Abstract
The RNA-binding protein AUF1 binds AU-rich elements in 3′-untranslated regions to regulate mRNA degradation and/or translation. Many of these mRNAs are predicted microRNA targets as well. An emerging theme in post-transcriptional control of gene expression is that RNA-binding proteins and microRNAs co-regulate mRNAs. Recent experiments and bioinformatic analyses suggest this type of co-regulation may be widespread across the transcriptome. Here, we identified mRNA targets of AUF1 from a complex pool of cellular mRNAs and examined a subset of these mRNAs to explore the links between RNA binding and mRNA degradation for both AUF1 and Argonaute 2 (AGO2), which is an essential effector of microRNA-induced gene silencing. Depending on the specific mRNA examined, AUF1 and AGO2 binding is proportional/cooperative, reciprocal/competitive or independent. For most mRNAs in which AUF1 affects their decay rates, mRNA degradation requires AGO2. Thus, AUF1 and AGO2 present mRNA-specific allosteric binding relationships for co-regulation of mRNA degradation.
INTRODUCTION
The discovery of microRNAs (miRNAs) has revolutionized the field of post-transcriptional gene control, as it has unveiled perhaps thousands of additional regulatory molecules (1–3). They are 21–23 nt in length and are encoded within the genome. They act through partially complementary binding sites within target mRNAs. Early studies identified them as translational suppressors, but later work showed they can promote degradation of many mRNAs as well (4). miRNAs assemble with a number of proteins, including the Argonaute proteins (AGO1–4) and glycine–tryptophan repeat protein 182 (GW182), to form the miRNA-induced silencing complex (miRISC) (5–8). miRISC controls translation via the ability of GW182 to recruit the carbon catabolite repressor 4-negative-on-transcription (CCR4-NOT) deadenylase complex to remove the poly(A) tail (9–11). Recent evidence also indicates that a complex of PUF (Pumilio/FBF) RNA-binding proteins, AGO proteins and translation elongation factor eEF1A can suppress translation by attenuating elongation steps (12). miRISC controls mRNA degradation via endonucleolytic cleavage of its mRNA targets (6–8,13) or by triggering deadenylation and decapping (8–11,14,15). The latter mechanism involves initial poly(A) tail trimming by the poly(A) binding protein-dependent, poly(A)-specific ribonuclease 2 (PAN2)-PAN3 ribonucleases, followed by full deadenylation catalysed by the CCR4-NOT deadenylase complex; decapping protein 1 (DCP1)-DCP2–dependent decapping and degradation of the mRNA body ensue. The GW182 subunit of miRISC coordinates these actions in three ways (9–11): (i) the N-terminal region of GW182 has an AGO-interacting domain; (ii) the C-terminal region of GW182, known as the silencing domain, contains motifs required for binding the cellular negative-on-transcription 1 (CNOT1) subunit of the CCR4-NOT deadenylase; and (iii) the silencing domain also binds the PAN3 subunit of the PAN2-PAN3 ribonuclease complex.
As predicted by Tenenbaum and colleagues several years ago (16), there are a growing number of examples of mRNA co-regulation by RNA-binding proteins and miRNAs (17). Indeed, a recent study coupling experiments and bioinformatic analyses revealed that mRNA sequences that contact RNA-binding proteins often couple to miRNA-binding sites (18). This is particularly true for AU-rich elements (AREs) and U-rich elements. AREs represent one of the best characterized families of elements that control mRNA stability and translation (17,19–21). They are located within the 3′-untranslated regions (UTRs) of many labile mRNAs encoding oncoproteins, cell cycle regulators, signalling proteins and regulators of immune responses. There are ∼20 ARE-binding proteins (AUBPs), although not all bind to all ARE-mRNAs. Nonetheless, the constellation of AUBPs bound to a particular ARE-mRNA somehow integrates their inputs to dictate an mRNA half-life or translation rate characteristic for that mRNA. As well, signalling events can alter the decoration patterns of AUBPs bound to the ARE-mRNP with concomitant effects on mRNA degradation and/or translation rates.
AU-rich element binding factor 1 (AUF1), also known as heterogeneous nuclear ribonucleoprotein D (hnRNP D), is a well characterized family of AUBPs (22). It consists of four isoforms of 37, 40, 42 and 45 kDa generated by alternative pre-mRNA splicing. The isoforms are differentially phosphorylated and shuttle between the nucleus and cytoplasm. The isoforms each present a range of ARE-binding affinities, RNA-induced protein oligomerization and effects on local RNA structure (23). Studies using auf1−/− mice or gene knockdowns in cell culture indicate that it controls ARE-mRNA degradation and/or translation depending on the particular ARE-mRNA examined (17,22,24).
In the work presented here, we used recombinant AUF1 protein to affinity purify and identify its mRNA targets from a complex pool of cellular mRNAs. Given the growing awareness of relationships between RNA-binding proteins and miRNAs for co-regulation of mRNAs, we examined binding of AUF1 and the RISC subunit AGO2 to a chosen subset of AUF1 target mRNAs by mRNP-immunoprecipitation and quantitative reverse transcriptase–polymerase chain reaction (RT-PCR). We examined the effects of AUF1 and/or AGO2 knockdown on decay kinetics of these mRNAs as well. The results indicate that depending on the mRNA examined, binding of AUF1 and AGO2 to mRNAs can be either reciprocal/competitive, proportional/cooperative or independent. For cases in which AUF1 affects mRNA decay, degradation requires AGO2. Thus, AUF1 and AGO2 can functionally couple by distinct mechanisms, perhaps programmed by the mRNA targets themselves, to control mRNA degradation.
MATERIALS AND METHODS
Preparation of RNA for affinity purification
Plasmid DNA was purified from a cDNA expression library prepared from adult human heart mRNA (Invitrogen). The vector backbone is pcDNA3, which permits in vitro synthesis of RNA from linearized library DNA. Twenty micrograms of library DNA was linearized in separate reactions with either NotI or XbaI and then purified with the High Pure PCR Product Purification Kit (Roche) per the manufacturer’s instructions. In initial experiments, 0.5 µg of NotI- and XbaI-digested DNAs were used as templates in separate in vitro transcription reactions with the RiboMAX kit (Promega). Reactions were supplemented with [α-32P] UTP (500 Ci/mmol) to permit trace labelling of RNA for quantification and estimations of recoveries during affinity chromatography. RNAs were labelled to ∼300 c.p.m./ng. After in vitro transcriptions, template DNAs were degraded with RQ1-DNase (Promega). RNAs were cleaned and purified with RNeasy columns (Qiagen), and RNA was quantified by liquid scintillation counting. Unlabelled RNA was generated exactly as previously described, except 2.5 µg of linearized DNA was used per reaction to maximize RNA production, and radiolabel was omitted. Unlabelled RNAs were quantified spectrophotometrically by A260.
Expression and purification of recombinant His6-p37AUF1
Recombinant protein was purified as previously described by Wilson et al. (25). Briefly, Escherichia coli TOP10 cells were transformed with plasmid pBAD/HisB-p37AUF1. Bacterial cells were pelleted and lysed by sonication, and recombinant protein was purified by Ni2+ affinity chromatography with a HiTrap Chelating affinity column (GE Healthcare). To determine protein concentration, sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed with purified recombinant His6-p37AUF1 and titrations of bovine serum albumin (BSA). Gels were stained with Coomassie Blue, and band intensities were quantified with the Kodak EDAS 120 imaging system. BSA band intensities were used to create a standard curve for determinations of His6-p37AUF1 protein concentration. Protein activity was confirmed by electrophoretic mobility shift assays.
Affinity purification of AUF1 target mRNAs
RNAs directly binding p37AUF1 were purified by the affinity selection approach of Bhattacharya et al. (26) with some modifications. Two hundred micrograms of library-derived mRNA was mixed with 18 µg of either His6-p37AUF1 protein or BSA (as a negative control) in a 400-μl reaction volume. As the cDNA inserts from the cardiac library range from ∼400–4000 nt, this yields a median size of 1800 nt. Using 1800 nt as the length of transcripts, the approximate molarity of mRNA in the reaction is 0.8 µM. p37AUF1 is ∼36 kDa and binds as a dimer, yielding a dimer concentration of 0.25 µM. Thus, binding reactions contained a molar excess of mRNA to limit p37AUF1 binding to the highest affinity mRNA targets. Protein–mRNA binding reactions were performed in 100 mM potassium acetate (KOAc), 50 mM Tris–HCl (pH 7.5), 10 mM dithiothreitol (DTT) and 1 μg/µl of heparin on ice for 30 min and were then transferred to room temperature for 3 min. A methylcellulose disc (Millipore, HAWP 0047) was soaked in binding buffer 1 [BB1: 100 mM KOAc, 50 mM Tris–HCl (pH 7.5) and 10 mM DTT]. After initial wetting of the disc, it was never allowed to dry during the experimental procedure. To isolate mRNA–protein complexes, the disc was filtered with the mRNA–AUF1 (or mRNA–BSA) mix. RNA not bound to protein was removed by washes with cold BB1. Protein bound RNA was eluted with 2 ml of binding buffer 2 [BB2: 200 mM KOAc, 50 mM Tris–HCl (pH 7.5), 500 mM NaCl and 10 mM DTT]. RNA was desalted and purified with the RNeasy kit (Qiagen) according to the manufacturer’s protocol. Affinity chromatography with His6-p37AUF1 protein and BSA was then repeated with mRNA recovered from the first round of affinity purification. Desalted and cleaned mRNA was then used for subsequent microarray analyses.
Similar to Bhattacharya et al. (26), pilot purification experiments were performed to assess the capacity of p37AUF1 to selectively isolate an ARE-RNA from a complex pool of RNAs. Cellular poly(A+) RNA was supplemented with 32P-labelled RNA containing or lacking a high affinity AUF1-binding site [a coding-region fragment of rabbit β-globin mRNA (Rβ) or Rβ linked to the FBJ murine osteocarcoma viral oncogene homolog (FOS) ARE, respectively]. Each mixture was fractionated by AUF1-affinity selection. After each round of selection, recovered 32P-labelled RNAs were quantified by scintillation counting.
Microarray analyses of affinity-purified RNA
The Human 1 A(V2) Oligo Microarray chip (Agilent), which contains probes for >18 000 known human genes, was used to identify affinity-purified mRNAs. Two pools of RNA were labelled: one was the experimental RNA (AUF1- or BSA-selected RNA); the other was total RNA from in vitro transcription reactions. One microgram of affinity-purified RNA and 1 μg of total RNA were reverse transcribed using random hexamers in the presence of Cy5-dUTP and Cy3-dUTP (Perkin-Elmer), respectively. Labelled cDNAs were mixed, fragmented, dried and resuspended in hybridization buffer. The mixture was incubated with microarray chips for 17 h at 60°C. To prevent data misinterpretation from dye bias, reciprocal labelling reactions and hybridizations were performed with a second chip. Arrays were visualized with a Perkin Elmer ScanArray microarray scanner, and raw data were analysed and normalized using ScanArray software (Perkin Elmer). For a given spot, the difference between the average log fluorescence intensities of the experimental and control pool yields M = log2(Cy3) − log2(Cy5) = log2(Cy3/Cy5). The t-statistic, t, corrects for poor spots in an array that can give rise to high M values by incorporating the standard deviation between replicates. Thus t = M/(s/n1/2), where s is the standard deviation and n is the number of replicates. However, the t-statistic can result in false–positives arising from spots with erroneously small standard deviations. To correct for these biases, B-statistics were determined. The B-statistic, B, corrects the t-statistic by applying a penalty to the standard deviation of every gene. The penalty is estimated using both the mean and standard deviation and yields a more accurate representation of selection (27). B = M/[(a + s2)/n)]1/2, where M is the difference between average log2 intensities (defined earlier in the text), s is the standard deviation, n is the number of replicates and a is the penalty. Genes with B > 0 have a >50–50 probability of being selected. Microarray data were analysed with the R-statistical program, version 1.8.1, to assign B-values to AUF1- and BSA-selected RNAs. To functionally classify proteins encoded by AUF1-associated RNAs, a computer program was designed (R. Muldowny and P. Kahn, Douglas College, Rutgers University) to import gene ontology data from the National Center for Biotechnology Information (NCBI) Entrez Gene database www.ncbi.nih.gov/entrez into a Microsoft excel file. Data were subsequently analysed with a local server.
Construction of reporter gene plasmids
Primers were designed to amplify the 3′-UTRs of selected AUF1 target genes for insertion into the 3′-UTR of the Rβ gene. The forward and reverse primers all contained a 5′-BglII site to allow insertion of PCR products into the unique BglII site just 3′ of the stop codon in the Rβ gene. The sequences of primer pairs (forward and reverse, respectively) used are:
| IFI16: | 5′-GCAGTCTAGATCTAGAAATCTGGATGTCATTGACGATAA-3′ and 5′-GCAGAATCTAGATCTTCACAAAAAAGATAATGTTTATTTAT-3′; |
| IL1RN: | 5′-GCAGAGATCTAGATCTTACTGCCCAGGCCTGCCT-3′ and 5′-GCAGAAGCTTAGATCTAGAAGGCATTTTCAAGATTTTATTGTA-3′; |
| FBXO24: | 5′-GCACAGATCTAGATGCAGAGGGCTGAAGGAGGC-3′ and 5′-GCACAGATCTAGACTCGCTGCAGCCTTTTATTCTATC-3′; |
| DOHH: | 5′-GCACAGATCTAGAGGCCCCACCCTCACCC-3′ and 5′-GCACAGATCTAGACCTTGGCAAAAATAATTTAATATTCC-3′; |
| GNLY: | 5′-GCACAGATCTAGAGCCCTCTCACCTTGTCCTGTG-3′ and 5′-GCACAGATCTAGATCTTGCTTGACACTTTATTCTCGTG-3′; |
| THRAP5: | 5′-GCACAGATCTAGACGGCCGGGGGTCCAGGCG-3′ and 5′-GCACAGATCTAGACTGGGCGCAGAGGGCGTTTATTGGA-3′. |
DNAs were amplified from the cDNA expression library described earlier in the text. PCR products were fractionated in a 1% agarose gel to confirm size and then gel purified with the QIAquick Gel Extraction kit (Qiagen) according to the manufacturer’s instructions. PCR products were digested with BglII and ligated with plasmid pTRE/Rβ-wt digested with BglII. pTRE/Rβ-wt contains the Rβ gene linked to the tetracycline-responsive promoter of plasmid pTRE (28). pTRE/Rβ-3′-UTR plasmids were used for transfection-based mRNA decay and mRNP-immunoprecipitation assays.
Cell culture and transfections of plasmids expressing shRNAs
HeLa/Tet-Off cells (human cervical carcinoma; BD Biosciences) (29), which express the tetracycline-responsive transcriptional activator tTA, were cultured at 37°C in 5% CO2 with Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS; Hyclone), 1% L-glutamine and 100 μg/ml of geneticin (GIBCO). Plasmids expressing shRNA directed against all four isoforms of AUF1 or a random sequence not found in the human genome (28) were linearized with XmnI and transfected into HeLa/Tet-Off cells with Effectene (Qiagen, Hilden, Germany). Stably transfected cells were selected with 250 U/ml of Hygromycin B (Calbiochem). To assess knockdown, cells were harvested, lysed in SDS gel loading buffer, fractionated by SDS–PAGE and analysed by western blotting with antibodies to AUF1 and α-tubulin (as an internal control; 1:8000 dilution; Sigma), exactly as described by Sinsimer et al. (28).
K562 cells (human chronic myeloid leukaemia; ATCC) (30) were cultured at 37°C in 5% (v/v) CO2 with RPMI-1640 medium supplemented with 10% (v/v) FBS (GIBCO) and 2 mM glutamine (Invitrogen). For RNAi-mediated gene knockdowns, 2 × 107 cells were transfected with 40 µg shRNA plasmid by electroporation with a Gene Pulser (Bio-Rad). Electroporation parameters were 360 V, 500 µF. Transfected cells were maintained in culture medium without antibiotics for 48 h to permit knockdown before assays. Medium was replaced with fresh culture medium lacking antibiotics 24 h post-transfection. The following plasmids were used: plasmids expressing shRNA directed against all four isoforms of AUF1 or a random sequence not found in the human genome; shAGO2-1-8 and shAGO2-2-6 against AGO2 (provided by Dr Shobha Vasudevan) (31). Knockdown efficiency was assessed by western blot analysis for AUF1, α-tubulin and AGO2 (1:1000 dilution, Millipore).
mRNP-immunoprecipitation
Immunoprecipitations of endogenous protein–mRNA complexes were used to assess association of AUF1 and/or AGO2 with endogenous target mRNAs. To pre-clear lysates in preparation for immunoprecipitation with AUF1 antibody, lysates of 2 × 107 K562 cells or HeLa/Tet-Off cells were incubated with rabbit non-immune serum (Sigma) for 45 min at 4°C, then magnetic Dynabeads coupled to protein A (Invitrogen) were added for 30 min at 4°C; beads were removed with a magnet. For immunoprecipitations, fresh beads were coated with anti-AUF1 or rabbit non-immune serum in NT-2 buffer [50 mM Tris–HCl (pH 7.4), 1 mM MgCl2, 150 mM NaCl and 0.05% Nonidet P-40] and washed. Pre-cleared cell lysates (1 mg protein) were incubated with 50 μl of coated beads in 200 μl of NT-2 buffer supplemented with 2.5 μl of RNase Out (Invitrogen) and 2 μl of 100 mM DTT for 3.5 h at 4°C with constant rocking. Beads were washed eight times with ice-cold NT-2 buffer and two times with NT-2 buffer supplemented with 0.5 M urea. Proteins were digested with proteinase K (Promega), and mRNAs were purified by phenol–chloroform extraction and ethanol precipitation. Immunoprecipitations were also performed to assess association of endogenous AGO2 in K562 cells with predicted target mRNAs using a similar protocol. Fresh beads were coated with anti-AGO2 IgG (Abcam) or control IgG (Cell Signaling). For AGO2 immunoprecipitations, pre-clearing of cell lysates was not necessary, and incubated beads were washed with NT-2 buffer (not supplemented with urea) eight times. For quantitations of mRNAs in precipitates, purified RNAs were reverse transcribed into cDNAs with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems), followed by SYBR green quantitative PCRs. The forward and reverse primer sequences, respectively, are as follows:
| MYC: | 5′-CCCTCAACGTTAGCTTCACC-3′ and 5′-GTAGTGGTGGTGCGAGATTC-3′; |
| IFI16: | 5′-ACAAACCCGAGAAACAATGACC-3′ and 5′-GCATCTGAGGAGTCCGAAGA-3′; |
| IL1RN: | 5′-CATTGAGCCTCATGCTCTGTT-3′ and 5′-CGCTGTCTGAGCGGATGAA-3′; |
| FBXO24: | 5′-GGACCCTTCAAGCCTTTGACC-3′ and 5′-GCTGCCCGTATCTGTTATTTCC-3′; |
| DOHH: | 5′-CCCTGCTCAAGCACGAGCTGG-3′ and 5′-GCCTCCCCTGCCTCATGGC-3′; |
| GNLY: | 5′-CCTGTCTGACGATAGTCCAAAAA-3′ and 5′-GACCTCCCCGTCCTACACA-3′; |
| THRAP5: | 5′-TGCTGGACATGAACACACTG-3′ and 5′-AGGGAACCCTGGTTGGGTA-3′; |
| NDOR1: | 5′-GACTCCTACCCGGTGGTGAAT-3′ and 5′-TTGGCCTGTAGTTGCACAAAC-3′; |
| HOXB8: | 5′-GTCCCTGCGCCCCAATTATTA-3′ and 5′-GCCCGTGGTAGAACTCCTG-3′; |
| GAPDH: | 5′-TTTAACTCTGGTAAAGTGGATATTGTTG-3′ and 5′-ATTTCCATTGATGACAA GCTTCC-3′. |
Immunoprecipitations were also performed to assess association of endogenous AUF1 specifically with 3′-UTRs of selected target mRNAs by transfecting HeLa/Tet-Off cells (1 × 107 cells) with 1.2 μg of reporter plasmid containing the rabbit β-globin (Rβ) gene linked to target 3′-UTRs (see reporter plasmid constructions earlier in the text). For quantitations of Rβ transgene-encoded mRNAs and glyceraldehyde-3-phosphte dehydrogenase (GAPDH) (GAPDH; as a control), purified RNAs were reverse transcribed into cDNAs with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems), followed by Taqman-based qPCR. The forward and reverse primer pairs, respectively, and probe sequences used are as follows:
| Rβ: | 5′-GTGAACTGCACTGTGACAAGC-3′ and 5′-ATGATGAGACAGCACAATAACCAG-3′ and probe 5′-FAM-CGTTGCCCAGGAGCCTGAAGTTCTCA-BHQ_1-3′; |
| GAPDH: | 5′-TTTAACTCTGGTAAAGTGGATATTGTTG-3′ and 5′-ATTTCCATTGA TGACAAGCTTCC-3′ and probe 5′-Cy3-CCATGGCACCGTCAAGGCTG-BHQ_2-3′. |
Determinations of mRNA decay kinetics
Half-lives of endogenous mRNAs in K562 cells were determined by culturing cells with actinomycin D (5 μg/ml) to inhibit transcription. Cells were harvested at times indicated in the figure panels. Time courses were limited to 3 h to avoid actinomycin D-induced apoptosis (32), which would complicate mRNA decay analyses. Total RNA was isolated with the RNeasy kit (Qiagen) following the manufacturer’s instructions. Purified RNAs were reverse transcribed into cDNAs with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) and analysed with SYBR green quantitative PCR. The forward and reverse primer pairs were described earlier in the text. Levels of each mRNA were normalized to GAPDH mRNA at each time point. Percent mRNA remaining was plotted versus time post-actinomycin D addition, and the first-order decay constant, k, was determined by non-linear regression analyses with Prism 5.0 software (GraphPad, San Diego, CA, USA). Messenger RNA half-life was calculated as t1/2 = ln2/k.
Assays of Rβ reporter mRNA half-lives were determined as described previously (28,33). Briefly, 1 × 105 HeLa/Tet-Off cells expressing shRNAs were transfected with Superfect (Qiagen) containing 50 ng of pTRE/Rβ or pTRE/Rβ-3′-UTR plasmids together with 200 ng of plasmid pEGFP-C2 (encoding internal control mRNA) per well of a six-well plate. After 48 h, doxycycline (Sigma) was added to 2-μg/ml final concentration to block transcription of the reporter gene. Cells were harvested at each time point, lysed with Qiagen QIAshredder cartridges and RNA was purified with the Qiagen RNeasy kit. Multiplex qRT–PCR reactions were assembled with the SuperScriptIII Platinum One-Step qRT-PCR kit (Invitrogen, Carisbad, CA, USA) together with each primer and probe specific for Rβ and EGFP as described earlier in the text. First-order decay constants and half-lives were determined as described earlier in the text for endogenous mRNAs.
Statistical analyses
Comparisons between data sets were performed with the unpaired two-tailed Student’s t-test (GraphPad, San Diego, CA, USA). P-values of <0.05 were considered significant.
RESULTS
Selection and identification of mRNA-binding targets of AUF1
To identify the repertoire of mRNAs that interact with AUF1 via direct protein–RNA association, we used the previously described affinity chromatography approach of Bhattacharya et al. (26) with purified recombinant p37AUF1 and introduced three modifications. First, a substrate pool transcribed in vitro rather than tissue-derived mRNA was used for selection. This modification permits affinity purification with an unlimited amount of starting mRNA material. This is critical when using tissues that are difficult to obtain. Secondly, in vitro transcribed RNA permits affinity purification under conditions of molar excess of mRNAs to protein. This modification increases stringency of the affinity selection process, as there is competition for protein binding among the different mRNA species present. Finally, microarray-based identification of purified mRNAs eliminates cloning bias towards abundant mRNAs rather than those transcripts that are enriched in the AUF1-selected pool. Based on the results of two replicate arrays, p37AUF1 selected 499 transcripts (Supplementary Table S1).
Proteins encoded by these 499 target mRNAs were classified into functional groups using the Database for Annotation, Visualization and Integrated Discovery (DAVID; Figure 1) (34,35). Almost 24% of the genes participate in cellular metabolic processes. Genes involved in gene expression (e.g. DNA-binding, RNA-binding, transcription and translation) collectively account for 21% of the targets. Cytokines and immunity/defence genes collectively represent 11% of genes, consistent with the importance of AUF1 in the immune system (24,28,36–40). Structural proteins account for 10% of expressed genes in human heart tissue (41) and for ∼50% of highly expressed genes (42). However, genes encoding structural proteins accounted for <10% of the AUF1-selected targets. Although ribosomal protein genes comprise 2.5% of the transcriptome in heart tissue (42), they accounted for only 0.6% of AUF1-selected transcripts. Thus, many of the p37AUF1 targets encode proteins involved in metabolism, transcription, RNA processing and immune responses. Some of these mRNAs had no apparent AREs in their 3′-UTR (data not shown); thus, AUF1 may have broader sequence recognition than previously thought. Detailed analyses of AUF1-binding motifs will be presented in forthcoming work (manuscript in preparation).
Figure 1.
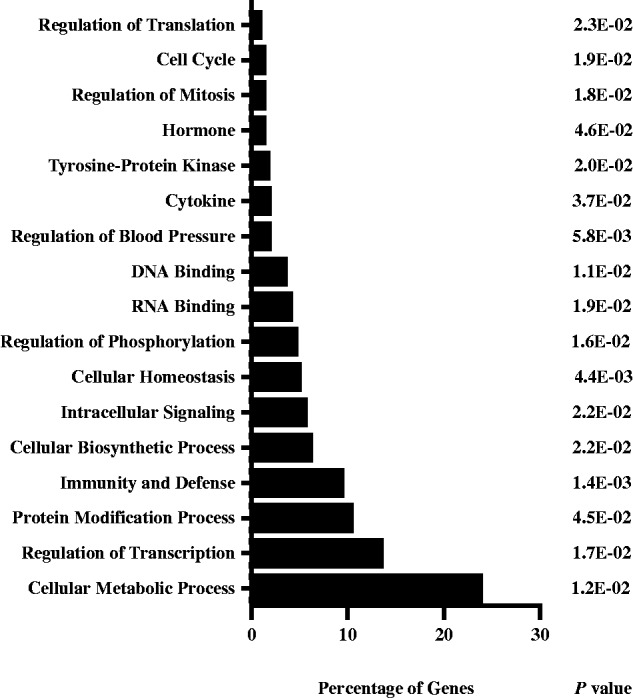
Gene ontogeny of p37AUF1 targets. The 499 identified target transcripts were analysed with DAVID. The percentage of genes in each category is shown along with P-values. The top percentages of targets regulate metabolic processes, transcription, protein modification and immunity/organism defence.
Binding of AUF1 to its target mRNAs in cells
Six transcripts were randomly chosen for validation of AUF1 binding: interferon γ-inducible protein 16 (IFI16; NM_001206567.1), interleukin-1 receptor antagonist (IL1RN; NM_173842.2), thyroid hormone receptor-associated protein (THRAP5/MED16; NM_005481.2), F-Box only protein 24 (FBXO24; NM_033506.2), granulysin (GNLY; NM_012483.2) and deoxyhypusine hydroxylase/monooxygenase (DOHH; NM_001145165.1). The association of these target mRNAs with AUF1 was tested by immunoprecipitation of cytoplasmic mRNP complexes from K562 cells, a human chronic myelogenous leukaemia cell line (30) and HeLa cells, a human cervical carcinoma cell line (29). Transcripts were quantified by real-time RT–PCR. Tumour necrosis factor-α (TNFα) and myelocytomatosis viral oncogene homolog (MYC) mRNAs served as positive controls for HeLa and K562 cells, respectively, as AUF1 binds the AREs within these mRNAs (28,43). Western blot analyses of aliquots of immunoprecipitates with anti-AUF1 antibody verified specific precipitation of AUF1 isoforms (Figure 2A and B, upper panels). All of the predicted target transcripts, as well as the positive control TNFα (for HeLa cells) and MYC (for K562 cells), were significantly enriched in AUF1-mRNP immunoprecipitations from extracts of both HeLa (Figure 2A) and K562 cells (Figure 2B) relative to GAPDH mRNA (P < 0.01 for all). These results indicate that AUF1 forms mRNP complexes with the transcripts in cells, consistent with the affinity purification results.
Figure 2.

Association of AUF1 with mRNA targets in cells. Lysates of HeLa/Tet-Off (A) and K562 cells (B) were prepared and fractionated by ribonucleoprotein immunoprecipitation (IP) with non-immune serum or AUF1 anti-serum. (A and B) Upper panels: aliquots of immunoprecipitated materials were analysed by western blot (WB) to verify anti-AUF1–dependent recovery of AUF1. The AUF1 isoforms are noted to the right of the panel. The p40/42 isoforms do not readily resolve by SDS–PAGE with mini-gels. The p37 isoform is the least abundant and is typically <10% of total AUF1 (our observations); it is not detected in the exposures shown. (A and B) Lower panels: both input and immunoprecipitated materials were analysed by qRT–PCR for each indicated endogenous mRNA and plotted as a percent of the input mRNA level. The means ± standard deviation (SD) from three independent experiments are shown. **P < 0.01 versus GAPDH mRNA.
AUF1 binding to and degradation of 3′-UTR reporter mRNAs
The next experiments determined whether AUF1 can associate with the mRNAs via their 3′-UTRs. mRNP-immunoprecipitation assays were performed for β-globin (Rβ) reporter transcripts engineered with full-length 3′-UTRs from the same mRNAs chosen earlier in the text; wild-type Rβ mRNA (i.e. lacking any inserted sequences) served as a negative control. Transgenes were transcribed from the minimal cytomegalovirus promoter linked to the tetracycline operator, and thus require the tetracycline-repressor–VP16 fusion protein, tTA (44). HeLa/Tet-Off cells, which express tTA, were transfected with reporter plasmids. Reporter transcripts containing the 3′-UTRs of IFI16, IL1RN, FBXO24, DOHH, GNLY, THRAP5 and the TNFα ARE were significantly enriched by antibody against AUF1 relative to the negative control Rβ mRNA (P < 0.01 for all; Figure 3A); again, GAPDH mRNA did not detectably immunoprecipitate. We note that the percentage of input reporter mRNAs immunoprecipitated was typically 1% of those for endogenous mRNAs (compare with Figure 2A). This is most likely because of the 50- to 100-fold higher cellular levels of reporter mRNAs compared with endogenous mRNAs (our observations). Nonetheless, the data in Figure 3A suggest that AUF1 can form mRNPs via the 3′-UTRs of the chosen transcripts. However, this does not exclude the possibility that AUF1 may bind other regions of the endogenous mRNAs as well.
Figure 3.
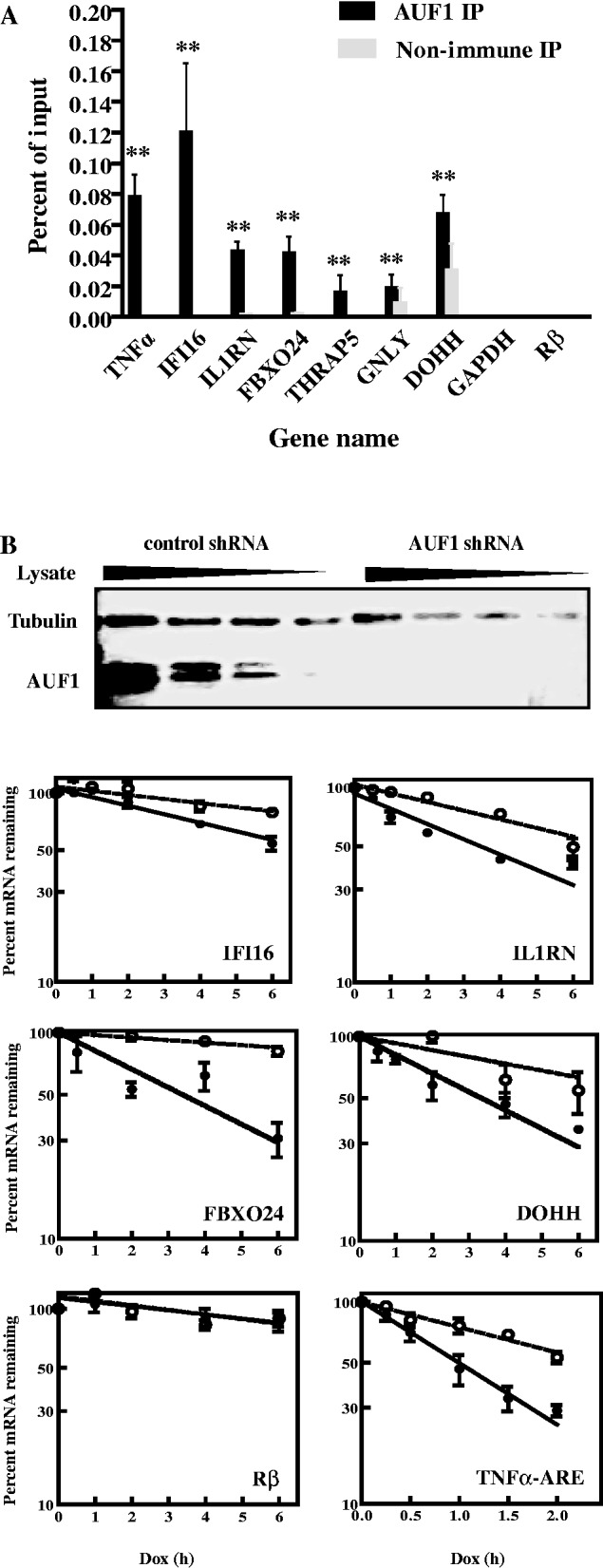
Analyses of AUF1 binding to and degradation of 3′-UTR reporter transcripts in HeLa/Tet-Off cells. (A) Association of AUF1 with 3′-UTR reporter transcripts in cells. A rabbit β-globin (Rβ) reporter plasmid containing the 3′-UTR from each indicated gene was transfected into HeLa/Tet-Off cells. After 2 days, cell lysates were immunoprecipitated with pre-immune serum or AUF1 anti-serum. Abundance of each indicated reporter mRNA in precipitates was determined by qRT–PCR and plotted as a percent of the input mRNA level. The means ± SD from three independent experiments are shown. **P < 0.01 versus Rβ mRNA (lacking any heterologous 3′-UTR insert). (B) Effects of AUF1 knockdown on degradation of Rβ-3′-UTR reporter mRNAs. HeLa/Tet-Off cells stably expressing control or AUF1-directed shRNA were prepared by transfection of plasmids and drug selection. Upper panel: western blot analysis of AUF1 knockdown. A 2-fold dilution series of lysate from cells expressing control shRNA was used to permit estimations of AUF1 knockdown efficiency. Tubulin served as a loading control. Lower panel: the Rβ-3′-UTR reporter genes were co-transfected with plasmid pEGFP-C2 into HeLa/Tet-Off cells expressing control (solid circles, solid lines) or AUF1 shRNAs (open circles, dashed lines). After 48 h, doxycycline was added to inhibit transcription of reporter genes. Levels of reporter mRNAs were normalized to that of EGFP mRNA and plotted as the percent reporter mRNA remaining as a function of time following doxycycline treatment. Each time point represents mean ± SD from n ≥ 3 independent experiments. Non-linear regression analyses yielded first-order decay constants (k) and associated mRNA half-lives. Representative graphs are shown, and all data are tabulated in Table 1. Dox, doxycycline.
The effects of AUF1 knockdown on 3′-UTR-mediated mRNA degradation were investigated next. Given the structure of mRNAs encoding each AUF1 isoform (45,46), it has not been possible to selectively reduce abundance of a single isoform by RNA interference. Thus, a plasmid expressing a shRNA that efficiently reduces all four AUF1 isoforms was used. This shRNA has been used extensively to examine the effects of AUF1 on mRNA degradation and translation (28,36,43,47–49). Two HeLa/Tet-Off cell lines were prepared: one stably expresses a plasmid encoding control shRNA, and the other stably expresses the shRNA against AUF1. Western blotting confirmed >95% knockdown of AUF1 with this shRNA, as expected (Figure 3B, top panel). Cells were transfected with the tetracycline-regulated reporter plasmids described earlier in the text. After 48 h, cells were cultured with doxycycline to block reporter gene transcription. RNA was harvested at various time points for analyses with qRT–PCR. First-order decay constants, k, were calculated from non-linear regression analyses of percent mRNA remaining as a function of time. Half-lives were calculated from k values and compared between cells expressing control versus AUF1 shRNA. AUF1 knockdown extended the half-lives of IFI16, IL1RN, FBXO24 and DOHH reporter mRNAs by 2- to 3-fold (Figure 3B and Table 1). Additionally, AUF1 knockdown stabilized Rβ mRNA linked to the TNFα ARE (Figure 3B and Table 1; 1.0 versus 2.0 h, P < 0.01). This result is consistent with its stabilization on AUF1 knockdown in THP-1 cells, a human monocyte cell line; likewise, AUF1 knockdown stabilizes endogenous TNFα mRNA in THP-1 cells (28,50). As expected, AUF1 knockdown had no effect on stability of wild-type Rβ mRNA (Figure 3B and Table 1). Lowering AUF1 abundance did not affect GNLY and THRAP5 reporter mRNAs, as they remained relatively stable with half-lives >10 h (Table 1). This suggests AUF1 might affect translation, rather than decay, of GNLY and THRAP5 mRNAs, just as it does for MYC mRNA (43) (see ‘Discussion’ section). Collectively, these findings indicate that a subset of 3′-UTRs from the chosen target transcripts can confer mRNA degradation to a reporter mRNA in an AUF1-dependent fashion.
Table 1.
Decay kinetics of β-globin reporter mRNAs in control versus AUF1-depleted HeLa/Tet-Off cells
| Reporter mRNA | Control shRNA |
AUF1 shRNA |
||||
|---|---|---|---|---|---|---|
| k (h−1)a | t1/2 (h)b | n | k (h−1)a | t1/2 (h)b | n | |
| Rβ | 0.05 ± 0.01 | >10 | 3 | 0.05 ± 0.01 | >10 | 3 |
| TNFα-ARE | 0.72 ± 0.16 | 1.0 | 5 | 0.34 ± 0.06** | 2.0 | 4 |
| IFI16 | 0.11 ± 0.01 | 6.3 | 3 | 0.05 ± 0.01* | >10 | 3 |
| IL1RN | 0.18 ± 0.01 | 3.8 | 3 | 0.10 ± 0.01* | 6.9 | 3 |
| FBXO24 | 0.20 ± 0.03 | 3.5 | 3 | 0.03 ± 0.01** | >10 | 3 |
| DOHH | 0.21 ± 0.01 | 3.3 | 3 | 0.08 ± 0.01* | 9 | 3 |
| GNLY | 0.04 ± 0.01 | >10 | 3 | 0.06 ± 0.01 | >10 | 3 |
| THRAP5 | 0.03 ± 0.01 | >10 | 3 | 0.03 ± 0.01 | >10 | 3 |
aFirst-order decay constants derived from plots of percent mRNA remaining versus time following Dox treatment. Values represent the means ± SD for n independent experiments.
bAverage mRNA half-lives were calculated as t1/2 = ln2/k.
*P < 0.05; **P < 0.01 versus control shRNA.
Degradation of endogenous mRNAs targeted by AUF1 in K562 cells
Figure 2B showed that AUF1 forms mRNP complexes in K562 cells with the six randomly chosen mRNAs—IFI16, IL1RN, THRAP5, FBXO24, GNLY, DOHH—and the positive control MYC. Two additional mRNAs were examined in extracts of K562 cells: homeobox B8 (HOXB8; NM_024016.3) and nicotinamide adenine dinucleotide phosphate (NADPH)-dependent diflavin oxidoreductase 1 (NDOR1; NM_001144026.1). These were chosen for two reasons: (i) AUF1 mRNP immunoprecipitation of K562 lysates and microarray analysis indicated AUF1 bound to both mRNAs (B. Liao, G. Brewer, unpublished observations) and (ii) analyses of the abundance of HOXB8 and NDOR1 mRNAs on AUF1 knockdown suggested that it might promote degradation of HOXB8, which is also a validated miRNA target (13), but promote stabilization of NDOR1 (B. Liao and G. Brewer, unpublished observations). Cytoplasmic mRNP complexes were immunoprecipitated with antibody against AUF1. Both HOXB8 and NDOR1 mRNAs were significantly enriched in AUF1-mRNP immunoprecipitations relative to GAPDH mRNA (P < 0.01 for all; Figure 2B), confirming that AUF1 formed mRNP complexes with these transcripts in K562 cells.
Knockdown of AUF1 was next used to examine the effects on decay kinetics of the endogenous mRNAs. Cells were transiently transfected with the plasmids expressing control shRNA or the shRNA against the four AUF1 isoforms, both described earlier in the text. Western blots confirmed >80% knockdown (Figure 4A). Cells were then cultured with actinomycin D (5 µg/ml), and RNA was harvested at various time points for determinations of mRNA half-lives as described earlier in the text. AUF1 knockdown in K562 cells extended the half-lives of IFI16, IL1RN, FBXO24 and DOHH mRNAs by 2- to 4-fold (Figure 4B and Table 2), consistent with the results of reporter mRNA analyses with HeLa/Tet-Off cells (Figure 3B and Table 1). Compared with control shRNA-transfected cells, AUF1 knockdown also extended the half-life of HOXB8 mRNA by 70% (Table 2; 3.5 versus 5.8 h, respectively; P < 0.05). However, AUF1 knockdown had no effect on the half-life of MYC mRNA, as expected, nor did AUF1 knockdown affect the half-lives of GNLY and THRAP5 mRNAs (Table 2), again consistent with results from 3′-UTR reporter mRNA analyses with HeLa/Tet-Off cells (Table 1). Thus, AUF1 might control translation of GNLY and THRAP5 mRNAs, as it does MYC mRNA (43). Together, these results indicated that AUF1 can promote degradation of subsets of its target mRNAs in K562 cells. NDOR1 mRNA was relatively stable with a half-life of 10 h in control shRNA-transfected cells, but as suspected, AUF1 knockdown reduced mRNA half-life >2-fold, to 4.3 h (Table 2; P < 0.05). Thus, AUF1 may act to promote stability of NDOR1 mRNA (see ‘Discussion’ section).
Figure 4.
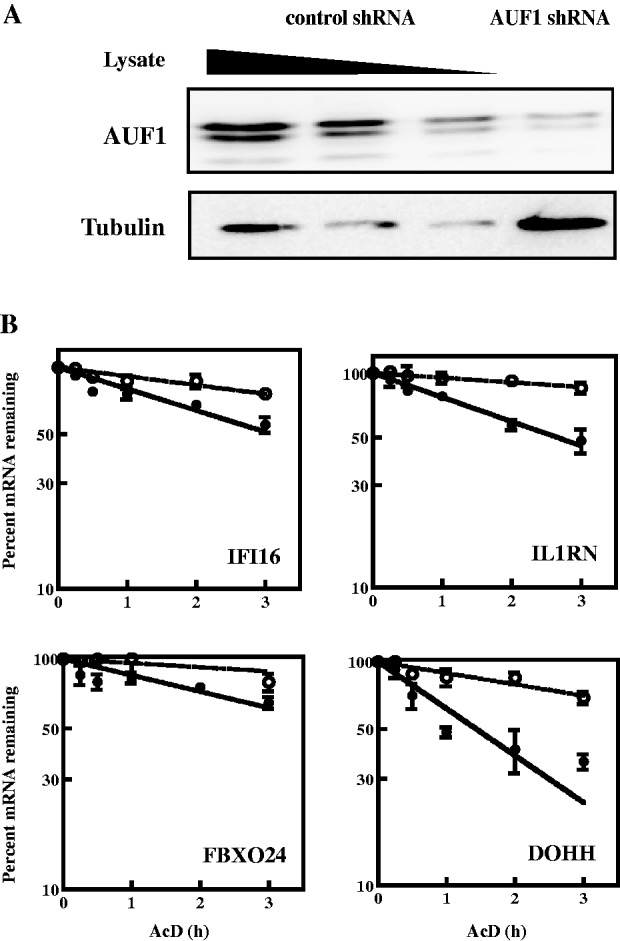
Effects of AUF1 knockdown on degradation of mRNAs in K562 cells. Cells were transiently transfected with plasmids expressing control or AUF1-directed shRNA. (A) Western blot analysis of AUF1 knockdown. A 2-fold dilution series of lysate from cells expressing control shRNA was used to estimate AUF1 knockdown efficiency. Tubulin served as a loading control. (B) Analyses of mRNA decay kinetics. Two days after transfection of shRNA-expressing plasmids, actinomycin D was added to inhibit transcription. For each indicated gene, levels of the endogenous mRNA were normalized to those of GAPDH mRNA and plotted as the percent mRNA remaining as a function of time following actinomycin D addition. Each time point represents mean ± SD from n ≥ 3 independent experiments for cells expressing control shRNA (solid circles, solid lines) or AUF1 shRNA (open circles, dotted lines). Non-linear regression analyses yielded first-order decay constants (k) and associated mRNA half-lives. Representative graphs are shown, and all data are tabulated in Table 2. AcD, actinomycin D.
Table 2.
Effects of AUF1 and/or AGO2 knockdown on endogenous mRNA decay kinetics in K562 cells
| mRNA | Control shRNA |
AUF1 shRNA |
AGO2 shRNA |
AUF1 + AGO2 shRNA |
||||
|---|---|---|---|---|---|---|---|---|
| k (h−1)a | t1/2 (h)b | k (h−1)a | t1/2 (h)b | k (h−1)a | t1/2 (h)b | k (h−1)a | t1/2 (h)b | |
| MYC | 1.46 ± 0.04 | 0.48 | 1.33 ± 0.04 | 0.52 | 0.90 ± 0.07** | 0.77 | 1.30 ± 0.03 | 0.53 |
| IFI16 | 0.23 ± 0.01 | 3.0 | 0.09 ± 0.01* | 8 | 0.05 ± 0.01** | >10 | 0.08 ± 0.01* | 8.7 |
| IL1RN | 0.26 ± 0.01 | 2.7 | 0.05 ± 0.01** | >10 | 0.34 ± 0.03 | 2.0 | 0.07 ± 0.01* | 10 |
| FBXO24 | 0.16 ± 0.01 | 4.3 | 0.04 ± 0.01** | >10 | 0.03 ± 0.01** | >10 | 0.06 ± 0.01* | >10 |
| HOXB8 | 0.20 ± 0.01 | 3.5 | 0.12 ± 0.01* | 5.8 | 0.03 ± 0.01* | >10 | 0.05 ± 0.01* | >10 |
| THRAP5 | 0.09 ± 0.01 | 8 | 0.09 ± 0.01 | 8 | 0.12 ± 0.01 | 5.8 | 0.06 ± 0.01 | >10 |
| NDOR1 | 0.07 ± 0.01 | 10 | 0.16 ± 0.01* | 4.3 | 0.06 ± 0.01 | >10 | 0.07 ± 0.01 | 10 |
| GNLY | 0.09 ± 0.01 | 8 | 0.07 ± 0.01 | 10 | 0.09 ± 0.01 | 8 | 0.08 ± 0.01 | 9 |
| DOHH | 0.51 ± 0.02 | 1.4 | 0.12 ± 0.01* | 5.8 | 0.20 ± 0.01* | 3.5 | 0.14 ± 0.01* | 5.0 |
aFirst-order decay constants derived from plots of percent mRNA remaining versus time following actinomycin D treatment, as described in ‘Materials and Methods’ section. Values represent the means ± SD for n ≥ 3 independent experiments.
bAverage mRNA half-lives were calculated as t1/2 = ln2/k.
*P < 0.05; **P < 0.01 versus control shRNA.
A subset of AUF1 target mRNAs binds AGO2 for their degradation
An emerging theme is that RNA-binding proteins and miRNAs can co-regulate mRNAs (18). Indeed, miRNA prediction algorithms suggest that, with the exception of THRAP5 mRNA, IFI16, IL1RN, FBXO24, NDOR1 and GNLY are miRNA targets. MYC, HOXB8 and DOHH are experimentally validated miRNA targets (13,51–54). For example, let-7, miR-34c and miR-185-3p control MYC; miR-196a controls HOXB8; and miR-331-3p and miR-642-5p control DOHH.
Interaction of AGO2, an integral subunit of RISC, with AUF1 mRNA targets was examined by performing AGO2 mRNP-immunoprecipitations with cytoplasmic lysates of K562 cells. Precipitates were examined for endogenous mRNA candidates with qRT–PCR. An AGO2 western blot of a sample immunoprecipitated with anti-AGO2 or control IgG showed specificity of the immunoprecipitation (Figure 5A, top panel). As expected, MYC, HOXB8 and DOHH mRNAs were immunoprecipitated. Except for IL1RN and GNLY, IFI16, FBXO24, THRAP5 and NDOR1 mRNAs also immunoprecipitated with AGO2 antibody, as compared with control IgG and the negative control GAPDH mRNA (Figure 5A, bottom panel, P < 0.01 for all). This result suggested that AGO2 associated with a subset of the AUF1 target mRNAs.
Figure 5.
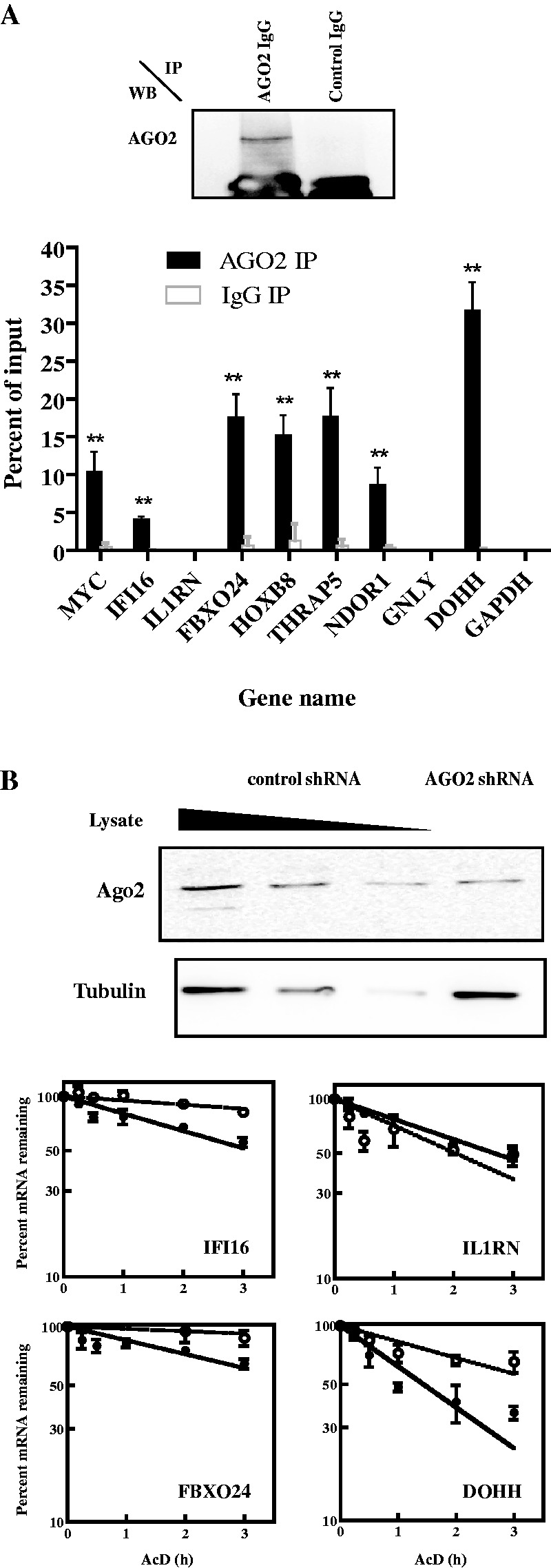
Analyses of AGO2 binding to and degradation of mRNAs in K562 cells. (A) Association of AGO2 with AUF1-target mRNAs in cells. Lysates of K562 cells were prepared and fractionated by ribonucleoprotein immunoprecipitation (IP) with IgG against AGO2 or control IgG. Upper panel: immunoprecipitated materials were analysed by western blot (WB) to verify anti-AGO2–dependent recovery of AGO2. Bottom panel: both input and immunoprecipitated materials were analysed by qRT–PCR, and abundances of the indicated mRNAs were plotted as a percent of the input mRNA level. The means ± SD from three independent experiments are shown. **P < 0.01 versus GAPDH mRNA. (B) Effects of AGO2 knockdown on mRNA decay kinetics. K562 cells were transiently transfected with two plasmids expressing distinct AGO2-directed shRNAs. Upper panel: western blot analysis of AGO2 knockdown. A 2-fold dilution series of lysate from cells expressing control shRNA was used to estimate AGO2 knockdown efficiency. Tubulin served as a loading control. Lower panel: 2 days after transfection of shRNA-expressing plasmids, actinomycin D was added to inhibit transcription. For each gene, levels of the endogenous mRNA were normalized to those of GAPDH mRNA and plotted as the percent mRNA remaining as a function of time following actinomycin D addition. Each time point represents mean ± SD from n ≥ 3 independent experiments for cells expressing control shRNA (solid circles, solid lines) or AGO2 shRNA (open circles, dotted lines). Non-linear regression analyses yielded first-order decay constants (k) and associated mRNA half-lives. Representative graphs are shown, and all data are tabulated in Table 2.
To examine whether AGO2 affects mRNA degradation, K562 cells were transiently co-transfected with two plasmids expressing distinct shRNAs against AGO2 (31). Western blot analysis indicated ∼75% knockdown of AGO2 (Figure 5B, top panel). Decay kinetics of endogenous mRNAs were then evaluated and compared with mRNA decay kinetics from cells transfected with control shRNA plasmid. AGO2 knockdown extended the half-lives of IFI16, FBXO24, HOXB8 and DOHH mRNAs by 2- to 3-fold (Figure 5B and Table 2), and it extended the half-life of MYC mRNA by 60% (Table 2; 0.48 versus 0.77 h; P < 0.01). AGO2 knockdown did not affect the half-lives of IL1RN, THRAP5, NDOR1 or GNLY mRNAs (Figure 3B and Table 2). This was not unexpected, as AGO2 did not associate with IL1RN or GNLY mRNAs (Figure 5A), and THRAP5, NDOR1 and GNLY mRNAs were stable, even when AGO2 was present (Table 2). However, the mRNA decay assays collectively indicated that rapid turnover of IFI16, FBXO24, HOXB8 and DOHH mRNAs required both AUF1 and AGO2. Thus, each is necessary but not sufficient for decay of these mRNAs.
If both AUF1 and AGO2 are required for degradation of at least some AUF1 target mRNAs, they may act as cofactors. The next experiments examined the effects of joint knockdown of AUF1 and AGO2 on mRNA decay. K562 cells were co-transfected with plasmids encoding shRNAs against AUF1 and AGO2. Western blot confirmed reductions in protein levels, although AGO2 reduction was less effective in cells co-transfected with shAUF1 + shAGO2 plasmids (Figure 6A); this type of effect has been observed before and may reflect competition of multiple shRNAs for processing and/or loading of the processed siRNA into RISC (55). Decay kinetics of endogenous mRNAs were then evaluated and compared with mRNA decay kinetics from cells transfected with control shRNA plasmid. Representative decay graphs are shown in Figure 6B. All data are tabulated in Table 2 to allow comparisons with single knockdown of AUF1 or AGO2. The conclusion from these experiments is that, for the majority of mRNAs subject to control of mRNA decay, joint knockdown of AUF1 and AGO2 had the same mRNA stabilizing effect as knockdown of either one singly, i.e. joint knockdown stabilized IFI16, IL1RN, FBXO24, HOXB8 and DOHH mRNAs 2- to 3-fold.
Figure 6.
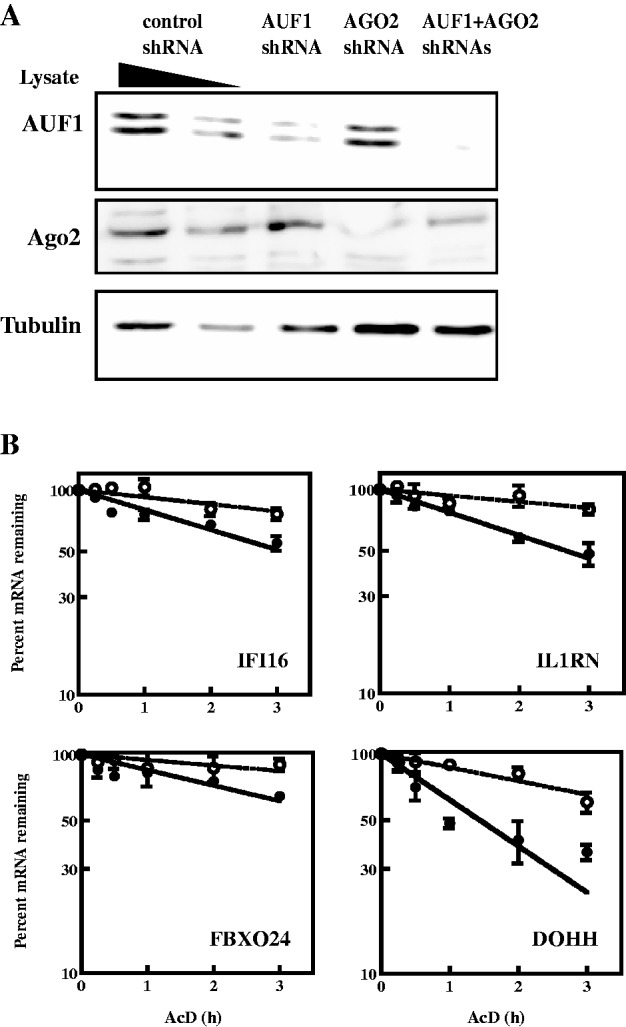
Effects of joint knockdown of AUF1 and AGO2 on degradation of mRNAs in K562 cells. Plasmids encoding shRNAs against AUF1 and/or AGO2 were transiently co-transfected. (A) Western blots of AUF1 and AGO2 protein levels in serially diluted lysates of cells expressing control shRNA were used to estimate knockdown efficiencies in cells expressing shRNA against AUF1 and/or AGO2, as indicated for each lane. Tubulin served as a loading control. (B) Two days after transfection of shRNA-expressing plasmids, actinomycin D was added to inhibit transcription. Levels of mRNAs were normalized to those of GAPDH mRNA and plotted as the percent mRNA remaining as a function of time following actinomycin D addition. Each time point represents mean ± SD from n ≥ 3 independent experiments for cells expressing control shRNA (solid circles, solid lines) or AUF1 + AGO2 shRNAs (open circles, dotted lines). Non-linear regression analyses yielded first-order decay constants (k) and associated mRNA half-lives. Representative graphs are shown, and all data are tabulated in Table 2.
There were two exceptions. One was MYC mRNA. As shown earlier, although knockdown of AUF1 had no effect on mRNA decay, knockdown of AGO2 increased mRNA half-life to 0.77 h from 0.48 h in control shRNA-transfected cells (Table 2; P < 0.01). However, knockdown of both AUF1 and AGO2 reduced mRNA half-life to a value statistically indistinguishable from that in cells transfected with control shRNA (Table 2; 0.53 versus 0.48 h, respectively; P > 0.05), i.e. joint knockdown restored rapid mRNA decay (see ‘Discussion’ section). The other exception was NDOR1 mRNA. Compared with decay of NDOR1 mRNA induced by knockdown of AUF1 alone, knockdown of both AUF1 and AGO2 stabilized the mRNA >2-fold (Table 2; 4.3 versus 10 h, respectively, P < 0.05). Thus, AUF1 may normally promote stabilization of NDOR1 mRNA by somehow inhibiting the ability of AGO2 to promote decay of the mRNA (see ‘Discussion’ section).
Effects of AUF1 and AGO2 abundance on their relative interactions with mRNAs
The results, so far, indicated the following: (i) AUF1 and AGO2 both bound a subset of the AUF1 target mRNAs examined; (ii) for some of these AUF1 targets, knockdown of either AUF1 or AGO2 stabilized the mRNA, suggesting they may act as cofactors; and (iii) AUF1 binding stabilized NDOR1 mRNA. These results suggest pleiotropic effects of AUF1 and AGO2 on decay of their target mRNAs, and perhaps, complex functional interactions between these two proteins. A precedent for this is the observation that AUF1 and the ARE-binding protein HuR act as cofactors to recruit AGO2-RISC to p16INK4a mRNA for its degradation (49). As such, the next series of experiments assessed the effect of suppressing AUF1 or AGO2 expression on mRNA target binding by the remaining factor.
K562 cells were transiently transfected with control shRNA or shRNA against either AUF1 or AGO2, and mRNP-immunoprecipitations were performed for the other protein. qRT–PCR was used to detect the nine AUF1 target mRNAs. MYC again served as a positive control, as it binds both AUF1 and AGO2 (43,51). AUF1 knockdown increased AGO2 association with MYC mRNA >2-fold compared with cells expressing control shRNA (Figure 7A, P < 0.05). Functionally, this is consistent with the observation that knockdown of AUF1 reduces translation of MYC mRNA (43). By contrast, AUF1 knockdown decreased AGO2 association with HOXB8 mRNA by 2-fold (Figure 7A, P < 0.05). This is consistent with the increase in half-life of HOXB8 mRNA on AUF1 knockdown (Table 2). Finally, knockdown of AUF1 had no effect on AGO2 association with IFI16, FBXO24, THRAP5, NDOR1 or DOHH mRNAs, suggesting that AGO2 binding to these mRNAs can occur independently of AUF1 (Figure 7A).
Figure 7.
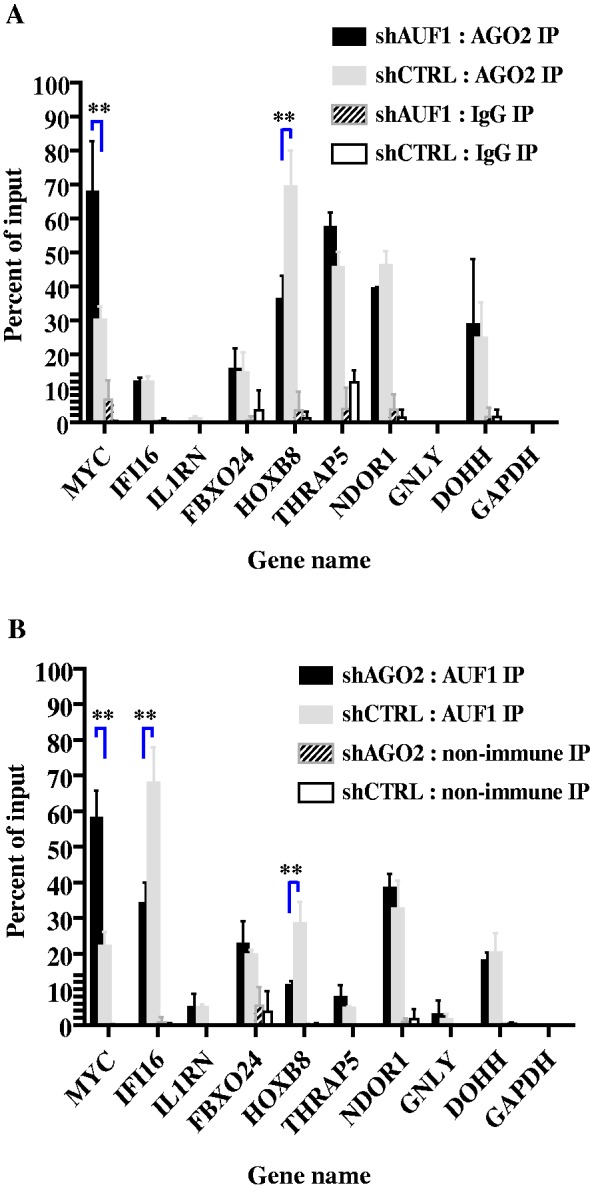
Effects of AUF1 and AGO2 abundance on mRNA binding. K562 cells were transiently transfected with plasmids encoding control shRNA or shRNA against either AUF1 or AGO2. Two days after transfection of shRNA-expressing plasmids, lysates were prepared and fractionated by mRNP-immunoprecipitation with control antibodies or anti-AUF1 antibody (for AGO2-knockdown cells) or anti-AGO2 antibody (for AUF1-knockdown cells). Both input and immunoprecipitated materials were analysed by qRT–PCR, and abundances of the indicated mRNAs were plotted as a percent of the input mRNA level. The means ± SD from three independent experiments are shown. **P < 0.01 versus control shRNA. (A) Knockdown of AUF1 followed by AGO2 mRNP-immunoprecipitation. (B) Knockdown of AGO2 followed by AUF1 mRNP-immunoprecipitation.
In the converse experiment, AGO2 knockdown increased AUF1 association with MYC mRNA ∼3-fold (Figure 7B, P < 0.05). Thus, AUF1 and AGO2 binding to MYC mRNA was inversely proportional, i.e. reciprocal. This again is consistent with a role for AUF1 in MYC translation, as earlier work showed that AUF1 knockdown reduced MYC translation (43) and raises a potential role for AGO2 in this event. By contrast, AGO2 knockdown reduced AUF1 association with IFI16 mRNA (Figure 7B, P < 0.05). However, AUF1 knockdown did not affect AGO2 association with IFI16 mRNA (Figure 7A). Thus, AGO2 may facilitate AUF1 association with IFI16 mRNA, but not vice versa. Likewise, AGO2 knockdown reduced AUF1 association with HOXB8 mRNA (Figure 7B, P < 0.05). AGO2 and AUF1 association with HOXB8 mRNA thus seems proportional, and perhaps binding of one protein facilitates mRNA association by the other. This is again consistent with the increase in half-life of HOXB8 mRNA on knockdown of either AUF1 or AGO2 (Table 2). Finally, knockdown of AGO2 had no effect on AUF1 binding to FBXO24, THRAP5, NDOR1 or DOHH mRNAs (Figure 7B). Thus, binding of AUF1 and AGO2 to these four mRNAs seemed independent. Taken together, the results of Figure 7 indicated that depending on the mRNA, AUF1 and AGO2 binding can be reciprocal with each other, proportional to each other or independent of each other.
DISCUSSION
Table 3 summarizes the relationships between mRNA binding by AUF1 and AGO2 and their roles in mRNA degradation. Binding of AUF1 and AGO2 to MYC mRNA is reciprocal (Figure 7). Earlier work showed that AUF1 promotes MYC translation, as its knockdown reduced MYC translation (43). This effect is in part because of enhanced mRNA binding by translation suppressor protein T cell intracellular antigen-1-related protein (TIAR) on AUF1 knockdown. However, the earlier work also showed that joint knockdown of AUF1 and TIAR did not restore translation, suggesting a second suppressor (43). Indeed, subsequent work showed that the AUBP HuR recruits AGO2 to MYC mRNA, which suppresses MYC translation (51). Perhaps AUF1 and HuR also present competitive binding effects on MYC mRNA as a mechanism to control AGO2 occupancy on the mRNA. Nonetheless, AGO2 also exerts modest effects on MYC mRNA decay, as its knockdown increased mRNA half-life by 60% (Table 2). By contrast, AUF1 knockdown in cells did not affect MYC mRNA decay (Table 2). This latter result is consistent with the observation that AUF1 knockdown leaves AGO2 bound to MYC mRNA (Figure 7), where it remains poised to promote decay (and translational suppression) of the mRNA. However, joint knockdown of AUF1 and AGO2 decreased mRNA half-life compared with knockdown of AGO2 alone (Table 2). The simplest interpretation is that knockdown of both AUF1 and AGO2 allows compensating AUBPs to bind MYC mRNA and thereby recruit decay enzymes other than AGO2. Clearly, downstream biochemical characterization experiments will be required to assess what combinations of bound AUBPs are possible and the allosteric relationships between them.
Table 3.
Effects of AUF1 and AGO2 on post-transcriptional control of mRNAs
| mRNA | AUF1 and AGO2 association | Function |
|---|---|---|
| MYC | Reciprocal | Translation (AUF1) |
| Decay/translation (AGO2) | ||
| IFI16 | AGO2 facilitates AUF1 binding | Decay requires AUF1 and AGO2 |
| HOXB8 | AGO2 facilitates AUF1 binding and vice versa | Decay requires AUF1 and AGO2 |
| FBXO24 | Independent | Decay requires AUF1 and AGO2 |
| DOHH | Independent | Decay requires AUF1 and AGO2 |
| NDOR1 | Independent | Stability requires AUF1 |
| THRAP5 | Independent | Stable mRNA |
| IL1RN | AUF1 binding only | Decay requires AUF1 |
| GNLY | AUF1 binding only | Stable mRNA |
AUF1 and AGO2 binding is proportional/cooperative for HOXB8 and IFI16 mRNAs (Figure 7). miR-196a degrades HOXB8 mRNA via endoribonucleolytic cleavage, as the miRNA fully base pairs with the mRNA (although there is one G:U base pair) (13). Nonetheless, AUF1 knockdown reduced AGO2 binding (Figure 7) and increased half-life of the mRNA (Table 2). Two possible explanations are that direct AUF1-AGO2 protein–protein associations facilitate their co-binding to the mRNA, or that AUF1 binding to HOXB8 mRNA may expose the miR-196a binding site, much like RNA-binding protein Pumilio exposes miR-221/222 sites in p27Kip1 mRNA by altering local RNA structure (56). Consistent with the latter idea, AUF1 possesses an RNA chaperone-like activity that can remodel local RNA structure (23,57). In the case of IFI16 mRNA, however, AGO2 knockdown reduced AUF1 association with the mRNA, but in the converse experiment, knockdown of AUF1 did not affect AGO–mRNA association (Figure 7). Thus, AGO2–IFI16 mRNA association does not require AUF1. Nonetheless, AGO2 does require AUF1 as a cofactor to effect mRNA degradation, as knockdown of either protein increased mRNA half-life (Table 2). Perhaps AGO2 requires AUF1 to assist its recruitment of the CCR4-NOT deadenylase complex to the IFI16 mRNP (8–11).
AUF1 and AGO2 binding is independent for FBXO24, THRAP5, NDOR1 and DOHH mRNAs (Figure 7). THRAP5 mRNA is stable, and knockdown of AUF1 and/or AGO2 had no effect on mRNA half-life (Table 2). We speculate these proteins control translation of the mRNA, as they do for MYC mRNA (43,51). Both FBXO24 and DOHH mRNAs were unstable, and knockdown of either AUF1 or AGO2 stabilized these mRNAs (Table 2). Thus, AUF1 and AGO2 may be cofactors for promoting degradation of some mRNAs. Consistent with this idea, a recent report (54) showed that miR-331-3p and miR-642-5p control abundance of DOHH mRNA and protein in prostate cancer cells. This observation is in line with our results showing that AGO2 knockdown increased mRNA half-life >2-fold (Table 2). NDOR1 mRNA presented a special case: it is normally a stable mRNA, but AUF1 knockdown reduced its half-life >2-fold (Table 2); in the absence of AUF1, mRNA degradation required AGO2, as joint knockdown of AUF1 and AGO2 stabilized the mRNA (Table 2). This raises the intriguing question as to how AUF1 could block mRNA degradation activity programmed by AGO2 bound to NDOR1 mRNA.
Finally, AUF1 binds IL1RN and GNLY mRNAs, whereas AGO2 does not (Figures 2, 5, and 7). AUF1 is essential for IL1RN mRNA decay, as its knockdown increased mRNA half-life >4-fold; by contrast, GNLY mRNA is relatively stable, and AUF1 knockdown had no effect on mRNA half-life (Table 2). Thus, how does AUF1 promote decay of some mRNAs and not others? This will require analyses of the network of proteins bound to each mRNA, dictated, in part perhaps, by the first protein with the highest affinity for (or access to) mRNA sequence(s) that trigger hierarchical binding of additional proteins that together allow mRNA decay, stability and/or translation.
Given the apparent complex relationships demonstrated here between AUF1 and AGO2 association with mRNAs, and their effect on mRNA decay, future work will address four areas: (i) to determine where on the mRNAs both AUF1 and AGO2 bind using techniques, such as photoactivatable ribonucleoside-enhanced–cross-linking and immunoprecipitation (PAR-CLIP) and related methods (58–61), and whether AUF1-AGO2 physically associate; (ii) to identify additional RNA-binding proteins associated with AUF1 target mRNAs and where they bind (as noted earlier in the text); (iii) to determine whether binding of one (or more) proteins alters RNA presentation to allow binding by others, including AGO2 and perhaps miRISC; and (iv) to consider whether there are ‘mRNA codes’ that programme the variety of effects observed here. In addition to its well characterized binding to AREs (17,22), AUF1 might bind a variety of sequences as well (manuscript in preparation). It is unlikely that one or another mRNA-binding mechanism (reciprocal/competitive, proportional/cooperative or independent) will be restricted to any one type of sequence element. Thus, it is likely that putative codes might reside in RNA secondary/tertiary structures or within mRNP structures that await identification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
National Institutes of Health (NIH) [T32 GM066706 to B.E.Z., AI057596 to Y.S., R25 GM055145 to P.I.B., CA091358 to J.W., CA102428 to G.M.W. and CA052443 to G.B.]; American Heart Association [11PRE6900008 to B.E.Z.]. Funding for open access charge: NIH [R01 CA052443].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Rob Muldowny and Peter Kahn, Department of Computer Science, Rutgers University, New Brunswick, NJ, USA, for their computer programming expertise, and Shobha Vasudevan, Massachusetts General Hospital, Boston, MA, USA, for the shAGO2 plasmids.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 3.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 12.Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 14.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell. Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George AD, Tenenbaum SA. MicroRNA modulation of RNA-binding protein regulatory elements. RNA Biol. 2006;3:57–59. doi: 10.4161/rna.3.2.3250. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–1019. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 20.Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:D137–D140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 22.Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley interdisciplinary reviews. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J. Biol. Chem. 2010;285:39127–39139. doi: 10.1074/jbc.M110.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson GM, Sutphen K, Chuang K, Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J. Biol. Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya S, Giordano T, Brewer G, Malter JS. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 1999;27:1464–1472. doi: 10.1093/nar/27.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonnstedt I, Britton T. Hierarchical Bayes models for cDNA microarray gene expression. Biostatistics. 2005;6:279–291. doi: 10.1093/biostatistics/kxi009. [DOI] [PubMed] [Google Scholar]
- 28.Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, Maher LR, Scrudato S, Rivera YM, Gupta S, et al. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol. Cell. Biol. 2008;28:5223–5237. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J. Exp. Med. 1953;97:695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 31.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene. 1998;17:931–939. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- 33.Ysla RM, Wilson GM, Brewer G. Chapter 3. Assays of adenylate uridylate-rich element-mediated mRNA decay in cells. Methods Enzymol. 2008;449:47–71. doi: 10.1016/S0076-6879(08)02403-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol. Cell. Biol. 2011;31:3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar S, Han J, Sinsimer KS, Liao B, Foster RL, Brewer G, Pestka S. RNA-binding protein AUF1 regulates lipopolysaccharide-induced IL10 expression by activating IkappaB kinase complex in monocytes. Mol. Cell. Biol. 2011;31:602–615. doi: 10.1128/MCB.00835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J. Interferon Cytokine Res. 2008;28:679–691. doi: 10.1089/jir.2008.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brewer G, Saccani S, Sarkar S, Lewis A, Pestka S. Increased interleukin-10 mRNA stability in melanoma cells is associated with decreased levels of A + U-rich element binding factor AUF1. J. Interferon Cytokine Res. 2003;23:553–564. doi: 10.1089/107999003322485053. [DOI] [PubMed] [Google Scholar]
- 39.Sadri N, Lu JY, Badura ML, Schneider RJ. AUF1 is involved in splenic follicular B cell maintenance. BMC Immunol. 2010;11:1. doi: 10.1186/1471-2172-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadri N, Schneider RJ. Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J. Invest. Dermatol. 2009;129:657–670. doi: 10.1038/jid.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steenman M, Chen YW, Le Cunff M, Lamirault G, Varro A, Hoffman E, Leger JJ. Transcriptomal analysis of failing and nonfailing human hearts. Physiol Genomics. 2003;12:97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 42.Hwang DM, Dempsey AA, Wang RX, Rezvani M, Barrans JD, Dai KS, Wang HY, Ma H, Cukerman E, Liu YQ, et al. A genome-based resource for molecular cardiovascular medicine: toward a compendium of cardiovascular genes. Circulation. 1997;96:4146–4203. doi: 10.1161/01.cir.96.12.4146. [DOI] [PubMed] [Google Scholar]
- 43.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 44.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 46.Dempsey LA, Li MJ, DePace A, Bray-Ward P, Maizels N. The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics. 1998;49:378–384. doi: 10.1006/geno.1998.5237. [DOI] [PubMed] [Google Scholar]
- 47.Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol. Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol. Cell. Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J. Biol. Chem. 2010;285:20940–20951. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, Elia A, Kress TR, Dickens M, Clemens MJ, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc. Natl. Acad. Sci. USA. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao JM, Lu H. Autoregulatory suppression of c-Myc by miR-185-3p. J. Biol. Chem. 2011;286:33901–33909. doi: 10.1074/jbc.M111.262030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Epis MR, Giles KM, Kalinowski FC, Barker A, Cohen RJ, Leedman PJ. Regulation of Expression of Deoxyhypusine Hydroxylase (DOHH), the Enzyme That Catalyzes the Activation of eIF5A, by miR-331-3p and miR-642-5p in Prostate Cancer Cells. J. Biol. Chem. 2012;287:35251–35259. doi: 10.1074/jbc.M112.374686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 57.Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 2003;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- 58.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP–transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J. Vis. Exp. 2011;pii:2638. doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Wen J, Parker BJ, Jacobsen A, Krogh A. MicroRNA transfection and AGO-bound CLIP-seq data sets reveal distinct determinants of miRNA action. RNA. 2011;17:820–834. doi: 10.1261/rna.2387911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


