloned into the 5′-untranslated region of plasmid pBSU0 (24) using an SpeI restriction site, leading to plasmid pBSU0_RS10. Plasmid constructs were isolated using GeneJETTM Plasmid Miniprep Kit (Fermentas). Correctness of individual assemblies was verified by sequencing.
Quantitative analysis for β-galactosidase activity and green fluorescent protein fluorescence
Enzymatic activity of the expressed β-galactosidase was determined at 40°C (25). All activity tests were performed in three independent experiments.
Green fluorescent protein (GFP) emission was measured in Sorensen’s phosphate buffer (pH 8.0) at 509 nm with excitation wavelength of 488 nm. Fluorescence intensity was normalized by OD600 and measured for all theophylline concentrations in three individual experiments.
Northern blot analysis
Escherichia coli Top10 cells were grown in the presence or absence of 2 mM theophylline until OD600 of 0.5–0.6 in 25 ml LB medium supplied with ampicillin and arabinose. Total RNA was extracted according to J. Lawrence (personal communication; http://rothlab.ucdavis.edu/protocols/D50.html).
Northern blot was performed using the NorthernMax formaldehyd-based system for northern blots (Ambion) according to manufacturer’s instructions. In all, 5 µg total RNA were separated on 1% agarose formaldehyde gel and transferred to BrightStarTM-Plus Membrane (Ambion).
DNA oligonucleotides (reverse complementary to target sequences) were radioactively labeled using γ-32P-ATP (Hartmann analytic). The primer sequences used are as follows:
bgaB: 5′-GTTCGATCTTGCTCCAACTG-3′
23S rRNA: 5′-ACGACGGACGTTAGCACCCG-3′
For quantitation of the mRNA levels, northern blot analysis was repeated twice. Intensities of bgaB signals were normalized using the 23S rRNA signals. The intensity of the mRNA level in the absence of theophylline was defined as 100%.
RESULTS
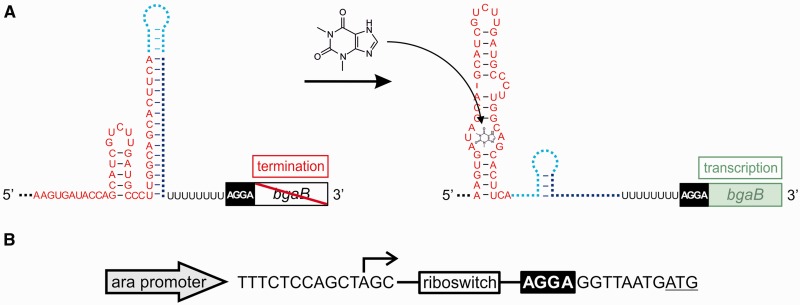
We devised an in silico selection pipeline based on the structure prediction methods implemented in the RNAfold program of the Vienna RNA package (22) to rationally design and select transcriptional riboswitches. Following the general paradigm of inverse folding algorithms (26–29), it operates by iteratively proposing candidate constructs and evaluating them with respect to the design goals. Each designed construct is composed of (i) the theophylline aptamer TCT8-4 that selectively binds its target at a Kd of 0.32 µM and strongly discriminates against the structurally closely related caffeine (23); (ii) a spacer sequence; (iii) a sequence complementary to the 3′-part of the aptamer; and (iv) a U stretch (Figure 1A). The rational model behind such a construct is to terminate transcription of a host gene if the target molecule theophylline is absent. This is achieved by the formation of a terminator structure composed of the aptamer 3′-part and elements (ii), (iii) and (iv). Ligand binding stabilizes the aptamer structure. Thus, the 3′-end of the aptamer sequence is unavailable for the formation of the terminator. As a consequence, the complete sequence of the host gene is transcribed and subsequently translated. Accordingly, a functional construct represents a transcriptional ON riboswitch.
Figure 1.
Riboswitch constructs. (A) Design strategy for theophylline-dependent riboswitches controlling transcription. The sequence of the TCT8-4 theophylline aptamer (red) was fused to a short spacer region (cyan) followed by a sequence complementary to the 3′-part of the aptamer (blue) and a U stretch (black). The Shine–Dalgarno sequence (black box) and the open reading frame of the reporter gene bgaB are located immediately downstream of this construct. On theophylline binding, terminator structure formation should be inhibited and transcription should proceed, resulting in expression of the reporter gene. (B) The final riboswitch constructs are transcribed from the arabinose promoter of plasmid pBAD. The transcription start point is indicated by the angled arrow. The Shine–Dalgarno sequence (AGGA) of the reporter gene for β-galactosidase is located immediately downstream of the riboswitch insert. The start codon of the reporter gene is underlined.
The functional role of the spacer sequence is 2-fold. First, it has to ensure that the binding-competent aptamer structure is formed and has sufficient time during transcription to sense the target molecule. Second, it has to provide a functional part within the terminator structure. These requirements translate to specific constraints on both composition and length of a functional spacer sequence. The spacer must not form base pairs with the aptamer sequence during transcription to avoid potential inhibition of target molecule sensing. This implies a structural constraint. The design algorithm starts from randomly generated spacer sequences with a length between 6 and 20 nt and uses folding simulations on the concatenation of aptamer and spacer to remove candidates that violate the described constraints. The reverse complement of the 3′-part of the aptamer followed by an 8 nt long U stretch is appended to the valid aptamer-spacer constructs, completing the possible intrinsic terminator structure. The co-transcriptional folding paths of these complete constructs are simulated using RNAfold predictions, and construct rejection is done based on predicted secondary structures that violate certain constraints of the riboswitch candidate.
Based on the design principles described earlier in the text, 12 theophylline sensing riboswitches have been constructed. From these predictions, six riboswitch constructs were selected, based on the following criteria of their computationally determined folding paths: (i) the unbound full-length transcript disrupts the aptamer and folds into a stable terminator structure; (ii) no stable secondary structure is formed downstream of the aptamer/theophylline complex; (iii) constructs with a calculated energy difference between bound and unbound state that deviates considerably from the binding energy of the aptamer/theophylline complex (−8.86 kcal/mol) were rejected. The designed constructs were assembled by overlap-extension polymerase chain reaction and cloned into plasmid pBAD2_bgaB between the ara promoter and the reporter gene for β-galactosidase from Bacillus stearothermophilus (24) (Figures 1B and 2A). Transcription was induced by arabinose in the presence or absence of 2 mM theophylline, and β-galactosidase activity of cell extracts was quantified using ortho-nitrophenyl-β-D-galactopyranoside. Three of the constructs turned out to stimulate reporter gene expression by 2-fold (RS1 and RS3) or 3-fold (RS10) (Figure 2B). Caffeine, as a control, could not activate the constructs. Fusing riboswitches RS2 and RS4 to the reporter gene resulted in a permanent active state of gene expression, independent of theophylline or caffeine. In contrast, in the RS8 construct, the reporter gene showed a rather low level of expression, resembling an inactive state under all tested conditions. As RS10 had the best theophylline-dependent activation ratio (Figure 2B), it was chosen for further investigations. To demonstrate the general functionality of such synthetic riboswitches in a different expression system, RS10 was inserted into plasmid pBSU0. It was positioned downstream of the constitutive promoter Prrn and upstream of the GFP-coding sequence (24). As in the original construct, reporter gene activity was induced on the addition of theophylline. The analysis of the resultant relative GFP fluorescence intensity showed a 3-fold increase in the presence of 1–2 mM theophylline, indicating a similar regulatory profile of RS10 as in the original construct with β-galactosidase expression (Figure 2C). The slight reduction in fluorescence at 2 mM theophylline likely results from the fact that high theophylline concentrations are detrimental to cell growth (7), leading to an increase of inactive cells that do not produce GFP anymore.
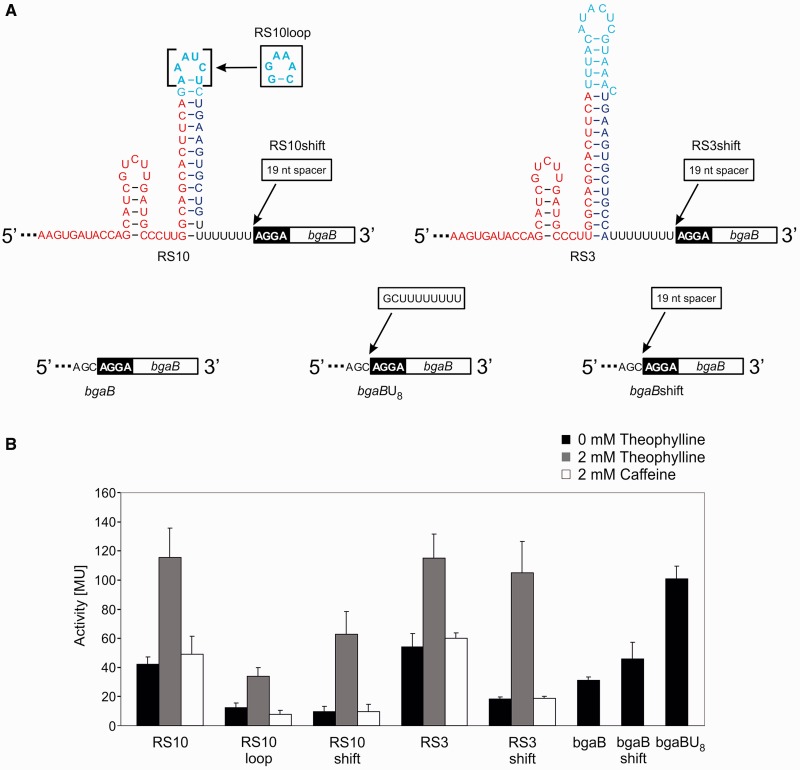
To optimize the functional riboswitches, we aimed for a reduction of the background expression level in the absence of theophylline. First, we replaced the tetraloop sequence of the original RS10 construct by GAAA, which is frequently found in natural terminators (30). Second, as a G-C base pair forms a stable basis for a tetraloop structure, we mutated the terminal A-U base pair in the terminator stem to G-C, leading to construct RS10loop (Figure 3A). This modification led to an increased efficiency in termination, as the background activity of the riboswitch was reduced from 40 to about 15 Miller units (MU) (Figure 3B). However, theophylline-induced gene expression activity was also lowered compared with the original construct. Obviously, this optimization of the terminator element stabilized its structure, resulting in an inhibition of a ligand-induced refolding of the upstream aptamer element required for ongoing transcription.
Figure 3.
Optimized RS10 and RS3 variants and control constructs. (A) In RS10loop, the tetraloop sequence of the terminator hairpin was adjusted to the frequently found sequence GAAA, including a closing G-C base pair. In RS10shift, a spacer sequence of 19 nt was inserted between the U8 stretch of the terminator and the Shine–Dalgarno sequence (black box). The same 19 nt spacer was inserted in RS3shift at the identical position. In control bgaB, the upstream untranslated region of the reporter gene did not carry any riboswitch or terminator inserts. Furthermore, the U8 stretch alone was placed immediately upstream of the ribosomal binding site, and the 19 nt spacer sequence region was inserted into the original bgaB control construct, leading to bgaBU8 and bgaBshift, respectively. (B) β-galactosidase activity test of the constructs described in (A). Activities are indicated in Miller units (MU). The tetraloop adaptation (RS10loop) in the terminator element led to a reduction of the background activity but lowered also the expression of the reporter gene. Increasing the distance between the riboswitch U8 stretch and the ribosomal binding site of the reporter gene in RS10shift and RS3shift increased the ON/OFF rate substantially, compared with the original constructs (for comparison, the regulation profiles of RS3 and RS10 shown in Figure 2B are indicated in this panel). In the control elements, the U8 stretch immediately upstream of the ribosomal binding site led to a rather high gene expression (bgaBU8), whereas increasing the distance between these sequences reduced the β-galactosidase activity (bgaBshift).
Furthermore, it was described that a stretch of eight U residues closely upstream of the Shine–Dalgarno sequence strongly enhances ribosome attachment and translation, whereas shorter U stretches have no such effect (31). As our riboswitches are located immediately upstream of the RBS, it is conceivable that the terminator U stretch of eight residues (U8) stimulates translation in a similar way, leading to an unspecific increase in background activity. Indeed, a construct bgaBU8, consisting of the U8 stretch and the reporter gene only (Figure 3A), showed a dramatic and constitutive increase in reporter gene activity up to 100 MU, compared with the original construct bgaB with 31 MU (Figure 3B). To prove that this effect depends on the close proximity of the U8 stretch and the ribosomal binding site, a 19 nt region from the original pBAD multiple cloning site was inserted upstream of the reporter gene without a riboswitch, leading to construct bgaBshift (Figure 3A). In contrast to the U8 insertion, this construct did not show a significant increase in gene activity compared with the original bgaB construct (Figure 3B). Accordingly, we inserted the 19 nt spacer element downstream of RS10, leading to construct RS10shift (Figure 3A). In the activity test, this riboswitch showed a strongly reduced background signal of ∼10 MU, resulting in a 6.5-fold activation of gene expression on theophylline addition (Figure 3B). To demonstrate the generality of this observation, we inserted the same 19 nt spacer region between RS3 and the ribosomal binding site. Similar to RS10shift, the RS3shift construct also showed an increased ON/OFF rate (5.7-fold), owing to a reduced background activity in the absence of the ligand (18 MU compared with 54 MU in RS3; Figure 3A and B).
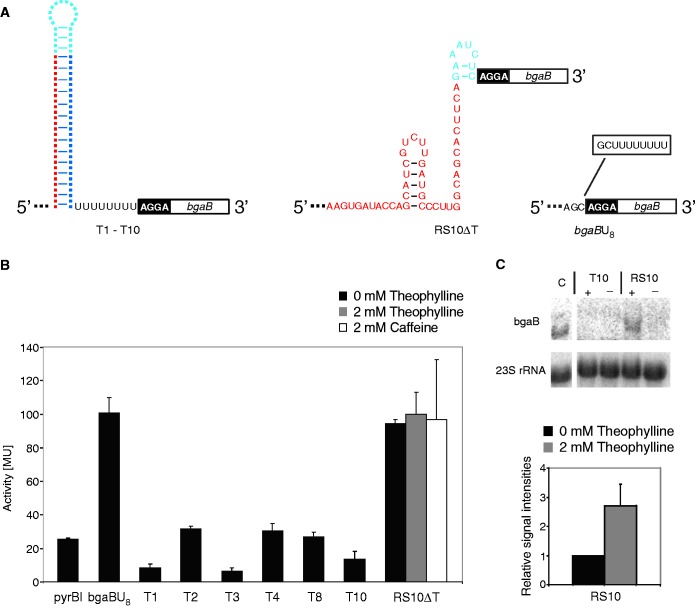
To further verify the functionality of the selected riboswitches, individual components were examined in the expression system. Deleting the 5′-part of the aptamer sequence led to isolated terminator structures T1–T10 that showed efficient repression of the reporter gene activity (Figure 4A and B). As a positive control, the terminator of the attenuator region of the pyrBI operon from E. coli was used (32). This natural regulatory element showed a termination efficiency comparable with that of the synthetic terminators, indicating that all generated terminator elements are functional (Figure 4B).
Figure 4.
Functional analysis of RS10 and synthetic terminator elements. (A) The isolated terminators of the tested riboswitch constructs were cloned upstream of the reporter gene. In a control construct, the 3′-complementary sequence of terminator 10 and the U8 stretch was deleted, leading to RS10ΔT. As shown in Figure 3, construct bgaBU8 represents the positive control. (B) β-galactosidase activity of terminator constructs. Although the positive control bgaBU8 (activation data from Figure 3B are presented) and the terminator deletion RS10ΔT show a strong enzyme activity, all terminators reduce the β-galactosidase activity at an efficiency comparable with that of the natural terminator from the pyrBI operon in E. coli (pyrBI). The terminator elements of the functional riboswitches 1, 3 and 10 show the strongest activity reduction. (C) Northern blot analysis of RS10. Left panel: Although the terminator-alone construct T10 showed no transcription of the reporter gene, RS10 allowed bgaB gene transcription exclusively in the presence (+) of theophylline. C, control expression of bgaB using pBAD2-bgaB without riboswitch inserts. 23S rRNA was used as an internal standard for normalizing the presented lanes. Right panel: Quantitation of bgaB transcription in RS10 construct with and without theophylline.
The observed terminator-dependent reduction of reporter gene activity is a first indication that the synthetic riboswitches control transcription, as terminator function is an essential prerequisite for a riboswitch-mediated transcriptional regulation. Hence, the integrity of the terminator structure should be essential for regulation, and a deletion of this element should lead to a constitutive active state. To test this, we removed the whole 3′-part of the terminator region of RS10, i.e. the complementary 3′-part of the hairpin and the U stretch, resulting in RS10ΔT (Figure 4A). This deletion led to a complete destruction of riboswitch function, and the construct exhibited permanent gene expression, independent of theophylline (Figure 4B).
Because the essential control elements for translation, in particular the ribosomal binding site, are still present in RS10ΔT and the corresponding terminator construct T10, these findings are a clear indication that the RS10 riboswitch regulates at the transcriptional level. To confirm this observation, northern blot analysis was performed for RS10 and the terminator-alone construct T10. Total RNA was prepared from corresponding E. coli TOP10 cells grown in the presence or absence of theophylline. Hybridization was performed using bgaB-specific oligonucleotide probes. To make sure that identical amounts of RNA were analysed, a probe for 23S rRNA was used as an internal control. Although the intensity of this control signal was identical in each preparation, a full-length mRNA of the RS10-bgaB construct was detected only in the presence of theophylline (Figure 4C, left panel). Quantitative analysis of the transcript of the RS10 construct revealed a 2.7-fold increase in mRNA levels in the presence of theophylline (Figure 4C, right panel). This is in good agreement with the levels of active protein measured in the β-galactosidase assay. The T10 construct, however, lacking the aptamer domain, completely inhibited transcription, regardless of theophylline. Taken together, this result is in agreement with the corresponding β-galactosidase activity tests and clearly shows that RS10 regulates at the transcriptional level.
DISCUSSION
We have successfully applied a combination of in silico, in vitro as well as invivo experiments to generate theophylline-dependent riboswitches that activate transcription on binding of the target molecule. This strategy is a promising alternative to riboswitch selection based on randomized RNA sequence libraries. The design of such libraries is crucial for the success of selection and requires features that differ depending on whether an ON or an OFF switch is to be designed (33,34). Furthermore, the screening approach with randomized RNA sequences was applied successfully only to riboswitches acting on translation (15,35–37). For such elements, the sole requirement is that the alternative structures lead to presentation or masking of the ribosomal-binding site, which comprises a short sequence of 4–6 nt (38). In transcription-controlling riboswitches, on the other hand, the alternative conformations have to promote or inhibit the formation of a larger and much more complex structure, i.e. an intrinsic terminator consisting of a hairpin element followed by a U-rich single-stranded region. Obviously, the screening of such a high sequence complexity with defined structural requirements is a difficult task for a selection approach, and corresponding efforts for designing transcriptional regulators failed so far (7). Hence, our rational design seems to be more promising for the design of such synthetic regulators.
The possibilities of comparing the synthetic riboswitches with their natural counterparts are limited, as almost all of those elements represent OFF-switches based on a different regulation principle. Here, ligand binding induces transcription termination, whereas our synthetic riboswitches cause ligand-induced transcription. Furthermore, regulated changes in expression vary dramatically (between 7 and 1.200-fold) in these riboswitches (39). The only well-characterized transcriptional ON-switch that activates gene expression on ligand recognition is the adenine-sensing ydhL riboswitch of B. subtilis (40). Here, the ligand is bound at a Kd value of 300 nM adenine [Kd of the theophylline aptamer: 320 nM theophylline (23)], and the ON/OFF rate is ∼10-fold compared with 6.5-fold as observed for RS10shift. For both natural (ydhL) and synthetic (RS10, RS3) riboswitches, similar external concentrations of the ligand molecules (0.5–2 mM theophylline versus 0.74 mM adenine) were used. The comparably lower regulation efficiency of the synthetic elements could be explained by the rather inefficient and slow theophylline uptake so that equilibrium is not reached (41). Altogether, the regulatory power of the synthetic ON switches is comparable with that of a natural ON switch.
In the case of other synthetic RNA-based regulators, theophylline-dependent translational riboswitches and the transcription-controlling elements presented here also show a similar efficiency in regulation (8-fold versus 6.5-fold) (15,17). However, these constructs are transcribed from different promoters (B. subtilis XylA, E. coli IS10) that can also influence termination efficiency (42). Furthermore, it was shown that sequences downstream of the U stretch can have an impact on termination efficiency (6,30), whereas additional U-rich elements enhance translation (43). Sequences immediately downstream of the start codon, described as downstream boxes, can also act as translational enhancers, as it was demonstrated for several E. coli and bacteriophage genes (44). Besides these and other sequence-specific features, there are further properties that greatly influence the expression of a given gene. Secondary structure elements and position effects of the aforementioned regulatory regions within a translational unit play a crucial role for productivity of protein synthesis (45,46). As these factors have a great impact on the efficiency of riboswitches in general, it is virtually impossible to compare the regulatory power of the elements. On the other hand, this plethora of features influencing bacterial gene expression allows a further optimization of the individual constructs, eventually leading to higher ON/OFF rates.
Although these data indicate that the nucleic acid sequence context can have a great impact on the efficiency of such synthetic riboswitches, RS10 shows a similar theophylline-dependent regulation in a different genetic background consisting of a constitutive promoter and GFP as a reporter (Figure 2C). A likely reason for the different ON/OFF rates of RS10 observed for the pBAD/bgaB system (6.5-fold) and the pBSU/GFP system (3-fold) are the specific properties of GFP as a reporter protein. After translation, GFP undergoes a series of maturation steps, leading to a considerable delay in fluorescence. Furthermore, in rapidly dividing cells, GFP will be diluted rather fast, leading to a reduced fluorescence signal in the growing culture (47). Hence, the absolute ON/OFF rates of the two reporter systems cannot be directly compared. Nevertheless, RS10 is active in both systems, indicating a functional generality of the design approach used in this study. Furthermore, the Prrn promoter-based transcription in the pBSU/GFP system indicates that arabinose used as an inducer in pBAD/bgaB has no influence on the conformational switch of the regulatory constructs but only activates transcription. This is further supported by the fact that in all individual assays based on the pBAD/bgaB system, arabinose was added for transcription induction, leading to a general normalization for the presence of arabinose in these experiments.
For transcription-controlling riboswitches, a kinetic control is discussed, where the riboswitch does not reach a thermodynamic equilibrium with its ligand, but it is driven by the kinetics of RNA folding, ligand interaction and RNA synthesis (48–50). Such a competition between ligand binding and speed of transcription seems to be a common regulatory feature. Several transcriptional riboswitches force the polymerase to pause downstream of the terminator element, giving the sensor sufficient time for ligand binding and, consequently, deciding whether to proceed or terminate transcription (51). In our riboswitches, sophisticated structural features such as transcriptional pause sites (49) or the energetic contribution of ligand binding to the stability of the riboswitch conformation (52) were not yet included in the design principles. The Kd value used to estimate the binding energy between target molecule and aptamer depends on experimental conditions, e.g. salt concentration and temperature, which are uncertain for the theophylline aptamer used here. Hence, the resulting ΔG value represents only a rough estimate and cannot be directly integrated into the pipeline at the moment. The current design strategy is based on MFE evaluations. This works as long as the unbound aptamer structure folds into the MFE conformation, as it is the case for the used theophylline aptamer. It needs to be replaced, however, if alternative structures are adapted preferentially. The usage of advanced co-transcriptional folding simulations and explicit predictions of alternative conformations will become necessary at that point.
As transcriptional riboswitches require an overlapping region between sensor (theophylline aptamer) and actuator (terminator), it is not possible to simply fuse a naturally occurring terminator to the aptamer. Natural intrinsic terminators consist of a GC-rich hairpin followed by a U-rich region that readily dissociates from the DNA template (30,53). As the hairpin element does not show a high conservation at the sequence level, this part of the terminator can be easily manipulated without destroying its regulatory function (54). As a consequence, it is possible to design artificial terminators simply by adjusting the 5′-half of the hairpin element to the sequence required for proper base pairing with the 3′-part of the aptamer. This is demonstrated by the fact that all designed terminator structures significantly shut down gene expression and represent therefore functional elements, comparable with natural terminators. Obviously, the simple descriptors used for our in silico construction are sufficient to predict efficient termination of the reporter system.
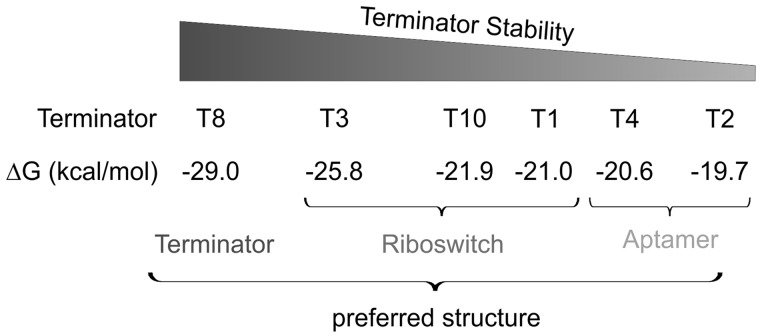
As shown in Figure 2, not every generated terminator led to a functional riboswitch. Nevertheless, the calculated free energy values of the individual terminators are in good agreement with functionality of the constructs (Figure 5). The terminator structure of RS8 showed the highest stability (−29.00 kcal/mol), suggesting that it is formed even in the presence of theophylline, and the bound aptamer structure cannot compete with the more stable terminator hairpin. Correspondingly, RS8 exhibits a constitutive low gene expression level. In contrast, terminators of RS2 and RS4, with constitutive gene expression, are the least stable structures with −19.70 kcal/mol and −20.60 kcal/mol, respectively. Here, the binding-competent structure of the aptamer is probably more stable and remains intact even in the absence of theophylline, interfering with terminator formation. The stabilities of terminators in functional riboswitches RS1, RS3 and RS10 were between −21.00 and −25.80 kcal/mol, allowing a stable formation of the ligand-bound aptamer structure in the presence of theophylline. In agreement with this correlation are the regulatory features observed for RS10loop, where the optimization of the terminator hairpin leads to an increased stability. Owing to this more robust terminator structure (ΔG = −24.8 kcal/mol), RS10loop has a reduced background activity compared with RS10. This is correlated with a lowered ON rate, as the ligand-bound aptamer structure now has to compete with a more stable terminator element. Yet, this interpretation is rather speculative, as the stability of a terminator structure is not necessarily correlated with termination efficiency.
Figure 5.
A possible correlation between terminator stability and riboswitch function. Among all tested terminators, T8 has the lowest free energy value and forms a stable structure that obviously cannot be disrupted on ligand binding. As a consequence, RS8 is locked in an OFF state. In contrast, T2 and T4 form a much weaker hairpin, shifting the equilibrium towards the aptamer structure that interferes with terminator formation. The functional riboswitches RS1, RS3 and RS10 show intermediate stabilities of their terminator elements that allow the desired ligand-dependent rearrangement of the constructs, switching between ON and OFF states. The stabilized terminator hairpin of RS10loop is located between the terminators of RS8 and the functional RS3. Although this stabilized hairpin reduces the background transcription rate considerably, it obviously impedes the structural rearrangement induced by ligand binding and allows only a moderate level of gene expression.
Although the adjusted loop sequences are found in most of the naturally occurring intrinsic terminators (30), one cannot transfer common features of stand-alone terminators into transcriptional riboswitches. Obviously, the requirements for structural rearrangements between ON and OFF states do not allow optimal terminator structures but favor a compromise between efficient termination and the possibility of riboswitch refolding on ligand binding. As energy values for most of the available aptamers are not known, it is not possible to predict a priori which of the designed terminators are suited for a certain aptamer domain to form a functional riboswitch. Here, biochemical analyses are required to verify a ligand-dependent regulation of transcription.
A great advantage compared with translational riboswitches is the fact that transcriptional riboswitches can in principle be combined as tandem copies in a single transcriptional unit, potentially leading to a rather tight regulation of gene expression or Boolean NOR gene control logic (7,55). Similar tandem arrangements are also described for several natural riboswitches, leading among other effects to an enhanced digital character of regulation (56). Furthermore, transcriptional riboswitches could be used as control elements for the release of non-coding regulatory RNAs that influence expression of certain target genes. However, to produce a functional regulatory RNA free of potentially interfering flanking sequences, additional cleavage sites in the primary transcript might be required.
Taken together, our in silico design to construct functional synthetic riboswitches acting at the level of transcription is a first promising approach to generate and optimize sophisticated regulatory RNA control elements. The implication of further parameters like transcriptional pause sites and ligand-binding behavior will lead to a more efficient regulatory power, comparable with that of the natural counterparts. Hence, fully understood synthetic elements have the potential to develop into useful and versatile building blocks in synthetic biology.
FUNDING
The Deutsche Forschungsgemeinschaft (DFG MO 634/8-1 and DFG STA 850/10-2); the Christiane-Nüsslein-Volhard-Stiftung (to M.W.). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank H. Betat, B. Klinkert and P. Kerpedjiev for valuable discussions and G. Domin for cloning RS8. They thank M. Mießler and S. Bonin for expert technical support.
REFERENCES
- 1.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotech. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 2.Wieland M, Hartig JS. Artificial riboswitches: synthetic mRNA-based regulators of gene expression. ChemBioChem. 2008;9:1873–1878. doi: 10.1002/cbic.200800154. [DOI] [PubMed] [Google Scholar]
- 3.Topp S, Gallivan JP. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2010;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. ChemBioChem. 2008;9:1906–1911. doi: 10.1002/cbic.200700713. [DOI] [PubMed] [Google Scholar]
- 8.Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson S, Bauer G, Fink B, Suess B. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005;11:503–511. doi: 10.1261/rna.7251305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa A. Rational design of artificial riboswitches based on ligand-dependent modulation of internal ribosome entry in wheat germ extract and their applications as label-free biosensors. RNA. 2011;17:478–488. doi: 10.1261/rna.2433111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput. Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Muranaka N, Abe K, Yokobayashi Y. Mechanism-guided library design and dual genetic selection of synthetic OFF riboswitches. ChemBioChem. 2009;10:2375–2381. doi: 10.1002/cbic.200900313. [DOI] [PubMed] [Google Scholar]
- 14.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 15.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 16.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babiskin AH, Smolke CD. Engineering ligand-responsive RNA controllers in yeast through the assembly of RNase III tuning modules. Nucleic Acids Res. 2011;39:5299–5311. doi: 10.1093/nar/gkr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D-S, Gusti V, Pillai SG, Gaur RK. An artificial riboswitch for controlling pre-mRNA splicing. RNA. 2005;11:1667–1677. doi: 10.1261/rna.2162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson KM, Syrett HA, Knudsen SM, Ellington AD. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002;2:21. doi: 10.1186/1472-6750-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP. Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic Acids Res. 2012;40:5775–5786. doi: 10.1093/nar/gks168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 24.Klinkert B, Cimdins A, Gaubig LC, Roßmanith J, Aschke-Sonnenborn U, Narberhaus F. Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J. Biotechnol. 2012;160:55–63. doi: 10.1016/j.jbiotec.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU. a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007;65:413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 26.Andronescu M, Fejes AP, Hutter F, Hoos HH, Condon A. A new algorithm for RNA secondary structure design. J. Mol. Biol. 2004;336:607–624. doi: 10.1016/j.jmb.2003.12.041. [DOI] [PubMed] [Google Scholar]
- 27.Busch A, Backofen R. INFO-RNA—a fast approach to inverse RNA folding. Bioinformatics. 2006;22:1823–1831. doi: 10.1093/bioinformatics/btl194. [DOI] [PubMed] [Google Scholar]
- 28.Hofacker I, Fontana W, Stadler P, Bonhoeffer L, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 29.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 30.d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 31.Buskirk AR, Kehayova PD, Landrigan A, Liu DR. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 32.Turnbough CL, Hicks KL, Donahue JP. Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA. 1983;80:368–372. doi: 10.1073/pnas.80.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muranaka N, Abe K, Yokobayashi Y. Mechanism-guided library design and dual genetic selection of synthetic OFF riboswitches. ChemBioChem. 2009;10:2375–2381. doi: 10.1002/cbic.200900313. [DOI] [PubMed] [Google Scholar]
- 34.Topp S, Gallivan JP. Riboswitches in unexpected places—a synthetic riboswitch in a protein coding region. RNA. 2008;14:2498–2503. doi: 10.1261/rna.1269008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topp S, Gallivan JP. Random walks to synthetic riboswitches—a high-throughput selection based on cell motility. ChemBioChem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 37.Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J. Am. Chem. Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- 38.Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1975;72:4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 40.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 41.Koch AL. The metabolism of methylpurines by Escherichia coli. I. Tracer studies. J. Biol. Chem. 1956;219:181–188. [PubMed] [Google Scholar]
- 42.Goliger JA, Yang XJ, Guo HC, Roberts JW. Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J. Mol. Biol. 1989;205:331–341. doi: 10.1016/0022-2836(89)90344-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Deutscher MP. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc. Natl Acad. Sci. USA. 1992;89:2605–2609. doi: 10.1073/pnas.89.7.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sprengart ML, Fuchs E, Porter AG. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 45.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 46.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leveau JH, Lindow SE. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J. Bacteriol. 2001;183:6752–6762. doi: 10.1128/JB.183.23.6752-6762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 49.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 50.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 51.Haller A, Souliere MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc. Chem. Res. 2011;44:1339–1348. doi: 10.1021/ar200035g. [DOI] [PubMed] [Google Scholar]
- 52.Gouda H, Kuntz ID, Case DA, Kollman PA. Free energy calculations for theophylline binding to an RNA aptamer: comparison of MM-PBSA and thermodynamic integration methods. Biopolymers. 2003;68:16–34. doi: 10.1002/bip.10270. [DOI] [PubMed] [Google Scholar]
- 53.Martin FH, Tinoco I. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl Acad. Sci. USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breaker RR. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]