Abstract
RNA editing by adenosine deaminases that act on RNA (ADARs) diversifies the transcriptome by changing adenosines to inosines. In mammals, editing levels vary in different tissues, during development, and also in pathogenic conditions. From a screen for repressors of editing we have isolated three proteins that repress ADAR2-mediated RNA editing. The three proteins RPS14, SFRS9 and DDX15 interact with RNA. Overexpression or depletion of these proteins can decrease or increase editing levels by 15%, thus allowing a modulation of RNA editing up to 30%. Interestingly, the three proteins alter RNA editing in a substrate-specific manner that correlates with their RNA binding preferences. In mammalian cells, SFRS9 significantly affects editing of the two substrates CFLAR and cyFIP2, while the ribosomal protein RPS14 mostly inhibits editing of cyFIP2 messenger RNA. The helicase DDX15, in turn, has a strong effect on editing in Caenorhabditis elegans.
Expression of the three factors decreases during mouse brain development. Moreover, expression levels of SFRS9 and DDX15 respond strongly to neuronal stimulation or repression, showing an inverse correlation with editing levels.
Colocalization and immunoprecipitation studies demonstrate a direct interaction of SFRS9 and RPS14 with ADAR2, while DDX15 associates with other helicases and splicing factors. Our data show that different editing sites can be specifically altered in their editing pattern by changing the local RNP landscape.
INTRODUCTION
Adenosine to inosine deamination of RNAs is the most prevalent RNA editing mechanism in metazoa. The enzymes responsible for this base recoding event are adenosine deaminases that act on double-stranded RNA (ADARs). Four different types of ADARs are found in the mammalian genome ADAR1, ADAR2, ADAR3 and TENR (1–4). ADAR1 and ADAR2 are expressed in all tissues but ADAR3 is predominantly expressed in the brain (5). ADAR1 and ADAR2 have proven deaminase activity, while no editing activity could be assigned to ADAR3 and TENR (3).
ADARs convert adenosines to inosines in double-stranded regions of RNAs. Inosines are recognized as guanosines by most cellular machineries. Therefore, this type of RNA editing can alter the base pairing, folding, localization, stability or transport of RNAs (6). If a codon is affected, the A-to-I conversion can lead to codon changes in messenger RNAs (mRNAs), resulting in the translation of proteins that differ from the genomically encoded ones.
The RNAs affected by ADAR-mediated editing frequently encode proteins involved in neurotransmission such as channels and receptors (6). The introduced amino acid exchanges can alter the assembly and transport of the affected proteins but also their kinetic properties (7). Interestingly, it could be shown that editing levels of the RNAs encoding these neuronally expressed proteins vary throughout development despite a relatively constant level of ADAR proteins (8). Moreover, global hypoediting has been reported in tumor tissues, indicating a correlation between editing and cancer (9–11). Similarly, alterations in editing patterns have been observed in association with mental disorders (11).
Until now, different mechanisms that can regulate ADAR activity have been proposed such as alternative splicing of ADAR pre-mRNAs, leading to the formation of less active variants of ADARs and post-translational modifications of the enzymes (12–16). For instance, peptidyl prolyl isomerase (Pin1) can stimulate ADAR2 activity by ensuring nuclear localization and stability of ADAR2. At the same time, E3 ubiquitin ligase WWP2 negatively regulates ADAR2 by catalysing its ubiquitination, thus stimulating its degradation (16). Similarly, ADAR1 activity has been shown to down-regulated by sumoylation (14).
At the transcriptional level, ADAR2 expression is stimulated by CREB1 (17). Thiamine deficiency, on the other hand, has been shown to down-regulate ADAR2 expression in vitro (18). Finally, subcellular sequestration of ADAR2 to nucleoli has also been suggested to regulate ADAR activity (19,20).
To identify potential cellular repressors of ADAR2 activity, we have used a yeast-based editing assay that allows for the unbiased identification of factors affecting editing. To this end we have isolated three RNA-binding proteins that repress RNA editing both in a heterologous yeast assay but also in mammalian cells. Most interestingly, the three candidates repress editing with site preferences. For one of the candidates, the RNA-helicase DDX15, conservation of the inhibitory activity on ADAR-mediated editing could be verified in Caenorhabditis elegans. Expression of the candidate proteins described here changes in the developing brain and on neuronal stimulation, making them excellent candidates as substrate-specific regulators of editing.
MATERIALS AND METHODS
Creation of a yeast strain to screen for repressors of editing
A stem-loop substrate based on the GluR-B R/G site was mutated by polymerase chain reaction (PCR) to replace the arginine codon at the editing site by an amber stop codon. On editing, the amber stop is converted into a tryptophan codon, allowing continuous translation of a downstream open reading frame. The stem-loop substrate was introduced in-frame 100 nucleotides downstream of the start codon of the his3 gene, using an artificially created XhoI restriction site. Downstream of the stem-loop the ura3 gene was introduced. The whole construct was fused to a neo resistance cassette and introduced via homologous ends into the leu2 gene of Saccharomyces cerevisiae strain W303 (21).
A Flag-tagged version of rat ADAR2 (a kind gift of R. Emeson, Vanderbilt University) was cloned into and expressed from the centromeric tetracycline inducible vector pCM251 (22).
Library transformation
To identify factors that interfere with editing a HeLA complementary DNA (cDNA) library cloned in pJG4 and bearing trp1, auxotrophy marker gene was transformed as described (23). Aliquots of the transformed cells were plated onto plates lacking tryptophan without further selection to determine the transformation efficiency. The remainder of the transformed cells was plated onto plates containing 5-fluoroorotic acid (5-FOA) to allow a negative selection against editing.
Plasmid isolation from yeast
Single colonies were picked and grown overnight in a selective medium. Plasmid DNA was prepared as described (24).
Tissue culture transfection
The open reading frames encoding candidate proteins were cloned into the tissue culture expression vector pCDNA3.1(−) (Invitrogen, CA, USA) N-terminally fused to 6xmyc tags and a NLS sequence. Transfection was performed using Nanofectin reagent (PAA, Austria) according to manufacturer’s instructions. After 32–72 h, cells were processed to obtain RNA, protein extracts, or were stained to visualize protein expression.
FACS analysis
For FACS analysis, vectors expressing repressors of editing were cotransfected with the substrate vector in a 4:1 ratio. After 72 h, red and green fluorescence was measured on a FACScalibur flow cytometer (BD Biosciences) using CellQuest 3.3 software. Statistical assessment was performed with FlowJo 6.3.1 software. For statistic evaluation, six different gates were taken above background for the red channel, while the green channel was wide open. The mean green florescence values from each gate were divided by the mean red fluorescence values. These normalized fluorescence values were plotted on a graph against the chosen gates. Statistical significance was measured by Student’s T-test.
Immunofluorescence analysis
Transfected cells were fixed (25), and myc-tagged proteins were detected with mAb 9E10 (26) and goat-anti mouse Alexa Fluor 568 (Invitrogen, CA, USA), and in case of Flag-tagged rADAR2 with either rabbit anti-Flag (Sigma) or anti-ADAR2 antibody (rabbit serum made in our lab) followed by goat Alexa Fluor 488 anti-rabbit antibody (Invitrogen, CA, USA).
Immunoprecipitation analysis
Cells were lysed by sonication in NET-2 buffer (150 mM NaCl, 80 mM Tris pH 7.4, 0.05% NP-40) (27). The extract (150–200 µg) was added to Protein A Sepharose beads (GE Healthcare) coupled with anti-myc mAb 9E10 or with anti-Flag antibody. After 2 h of incubation on a rotating wheel, beads were washed with NET-2 and bound proteins were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and western blotting. Myc-tagged proteins were detected with mAb 9E10 and alkaline phosphatase–coupled goat anti-mouse antibody (Sigma). Flag-tagged rADAR2 was detected with either rabbit anti-Flag (Sigma, St. Louis MO, USA) or a home-made anti-ADAR2 antibody followed by goat alkaline phosphatase anti-rabbit antibody (Pierce, Rockford, IL, USA). Candidate proteins were detected with monoclonal mouse anti-myc antibody and a secondary goat anti-mouse antibody coupled to alkaline phosphatase. Phosphatase activity was detected with the chromogenic substrate NBT/BCIP.
RNA isolation
RNA was idirectly isolated from cells growing in a tissue culture dish using guanidinium thiocyanate–phenol–chloroform extraction with TriFast reagent (Peqlab, Germany) according to manufacturer’s protocols.
RNA-IP
To determine whether substrate RNAs show preferential binding to any of the candidate proteins, RNA-IPs were performed. To do this, HeK293 cells stably transfected with ADAR2 were transiently transfected with plasmids expressing any of the three candidates as myc-tagged fusion proteins. Immunoprecipitations were performed as above, and RNAs were extracted using Trifast (Peqlab, Germany) from IP reactions with myc-coupled protein-A sepharose or uncoupled beads (mock). RNAs were precipitated, reverse transcribed and cDNAs were used for qPCRs. Enrichment of the specific RNAs over mock IPs was calculated by comparing the amount of RNA in the input versus RNA in the IP (delta CT). This value was compared for each specific IP over the corresponding Mock IP (delta-delta CT). The resulting value was used to calculate the fold-enrichment of each RNA compared with a mock IP.
Reverse transcription
cDNA was synthesized with RevertAid M-MuLV Reverse Transcriptase (Fermentas) according to the manufacturer’s instructions using random hexamers. Aliquots of the heat inactivated RT-reactions were used as templates for PCR reactions.
Quantification of editing by sequencing
Editing was quantified by dividing the height of G peak to the sum of A and G peak. All measurements were made in Adobe Photoshop CS5. For statistical evaluation, three or more measurements were compared by a non-paired Student’s T-test.
Construction of shRNA vectors
To generate shRNA expression cassettes targeting rps14, ddx15 or srsf9, the pLKO.1 lentiviral vector system was used (28) as described by Addgene (http://www.addgene.org/plko). Briefly, specific oligonucleotides (Sigma) corresponding to the following Broad TRC RNAi shRNA library (The RNAi Consortium) sequences were introduced into the Age I–EcoR I sites of pLKO.1 (Addgene plasmid # 10878).
shRNA sequences can be found in Supplementary Table S2.
Cells transfected with non-targeting shRNA vectors NT-2 or anti luciferase, activating RISC and the RNAi pathway, but not targeting any human gene, were used as negative controls in knockdown experiments. All shRNA expression cassettes were sequence verified.
Viral particle production and target cell infection
Described shRNA-pLKO.1 constructs were cotransfected with the packaging plasmid pPax2 (Addgene plasmid # 12260) and the envelop plasmid pMD2.G (Addgene plasmid #12259) into human embryonic kidney 293FT cells using Lipofectamine 2000 (Invitrogen). Virus was harvested 72 h post-transfection and concentrated using a PEG virus precipitation kit (BioVision). Infections of HeK293 cells either stably expressing the target protein, or ADAR2 were carried out in the presence of 10 μg/ml hexadimethrine bromide (a.k.a. polybrene, Sigma). Following transduction, cells were selected with 1 μg/ml puromycin.
RNA isolation from C. elegans
C. elegans strains were obtained from CGC. VC2277 (ddx-15+/−) with pharyngeal GFP on a balancer were selfed, and progeny was selected for heterozygous ddx-15+/− GFP, and homozygous non-GFP ddx-15−/− adults. Homozygous glh-2−/− worms were also obtained from CGC. For RNA extraction, 24 animals were collected in 100 μl water. After the addition of 100 µl of glass beads and 500 µl of TriFast (Peqlab, Germany), the worms were vortexed at 4°C for 30 min. After the addition of 200 µl of chloroform, the extract was vortexed for 10 min at 55° C. Nucleic acids were precipitated from the aqueous phase with 2-propanol. Contaminating DNA was removed by DNase I digestion.
RT PCR and qPCR
cDNA synthesis was done with random hexamers and RevertAid RNAseH minus mMuLV reverse transcriptase following the manufacturers instruction (Fermentas, Lithuania). As a control, MOCK reactions without RTase were set up.
For qPCR, a GoTaq qPCR master mix was used (Promega, Madison, WI) on a BioRad iQ5 cycler (BioRad, Hercules, CA, USA). At least three biological and two technical replicates were done for each qPCR assay.
Relative differences in RNA levels were determined by using the delta delta CT method (29). Statistical significance of differences was calculated using a Student’s T-test.
Rat brain slice preparation and treatments
All procedures were carried out in accordance with UK Home Office regulations. Sprague–Dawley rat hippocampi were dissected from pups (postnatal age 5 days) in a sucrose-modified Gey’s balanced salt solution, which was as follows: 175 mM sucrose, 50 mM NaCl, 2.5 mM KCl, 0.85 mM Na2HPO4, 0.65 mM KH2PO4, 28 mM MgSO4, 2 mM MgCl2, 0.5 mM CaCl2, 25 mM glucose and 10 mg/ml phenol red (∼330 mOsm, pH 7.3). Transverse hippocampal slices (350 µm thick) were cultured using the roller-tube method on collagen-coated coverslips in an incubator at 36°C without humidity or CO2 control (30,31). Cultures were held as described (32).
For interface cultures, Millicell cell culture inserts of 0.4 µm pore size (PICM0RG50, Millipore) were placed in six-well plates containing 1 ml of pre-warmed culture medium (as above). Three slices were individually transferred and positioned on the membrane of each insert and gently washed with culture medium. Slices were maintained in an incubator at 37°C and supplied 5% CO2 (32).
Slices were cultured for at least 3 weeks before treatments. For treatments, slices were fed with culture medium containing tetrodotoxin (TTX, 1 µM), bicuculline (BIC, 20 µM) or no drug (CTRL). Slices were returned to the incubator for a duration of 48 h, after which hippocampal subfields were dissected for RNA extraction.
qPCR on cDNA samples was performed with primers specific for the three candidates and primers for GAPDH as a control.
Analysis of gene expression atlas data
Affymetrix gene expression data was downloaded from http://www.ebi.ac.uk/gxa/ for experiment E-GEOD-35366. Data for six wild-type mouse brains each at e14, p0 and p14 was extracted for the three genes SRSF9, DDX15 and RPS14 as well as tubulin and actin for normalization. Normalization to actin was performed (the essentially identical data was obtained when tubulin was used for normalization).
RESULTS
Creation of a yeast strain for a heterologous editing assay
To identify cellular RNAs and proteins that can repress editing, we have modified a yeast reporter strain that was originally used to identify factors that stimulate ADAR2 activity (Garncarz, Tariq, Handl, Pusch, and Jantsch, RNA Biology, in press). Briefly, a hairpin derived from the human GluA2 glutamate receptor gene that harbors an amber stop codon surrounding a bone fide editing site was cloned into the 5′ region of a his3::ura3 fusion construct (Supplementary Figures S1 and S2). On editing, the stop codon is converted into a tryptophan (W) codon, allowing for the expression of the his3::ura3 fusion. The entire cassette, driven by the his3 promoter, is integrated into the genome.
A rat ADAR2 cDNA was introduced on a tetracycline-inducible centromeric vector to confer editing to yeast cells. As a positive control, a ‘pre-edited’ version of the reporter construct was prepared as well that allows constitutive expression of ura3. On minimal media lacking uracil only strains harboring the pre-edited stem loop, or strains showing successful editing of the amber codon can grow. In the presence of uracil and the drug 5-FOA, however, only cells that show no editing and thus no expression of ura3 can grow. URA3 converts FOA to 5-fluoro-uracil, which is toxic to cells. Thus, only cells that are inhibited in their editing activity and therefore fail to express URA3 are able to grow on this selection medium (Supplementary Figures S3 and S4). This strain therefore allows to screen for factors or cDNAs that express proteins that will repress editing, therefore allowing growth on plates harboring 5-FOA.
Screening for inhibitors of editing in a yeast reporter strain
To screen for proteins that interfere with editing, a HeLa cDNA library cloned into a yeast expression plasmid was transformed into the screening strain described above. Transformants were plated on selective media containing FOA to select against URA3 expression, and thus against editing. From a total of 1 × 106 colonies screened, about 140 showed growth under FOA selection indicating repression of editing or successful prevention of 5-fluoro-uracil accumulation.
On retransformation into the original screening strain, to eliminate false positive clones, and to compare positive hits with each other, 12 clones could clearly and reproducibly support growth on FOA plates (see Supplementary Table S1). The remaining cDNAs either failed to reproducibly support growth on FOA plates or also supported growth of a strain constitutively expressing Ura3 from a pre-edited stem loop, independent of ADAR2 editing.
Of the 12 positive clones, four did encode RNA-interacting proteins (see Supplementary Table S1).
Validation of candidates in mammalian cells
To test whether the clones isolated in the yeast screen are also able to inhibit editing in a more natural surrounding, the cDNAs were cloned in frame into a mammalian expression plasmid harboring a myc-tag for easier detection.
The resulting plasmids were transfected with a reporter plasmid into HeK293 cells that were stably expressing ratADAR2. The reporter plasmid allows to quantify editing with the aid of a fluorescent reporter; the open reading frames of RFP and GFP are separated by the GluA2 stem loop harboring a stop codon at an editing site (33). An increase in editing leads to an increase in green fluorescence, while red fluorescence remains constant. Similarly, inhibition of editing reduces green fluorescence (Supplementary Figure S5). The impact of expression of the candidate cDNAs on red and green fluorescence was measured by flow cytometry (FACS) (data not shown). Three out of the 12 candidates did show a significant reduction in green fluorescence relative to red fluorescence, these clones encoded fragments of the RNA helicase DDX15, the splicing factor SFRS9 and the full-length cDNA ribosomal protein RPS14. All other clones only mildly affected editing in the mammalian reporter assay.
Next, the full-length versions of these cDNAs were cloned and tested for their effect on editing of the stem-loop reporter (Figure 1). Clearly, full-length DDX15, SFRS9 and RPS14 led to a strong reduction in green fluorescence (shift to the left) when compared with red fluorescence. A scheme of the three proteins and the fragments recovered from the primary yeast screen is given in Supplementary Figure S6.
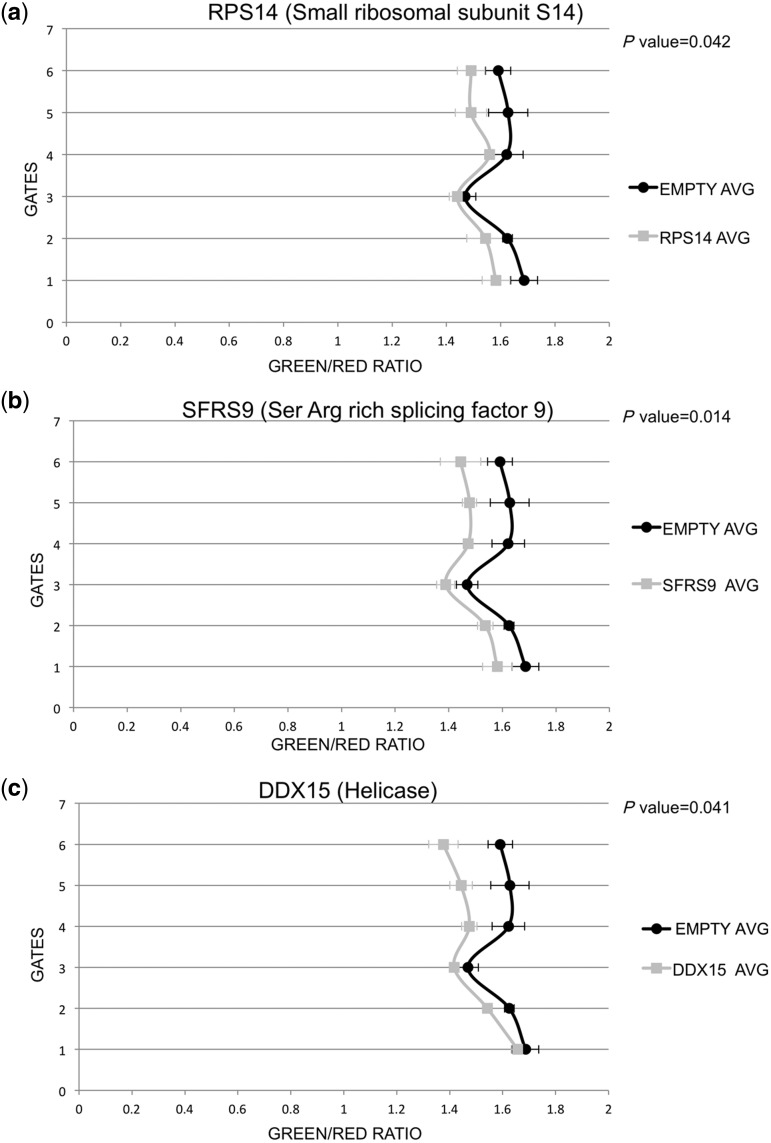
Figure 1.
Fluorescence activated cell sorting demonstrates inhibition of editing by the three candidates isolated. Cells stably transfected with ratADAR2 were cotransfected with two plasmids. One plasmid expresses the editing reporter RFP and GFP separated by an editable stem loop. Editing removes a stop codon and allows GFP expression to occur. Red and green fluorescence ratios are calculated for each cell. Values are then averaged for 6 different intensity windows. A shift to the left indicates a lower editing level induced by coexpression of an editing repressor. (a) RPS14 (grey) is compared with control empty vector (black). Expression of RPS14 leads to a significant decrease in editing. (b) Similarly, also expression of SFRS9 (grey) or DDX15 (c) leads to a decrease in editing. P values were calculated for gate #5 using Student’s T-test.
The FACS-based reporter system only indirectly determines the impact of cellular proteins on RNA editing. Therefore, to directly compare editing levels of the reporter stem loop, A to I conversions were measured by sequencing of RT-PCR products covering the stem loop. As expected, coexpression of DDX15, RPS14 and SFRS9 led to a slight, but reproducible, decrease in editing of the reporter stem loop (Figure 2).
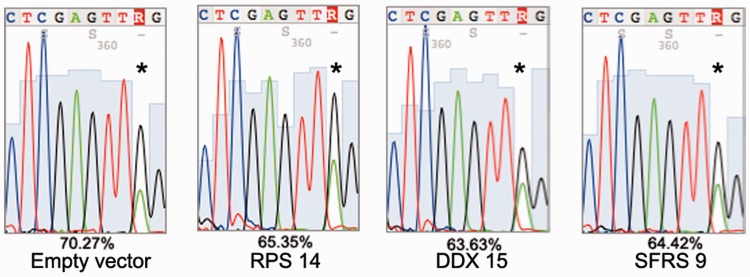
Figure 2.
Direct comparison of editing levels in the RFP–stem-loop–GFP construct. RNA from cells expressing rat ADAR2, the RFP-GFP reporter construct and the candidate repressors of editing was isolated. The stem-loop sequence of the editing reporter construct was reverse transcribed and amplified by PCR. An RT (−) control was included for all reactions. Editing levels were compared by direct sequencing and measuring of A and G peaks of forward and reverse sequencing tracks. Editing efficiency was calculated by dividing G peak height versus sum of A and G peak height. Presence of an empty vector leads to 70.3% editing. Coexpression of RPS14, DDX15 and SFRS9 leads to a reduction of editing by 5–6%. The asterisk marks the edited nucleotide.
Candidate proteins exhibit a substrate-specific inhibition of editing
At first sight, the effect of overexpression of the candidate proteins seems low. However, the mammalian editing reporter system relies on the successful cotransfection of two plasmids, the RFP-GFP reporter and the candidate cDNA in the same cell. Cotransfection of two plasmids hardly ever exceeds 30%; therefore, only a small fraction of cells expressing the reporter will be affected by the simultaneous expression of the candidate protein (A. Tariq and M. Jantsch, unpublished). Moreover, the GluA2-derived stem loop is taken out of its natural context and may therefore not reflect an endogenous substrate.
We therefore tested how editing of endogenous substrates is influenced by the candidates. To this end, three different substrates were chosen: the pre-mRNA encoding CyFip2 harbors an editing site in exon 10 about 50 nucleotides away from the next splice site, the pre-mRNA of the actin-crosslinking protein FLNA, which has an editing site just 2 nucleotides upstream of the next 5′ splice site and the pre-mRNA encoding the CFLAR protein that harbors multiple editing sites within its 3′ UTR.
Cotransfection of any of the three cDNAs encoding the candidate proteins did lead to a significant inhibition in editing of CyFIP2 ranging from 8% to 16% (Figure 3a). Editing of FLNA, in contrast, was barely affected by the cotransfected proteins (Figure 3b). Editing of the 3′ UTR located in CFLAR, finally, was inhibited by SFRS9 by up to 12% and to a minor extent by RPS14 (Figure 3c). Direct quantification of editing levels in the substrate stem loop, confirmed a reduction of editing by all three candidates (Figure 3d). Thus, the three factors identified in the screen inhibit editing to different, substrate-specific extents. Quantification of three biological replicates also indicates that some of the induced changes in editing levels are highly significant.
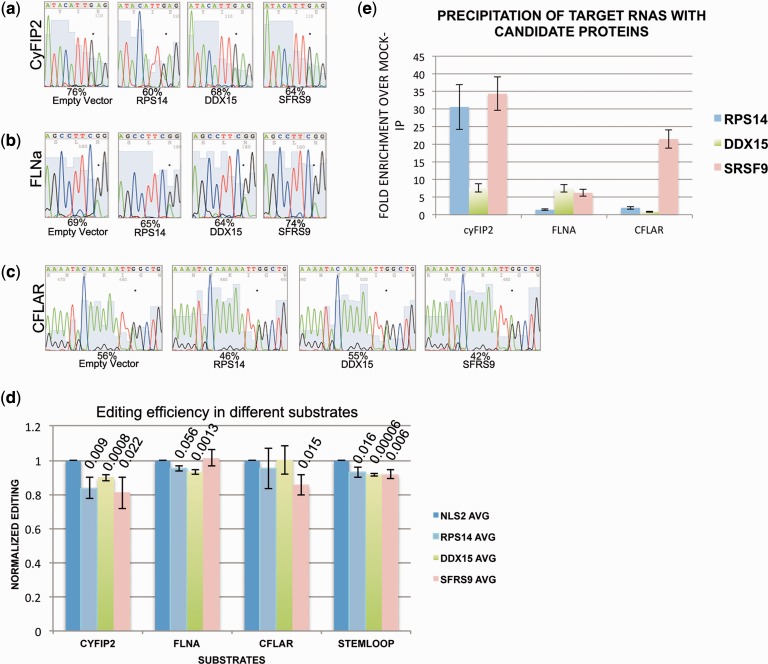
Figure 3.
Editing of endogenous substrates is selectively inhibited by repressors of editing. Editing of endogenous (a) CyFip2, (b) FLNa and (c) CFLAR RNA was determined on transfection of cDNAs encoding the three candidate repressors of editing RPS14, DDX15 and SFRS9. CyFIP2 editing is most strongly affected by expression RPS14 and SFRS9. FLNA, in contrast, is barely affected by any of the three candidates. CFLAR editing, finally, is only affected by SFRS9. Editing efficiency was calculated by dividing A and G peak height by G peak height. The edited adenosine is marked by an asterisk. (d) The average editing levels of three biological replicates are calculated and normalized to mock-transfected samples. P-values are calculated and indicated where significant. (e) myc tagged candidates were precipitated and the associated RNAs isolated. The presence of substrate RNAs in the precipitate was quantified by quantitative RT-PCR and normalized to the amount of RNA present in the cellular lysate. Fold enrichment was calculated by comparison to a mock IP with protein A sepharose beads only. cyFIP2 RNA is strongly enriched in IPs with SRSF9 and RPS14, while CFLAR RNA is strongly enriched with SFRS9 only.
Repressors of editing show distinct affinities for substrate RNAs
Substrate-specific inhibition of RNA editing might be the consequence of binding preferences of the inhibitors to a particular subset of RNAs. Therefore, to determine whether the candidate proteins RPS14, DDX15 and SRSF9 would discriminate amongst the three substrates, we tested for the association of the substrate RNAs with the candidates using RNA co-IPs. To do so, HeK293 cells expressing ADAR2 were transfected with plasmids expressing the myc-tagged candidates. After 24 h, cells were lysed and immunoprecipitations were performed using protein A beads coupled with the myc-specific monoclonal antibody 9E10 or with uncoupled protein A beads as a control. RNAs were extracted from the immunoprecipitated material and from the input material. Using primers specific for the three substrate RNAs, the amount of RNA that was copurified with any of the three candidate proteins was normalized to the amount of RNA found in the input material. By comparing these values with those obtained from IPs performed with uncoupled protein A beads (mock), the fold enrichment of each RNA bound to any of the three candidate proteins was calculated (Figure 3e). Most interestingly, the different candidates displayed clear binding preferences for the three substrate RNAs. The binding preferences also directly correlated with the negative impact on RNA editing that was exhibited by the candidates. For instance, editing of FLNA was the least affected by any of the three candidates. Consistently, FLNA RNA was also the least bound by the candidates. Editing of cyFIP2 RNA, on the other hand, was most strongly affected by SRSF9 and RPS14, which were also the two proteins to which cyFIP2 RNA bound most strongly. CFLAR RNA, finally, was most strongly bound by SRSF9, which also had the strongest negative influence on editing of this RNA. Taken together, it seems that the RNA-binding preferences of the candidates isolated can explain the substrate-specific inhibition of editing displayed by them (Figure 3d and e).
Knockdown of candidate proteins stimulates editing
Overexpression of the candidate proteins by transient transfection only affects a fraction of cells, whereas editing levels are measured on all cells. Therefore, to get a better impression on the impact of the three candidates on RNA editing, we tested whether repression of the candidates would lead to a stimulation of editing. To achieve this, we tried to silence the three candidate proteins using lentiviral-delivered shRNAs, which normally gives rise to a homogenous population of cells, all selected for the stable expression of a shRNA (see Supplementary Table S2). The efficiency of silencing was monitored in two ways. On the one hand, HeK293 cells stably expressing myc-tagged versions of the candidate proteins were treated with shRNA expressing lentivirus and subsequently stained with an anti-myc antibody. On the other hand, reduction of endogenous RNA levels were monitored by qRT-PCR. Silencing was successful for RPS14 and SFRS9, while DDX15 expression was only moderately affected (see Supplementary Figure S7).
The effect of silencing of the candidate proteins was determined by monitoring editing levels of the RNA encoding cyFIP2. Silencing of SFRS9 and RPS14 indeed led to an increase in editing of cyFIP2 by 15% and 12%, respectively. Treatment with shRNAs against DDX15 had no effect on the editing levels of cyFIP2, consistent with the inefficiency to knockdown this protein (see Figure 4). Clearly, RPS14 and SRSF9 display the strongest effect on the three mammalian substrates. Overexpression or repression of RPS14 can down- or up-regulate editing of cyFIP2 pre-mRNA by up to 15%.
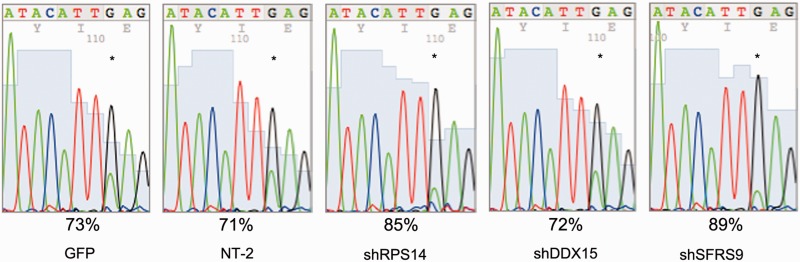
Figure 4.
Silencing of inhibitory factors leads to an increase in editing. The candidate repressors of editing were silenced using lentiviral-delivered shRNAs. Silencing was monitored by immunofluorescence and qRT-PCR. Silencing of SFRS9 and of RPS14 leads to a boost in editing by >15%. Silencing of DDX15, in contrast, has no effect on cyFIP2 editing. Asterisks mark the edited nucleotides. A non-target shRNA (NT-2) and a shRNA against GFP were used as a control. Because the cells used were stably transfected clones, no biological replicates were performed.
C. elegans DDX15 represses RNA editing
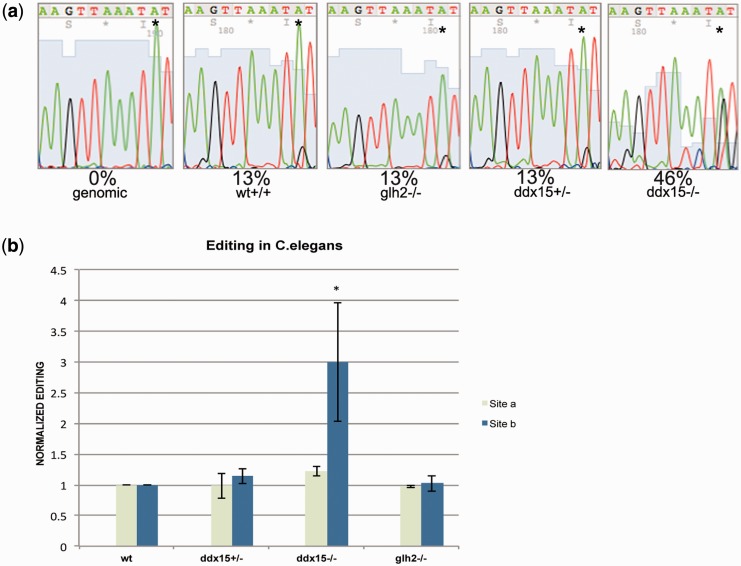
A well conserved DDX15 ortholog can be found in C. elegans. Moreover, a ddx-15 deletion strain is available from the CGC (vc2277). ddx-15−/− worms are homozygous sterile but can be kept as heterozygotes. To test whether DDX15 would also inhibit RNA editing in C. elegans, we tested editing levels in wild-type worms, and heterozygous and homozygous deletions of ddx-15. To test whether any RNA-helicase would have a similar impact on editing, a strain carrying a homozygous deletion for the helicase glh-2 was used for comparison (KB1). In C. elegans, RNA editing had been shown to occur mainly in structured 3′ UTRs. We therefore determined the impact of the above-mentioned deletions on the editing of the 3′ UTR of the C. elegans gene zc239.6, which had been shown to be edited at several sites (34). As editing levels vary between different sites, we focused on two sites that show weak basal editing levels (site a & b). Site a had previously been identified as an editing site, while site b was only identified to be edited in this study (34). Wild-type C. elegans as well as worms carrying a homozygous glh-2 deletion but also heterozygous ddx-15+/− worms displayed no change in editing at either site with a basal editing level of ∼13% at site b. A homozygous deletion of ddx-15−/−, in contrast, led to a dramatic increase in editing, boosting editing to 50% at site b (Figure 5a and b: dark blue bars). Most interestingly, the closely spaced site a is unaffected by the presence or absence of DDX15 (light green bars in Figure 5b and data not shown). Together, this shows once more that the candidates isolated in the primary yeast screen can affect editing to different extents, depending on the target RNA and the cellular surrounding.
Figure 5.
The helicase DDX15 but not GLH-2 strongly affects editing in C. elegans. Wild-type C. elegans, worms homozygous for the helicase glh-2, or heterozygous and homozygous ddx15 individuals were picked and editing levels were determined in the 3′ UTR of transcript zc239.6.This 3′ UTR had been reported to be edited at several positions. (a) Electropherogram shows that editing is strongly affected at site b (asterisks). (b) Graphical representation of editing levels of three biological replicates. Editing at site b (dark blue) corresponding to the electropherograms in (a) is strongly affected. Another nearby site that is known to be edited (site a, light green bar) is hardly affected by the presence or absence of DDX15.
RPS14 and SFRS9 interact with ADAR2
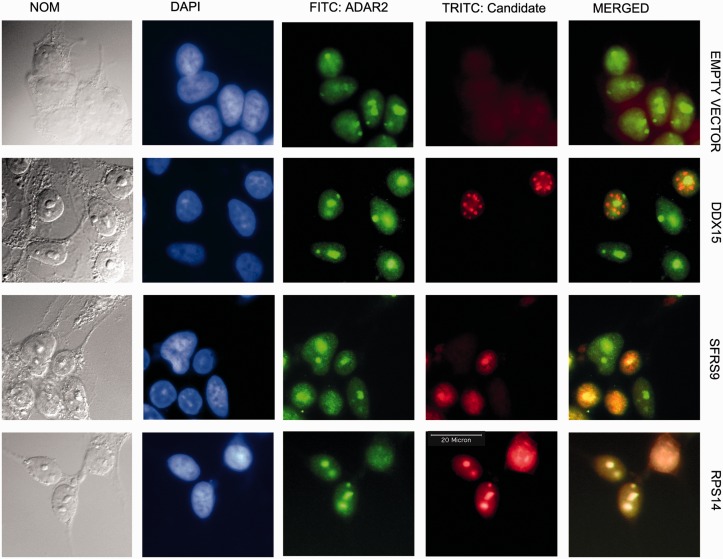
To determine whether the inhibitors of editing can form a complex with ADAR2, we first performed colocalization studies. Myc-tagged RPS14, SFRS9 and DDX15 were transfected in HeK293 cells stably expressing Flag-tagged rat ADAR2. Simultaneous staining showed that coexpression of both proteins led to a strong colocalization of ADAR2 with RPS14 and SFRS9, which was most prominent in nucleoli. RPS14 is primarily localized to nucleoli and therefore a colocalization with the nucleolar localized ADAR2 is not surprising. SRSF9, however, is a splicing factor that normally localizes in a diffuse nuclear speckled pattern (see Supplementary Figure S7). Overexpression of ADAR2 apparently forces SRSF9 to colocalize to nucleoli, already suggesting a strong interaction between the two factors (Figure 6). DDX15, in contrast, localizes to nuclear foci and does not interact with ADAR2 (Figure 6).
Figure 6.
ADAR2 colocalizes with SFRS9 and RPS14. Cells stably transfected with FLAG-tagged rat ADAR2 were cotransfected with an empty vector or myc-tagged DDX15, SFRS9 or RPS14. The green FITC channel shows the localization of ADAR2, while the candidate proteins are shown in the red TRITC channel. Immunofluorescence staining indicates that SFRS9 and RPS14 colocalize with ADAR2 to nucleoli, whereas DDX15 localizes in nuclear dots that do not overlap with nucleoli. In normal cells that do not overexpress ADAR2, SFRS9 is localized to the nucleoplasm (see Supplementary Figure S7). The colocalization of SFRS9 to nucleoli therefore depends on the presence of ADAR2. The colocalization also reflects the coimmunoprecipitation of ADAR2 with RPS14 and SFRS9 shown in Figure 7.
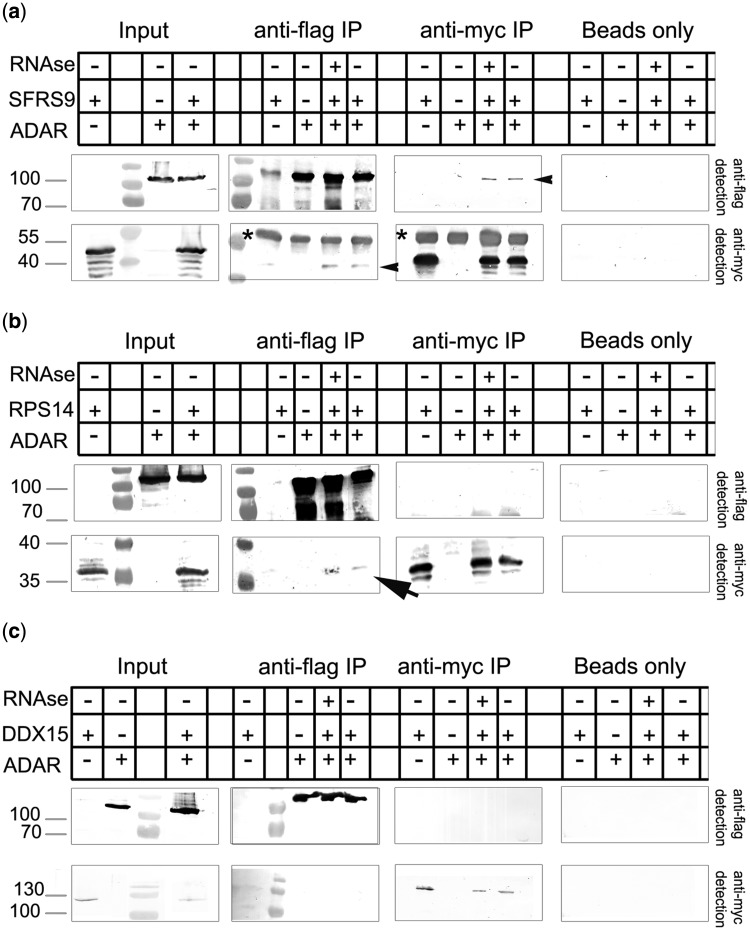
To further verify the presence of the candidate proteins and ADAR2 in the same complex, coimmunoprecipitation experiments were performed. Because the candidate proteins were myc-tagged and rADAR2 was flag-tagged, the precipitation experiments were done in both directions, one pulling on ADAR2 and detecting the candidate proteins, and the other pulling on the candidate proteins and detecting ADAR2. SFRS9 and ADAR2 showed an RNA-independent interaction with ADAR2 by coimmunoprecipitation (Figure 7a). ADAR2 could be detected in immunoprecipitates of SRFS9 and vice versa, proving the colocalization pattern mentioned above. An interaction between RPS14 and ADAR2 was less prominent but also RNA independent (Figure 7b). Moreover, while myc-RPS14 could be detected when FLAG-ADAR2 was precipitated, the opposite was not the case. This may, however, reflect different expression levels of both tagged and endogenous proteins that may give rise to non-stoichometric complex formation. DDX15, finally, failed to interact with rADAR2, confirming the colocalization experiments (Figure 7c). Also, immunoprecipitation of TAP-tagged DDX15 followed by mass spectrometric analysis of associated proteins failed to detect an interaction with ADAR2 but verified its interaction with spliceosomal components, in agreement with its function in pre-mRNA splicing (35) (Supplementary Figure S8).
Figure 7.
ADAR2 interacts with SFRS9 and RPS14 but not with DDX15. RPS14, SFRS9 and DDX15 were expressed as myc-tagged proteins together with Flag-tagged ADAR2 in transfected cells. Immunoprecipitations were done either with an anti-myc antibody or an anti-FLAG antibody, and the precipitate was detected for the presence of either protein. (a) SFRS9 can be pulled down with ADAR2 and vice versa (black arrowheads, top panel). (b) RPS14 shows a weak interaction with ADAR2 (black arrow). RPS14 can be copurified with ADAR2, while no RPS14 can be detected in the ADAR2 precipitate. This discrepancy may reflect different expression levels of the two proteins. No interaction can be detected between DDX15 and ADAR2 (c). Asterisks indicate immunoglobulin bands originating from the immunoprecipitation.
ADAR2 transcription is unaffected by inhibitors of RNA editing
A simple mechanism by which the isolated inhibitors of editing might act would be an alteration of ADAR2 expression. To test this possibility, we determined ADAR2 expression by RT-qPCR in cells transfected with the isolated candidate proteins. However, no change in ADAR2 mRNA levels could be identified, indicating that expression levels of ADAR2 are unaffected, at least at the RNA level (Supplementary Figure S9).
Expression of candidates changes during development and on neuronal stimulation
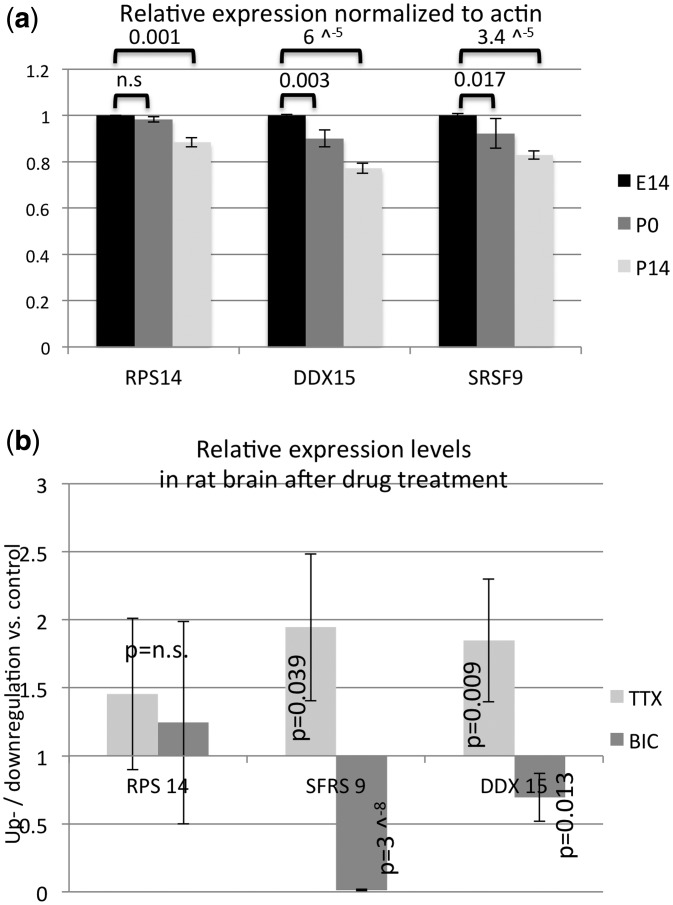
To determine the involvement of the isolated candidates in the regulation of editing in vivo, we first screened the mouse expression atlas for changes in expression levels of RPS14, DDX15 and SRSF9 during brain development. In particular, experiment E-GEOD-35366 screens brains of three different wild-type lines in two duplicates each at e14, p0 and p14 on an Affymetrix array (http://www.ebi.ac.uk/gxa/experiment/E-GEOD-35366). Expression levels of the three candidates were normalized to actin expression and compared with each other. Most interestingly, a significant drop in expression was observed for all three candidates during development being consistent with the reported increase in editing in the brain during these stages (8) (Figure 8a).
Figure 8.
Expression levels of inhibitors of editing vary during development and on neuronal stimulation. (a) Expression data for experiment E-GEOD-35366 was downloaded for six biological replicates of mouse brain RNAs at stage E14, p0 and p14. Expression profiles for RPS14, DDX15 and SRSF9 were calculated by normalization to actin. A decrease in expression throughout development was observed for all three candidates. P values calculated by Student’s T-test are indicated. (b) Expression of RPS14, DDX15 and SRSF9 was determined by qPCR of cDNAs prepared from rat brain slices cultured with BIC or TTX. TTX treatment leads to an ∼2-fold up-regulation of DDX15 and SRSF9, while BIC treatment leads to an almost 80-fold drop in SRSF9 RNA levels and to a 25% drop in DDX15 RNA. RPS14 levels were unaffected by the treatment. P values calculated by Student’s T-test are indicated (calculated for difference to untreated samples).
Moreover, a recent report has demonstrated a change in hippocampal CA1 editing patterns that correlated with neuronal stimulation. In particular, treatment of brain slices with the sodium channel inhibitor TTX or the GABA-A channel blocker BIC leads to a decrease or increase in GluA2 editing at the R/G site, respectively (36,37). We therefore tested cDNA samples of treated hippocampal rat brain slices for a change in expression of the three candidates via quantitative RT-PCR. Most interestingly, while RPS14 showed no change in expression patterns on TTX or BIC treatment, SFRS9 showed an increased expression on TTX treatment and a strong (<80-fold) and highly significant reduction on BIC treatment. Also DDX 15 showed increased expression on TTX treatment and decreased expression on BIC treatment (Figure 8b).
Together, these data indicate that the candidates identified in this screen show variable expression patterns during brain development but also on electrophysiological stimulation of neuronal tissue, and may thereby well contribute to the observed changes in editing in these conditions (8).
DISCUSSION
Adenosine deamination by ADARs diversify the transcriptome, both at the primary nucleotide level and at the structural level. Deregulation of editing is associated with several neurological disorders (9). Recently, some mechanisms that alter editing activity have been proposed such as the formation of ADAR isoforms, post-translational modification or specific subcellular localization patterns (19,38,39).
Here we have aimed to take an unbiased approach to identify factors that modulate ADAR2 activity. To this end, three factors that inhibit editing activity could be identified. All three proteins, RPS14, SFRS9 and DDX15, are RNA binding proteins. Surprisingly, however, the candidates were showing different levels of inhibition on different substrates. This indicates substrate specificity of the inhibitors and may reflect the position of the editing site relative to adjacent introns but also differences in local sequence context. In fact, we could show that the candidates exhibit clear substrate-specific RNA binding that reflects their ability to inhibit editing in the substrates tested. Thus, the most likely explanation for the substrate-specific inhibition of editing would be direct competitive binding at or near the editing site. Our finding also implies that editing levels may vary from substrate to substrate even within one tissue. Therefore, while global editing patterns may change during development or disease progression, editing of an individual substrate may still fail to follow the general trend observed in this tissue depending on the RNA-binding proteins associated (8,40). However, whether this prediction holds true still needs to be determined.
Besides competitive binding to substrates, the candidates identified here may also act at another level: a direct interaction and colocalization of ADAR2 with SFRS9 and to some extent also with RPS14 was observed, also leaving the possibility that the inhibition of editing is mediated by this interaction. In this scenario, complexes formed between ADAR2 and RPS14 or SRSF9, respectively, would be targeted to specific editing sites or assembled at these sites where editing is then repressed by the protein–protein interaction between the two factors. However, to separate these two possibilities, specific mutations in SFRS6 and RPS14 would need to be generated that selectively inhibit either RNA-binding or ADAR2 binding.
While one might get the impression that all RNA binding proteins may inhibit RNA binding, it should be mentioned that this is not the case. In a similar screen, aimed at the identification of factors that stimulate RNA editing, the hnRNP A2B1 protein could be isolated and verified as an enhancer of editing (Garncarz, W., Tariq, A., Handl, C., Pusch, O., and Jantsch, M.F., RNA Biology, in press). Thus, it is likely that changes in the global RNP landscape can both stimulate and inhibit binding, depending on the substrate site investigated and its overall accessibility. In this context, it is also worth mentioning that ADARs have been reported to be part of lnRNPs, large ribonucleoprotein particles that have been suggested to serve as coordination platforms of several post-transcriptional processing events (41).
Not all factors had the same impact on editing levels. While SFRS9 had the strongest effect in tissue culture cells, the helicase DDX15 has almost no effect on editing in this cellular surrounding. Instead, DDX15 did slightly alter editing in the yeast but had a dramatic effect on the editing of C. elegans 3′ UTRs. Worm DDX15 (F56D2.6) is 74.28% similar to human DDX15 at the protein level. This finding also indicates that the yeast screen used here, has the capacity to be used to identify modulators of editing from several species. The ATP-dependent RNA helicase DDX15 is involved in the disassembly of spliceosomes after release of the spliced product. As DDX15 showed the weakest substrate binding and also failed to associate with ADAR2, it is most likely that the helicase unwinds the double stranded RNA, hence dismantling the ADAR2 substrate as reported earlier for another RNA helicase (RNA helicase A RHA) (42). However, glh-2 another helicase, had no effect on editing in C. elegans, leaving the possiblitly that DDX15 may specifically interact with the 3′ UTR investigated here. Clearly, the series of molecular events that lead to the inhibition of ADAR2 activity still needs further investigation.
Most interestingly, analysis of expression profile data clearly shows that expression of all three candidates decreases during mouse brain development, therefore also linking them to changes in expression patterns observed in vivo (8). Moreover, recent data have shown that editing levels can vary with neuronal activity (36,37). Expression levels of SRSF9 and DDX strongly respond to treatment with neuronal stimulators or inhibitors that correlates nicely with the observed changes in editing patterns on treatment. SFRS9 RNA levels even drop 80-fold on bicucullin treatment, making this an interesting target for future studies. The fact that not all candidates isolated in our screen respond to neuronal stimulation or inhibition (eg. RPS14) also demonstrates that BIC or TTX does not lead to a global change in transcription but may selectively affect transcription of a few genes. It is also worth mentioning that the reported change in GluA2 editing levels on BIC and TTX treatment was accompanied by a change in ADAR2 levels (37). The finding that ADAR2 levels do not change on overexpression or depletion of any of the three candidates (see Supplementary Figure S9) suggests further that BIC and TTX treatment do not regulate ADAR2 levels via the candidates described here but rather affects expression of the candidates and ADAR2 independently.
Taken together, the candidates presented here, qualify nicely as potential regulators of editing under physiological conditions, while the mechanism by which their expression is regulated on neuronal stimulation still requires further research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–9 and Supplementary Material and Supplementary Reference [43].
FUNDING
Austrian Science Foundation (SFB 4313). Pakistani Higher Education Commission (to A.T.); Czech Academy of Sciences (RVO:67985823 to A.B.). Funding for open access charge: Austrian Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Ronald Emeson (Vanderbild University) for the kind gift of a rat ADAR2 plasmid. Dr. Ingo Greger (LMB, Cambridge) is kindly acknowledged for providing us with cDNA samples of rat brain slices treated with BIC and TTX. The authors would also like to thank the MFPL mass spectrometry unit for their excellent help and support in analysing immunoprecipitated proteins.
REFERENCES
- 1.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci. USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Connell MA, Gerber A, Keller W. Purification of human double-stranded RNA-specific editase 1 (hRED1) involved in editing of brain glutamate receptor B pre-mRNA. J. Biol. Chem. 1997;272:473–478. doi: 10.1074/jbc.272.1.473. [DOI] [PubMed] [Google Scholar]
- 3.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 4.Connolly CM, Dearth AT, Braun RE. Disruption of murine Tenr results in teratospermia and male infertility. Dev. Biol. 2005;278:13–21. doi: 10.1016/j.ydbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmauss C, Howe JR. RNA editing of neurotransmitter receptors in the mammalian brain. Sci. STKE. 2002 doi: 10.1126/stke.2002.133.pe26. 2002, p e26. [DOI] [PubMed] [Google Scholar]
- 8.Wahlstedt H, Daniel C, Enstero M, Öhman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galeano F, Tomaselli S, Locatelli F, Gallo A. A-to-I RNA editing: the “ADAR” side of human cancer. Semin. Cell Dev. Biol. 2012;23:244–250. doi: 10.1016/j.semcdb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl Acad. Sci. USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maas S, Kawahara Y, Tamburro K, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol. Cell. Biol. 2006;26:480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agranat L, Sperling J, Sperling R. A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2. RNA Biol. 2010;7:253–262. doi: 10.4161/rna.7.2.11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto Desterro JM, Keegan LP, Jaffray E, Hay RT, O'Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber A, O'Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 16.Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, Bisso A, Keegan LP, Del Sal G, O'Connell MA. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, Lau L, Liu S, Liu F, Lu Y. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Yang G, Yong Y, Liu Y, Zhao L, Xu J, Zhang X, Wan Y, Feng C, Fan Z, et al. ADAR2-dependent RNA editing of GluR2 is involved in thiamine deficiency-induced alteration of calcium dynamics. Mol. Neurodegener. 2010;5:54. doi: 10.1186/1750-1326-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl Acad. Sci. USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 21.Baudin-Baillieu A, Guillemet E, Cullin C, Lacroute F. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast. 1997;13:353–356. doi: 10.1002/(SICI)1097-0061(19970330)13:4<353::AID-YEA86>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 22.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 23.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh MV, Weil PA. A method for plasmid purification directly from yeast. Anal. Biochem. 2002;307:13–17. doi: 10.1016/s0003-2697(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 25.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 26.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto- oncogene product. Mol. Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steitz JA. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- 28.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 29.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gahwiler BH. Organotypic monolayer cultures of nervous tissue. J. Neurosci. Methods. 1981;4:329–342. doi: 10.1016/0165-0270(81)90003-0. [DOI] [PubMed] [Google Scholar]
- 31.Gahwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends Neurosci. 1997;20:471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- 32.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res. 2007;35:3723–3732. doi: 10.1093/nar/gkm314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl Acad. Sci. USA. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fouraux MA, Kolkman MJ, Van der Heijden A, De Jong AS, Van Venrooij WJ, Pruijn GJ. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA. 2002;8:1428–1443. doi: 10.1017/s1355838202021076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Levanon EY, Hueske EA, Ambrose JM, Li JB. Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics. 2012;192:281–287. doi: 10.1534/genetics.112.141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balik A, Penn AC, Nemoda Z, Greger IH. Activity-regulated RNA editing in select neuronal subfields in hippocampus. Nucleic Acids Res. 2013;41:1124–1134. doi: 10.1093/nar/gks1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maas S, Gommans WM. Novel exon of mammalian ADAR2 extends open reading frame. PLoS One. 2009;4:e4225. doi: 10.1371/journal.pone.0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slavov D, Gardiner K. Phylogenetic comparison of the pre–mRNA adenosine deaminase ADAR2 genes and transcripts: conservation and diversity in editing site sequence and alternative splicing patterns. Gene. 2002;299:83–94. doi: 10.1016/s0378-1119(02)01016-8. [DOI] [PubMed] [Google Scholar]
- 40.Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008;283:7251–7260. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- 41.Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc. Natl Acad. Sci. USA. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bratt E, Ohman M. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003;9:309–318. doi: 10.1261/rna.2750803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helmut B, Hildburg B, Hans JG. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.