Abstract
Single-molecule techniques facilitate analysis of mechanical transitions within nucleic acids and proteins. Here, we describe an integrated fluorescence and magnetic tweezers instrument that permits detection of nanometer-scale DNA structural rearrangements together with the application of a wide range of stretching forces to individual DNA molecules. We have analyzed the force-dependent equilibrium and rate constants for telomere DNA G-quadruplex (GQ) folding and unfolding, and have determined the location of the transition state barrier along the well-defined DNA-stretching reaction coordinate. Our results reveal the mechanical unfolding pathway of the telomere DNA GQ is characterized by a short distance (<1 nm) to the transition state for the unfolding reaction. This mechanical unfolding response reflects a critical contribution of long-range interactions to the global stability of the GQ fold, and suggests that telomere-associated proteins need only disrupt a few base pairs to destabilize GQ structures. Comparison of the GQ unfolded state with a single-stranded polyT DNA revealed the unfolded GQ exhibits a compacted non-native conformation reminiscent of the protein molten globule. We expect the capacity to interrogate macromolecular structural transitions with high spatial resolution under conditions of low forces will have broad application in analyses of nucleic acid and protein folding.
INTRODUCTION
Model RNA and DNA hairpins have been extensively characterized by optical trapping methods, providing a detailed view of the folding energy landscapes of these fundamental nucleic acid structures (1–5). However, the ability to achieve high spatial resolution using force spectroscopy relies on the application of relatively large stretching forces (>10 pN) to suppress the measurement noise introduced by the long flexible DNA handles used to attach the structure of interest to micron-scale beads held in the optical trap. Recently, a study of the force-dependent structural dynamics of single Holliday junctions was reported, which paired an optical trap to apply precisely calibrated stretching forces with single-molecule Förster resonance energy transfer (smFRET) to monitor DNA structural dynamics (6). This fluorescence–force method provided a powerful tool for probing sub-nanometer–scale structural rearrangements within single Holliday junctions at very low stretching forces (<1 pN) over short periods. In addition, several groups have reported measurements that combine smFRET with a magnetic tweezers apparatus (7,8). The use of magnets to apply mechanical loads to individual DNA molecules has several potential advantages over optical traps. For example, the combination of a high-power optical trapping laser with single-molecule fluorescence is technically difficult to implement, typically requiring interlacing of the FRET excitation and optical trapping beams to avoid rapid photo damage of the FRET probes by the high-power trapping laser (9,10). Moreover, the sensitivity of optical traps to mechanical drift makes the application of low stretching forces (<1 pN) over extended periods far more challenging than with a simple magnetic tweezers system. Here we describe an integrated fluorescence and magnetic tweezers microscope capable of measuring nanometer-scale structural transitions in single DNA molecules at low stretching forces. We demonstrate the utility of this approach by analyzing the mechanical unfolding pathway of a model human telomere DNA substrate.
Telomeres are specialized chromatin structures that protect linear ends of eukaryotic chromosomes from aberrant DNA processing by DNA damage repair machinery (11). The foundation of human telomere structure is a long stretch of double-stranded DNA composed of a hexanucleotide DNA repeat sequence (T2AG3). In addition, all telomeres terminate with a 3′ single-stranded G-rich DNA tail, which has the capacity to fold into a unique secondary structure called a G-quadruplex (GQ). Human telomere DNA GQs are proposed to play a central role in telomere homeostasis, and small-molecule ligands that selectively bind and stabilize telomere DNA GQs have shown promise as anti-cancer drugs (12,13). Thus, intensive efforts have been made to better understand the structure and function of telomere DNA GQs. The first solution structure of a human telomere GQ revaled a fundamental structural architecture in which guanine bases are hydrogen bonded in a planar quartet geometry and may coordinate a single centrally located monovalent metal ion (Figure 1A, top left) (14). Three adjacent intra-molecular G-quartets may interact via stacking interactions and are topologically linked by short intervening DNA loop sequences (Figure 1A, bottom left). Moreover, the folding properties of telomere DNA GQs vary with the presence of different monovalent cations. Na+ ions predominantly promote the formation of an anti-parallel GQ conformation (14), whereas GQ DNA crystals formed in the presence of K+ ions revealed a distinct parallel GQ folding topology (15). More recent solution studies have demonstrated that multiple GQ topologies coexist in the presence of K+, including the anti-parallel, parallel and several hybrid forms (16–19).
Figure 1.
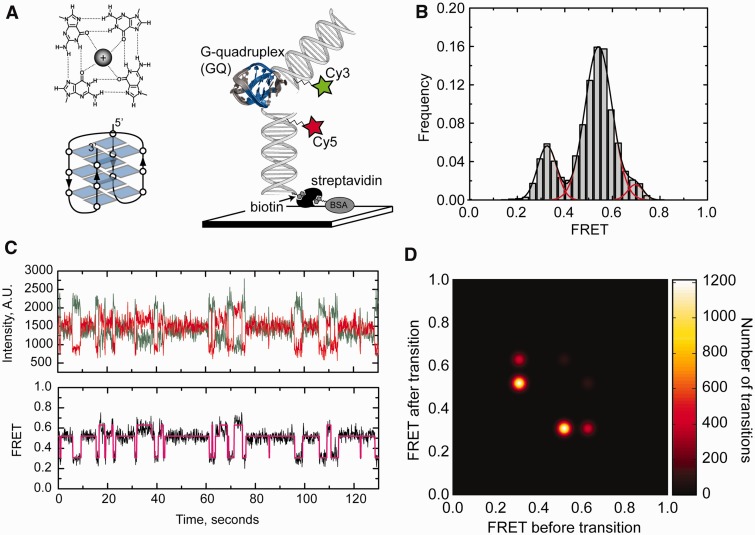
Single-molecule FRET analysis of Na+-induced telomere DNA G-quadruplex folding in the absence of force. (A) Top left: Diagram of H-bonding network within a single G-quartet with a monovalent metal ion coordinated at its center. Bottom left: Schematic illustration of the anti-parallel G-quadruplex folding topology, with each planar G-quartet represented as a quartet of blue rectangles. Right: Schematic diagram of smFRET experimental setup [a model telomere DNA sequence ((TTAGGG)4, Tel24]. G-quartets highlighted in blue are flanked by two duplex DNA handles harboring a FRET donor (Cy3) and acceptor (Cy5) dye, respectively. Tel24 molecules were surface immobilized via a biotin–streptavidin linkage and imaged using prism-type total internal reflection microscopy. The antiparallel G-quadruplex structure solved by NMR is shown in the illustration (PDB code 143D). (B) Single-molecule FRET histogram derived from data collected on >100 molecules is fit with three Gaussian functions centered at FRET = 0.33, 0.54 and 0.70. (C) A representative smFRET trajectory for Tel24 in the presence of 100 mM NaCl. Individual donor (green) and acceptor (red) dye intensities (top) and calculated FRET ratios (bottom) are plotted as a function of time. FRET trajectories were fit with a hidden Markov model yielding idealized FRET traces (magenta). (D) Idealized FRET traces from HaMMy fitting were used to generate a transition density plot indicating the frequency of each observed transition. Color bar on right indicates the number of transitions.
The structural and dynamic properties of telomere DNA GQs have been studied using smFRET (20–22). These experiments revealed that a particular GQ topological fold must transit through an obligatory unfolded intermediate to isomerize into a distinct GQ fold. More recently, the rupture force distribution of single telomere DNA GQs has been analyzed using atomic force microscopy and optical trapping, providing a direct measurement of telomere DNA GQ mechanical stability (23–26); however, these force spectroscopy studies did not analyze the force dependence of telomere DNA GQ folding/unfolding at equilibrium. Here, we report a detailed analysis of the force dependence of telomere DNA GQ folding and unfolding at equilibrium under Na+ folding conditions. Our results demonstrate that the structural equilibrium between the unfolded and Na+-induced telomere DNA GQ structure is highly sensitive to forces between ∼1 and 8 pN. Analysis of the force-dependent rate constants for folding and unfolding provides a direct measurement of the position of the transition state barrier for telomere DNA GQ unfolding along the DNA-stretching reaction coordinate. Interestingly, in contrast to other DNA secondary structures characterized by force spectroscopy, we find telomere DNA GQs exhibit a very short distance (<1 nm) to the transition state barrier for unfolding. This unfolding behavior indicates telomere DNA GQ structure is significantly stabilized by long-range contacts, and once these contacts are disrupted, the entire GQ fold readily dissolves.
MATERIALS AND METHODS
DNA oligonucleotides
All DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. The sequences of all DNAs used in the study are listed in Supplementary Table S1.
Dye labeling of DNA oligonucleotides
The biotinylated DNA handle was labeled at the amino modification on C6 of T3 with monoreactive Cy5 (GE Healthcare), and the EcoRI DNA Handle was labeled at the amino modification on C6 of T16 with monoreactive Cy3 (GE Healthcare). Dye-labeled fragments were EtOH precipitated and purified by reverse-phase chromatography using a C8 column (Agilent, Eclipse XDB-C8) on an AKTA purifier. After HPLC purification samples were EtOH precipitated, and resuspended in ddH2O. DNA concentrations were determined using a NanoDrop 2000C (Thermo Scientific).
DNA annealing reactions
The biotinylated, Cy5-labeled and Cy3-labeled strands were annealed to the Tel24 abasic, 15R60/T8 hairpin abasic, or polyT abasic sequence by heating to 95°C for 4 min, followed by slow cooling to room temperature (over several hours) in the presence of a buffer (50 mM Tris–HCl, pH 8) containing either 100 mM NaCl or 100 mM KCl.
DNA molecule for integrated fluorescence and magnetic tweezers measurements
DNA molecules were constructed by ligation of three precursor DNA fragments: the annealed biotinylated smFRET fragment, a digoxigenin-modified DNA linker fragment and lambda DNA handle. The preparation of desired Cy5- and Cy3-labeled DNA insert is described earlier in the text. The digoxigenin-labeled DNA linker is synthesized by PCR reaction with a pUC19 template using primers flanking the multiple cloning site. PCR reactions were set up using a dNTP mixture containing a 1:4 molar ratio of digoxigenin-11-dUTP (Roche): dTTP and digested with BamHI. The 15 721-bp lambda DNA handle was prepared by BamHI and EcoRI enzymatic digestion and agarose gel purification. Ligation reactions were set up with a 1:1:1 molar ratio of the desired Cy5- and Cy3-labeled DNA insert harboring an EcoRI sticky end, the purified EcoRI/BamHI lambda DNA handle and the BamHI digoxigenin-modified DNA linker fragment. Ligation reactions were run overnight at 16°C in the presence of T4 DNA ligase (NEB). The final ligation product was heat inactivated at 65°C to ensure the T4 DNA ligase did not remain associated with the DNA.
Single-molecule FRET measurements in the absence of force
For these experiments, quartz slides (Finkenbeiner Inc.) were cleaned by sonicating for 20 min in 10% w/v Alconox, 20 min in ddH2O, 20 min in acetone, 20 min in 1 M KOH and then 20 min in fresh 1 M KOH. Slides are then rinsed with ddH2O and dried under nitrogen, followed by flame cleaning with a propane torch for ∼2 min. Sample chambers were prepared by sandwiching pieces of parafilm between the quartz slide and a plasma-cleaned cover glass (Harrik Plasma Cleaner), cover glasses from TED Pella, Inc. (Prod. No. 260146). Chambers were heated on a hot block (95°C) for 1 min to seal the parafilm to the glass. Channels (∼10 µL total volume) were treated with 35 µL of 1 mg/mL of biotinylated bovine serum albumin (Sigma Cat. No.A8549) for 5 min, washed with 100 µL of T50 buffer [10 mM Tris–HCl (pH 8), 50 mM NaCl], incubated with 50 µL of 0.2 mg/mL streptavidin (Invitrogen Cat. #S888) and then washed with 100 µL of T50. Channels were then equilibrated with buffer matching the desired experimental condition (see main text).
Next, 100 µL of 5–10 pM annealed fluorescent and biotin-labeled smFRET DNA was deposited onto the streptavidin-coated quartz slide in the desired buffer condition. After 5-min incubation, desired buffer was flushed to the slide to remove the unbound DNA. Data were collected in imaging buffer containing 50 mM Tris (pH 8), 100 mM KCl or NaCl, 0.4% (w/v) d-glucose, 0.1 mg/ml glucose oxidase (Sigma Cat.# G2133-250KU), and 0.02 mg/ml catalase (CalBiochem Cat.#219001) and saturated with Trolox (Aldrich Cat.#23,881-3). Data were acquired using a green laser (532 nm, Laserglow, Inc.) and prism-type total internal reflection microscopy on an inverted Olympus IX71 microscope equipped with an Andor IXON (897) CCD camera with 100-ms integration time.
Single-molecule FRET analysis
Raw movie files were analyzed using in-house–written software available on request (IDL and MATLAB). FRET is defined as the efficiency of energy transfer between acceptor and donor dye, IA/(IA + ID). IA is the acceptor intensity, and ID is the donor intensity. FRET histograms were compiled by combining data from ∼100 individual single-molecule trajectories. The hidden Markov modeling was performed using the HaMMy (27) software program. First, dye intensity traces used in the HaMMy analysis were normalized after photo bleaching, and then the period before photo bleaching was cropped to be used in the analysis. Because individual traces did not typically possess a sufficient number of transitions to produce a reliable HaMMy fit, we opted to stitch all the normalized and cropped trajectories into a single trace using in-house–written MATLAB software. This compiled trace was then fit using the HaMMy software. The idealized FRET trace that was produced by the HaMMy fitting was then parsed back into individual traces using in-house MATLAB software to prevent the inclusion of artificial FRET transitions at the trace stitch points. The individual HaMMy fits were then used to generate a transition density plot using in-house–written MATLAB software. To fit the smFRET distributions, data were binned (bin size was FRET = 0.03) and fit with multiple Gaussian functions using Origin software. For dwell time analysis, idealized traces produced by fitting the single-molecule trajectories using the HaMMy software package were used to calculate the average lifetime of the folded or unfolded states. The smFRET histograms for the Na+ and K+ titrations, as well as in the presence of Mg2+, were constructed by averaging the observed FRET for each molecule over 2 s (20 frames at 100-ms integration time) and binning the data with a bin size of FRET = 0.03.
Integrated FRET and magnetic tweezers measurements
The DNA tethers for FRET–magnetic tweezers measurements were attached to anti-digoxigenin–coated magnetic beads. Anti-digoxigenin beads were made using EDC coupling chemistry (Pierce CAS# 22980) using carboxylated magnetic beads [Dynal, either 1-µm (Invitrogen Cat.#650.11) or 2.8-µm (Invitrogen Cat.#143.05D) diameter]. After incubating 10 min at 4°C, the DNA–magnetic bead mixture was washed to remove the excess DNA fragments before being deposited onto the streptavidin-coated cover glass (#1.5, Ted Pella) prepared using the same cleaning procedure described earlier in the text. After incubating 30 min at room temperature to allow the DNA–magnetic bead complexes to settle to the surface, 100 µL of 50 mM Tris with 100 mM Na+ (for GQ experiments) or 200 mM K+ (for control DNA hairpin experiments) was flowed onto the slide by gravity to remove the unattached DNA-linked magnetic beads.
To calibrate the stretching forces being applied to each DNA tether, forces were measured across a wide range of positions of the magnet assembly held above the sample chamber. Force measurements were determined using the expression F = LkBT/<χ2>, where F is the force in pico-Newtons (pN), L is the DNA tether length at a particular magnet height, kBT is thermal energy (4.1 pN × nm) at room temperature and <χ2> is the variance in the bead position in x-axis. The DNA tether length was measured by calibrating the diffraction ring pattern of the magnetic bead using a piezo-controlled objective positioning device (Mad City Labs) as previously described (28). Separate calibrations were made for the 1-µm and 2.8-µm magnetic beads. The force was found to decrease exponentially as the distance of the magnets was increased from the sample chamber as described (28). The magnet height versus force plots were fit with single exponential decay functions, and the parameters from these fits were then used to calculate the forces applied to subsequent experimental setups using the same-size bead. The bead manufacturer estimates a deviation in the radius (r) of the beads on the order of 1–2%, and the force scales with the volume of the bead (r3); thus, we estimate the systematic error of our force measurements based on an imperfect calibration to be on the order of 10%.
Data were collected in imaging buffer containing 100 mM Na+ or 200 mM K+ salt, 0.4% (w/v) d-glucose, 0.1 mg/ml glucose oxidase (Sigma Cat.# G2133-250KU), and 0.02 mg/ml catalase (CalBiochem Cat.#219001) and saturated with Trolox (Aldrich Cat.#23,881-3). Data were acquired using objective-type total internal reflection microscopy with a green laser (532 nm, Laserglow, Inc.) on an Olympus IX71 microscope equipped with an Andor IXON (860) CCD camera. A 100-ms integration time was used for telomere DNA GQ and polyT measurements, and a 33-ms integration time was used for hairpin measurement. The stretching force applied to individual DNA tethers was altered by changing the linear position of a pair of rare-earth magnets mounted on a computer-controlled translation stage above the sample. The DNA extension and applied stretching force were determined by visualizing the magnetic bead with a blue LED (Thorlabs) mounted above the magnet assembly as described previously (28). Before collecting smFRET data, the length of each DNA tether was monitored in real time as a function of applied twist to ensure that there was only a single DNA molecule attached between the bead and the surface. Molecules for which the extension changed as a function of twist represented DNA braids and were not used in the measurements.
Raw movie files were first analyzed using in-house–written software (Labview and MATLAB), and then fit with the HaMMy (27) software to generate idealized FRET traces used for dwell time analysis. Single-molecule FRET histograms generated from combined FRET–magnetic tweezers data were compiled from all molecules analyzed at a particular force set point. Traces were first normalized for photo bleaching and then cropped before the photo bleaching event for HaMMy analysis. We focused our analysis on the equilibrium between the predominant mid-FRET (folded) and the low-FRET (unfolded) state. To this end, for our analysis, we used the idealized FRET traces generated by the HaMMy (27) fitting to remove the minority of time spent in the alternative high-FRET (folded) state. For dwell time analysis, idealized traces produced by fitting the smFRET trajectories using the HaMMy software package were used to calculate the average lifetime of the folded or unfolded states at each of the indicated forces.
RESULTS
Single-molecule FRET analysis of telomere DNA GQ structure in the absence of force
The experimental geometry used in the present study of the force-dependent folding/unfolding of telomere DNA GQ structure includes attachment of duplex DNA handles to each end of a single GQ forming sequence (Figure 1A, right). A similar design has been used in recent optical trapping experiments (24–26), but it has not been analyzed using smFRET; therefore, we first set out to characterize the structure and dynamics of our model human single-stranded telomere DNA construct [Tel24, (TTAGGG)4] embedded within a duplex DNA molecule. For these experiments, the Tel24 sequence was flanked by non-telomeric DNA extensions, which were hybridized to their respective complementary DNA strands harboring either a FRET donor (Cy3) or acceptor (Cy5) dye (Figure 1A, right). To minimize the possibility of the adjacent duplex altering the folding properties of the GQ forming sequence, abasic sites were introduced between the Tel24 and the non-telomere sequence. Experiments investigating the structure of Tel24 in the absence of force were conducted on a prism-type total internal reflection fluorescence (TIRF) microscope (29). Throughout this study, we define FRET = IA/(IA + ID), where IA and ID are the background-corrected intensities of the acceptor and donor dyes, respectively.
We first measured the folding properties of Tel24 over a wide range of NaCl concentrations (Supplementary Figure S1). As the NaCl concentration was gradually increased to 100 mM, we observed the emergence of a predominant FRET population centered at FRET = 0.54, a second population centered at FRET = 0.33 and a minor population centered at FRET = 0.70 (Figure 1B). Analysis of single-molecule trajectories revealed the Tel24 construct is in a dynamic equilibrium between each of these three FRET states (Figure 1C). Conversion of the predominant mid-FRET state to the high-FRET state required formation of the transient low-FRET state, as has been previously reported (21). Single-molecule trajectories were fit to a hidden Markov model, which yielded idealized FRET trajectories (Figure 1C) (27). The idealized traces were used to generate transition density plots, which clearly indicated the majority of FRET transitions (70%, n = 2426/3475 transitions) occurred between the mid-FRET and the low-FRET states (Figure 1D). The ratio of time spent in the unfolded (FRET = 0.33) and the predominant folded (FRET = 0.54) states yielded a Keq = 0.25, corresponding to a ΔGunfold = 0.81 kcal mol−1, a value that is in good agreement with previously reported calorimetric studies of Tel24 unfolding in NaCl (30). Dwell time analysis yielded rate constants of 0.87 s−1 and 0.29 s-1 for the folding and unfolding reactions, respectively, giving a Keq = 0.33 in reasonable agreement with the value obtained by fitting the smFRET histograms. Nuclear magnetic resonance and CD spectroscopy experiments on telomere DNA substrates in the presence of NaCl have demonstrated the strong preference of single-stranded telomere DNA to form the anti-parrallel GQ conformation under this folding condition (14,31). Thus, while smFRET data alone are not sufficient to conclusively determine which GQ structure is present, it is likely the predominant FRET = 0.54 conformation observed in the presence of 100 mM Na+ represents the anti-parallel conformation and the FRET = 0.33 state represents the unfolded conformation.
In contrast to the Na+ folding condition, the presence of KCl produced a considerably broader distribution of FRET states, with increasing concentrations of K+ stabilizing several distinct high-FRET GQ conformations (Supplementary Figure S2). The smFRET distribution for Tel24 folded in 100 mM KCl is well fit by two Gaussian functions, with populations centered at FRET = 0.61 and FRET = 0.76. Consistent with this result, analysis of single-molecule trajectories in the presence of 100 mM KCl revealed two distinct long-lived FRET states, which occasionally inter-converted via the obligatory low-FRET intermediate (Supplementary Figure S3). In addition, the presence of Mg2+ in the K+ folding reaction appeared to have a slightly stabilizing effect on GQ structure, but did not significantly alter the smFRET histograms in the presence of 100 mM K+ (Supplementary Figure S4). Taken together, our smFRET results are in good agreement with previous findings that demonstrated Na+ ions promote homogeneous folding of telomere DNA GQs, while K+ ions induced a more complex distribution of FRET states that are more thermodynamically stable than the Na+-induced fold. Based on these results, we conclude that the presence of duplex DNA flanking the Tel24 sequence does not significantly alter telomere DNA GQ folding and stability.
Integrated fluorescence and magnetic tweezers spectroscopy
Previously reported force spectroscopy experiments using optical traps achieved sub-nanometer spatial resolution by applying relatively large stretching forces (>20 pN) to the system of study (32,33). Because many biologically important structural transitions in nucleic acids and proteins are induced by much smaller forces, we have developed a simple method for measuring sub-nanometer structural rearrangements within individual DNA molecules held under a wide range of stretching forces (0.1–50 pN). In our integrated fluorescence and magnetic tweezers system, a single Tel24 sequence is embedded within a duplex DNA molecule specifically attached between a microscope slide and a magnetic bead (Figure 2A). The length of the DNA handle attached to the microscope slide was 29 bp (∼10 nm), while the length of the DNA handle attached to the bead was made considerably longer (15.7 kb, 5.4-µm contour length) to prevent background signal introduced by the large magnetic bead entering the evanescent field produced near the surface of the glass slide by total internal reflection of the excitation laser. The DNA construct is further modified with FRET donor (Cy3) and acceptor (Cy5) dyes on the DNA handles flanking the Tel24 sequence, as well as abasic sites between the duplex DNA handles and the Tel24 sequence. Stretching force was applied to the tethered DNA molecule with a pair of permanent rare-earth magnets mounted on a computer-controlled translation stage. Integrated fluorescence and magnetic tweezers experiments used objective-type TIRF, which is readily compatible with the presence of the magnet assembly held above the sample chamber. We note that owing to slight differences in the optical components in the microscopes, smFRET values made by objective-type TIRF vary slightly from those measured using the prism-type TIRF setup described in Figure 1 [ie. mid-FRET (folded state) is 0.64 in low-force objective-type TIRF experiments versus 0.54 in prism-type TIRF microscope under zero force]; however, this variation does not impact our assignment of the mid-FRET and low-FRET states to the folded and unfolded telomere DNA GQ conformations, respectively.
Figure 2.
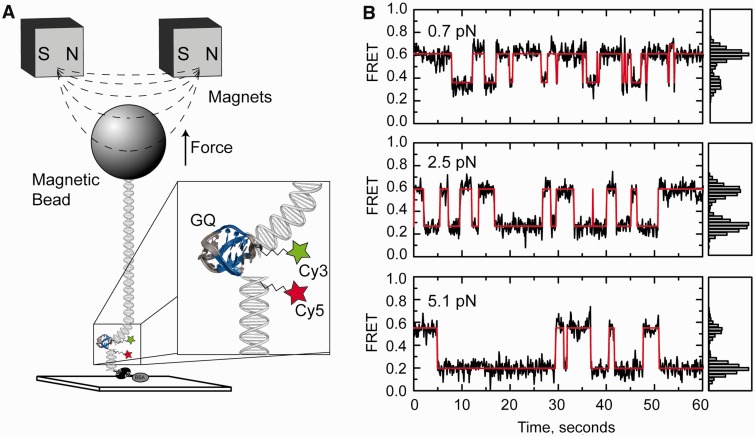
Force biases the telomere DNA G-quadruplex folding/unfolding equilibrium. (A) Schematic diagram of the experimental setup for integrated fluorescence and magnetic tweezers measurements. A FRET-labeled Tel24 molecule is flanked by a short biotinylated duplex DNA handle on one side, and a second longer duplex DNA handle with terminal digoxigenin modifications on the other side. Individual Tel24 constructs were attached between a streptavidin-coated microscope slide and an anti-digoxigenin–coated magnetic bead in a magnetic tweezers setup and imaged using objective-type total internal reflection microscopy. Variable stretching force was applied to individual Tel24 molecules by translating a pair of rare-earth magnets held above the sample chamber. (B) Representative smFRET trajectories of a single Tel24 molecule held under three different forces (0.7, 2.5 and 5.1 pN). Histograms of the FRET values obtained at each force set point are shown on the right side of the FRET trajectories.
In Figure 2B, representative smFRET trajectories are plotted at three different stretching forces for a single telomere DNA GQ in the presence of 100 mM NaCl. It is evident that at low forces (0.7 pN), the molecule spends the majority of the time in the high-FRET (folded) state consistent with our zero force measurements described earlier in the text. In contrast, a higher force (5.1 pN) substantially shifts the telomere DNA GQ folding equilibrium toward the low-FRET (unfolded) state. Notably, the application of small degrees of tension appeared to substantially reduce the number of transitions into the minority high-FRET (FRET = 0.70) conformation when compared with experiments performed in the absence of force; thus, we focused our analysis on the structural equilibrium between the low-FRET (unfolded) and predominant mid-FRET (folded) conformation. These data qualitatively demonstrate the capacity of our technique to detect nanometer-scale structural transitions within single telomere DNA GQs at low forces. However, to determine whether this approach can be used to extract quantitative information about force-induced structural transitions in DNA, we next performed control experiments on a model DNA hairpin construct (Supplementary Figure S5). Data taken on the model DNA hairpin were analyzed as a simple two-state system separated by a single energetic barrier, yielding an unfolding force F1/2 = 12.1 pN (defined as the force at which the hairpin folded and unfolded states are equally populated) and a distance between the folded and unfolded state Δx = 16.2 nm. Furthermore, analysis of the force-dependent rate constants for folding and unfolding placed the transition state barrier 8.9 nm from the folded state and 6.1 nm from the unfolded state. These results are in good quantitative agreement with previously reported optical trapping measurements on the same model DNA hairpin (2), demonstrating the ability of our integrated fluorescence and magnetic tweezers system to extract quantitative information about DNA folding processes.
Force dependence of telomere DNA GQ folding and unfolding
We next characterized in greater detail the force dependence of the telomere DNA GQ folding/unfolding equilibrium. For these experiments, we performed force titrations for 75 individual telomere DNA GQ molecules (for each force, a minimum of five different molecules were analyzed), and compiled the results into FRET histograms (Figure 3A, see Supplementary Figure S6 for complete data set). Analysis of the amount of time spent in the mid-FRET (folded) state versus the low-FRET (unfolded) state as a function of force provided a direct measurement of the effect of stretching force on the telomere DNA GQ folding equilibrium. We fit each histogram with two Gaussian functions and calculated Keq for the unfolding reaction as the area under the low-FRET peak divided by the area under the mid-FRET peak. From the slope of the plot of ln(Keq) as a function of stretching force, we determined the unfolding distance Δx = 2.7 nm and F1/2 = 2.5 pN (Figure 3B).
Figure 3.
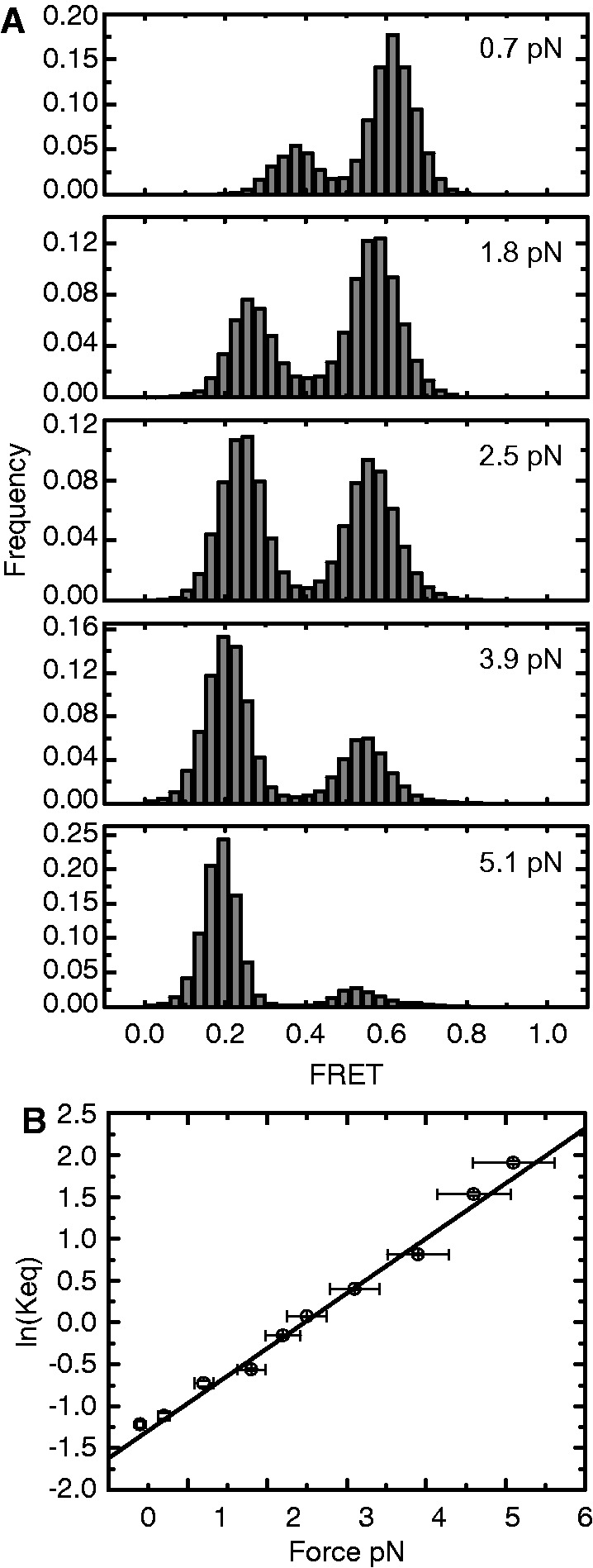
Telomere DNA G-quadruplex folding equilibrium as a function of applied force. (A) Compiled single-molecule FRET histograms from data taken on 75 individual Tel24 molecules. For each force, a minimum of five different molecules were analyzed. Data for five representative forces ranging from 0.7 to 5.1 pN are shown (see Supplementary Figure S6 for complete data set). (B) Plot of the ln(Keq) versus force. For each force set point at which folding/unfolding transitions were detected, smFRET histograms were fit with two Gaussian functions, and Keq for unfolding was calculated as the area under the low-FRET (unfolded) state divided by the area under the high-FRET (folded) state. Plot was fit with the expression ln(Keq) = FΔx/kBT − ΔGo/kBT, yielding an unfolding distance Δx = 2.7 nm and a F1/2 = 2.5 pN. Y-axis error bars are estimates of the standard error of the mean determined by bootstrapping analysis of smFRET data, and x-axis error bars reflect an ∼10% systematic error in determination of stretching force.
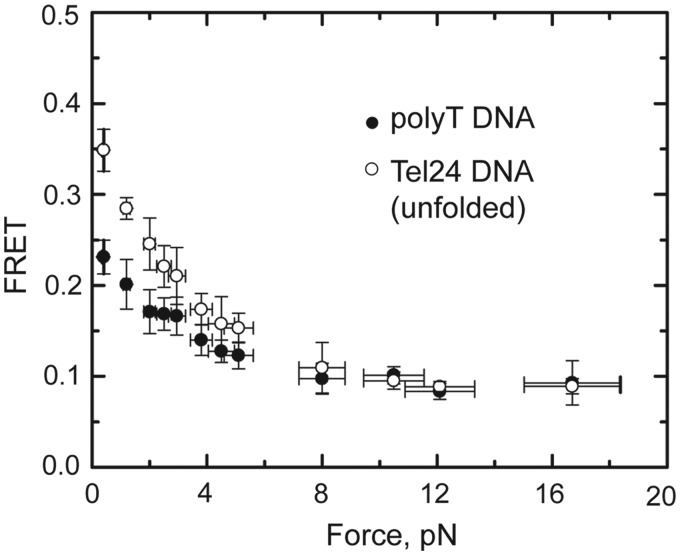
We observed that the centers of the Tel24 low-FRET (unfolded) and mid-FRET (folded) populations shifted toward lower values as the applied stretching force was increased (Figure 3A). The two different Tel24 conformations fall within significantly different regions of the FRET response curve; thus, the magnitude of the force-induced ΔFRET for the low-FRET population reflects a substantially larger change in distance between the FRET probes than the ΔFRET of the mid-FRET population (Supplementary Figure S7). The low-FRET Tel24 conformation, previously identified as an obligatory folding intermediate during inter-conversion between distinct topological isomers (21), was suggested to reflect an unfolded DNA conformation that would be expected to respond to stretching force by gradually increasing its end-to-end distance, as observed in our experiments. To further explore the structural properties of this obligatory unfolded intermediate, we compared the force response of the low-FRET (unfolded) telomere DNA GQ conformation with a 24-nucleotide polyT DNA strand, which has been used as a model for unstructured single-stranded DNA in previous smFRET experiments (34). We find that under low stretching force conditions, the unfolded conformation of the Tel24 construct is significantly more compact (higher FRET) than the polyT construct (Figure 4). This result demonstrates the G-rich Tel24 unfolded state experiences an attractive force, perhaps derived from increased stacking interactions between purine bases or transient H-bonding, which promotes telomere DNA GQ folding. Interestingly, the elastic properties of the unfolded Tel24 and polyT constructs converge within the force range (∼8 pN) that completely inhibits telomere DNA GQ folding within the 100-ms time resolution of these measurements (Supplementary Figure S6). We note that the gradual increase in extension of the unfolded Tel24 state with increasing force is not consistent with the presence of any long-lived structured GQ folding intermediates such as a DNA hairpin or triplex structure. These structured intermediates have been previously suggested to occur during telomere DNA GQ folding (35–37), but may be too transient to detect in our experiments.
Figure 4.
Force response of Tel24 low-FRET (unfolded) state and polyT DNA. The average FRET values of the Tel24 low-FRET (unfolded) state (open circles) and a 24-nucleotide polyT DNA construct (closed circles) are plotted as a function of force. Y-axis error bars are the standard deviation of the average FRET value measured for at least five individual molecules at each force set point, and x-axis error bars reflect an ∼10% systematic error in the calibration of the applied stretching force.
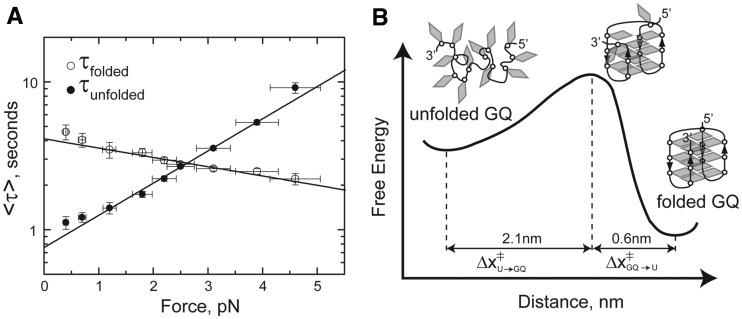
The force dependence of the rate constants for telomere DNA GQ folding and unfolding can also be analyzed from our single-molecule trajectories. Making the simplifying assumptions that along the reaction coordinate defined by the DNA-stretching axis, the position of the energy barrier is independent of the stretching force and the positions of the folded and unfolded energy wells are not substantially altered by the applied force, we modeled the lifetime of the folded state, τfolded, as given by τfolded(F) = τfolded,0 × exp(FΔx‡folded/kBT), where τfolded,0 is the folded lifetime at zero force, Δx‡folded is the distance from the folded to the transition state, kB is Boltzman’s constant and T is temperature. Similarly, the lifetime of the unfolded state as a function of force can be expressed as τunfolded(F) = τunfolded,0 × exp(FΔx‡unfolded/kBT). Fitting these expressions to a plot of the average lifetimes of the folded or unfolded states as a function of force yielded Δx‡unfolded = 2.1 nm, Δx‡folded = 0.6 nm, τfolded,0 = 4.1 s and τfolded,0 = 0.76 s (Figure 5A). The results of our kinetic analysis are in good agreement with the Δx value derived from the equilibrium data (Δx = Δx‡folded + Δx‡unfolded = 2.7 nm). In addition, the extrapolated zero force rate constants of kfold = 1.3 s−1 and kunfold = 0.24 s−1 are in good agreement with our smFRET measurements in the absence of force (see earlier in the text). The short distance (0.6 nm) from the folded state to the transition state barrier suggests the telomere DNA GQ structure is stabilized by long-range contacts, and that disruption of just a few of the terminal base pairing contacts is sufficient to promote the unfolding of GQ structure (Figure 5B).
Figure 5.
Force-dependent rate constants of telomere DNA G-quadruplex folding and unfolding. (A) Average lifetimes (τ) of the folded (high FRET) and unfolded (low FRET) states are plotted as a function of force. Plots are fit with exponential functions described in the main text, yielding Δx‡unfolded = 2.1 nm and Δx‡folded = 0.6 nm. (B) A schematic folding energy landscape for Tel24 in the presence of 100 mM NaCl. Cartoon model for the folded, transition-state and unfolded structures are superimposed on the folding energy landscape. The short distance between the transition-state barrier and the folded conformation suggests just a few base pairs are disrupted in the transition state structure.
DISCUSSION
Here, we describe an integrated fluorescence and magnetic tweezers technique that combines objective-type total internal reflection microscopy with a ‘classical’ magnetic tweezers setup (28,38). Although several groups have described combined fluorescence–force instruments using optical traps to apply stretching forces (6,9), these tools remain inaccessible to non-specialists, owing to the technical challenges associated with aligning and calibrating these advanced optical trapping systems. Furthermore, the integration of a high-power trapping laser with single-molecule fluorescence detection has the potential to negatively impact the photo physical stability of the fluorescence probes, thereby limiting the duration of the FRET measurement. By comparison, the technique described in the present study is a simple instrument to construct and maintain, and should therefore be a generally accessible tool for a wide range of biochemists and structural biologists. Moreover, the use of permanent magnets to apply stretching forces obviates concerns of photo damage induced by a high-power trapping laser and provides a means to stably apply low forces for arbitrarily long periods. Indeed, smFRET trajectories acquired using our approach routinely last for tens of minutes, facilitating measurement of a single molecule under a variety of experimental conditions. An additional advantage of hybrid force–fluorescence methods using magnetic tweezers is the ability to monitor torque-induced structural transitions by smFRET, as has recently been reported in studies of the torque-induced B-DNA to Z-DNA transition (7). We expect the capability of integrated fluorescence and magnetic tweezers systems to detect nanometer-scale structural rearrangements under conditions of low tensions and torques will be broadly applicable to a wide range of macromolecular folding studies.
In the present work, we have analyzed the force-dependent folding of a model human telomere DNA GQ in the presence of Na+. We find that the dynamic equilibrium between the Na+-induced GQ folded and unfolded states is extremely sensitive to applied stretching forces between ∼1 and 8 pN. By analyzing the effect of stretching force on the rate constants for telomere DNA GQ folding and unfolding, we have directly measured the position of the transition state barrier for GQ unfolding along the well-defined DNA-stretching reaction coordinate. Interestingly, we find the distance between the telomere DNA GQ folded state and the transition state barrier for unfolding is short (∼0.6 nm). Our finding is consistent with a recent report that used dynamic force spectroscopy (DFS) to reconstruct the distribution of rupture forces for a DNA GQ formed by a sequence upstream of the human insulin gene promoter (26). Fitting the DFS results to theoretical models that predict the distribution of rupture forces assuming a single transition state barrier suggested the barrier for folding is within ∼1 nm of the GQ folded state, in close accord with our present results. However, as with previous optical trapping experiments, which analyzed the mechanical stability of GQ DNA (24,25), the DFS measurements did not directly observe a reversible folding/unfolding equilibrium in the low force regimen as reported here. At present, it is unclear why the low-force ‘hopping’ behavior we report for the telomere DNA GQ in the present study is not observed in the optical trapping experiments, but the discrepancy may be due to differences in the sequences being used or challenges associated with detecting small extension changes at low stretching forces in an optical tweezers instrument.
Force spectroscopy experiments with RNA and DNA hairpins revealed the transition state barrier for unfolding lies closer to the unfolded state than the folded state (1,2). This long distance to the transition state barrier for unfolding is expected for compliant structures the folding stability of which is derived primarily from local interactions (i.e. zipping of adjacent base pairs within a DNA hairpin). However, a short distance to the transition state barrier for unfolding, as measured here for the telomere DNA GQ, is indicative of a more brittle structure the stability of which is reliant on critical contributions of long-range interactions. This result can be explained within the framework of the nuclear magnetic resonance structure for the Na+-induced anti-parallel GQ fold (14), wherein guanine bases separated by as many as 20 nucleotides must be brought together within a planar G-quartet configuration. Thus, disruption of just a few H-bonding interactions by mechanically pulling on the terminal bases of the telomere DNA GQ structure is sufficient to effectively destabilize the entire fold (Figure 5B), a finding that has direct implications on the molecular mechanisms of telomere-associated proteins and enzymes, which must resolve DNA GQs during telomere maintenance. Telomere DNA GQs are typically classified as DNA secondary structure; however, our studies reveal these unique DNA folds share important structural properties with RNA and protein tertiary structures, which have also been characterized by sub-nanometer distances to the transition state barrier for unfolding (1,39–41). Finally, understanding the critical contribution of long-range interactions in promoting telomere GQ folding stability should aid efforts in designing small-molecule drugs that target and stabilize GQ structure to disrupt telomere homeostasis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–7.
FUNDING
National Institutes of Health (NIH) [GM095850-02 to M.D.S.] and the National Science Foundation [DGE 0809125 to J.W.P.]. Funding for open access charge: NIH [GM095850-02].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 2.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl Acad. Sci. USA. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harlepp S, Marchal T, Robert J, Leger JF, Xayaphoummine A, Isambert H, Chatenay D. Probing complex RNA structures by mechanical force. Eur. Phys. J. E Soft Matter. 2003;12:605–615. doi: 10.1140/epje/e2004-00033-4. [DOI] [PubMed] [Google Scholar]
- 4.Li PT, Collin D, Smith SB, Bustamante C, Tinoco I., Jr Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump, and force-ramp methods. Biophys. J. 2006;90:250–260. doi: 10.1529/biophysj.105.068049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Jr, Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DM, Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee M, Kim SH, Hong SC. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl Acad. Sci. USA. 2010;107:4985–4990. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, Liphardt J. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Lett. 2005;5:1509–1514. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- 9.Brau RR, Tarsa PB, Ferrer JM, Lee P, Lang MJ. Interlaced optical force-fluorescence measurements for single molecule biophysics. Biophys. J. 2006;91:1069–1077. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarsa PB, Brau RR, Barch M, Ferrer JM, Freyzon Y, Matsudaira P, Lang MJ. Detecting force-induced molecular transitions with fluorescence resonant energy transfer. Angew. Chem. 2007;46:1999–2001. doi: 10.1002/anie.200604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 12.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 13.Rezler EM, Bearss DJ, Hurley LH. Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 15.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 16.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jena PV, Shirude PS, Okumus B, Laxmi-Reddy K, Godde F, Huc I, Balasubramanian S, Ha T. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J. Am. Chem. Soc. 2009;131:12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch S, Baker H, Byker SG, Zhou D, Sinniah K. Single molecule force spectroscopy on G-quadruplex DNA. Chemistry. 2009;15:8113–8116. doi: 10.1002/chem.200901390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koirala D, Dhakal S, Ashbridge B, Sannohe Y, Rodriguez R, Sugiyama H, Balasubramanian S, Mao H. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011;3:782–787. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Z, Koirala D, Cui Y, Easterling LF, Zhao Y, Mao H. Click chemistry assisted single-molecule fingerprinting reveals a 3D biomolecular folding funnel. J. Am. Chem. Soc. 2012;134:12338–12341. doi: 10.1021/ja303218s. [DOI] [PubMed] [Google Scholar]
- 26.de Messieres M, Chang JC, Brawn-Cinani B, La Porta A. Single-molecule study of g-quadruplex disruption using dynamic force spectroscopy. Phys. Rev. Lett. 2012;109:058101. doi: 10.1103/PhysRevLett.109.058101. [DOI] [PubMed] [Google Scholar]
- 27.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipfert J, Hao X, Dekker NH. Quantitative modeling and optimization of magnetic tweezers. Biophys. J. 2009;96:5040–5049. doi: 10.1016/j.bpj.2009.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 32.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 34.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mashimo T, Yagi H, Sannohe Y, Rajendran A, Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Dai J, Veliath E, Jones RA, Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrion-Vazquez M, Li H, Lu H, Marszalek PE, Oberhauser AF, Fernandez JM. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 40.Li PT, Bustamante C, Tinoco I., Jr Unusual mechanical stability of a minimal RNA kissing complex. Proc. Natl. Acad. Sci. USA. 2006;103:15847–15852. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams PM, Fowler SB, Best RB, Toca-Herrera JL, Scott KA, Steward A, Clarke J. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.