Abstract
Messenger RNA is a key component of an intricate regulatory network of its own. It accommodates numerous nucleotide signals that overlap protein coding sequences and are responsible for multiple levels of regulation and generation of biological complexity. A wealth of structural and regulatory information, which mRNA carries in addition to the encoded amino acid sequence, raises the question of how these signals and overlapping codes are delineated along non-synonymous and synonymous positions in protein coding regions, especially in eukaryotes. Silent or synonymous codon positions, which do not determine amino acid sequences of the encoded proteins, define mRNA secondary structure and stability and affect the rate of translation, folding and post-translational modifications of nascent polypeptides. The RNA level selection is acting on synonymous sites in both prokaryotes and eukaryotes and is more common than previously thought. Selection pressure on the coding gene regions follows three-nucleotide periodic pattern of nucleotide base-pairing in mRNA, which is imposed by the genetic code. Synonymous positions of the coding regions have a higher level of hybridization potential relative to non-synonymous positions, and are multifunctional in their regulatory and structural roles. Recent experimental evidence and analysis of mRNA structure and interspecies conservation suggest that there is an evolutionary tradeoff between selective pressure acting at the RNA and protein levels. Here we provide a comprehensive overview of the studies that define the role of silent positions in regulating RNA structure and processing that exert downstream effects on proteins and their functions.
INTRODUCTION
Sequencing of multiple genomes in recent decades revealed that the number of protein-coding genes in multicellular organisms is surprisingly low compared with the variety of biological functions performed by these proteins and the resulting physiological and morphological complexity of higher eukaryotic species (1–4). Such a major increase in functional complexity is largely generated at two fundamental levels: (i) transcriptional and post-transcriptional control that regulates differential gene expression, alternative transcription and splicing, and (ii) post-translational modifications that affect protein structure, function and metabolic fate, and facilitate a large variety of functions performed by these proteins in vivo (5–8). Prominently, events that occur in between these two levels of regulation and involve all the steps that lead from mRNA to protein have not been factored into this complexity in earlier studies.
Until recently, mRNA has been viewed solely as a carrier of the genetic code, transmitting information about the primary amino acid sequence from genes to proteins. Recent studies reveal a surprisingly important role of mRNA in the regulation of biological complexity. As we now know, mRNA is a key component of an intricate regulatory network of its own, which is different from the networks and pathways involved in DNA and protein regulation. Eukaryotic organisms carry multiple regulatory and structural signals in mature mRNA and pre-mRNA, delineated along the protein-coding and non-coding regions in complex overlapping manner. The key provision that enables mRNA to carry these regulatory functions is the redundancy of the genetic code that allows for many synonymous nucleotide substitutions that do not change amino acid sequences of the encoded proteins and therefore often called ‘silent’ mutations. Synonymous nucleotide substitutions due to mutagenesis, errors in splicing and RNA editing can confer dramatic differences to the structure and function of mRNA itself that provide diverse possibilities for the regulation of gene expression patterns (7–10). Within the protein-coding regions (CDSs), the redundancy of the genetic code allows for the overlap in encoding amino acid sequences and RNA functional and structural signals, especially at the key structural and reference sites, such as the vicinity of the start and stop codons (10) as well as the exon–intron boundaries (11). The question to what purpose and extent do the genomes exploit their non-coding potential is still open (4,12).
There are several well-documented ways in which synonymous sites exert their impact on gene functions: effect on mRNA splicing, mRNA folding, stability and regulation of translation through utilization of preferred synonymous codons that translate more efficiently and accurately. Additional and sometimes opposing selective forces appear to affect codon frequency as well. Previous findings show roles for synonymous positions in RNA–RNA interactions, which influence the translation efficiency, and in RNA–RNA cross-talk, which is a key to biological regulation of expression and transcriptome complexity (13–15). Emerging evidence shows that ‘silent’ substitutions carry a wealth of information, which is written over the encoded amino acid sequence, and that this information can be used to regulate translation speed, protein homeostasis, metabolic fate and even post-translational modifications, which will be discussed in this review. Here we will focus on the RNA level of regulation and the role of synonymous sites and mRNA structure in generating biological complexity.
SYNONYMOUS SITES AND CODON USAGE AFFECT GENE EXPRESSION
Although the genetic code is generally conserved among organisms, synonymous codons in different species are used with different frequencies—a trend commonly defined as codon usage bias. Codon usage bias reflects selection for optimization of the translation process by tRNA abundance in many organisms. However, other different factors such as GC nucleotide composition (16), RNA stability and folding (10), local RNA secondary structures (17), mRNA longevity (18), protein structure (19), compositional strand bias (20) and strand asymmetry induced by transcription-coupled repair (21) have also been proposed to affect nucleotide preferences at synonymous sites (9,22). Some of these factors are universal, whereas other factors act at specific levels of biological organization or under specific conditions.
Synonymous sites are not neutral
The neutral theory maintains that codon preferences exist because of the differences in codon mutability and most synonymous mutations spread to fixation by chance, and, therefore, have no effect on the fitness of organisms (23). However, a new wave of evidence for widespread selection pressure on the nucleotide level in the eukaryotic genomes and demonstration of the importance of synonymous positions for regulation of translation and splicing (8–10,24–29) cast doubt on the statement of the neutral theory (23). These observations support the theory that synonymous positions are under selection and codon bias is maintained by a balance between selection, mutation and genetic drift (30–32).
GC content is a significant feature affecting codon preferences in different organisms (11,25,28). Across many species (675 bacteria, 52 archea and 10 fungi), the differences in codon usage can be predicted from the nucleotide content of their non-coding sequences (33). However, GC content is determined not solely by genome-wide requirements, but also by selective forces that act on the coding regions (22). Indications of selection on synonymous positions were noted in Drosophila melanogaster and Caenorhabditis elegans, where most of the third positions in optimal codons contain a cytosine or guanine (32). Similarly, codon usage in mammals is obviously non-random due to elevated frequencies of G and C at synonymous sites (9,34). In different species, greater GC content at synonymous positions in the coding regions compared with the flanking introns could indicate selection at synonymous sites (34,35) and could be considered as a major factor of evolution (see ‘Evolution’ section). A pattern of polymorphism in GC-rich human genes, which is unexplained in the framework of the mutation bias hypothesis, is consistent with the action of natural selection or biased gene conversion (36). In mammals, synonymous sites within the first exons are more GC-rich than within the last exons of the genes, a feature, likely, relevant to translation regulation, whereas there is no difference between GC contents of first and last introns of genes (34). Different patterns in codon bias have also been observed at the beginning and at the end of bacterial genes (37).
At the mRNA level, synonymous positions were found to control folding, stability and secondary structures of mRNAs in different organisms and affect translation efficiency and post-translational regulation through mRNA–RNA and mRNA–protein interactions. Some of these structural and regulatory RNA features are defined by local nucleotide content, and codon preferences within specific genes, as well as across genes within the genome (9,10).
It is well established that synonymous codons are used non-randomly and can drive translational selection and affect codon preference in many organisms (22). It is difficult to explain by mutational pressure alone why preferred codons are recognized by more abundant tRNA molecules, or how the strong variability of codon bias across genes within the genome is maintained, where more pronounced codon usage bias is characteristic for highly expressed genes. The level of gene expression correlates strongly with codon bias in many prokaryotes and eukaryotes, while co-expressed genes have similar synonymous codon usages within the genomes of human, yeast, worms and bacteria (38). These observations suggest a role for synonymous position in the regulation of translation and support the notion that synonymous positions are not neutral.
Codon usage and selection for translation efficiency and accuracy
Expression level is an important determinant of protein evolution rates (39,40), and translational selection is one of the most important driving forces in evolution (22). Earlier studies considered codon selection for maximization of the translational efficiency under conditions when selection favors rapid translation and the relevant iso-acceptor tRNAs might not be equally abundant (22). Under such conditions, a pressure exists to use the codons that match the most abundant tRNAs to facilitate translation. Utilization of common or rare codons can significantly affect the rate of ribosome translocation through mRNA, as the limited availability of the corresponding aminoacyl-tRNAs is expected to cause delays and ribosome stalling at the rare codon sites. Differential codon usage is associated with varying expression rates in many organisms (9). Positive correlation between codon usage bias and gene expression level was established in bacteria (41,42) yeast (41), nematode (43) and insect (44). As expected, bias in favor of preferred codons is more pronounced in highly expressed genes and mostly observed in prokaryotic species with large populations, although some prokaryotes do not show any clear signs of selection for translation efficiency (45).
Recent experiments led to the conclusion that redundancy in the genetic code allows translation of synonymous but differentially coded mRNAs at different rates, even with fixed tRNA usage (46–48). Codon usage can significantly affect the speed of translation elongation in bacteria. In Escherichia coli, the rate for aminoacyl-tRNA association with different codons spans a 25-fold range and preferred codons accept aminoacyl-tRNAs faster than more rarely used codons (49). The use of common codons can increase the rate of translation elongation several folds, compared with the rare ones (50). In bacteria, codon usage represents an adaptation in those species that undergo rapid environmental changes and has been directly linked to changes in protein expression (38). In some fungi, natural selection also generally favors optimal codon variants, but fixation of optimal codons is reduced in rapidly evolving long genes (51).
A more complex picture emerges in mammalian species, where evidence supporting translational selection of codon choice is arguable (9,52,53). Experimental evidence was reported that tRNA content in rabbit reticulocytes is specialized for the synthesis of hemoglobin, which constitutes >80% of total protein expression in these cells (54,55). However, no correspondence between the usage of a codon in human protein-coding sequences and the abundance of iso-accepting tRNA has been found in several studies on the genome level (32,56–59). It was shown that translation selection, when co-adaptation of specific tRNA gene copy numbers and codon usage across genomes considered, is more than 10 times lower for mammalian than for non-mammalian organisms, such as E. coli, yeast and worms (52). Only a weak correlation was found between expression level and frequency of optimal codons for human genes (60). Similarly, a weak correlation between levels of gene expression and amino acid composition, accountable for ∼10% of the variation in expression levels, was reported recently for mouse protein-coding genes (61). This is not surprising, as the identity and diversity of the optimal codons in mammalian genomes is determined largely by the majority of genes, on which selection is much weaker, whereas selection for the use of optimal codons is strongest in highly expressed genes (33).
When most of the genes seem to be under selection to increase usage of the preferred codons, some genes undergo opposite selection (62). There is an advantage to use rare codons in certain positions where they have a potential to slow down translation rate, especially at the elongation stage, because of the relatively longer time of rare tRNA delivery (46). Rare codons are biased in lowly expressed genes in several genomes, including humans (60). In line with this, different protein structural elements are associated with specific codon usage: α-helical regions are enriched by common (fast translated) codons, whereas disordered and β-sheets structures are mostly encoded by rare codons (63). Thus, rare codons likely provide an opportunity for translation pause and allow the translated segment of the protein to be folded properly without potentially interfering with the downstream segments that have not been translated yet (64).
Selection on codon bias may also increase translation accuracy (65) because selection favors optimal codons at sites where changes are most likely to disrupt protein functions (44). Significant association of evolutionary conserved regions with optimal codons was found in many different species on the transcriptome level (65–67). Some studies suggest that selection for translation accuracy might be required to prevent protein misfolding errors leading to the loss of functional protein molecules (65). This idea is supported by the observation that buried amino acids, responsible for protein folding, are preferentially encoded by more optimal codons, compared with surface residues, which participate in intermolecular interactions (68).
Determination of the roles of synonymous positions on the multiple levels of protein regulation is a highly dynamic rapidly emerging field. Notably, these roles appear to be different in prokaryotes and eukaryotes. It is clear that protein-coding sequences in higher eukaryotes require diversification for functional integrity, and this is achieved by the use of different codons in their variable and constitutive regions through different selection mechanisms (69). Thus, a vast body of recent evidence demonstrates that nucleotide preferences in synonymous positions contribute to the efficiency and accuracy of protein expression, and a bias for preferred synonymous nucleotides is generated and maintained by selection (22,31,32,70).
More than codon usage
A recent study reviewing codon usage bias in hundreds of prokaryotic genomes revealed that this bias is highly variable in different prokaryotes, ranging from high degrees of differential use of synonymous codons among different genes to virtually none (71). As mentioned previously, this parameter was found to correlate with the range of habitats for particular organisms: those with the necessity to adapt to a variety of environments (including pathogens) demonstrated a higher extent of codon usage bias compared with those organisms that live only in a particular habitat. Thus, in prokaryotes, codon usage appears to represent an adaptation measure that can regulate the overall ability of the organism to undergo rapid changes under the pressure of each particular environment (71). Perturbing the codon usage directly affects the level or even direction of changes in protein expression in response to environmental stimuli. It has been shown for different prokaryotic and eukaryotic species that codon usage is universally function-specific and cells may need to dynamically alter their intracellular tRNA composition to adapt to their new environment or adopt a novel physiological role (38).
Apart from mRNA, translation efficiency depends on another essential player: tRNA. Transport RNA gene content is a key factor that defines the efficiency of the translation machinery. Remarkably, repertoire of tRNA genes varies greatly between different organisms (72–75). Certain tRNA species are absent in entire branches of the phylogenetic tree, whereas others are clearly predominant. For example, in Homo sapiens, 29 of the 43 tRNAAla genes (68%) correspond to the iso-acceptor tRNAAlaAGC. Similar relationships were reported for bacterial species, and the underlying reasons are poorly understood. A recent study tracing the correlation between two tRNA modifications in base 34 of the anticodon that increase codon-pairing ability, mediated by tRNA-dependent adenosine deaminases and uridine methyltransferases (76), found that the emergence of these modifications likely played a role in shaping of genomes and directing evolution of many species (77). Comparison of more than 500 different genomes showed that these two modifications likely define patterns of gene expression that correlate with the separation of living organisms into archaea, bacteria and eukaryotes (77). This study presents an entirely different angle in viewing the relationship between coding sequence and gene expression, and defines a novel feature of pro- and eukaryotic codon usage driven by tRNA modifications.
Moreover, not only codon usage, but also codon context or the positioning of the particular codons in relation to their neighbors (i.e. codon pair usage) is subject to evolutionary pressure and apparently plays an important role in mRNA translation. Comparison of codon context for multiple genes in several eukaryotic species showed that both synonymous and non-synonymous mutations are selected to maintain context biases (78). These data are in agreement with an observation that the amino acid replacement changes can disrupt the codon context sufficiently to increase the probability of fixation of subsequent silent changes in adjacent codons (79). In vivo studies provided evidence for the role of codon context in decoding fidelity and efficiency in different organisms, suggesting that codon context modulates evolution of the primary nucleotide sequence in the protein-coding genes and fine-tunes the structure of the open reading frames to ensure fidelity and efficiency of genome architecture (10,80).
In summary, many factors determine the choice of codons, and selection on the codon bias likely acts at both the transcriptional and translational level. tRNA relative abundance, modifications and codon usage could drive each other to synergistically optimize the efficiency of gene expression. Elevated GC content of synonymous positions in many eukaryotic and prokaryotic genomes suggests that the RNA-level selection pressure contributes to codon preferences. Local codon context or positioning of particular codons in relation to their neighbors also might help to accommodate diverse regulatory signals and RNA structural elements in the protein-coding regions.
Unlike prokaryotes, eukaryotic organisms appear not to use codon usage bias as a dominant mechanism of regulation of protein expression. Instead, codon preferences are used to accommodate diverse regulatory elements responsible for the variability of molecular and cellular mechanisms and to provide new level of the biological complexity, especially in protein-coding regions of higher eukaryotes.
ROLE OF SYNONYMOUS POSITIONS IN mRNA FOLDING, STABILITY AND PROTEIN FATE
mRNA secondary structure and regulation of translation
In 1972, White et al. (81) suggested that redundancy in the genetic code permits extensive variation of the nucleotide sequence and satisfies the requirements for both protein and RNA structure. Fitch (82) found first evidence that degeneracy of the genetic code is used to optimize base-pairing in mRNA molecules. Since then, the idea that redundancy of the genetic code allows preservation of mRNA folding has been supported by several lines of evidence that are discussed in this and following sections.
Single-stranded mRNA molecules form secondary structures through complementary self-interactions. Formation of RNA structures is dependent on the primary nucleotide sequence and folding environment, and is often defined by the longer-range interactions between the nucleotides. Evolutionarily conserved local secondary structures were described in eukaryotic and mammalian mRNAs and pre-mRNAs (83). Synonymous substitutions affect mRNA translation in different organisms (41,50,84). They can induce significant changes in the mRNA folding, causing formation of new stable hairpin loops and elements of higher-order folding. Recent studies suggest that the placement of stable structural elements within the mRNA sequence is far from random, and propose that transient ribosome stalling at key mRNA regulatory sites can affect protein abundance, folding and even post-translational modifications, as is discussed in the following sections. Stable structural elements can significantly affect translation initiation and ribosome translocation, inducing ribosome pausing and stalling that could considerably delay the overall progress of protein synthesis and folding of nascent polypeptides. Strong mRNA secondary structures formed due to gene-specific codon usage have been implicated in discontinuous translation and pauses in synthesis of insect silk fibroin, chicken collagen and other proteins (85,86). Although stable secondary structures capable of interfering with translation are generally avoided in mRNA coding regions (87), significant biases in favor of local RNA structures have been found in several bacterial species and yeast (17). Native mRNAs have a lower calculated folding free energy than random sequences (88), and correlations between mRNA and protein secondary structures have been noted (19). It was suggested that elevated C content at the third synonymous sites that stabilize RNA secondary structures (89) creating translational pauses is driven by usage of different encoded amino acids in alpha-helices, beta-sheets and disordered structures, which require different folding time. This phenomenon is associated with differential codon usage, as discussed in the previous section.
Periodic pattern of mRNA folding in protein-coding regions
Pronounced periodic pattern of mRNA secondary structure, stability and nucleotide base-pairing was found in the mammalian coding regions (Figure 1). This pattern is created by the structure of the genetic code, and the relative abundance of dinucleotides is important for its maintenance (10). Although synonymous codon usage contributes to this pattern, even in the absence of codon bias, such pattern can be observed at the degenerate codon sites. While all codon sites are important for the maintenance of mRNA secondary structure, degeneracy of the code allows regulation of stability and periodicity of mRNA secondary structure. Synonymous codon sites contribute most strongly to mRNA stability, and base-pairing at the third codon positions is significantly higher than at other codon sites in mammalian transcriptomes (Figure 1). Similar periodicities of mRNA stability were theoretically predicted in bacterial, yeast, worm and fly transcripts (90). These results convincingly support the hypothesis that redundancies in the genetic code allow transcripts to satisfy the requirements for both protein and RNA structure. The RNA-level selection on synonymous positions maintains a more stable and ordered mRNA secondary structure, which is likely to be important for the transcript stability and translation (10).
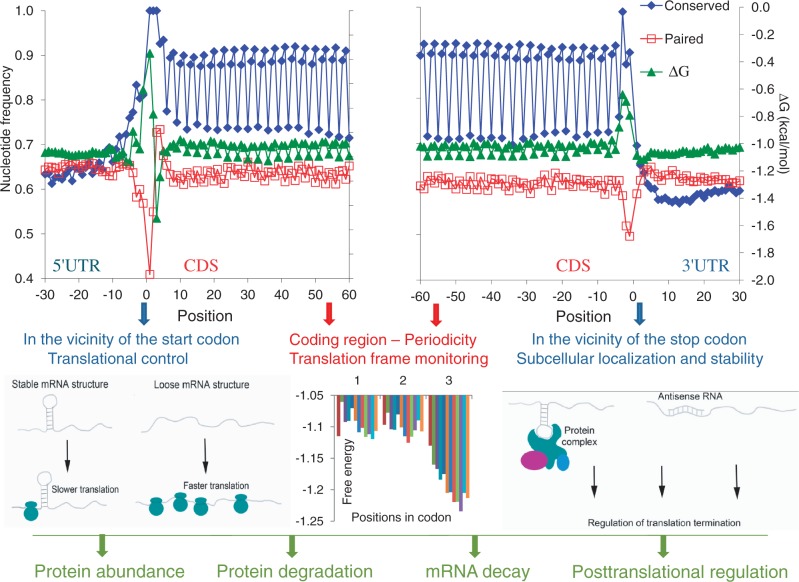
Figure 1.
Periodic pattern of nucleotide involvement in secondary structure formation and sequence conservation around the start and the stop codons in human mRNAs. Positions from −30 to −1 correspond to 5′-UTRs and positions from 1 to 60 correspond to CDSs (upper left panel). Positions from −60 to −1 correspond to CDSs and positions from 1 to 30 correspond to 3′-UTRs (upper right panel). Blue, sequence conservation in 6919 orthologous human and mouse mRNAs. Red, base-paired nucleotides in 19 317 human mRNAs. Green, free Gibbs energy of base-pairing in 19 317 human mRNAs. Structural features of the untranslated regions (UTRs) and coding sequences (CDSs) have a major role in the control of mRNA translation. The relaxed secondary structures in UTRs are common for many mRNAs and involved in regulation of initiation (low left panel) and termination (low right panel) of translation. Periodic pattern in the CDS is likely responsible for translation frame monitoring (low center panel). Groups of genes with distinct levels of expression (114) are presented in different colors. This figure is adapted from (10).
Recent application of Parallel Analysis of RNA Structure (PARS) at single-nucleotide resolution to profiling of mRNA secondary structure in budding yeast Saccharomyces cerevisiae confirmed in silico predictions of the three-nucleotide periodicity of secondary structure across the coding regions and the existence of a more stable secondary structure in the coding versus untranslated regions (91,92).
mRNA secondary structure in the vicinity of the start and stop codons
Genome-wide analysis of eukaryotic mRNAs revealed distinct patterns of evolutionary conservation at the boundaries of the untranslated and coding regions. Conservation patterns at the synonymous positions in eukaryotes are more pronounced at the ends of the CDS, in the vicinity of the start and stop codons [Figure 1, (93)]. Elevated sequence conservation at synonymous positions likely reflects increased selection pressure on the structural features in these regions. The start and stop codons of mammalian transcripts mostly reside in the unpaired regions of evolutionary conserved mRNA stem-loop structures (10). At the same time, functional mRNA domains (5′-UTRs, CDSs and 3′-UTRs) preferentially fold onto themselves, with likely cross-domain (UTR-CDS) interactions in their vicinity. Such distinct folding patterns and placement of the start and stop codons into relaxed structures likely facilitate efficient initiation and termination of translation (10). This trend of relaxed mRNA secondary structure near translation start codon was confirmed in other eukaryotic and prokaryotic species (93,94). This is a characteristic feature of highly expressed secretory proteins that tend to have relaxed secondary structure within the first 30 bases of their open reading frames (92). An anti-correlation between the mRNA translation efficiency and the stability of the structure in the vicinity of the translation start site was experimentally confirmed in yeast (92).
The effect of mRNA folding on the rates of translation initiation and protein expression level was studied in E. coli. Expression of coding variants of the green fluorescent protein in a synthetic library of 154 genes that varied randomly at the synonymous sites, but had the same amino acid sequence, showed 250-fold variations in protein expression levels (95). Stability of mRNA folding near the ribosomal binding site appeared to be the defining factor that could explain more than half of the variation in the protein levels, whereas codon usage bias did not correlate with gene expression. The results of this analysis suggest that mRNA folding and associated rates of translation initiation play an important role in shaping protein expression levels. Experimental studies of individual genes support in silico predictions and demonstrate the importance of the mRNA folding in the vicinity of the start codon. An interesting example involves catechol-O-methyltransferase (COMT) (96), a major enzyme controlling catecholamine levels that plays a central role in pain perception and cognition (97). One of the common, in the human population, COMT haplotypes carries the non-synonymous variation C(166)T within the upstream coding region of the RNA transcript. This haplotype codes for a less stable protein that exhibits an elevated protein expression in vitro (97), which would compensate for lower protein stability. It appears that structural destabilization near the start codon in the T allele mRNA could be related to the observed increase in the COMT expression. Folding simulations of the tertiary mRNA structures demonstrate that this destabilization lowers the folding transition barrier, thus decreasing the probability of occupying its native state. These data suggest a structural mechanism whereby functional synonymous variations near the translation initiation site affect translation efficiency through entropy-driven changes in mRNA dynamics and present an example of stable compensatory genetic variations in the human population.
Another case of the structure-dependent regulation involves mRNA sequences encoding leader peptides. Although traditionally it has been believed that the sole purpose of the leader sequences is to target proteins to the appropriate intracellular destinations, recent studies suggest that the leader sequence carries information on RNA secondary structure in the translation initiation region that may help to control the rate and speed of translation initiation. This is illustrated with yeast cytochrome oxidase subunit II (Cox2p) mRNA, whose upstream codons contain antagonistic control elements fine-tuning the translation: the positive control element includes the first 14 codons specifying the leader peptide, whereas the negative control element is contained within codons 15 to 91. These regulatory elements embedded in the translated COX2 mRNA sequence, together with trans-acting factors, could play a role in the coupling of regulated synthesis of nascent pre-Cox2p polypeptide to its insertion in the mitochondrial inner membrane (98,99). We expect that such mechanisms of translational control may be common, and other interesting cases will be reported in future studies to encompass a wide variety of proteins containing leader peptides.
RNA stability and protein abundance
Synonymous substitutions may affect translation by facilitating stable loops that can significantly delay translation initiation and/or ribosome translocation, or by loosening mRNA secondary structures and eliminating obstacles to speedy translation (8,29,95). Such mRNA-structure-dependent changes in translation rates can have dramatic effects on protein abundance and predispose to disease development. For example, a correlation was found between the vulnerability to myogenous temporomandibular joint disorder and synonymous mutations in the human COMT gene, which has been discussed in the previous section (11,29). Synonymous substitutions in three common COMT haplotypes result in the formation of different stem-loop structures in the middle of the protein-coding region, and the stability of these structures inversely correlates with the amount of translated protein, leading to significant differences in the level of COMT enzymatic activity in vivo. Synonymous substitutions in the COMT coding sequence substantially influence pain sensitivity and the risk of developing temporomandibular joint disorder by affecting expression of this key protein regulator of pain perception.
Another example of naturally occurring synonymous mutations that affect mRNA stability and protein synthesis was described for the human dopamine receptor D2 (DRD2) gene (100). Synonymous variant C957T, rather than being ‘silent’, altered the predicted mRNA folding, led to a decrease in mRNA stability and translation and dramatically changed dopamine-induced upregulation of DRD2 expression. Variant G1101A did not show an effect by itself but annulled the aforementioned effects of C957T, demonstrating that combinations of synonymous mutations can have functional consequences drastically different from those of each isolated mutation. These results provide insights into mechanisms of molecular population genetics of diseases with complex inheritance and indicate that synonymous variation can have effects of potential pathophysiological and pharmacogenetic importance. Doubtless, these enzymes are only several examples among the potential many (101) that may be regulated through this mechanism. Other examples for many proteins are emerging in some of the ongoing studies partially discussed elsewhere in this article.
Native mRNAs have a lower calculated folding free energy than random sequences, and the average folding energy and ΔG of dinucleotide interaction are significantly lower for abundant transcripts relative to rare ones (10). There is no direct link, however, between the thermodynamic stability of transcripts and their decay rates that are controlled by complex cellular mRNA decay systems using arrays of RNA-binding proteins and specific nucleases. There is abundant experimental evidence that the steady-state levels and decay rates of bacterial and mammalian mRNA strongly depend on the usage of synonymous nucleotides. Certain dinucleotides, for example, the across-codon dinucleotide T|A, are strongly avoided in both prokaryotes and eukaryotes, owing to fast enzymatic degradation of UA-rich mRNA species [reviewed in (102)].
mRNA structure, post-translational modifications and regulation of protein folding
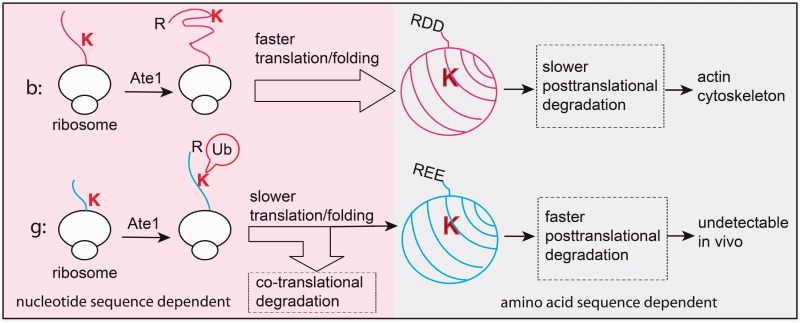
Recent studies demonstrate that variations in translation speed induced by mRNA secondary structures can lead to changes in post-translational modifications of the nascent polypeptide, a level of protein regulation that was previously believed to be unconnected with the RNA level regulation. An example of translation-dependent regulation of post-translational arginylation was recently shown for actins (103), abundant proteins represented by six gene copies in higher vertebrates that are nearly identical at the amino acid level but are encoded by different synonymous codons. It has been a subject of long-term debates in the actin field why mammalian genomes encode six highly similar actin proteins, and why do all these proteins appear to be only minimally redundant despite their near identity at the amino acid level. Non-muscle beta- and gamma-actin, two prevalent non-muscle actin forms that often coexist in the same cell in nearly equal levels, are differentially modified by post-translational arginylation that affects only beta-actin and regulates its function in the cell motility (104). Surprisingly, this difference in post-translational modifications appeared to be regulated entirely through mRNA, which differs by ∼12% between beta- and gamma-actin (103). Gamma-actin mRNA forms a stable secondary structure at the translation initiation site, whereas beta-actin mRNA is relatively unstructured in that region, resulting in a significant reduction in the translation speeds for gamma-actin compared with beta-. Although this difference does not significantly affect the overall protein abundance, it appears to selectively affect post-translationally modified states, causing slower folding of gamma-actin due to ribosome pausing and thus making it vulnerable to ubiquitin conjugation machinery attracted by co-translational arginylation. As a result, arginylated gamma-actin is selectively removed and never found in cells, whereas arginylated beta-actin, which escapes this degradation due to rapid synthesis and folding (103), accumulates in the cell (Figure 2). Thus, in the case of actin, synonymous codon-mediated changes in the mRNA secondary structure can lead to significant differences in protein translation rates and thus affect not only protein homeostasis but also post-translational modifications. It appears likely that such mechanism can also be involved in achieving selectivity in post-translational modifications of otherwise similar proteins.
Figure 2.
Differential arginylation of actin isoforms is regulated by a novel degradation mechanism coupled to the translation and folding dynamics in vivo. Top, faster translation and folding of beta-actin protects the Lys18 residue from potential co-translational ubiquitination and degradation on N-terminal arginylation. After emerging from the ribosome, arginylated beta-actin remains relatively stable and incorporates into actin cytoskeleton. Bottom, slower translation and folding of gamma-actin coupled with co-translational arginylation exposes arginylated gamma-actin for ubiquitination and ensures effective removal of 60–80% of arginylated gamma-actin protein. The fraction of arginylated gamma-actin that escapes the co-translational ‘check point’ is still degraded faster, with half-life of only 1 h, so that no arginylated gamma-actin can be detected in vivo. Image courtesy of Dr Fangliang Zhang.
Synonymous single-nucleotide polymorphisms within the same gene can create individual variations in translation speeds, leading to dramatic effects on protein folding between individuals. A striking example of this kind concerns multidrug resistance 1 (MDR1 or ABCB1) gene (105,106). In this gene, frequent-to-rare codon synonymous substitutions lead to the synthesis of proteins with identical primary structures but distinctly different folding patterns and varied intracellular functions. These differences are believed to be generated by ribosome stalling that, if it lasts long enough, can affect the protein folding and lead to alternate folding patterns. Although the conformational and functional differences between the native and alternate states may be minor, the MDR1 case illustrates that the protein folding barriers may nevertheless constitute sufficiently high hurdles on the physiological time scales, leading to kinetically trapped states with altered structures and functions. Other related examples have been identified in disease and discussed elsewhere. Overall, like with other effects of synonymous positions on protein functions, these cases are likely to be the first of many. Considering the possibility of selection against protein misfolding supported by recent studies (63,64,68), it is likely that additional experimental evidence of the role of mRNA structure in determination of protein fate may be found in the near future.
REGULATION OF TRANSLATION THROUGH RNA–RNA CROSS-TALK
It has been long assumed that RNA–RNA interactions in the course of translation are limited to the classical codon–anticodon base-pairing between mRNAs and tRNAs, as well as to interaction of ribosomal RNA (rRNA) with ribosome binding sites (RBS) on mRNAs in prokaryotes. Recent evidence suggests that interactions between clinger elements on rRNA molecules and complementary sites scattered along mRNAs are important factors in regulation of translation in both prokaryotes and eukaryotes. In prokaryotes, internal Shine-Dalgarno-like sites in the coding mRNA regions may function as translation delay signals. In addition to better known factors, such as codon usage and mRNA secondary structure, the complementary base-pairing between mRNA and rRNA molecules may play an important role in controlling protein synthesis (14,107). It was proposed that mRNA–rRNA cross-talk follows the multiple contact model (Figure 3A and B) through formation of duplexes between short complementary sites scattered over sequences (14,107,108). Universal occurrence of rRNA clingers in prokaryotes and eukaryotes suggests that this level of regulation was likely established early in evolution. Strong G/C asymmetry of the coding strands, as well as C-rich content of synonymous positions and 5′-UTRs in the vicinity of the start codon, might represent regulatory adaptations for a more efficient and fast translation.
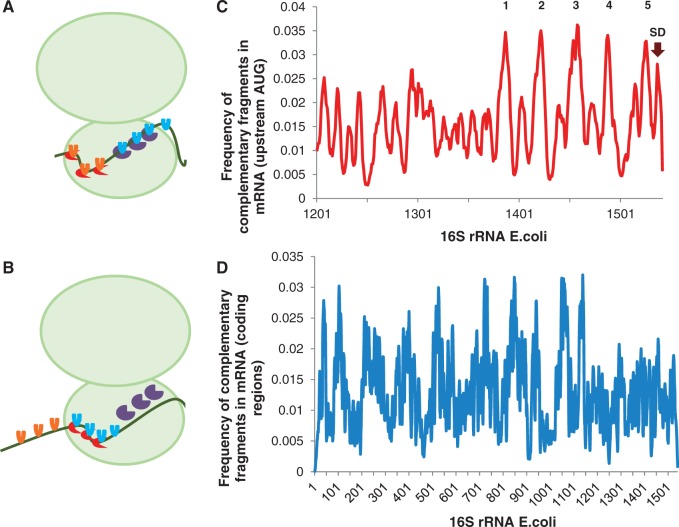
Figure 3.
The multiple contact model of mRNA–rRNA interactions. Hybridization affinity of 16S rRNA to mRNAs in E. coli. mRNA–rRNA interactions follow ‘the multiple contact model’ and occur due to the formation of multiple duplexes between short complementary sites scattered over the sequences (107,108). (A) mRNA–16S rRNA interactions affect translation efficiency. Efficient translation: rRNA clinger elements (red: located at 3′-end of 16S rRNA; brown: other) interact with the mRNA at complementary sites, which are indicated schematically as orange shapes in 5′-UTRs and blue shapes in CDSs. (B) Translation pausing: rRNA clinger elements located at 3′-end of 16S rRNA interact with mRNA sites located in CDSs. (C) The profile of complementarity of the 3′-end domain of 16S rRNA to the 5′ untranslated regions of mRNAs (30 nucleotides upstream AUG for ∼4200 sequences). The anti-Shine-Dalgarno site (UCACCUCC) is marked by arrow. Other clinger elements in the 3′-end of 16S rRNA: 1 (CCCGGGCCC), 2 (GGGAGUGGU), 3 (UCGGGAGGGC), 4 (UGGGGUGAA), 5 (AGGGGAACCUGCGG). (D) The profile of complementarity of 16S rRNA to the coding regions of mRNAs (∼4200 transcripts). Note significantly higher hybridization affinity of the 3′-end domain of 16S rRNA to UTRs (panel C) relatively to CDSs (panel D). Complementary targets to clinger elements in the 3′-end of 16S rRNA are avoided in the coding regions (107), in good agreement with recently published experimental data on the translation pausing driven by the Shine-Dalgarno-like sequences in the coding region of mRNAs (109). Panels C and D are adapted from 107.
mRNA–rRNA cross-talk in prokaryotes
Sequence analysis of 16S rRNA of E. coli identified multiple sites termed clinger elements or clingers that are complementary to the sites frequently occurring in mRNAs and tRNAs and represent potential regions of intermolecular hybridization. Clinger sites and their complementary mRNA partners are highly conserved in E. coli and might also operate in other prokaryotes by base-pairing of the 16S rRNA in the 30S ribosomal subunit with mRNAs (107). Major clingers on 16S rRNA pair with abundant mRNA motifs and represent universal binding sites for transcripts that belong to different functional groups (Figure 3C and D). Notably, clingers with pronounced hybridization affinity to 5′-UTRs of mRNAs are located in the 3′-end of 16S rRNA, where several G-rich high affinity clingers exist in addition to the classic anti-Shine-Dalgarno C-rich site (Figure 3C). Contrary, clingers complementary to mRNA coding regions are mainly located in the 5′ and core regions of 16S rRNA, whereas hybridization affinity of the 3′-end of 16S rRNA to mRNA coding regions is relatively low [(107), Figure 3D]. These results suggest an adaptation of structural organization of the 16S rRNA molecule to mRNA sequences, and support the idea that RNA interactions with clingers may contribute to upregulation of the translation process through increase in local concentration of mRNAs and tRNAs in the vicinity of the ribosome and their proper positioning, or reduction in efficiency of translation through non-specific mRNA–16S rRNA interactions (107) or transient pausing of ribosomes during translation (109).
This concept is supported by recent experimental study where a minimal reconstituted E. coli translation system was used to identify efficient RBSs in an unbiased high-throughput manner (110). The authors applied ribosome display, a powerful in vitro selection method, to enrich only those mRNA sequences that could direct rapid protein translation. In addition to canonical Shine-Dalgarno motifs, they recovered highly efficient C-rich sequences in mRNA coding regions that exhibit unmistakable complementarity to the 16S rRNA of the small subunit of the ribosome (Figure 3C), indicating that broad-specificity base-pairing may be an inherent general mechanism of efficient translation. Furthermore, given the conservation of ribosomal structure and function across species, the broader relevance of C-rich RBS sequences is supported by multiple diverse examples in nature, including C-rich RBSs in several bacteriophages and plants, a poly-C consensus before the start codon in lower eukaryotes and Kozak-like sequences in vertebrates (111).
Recently Weismann and colleagues (109) reported a genome-wide study of ribosome pausing in E. coli and Bacillus subtilis by ribosome profiling, a technique that allows the identification of ribosome-protected mRNA by high-throughput sequencing. Results of the study suggest that under nutrient-rich conditions, usage of rare codons does not lead to significant delays in translation. Rather, Shine-Dalgarno-like sites within the coding sequences cause pervasive translational pausing, due to hybridization between the mRNA and the 16S rRNA of the translating ribosome. To avoid excessive pausing, internal Shine-Dalgarno sequences are disfavored in the protein-coding sequences, avoiding codons and codon pairs that resemble canonical Shine-Dalgarno sites. Such disfavor creates an inadvertent bias in codon usage and also contributes to elevated C-content in highly translated mRNAs. As natural environments, unlike experimental conditions, often involve insufficient nutrient supplies, it appears likely that nutrient starvation and/or specific nutrient deficiencies induce evolutionary adaptations to cause a downstream effect of ribosome pausing in the content-dependent manner, and thus, redundancy in the genetic code likely constitutes a genuine evolutionary tool that controls translation rates. Internal Shine-Dalgarno-like sequences and C-rich RBSs are likely major determinants of translation rates and a global driving force for the coding of bacterial genomes.
mRNA–rRNA cross-talk in eukaryotes
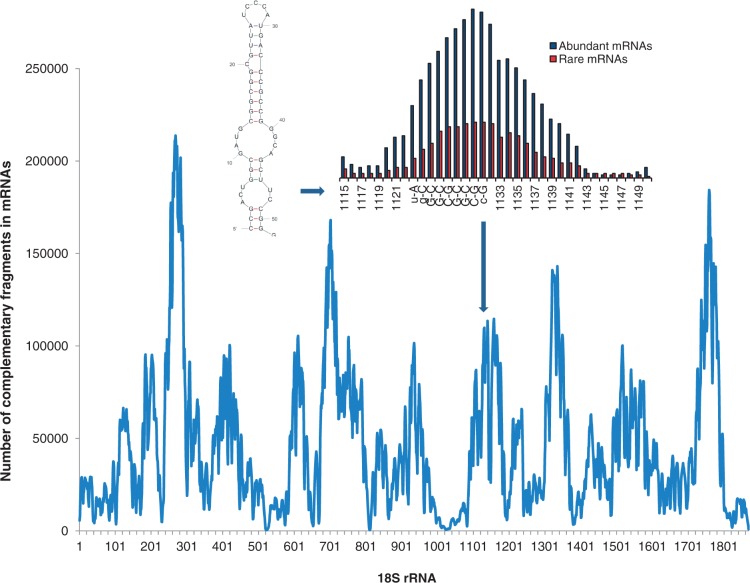
Intermolecular hybridization experiments demonstrated that human 5S rRNA and 18S rRNA molecules can hybridize with mRNAs during translation (112). Similarly, murine 18S rRNA and 28S rRNA form stable hybrid structures with mouse mRNAs, suggesting that such interactions could play a role in regulating translation speed. As discussed previously, mRNA may interact with rRNA through formation of duplexes between short complementary sites scattered over sequences to position mRNA properly for efficient translation (14,108). Sequence analysis identified multiple 18S rRNA clingers complementary to oligonucleotides that frequently occur in both 5′-UTR and coding regions of mRNA and represent potential hybridization regions (14). Many eukaryotic mRNAs contain sequences that resemble segments of 28S and 18S rRNAs, and these rRNA-like sequences are present in both sense and antisense orientations. For example, four potential 18S rRNA-interacting sequences were found in hundreds of different mRNAs, and the location of these sequences within the various genes was not random (113). The distribution of clingers along 18S rRNA sequence is universal for different mRNAs (Figure 4), and the affinity of clingers for mRNAs is 2-3 times higher than for intron sequences and for randomly generated sequences with the same nucleotide content. There is a significant variability in the hybridization affinity between different mRNAs that suggests a possible role of rRNA clingers in translation processes as universal regions of mRNA binding that can affect translation rates (14). Notable differences were found in the affinity of rRNA to the groups of abundant and rare mammalian mRNAs, as well as the prevalence of C-rich synonymous positions in the abundant mRNAs (9,93,114). Elevated C-content in mRNA synonymous sites likely represents an adaptation mechanism that adds to upregulation of translation rates of abundant high-expression mRNA species. For example, the hybridization affinity of 18S rRNA clingers to abundant protein kinase transcripts was ∼four-fold higher than for rare kinase transcripts [Figure 4, (114)].
Figure 4.
mRNA–rRNA intermolecular hybridization affinity. Distribution of complementarity of mouse 18S rRNA to several thousand mRNAs. Peaks represent potential clingers on mouse 18S rRNA. Hybridization affinity of abundant and rare protein kinase transcripts to verified 18S rRNA clinger (right insert box). Predicted secondary structure of verified 18S rRNA clinger (left insert box). This figure is adapted from 14 and 114.
The ability of several predicted clingers to interact with mRNA during translation was experimentally confirmed. There is evidence that mRNA sites interacting with rRNA may facilitate translation. A 9-nucleotide sequence from the 5′ leader of the mouse Gtx homeodomain mRNA facilitates translation initiation by base-pairing to 18S rRNA. Role played by the Gtx element in translation in eukaryotes to some extent resembles the function of Shine-Dalgarno sequences in translation in prokaryotic organisms (113,115,116). The presence of the Gtx element in various mRNAs suggests that this element may affect translation of different transcripts. Another sequence complementary to 18S rRNA is preferentially located within coding regions in multiple rodent genes immediately upstream of the termination codon. The effects of the sequence complementarity to 18S rRNA on translation were assessed using rodent mRNA encoding ribosomal protein S15. Mutations that decrease this complementarity without changing the amino acid sequence or affecting codon preference increase translation ∼1.5 fold (13). It is likely that direct base-pairing of particular mRNAs to rRNAs within ribosomes may provide a mechanism of translational control that works in both directions, and clinger sites may function both as upregulating and downregulating elements.
These and other studies allow a better understanding of the role of intermolecular RNA interactions in regulation of protein expression, and suggest that selection pressure on synonymous sites could be imposed by requirements to accommodate or avoid placement of RNA–RNA interaction sites within protein-coding sequences, which may contribute to upregulation of the translation process through increase in the local concentration of mRNAs in the vicinity of the ribosome and their proper positioning, or reduce the efficiency of translation through transient pausing of ribosomes during translation (14,107–109).
ROLE OF SYNONYMOUS POSITIONS IN THE OVERLAPPING CODES: EUKARYOTIC REGULATORY SIGNALS
Messenger RNA carries numerous short regulatory sequences, such as transcription factor binding sites, RNA editing and localization elements, splicing and translation initiation signals that often overlap with protein-coding regions. The repertoire of overlapping codes is particularly rich in eukaryotic coding regions that harbor regulatory signals involved in alternative transcription, splicing and nucleosome positioning (6,24), binding sites for diverse mRNA-associated proteins, microRNA (miRNA) target sites and other elements of RNA–RNA cross-talk (14,15,24). Selection pressure exerted on synonymous codon positions at such sites allows many degrees of freedom for evolution that might be used for achieving changes in regulation of biological function without modifications of protein sequences. Single-nucleotide changes at synonymous positions dramatically influence transcriptome repertoire and enrich structures of alternative isoforms expressed in different tissues and under different conditions (6,9,40,67). In this section, we will discuss the diversity of overlapping codes and regulatory signals in higher eukaryotes and their contribution to the complexity of transcriptome.
Given the key roles RNA signals and structural elements play in multiple aspects of normal physiology and regulation of protein function, it is not surprising that aberrations and alterations in RNA signals and structures at the primary and secondary levels can lead to dramatic consequences to health and has been implicated in a number of human diseases through various mechanisms (117).
miRNA–mRNA interaction and silencing influence synonymous codon choices
Synonymous codons are widely selected for the needs of various biological mechanisms in transcription regulation in eukaryotes. Recent evidence suggests that miRNA function may affect synonymous codon choices in the vicinity of miRNA target sites that are commonly located in the coding regions of plant genes. A general trend of relieved structural accessibility around miRNA target sites was observed in four plant genomes (118). It was found that G- and C-rich codons are avoided in the regions flanking miRNA target sites, and this selection is stronger for GC-rich genes compared with the genes located in the GC-poor regions. The authors suggest that synonymous codons near miRNA targets are selected for efficient miRNA binding, and natural selection on synonymous positions around miRNA target sites might, therefore, influence evolution of the coding regions. Similar selection may act on the coding regions in mammals and insects (119–122). Although the majority of characterized mammalian miRNA target sites are located in the 3′-UTRs, the large-scale studies show that they are also present and functional in coding regions and 5′-UTRs (123–127). Targeting of sites harbored by the coding regions is generally less effective. However, they contain representative numbers of miRNA target sites that mediate notable repression, as demonstrated by genome functional studies (128–131). This conclusion is also confirmed by several research groups in experiments using reporter assays (132–134).
Many of CDS-located target sites are conserved between closely related animal species (119,121,134). Coding regions of repeat-rich genes contain numerous potential target sites for particular miRNAs, and such genes are often strongly repressed. Such sequence repeats arise through evolutionary duplications and occur particularly frequently within families of C2H2 class of zinc-finger genes (127). Efficient targeting of coding-region repeats is highly predictable, and due to the large number of target sites within a single CDS, downregulation observed in reporter assays can be stronger than for many genes with 3′-UTR targets.
Synonymous mutations at the miRNA-binding sites disrupt target recognition and may be implicated in disease development. For example, synonymous polymorphism in the human IRGM gene affects binding site for miR-196 and leads to tissue-specific deregulation of the IRGM-dependent xenophagy that causes a predisposition to Crohn’s disease (135). Thus, recent studies suggest a role of the coding regions, and, specifically, the coding-sequence repeats, in post-transcriptional regulation. Selection pressure on the synonymous positions might affect synonymous codon choices in and around miRNA target sites in favor of higher accessibility of miRNA binding.
Splicing control imposed by mRNA folding and intermolecular interactions at exon-intron boundaries
The majority of protein-coding genes in mammals undergo alternative splicing, whereby the same sequence belongs to an exon in one subset of transcripts of a given gene locus and to an intron in another subset of transcripts. Indeed, the latest estimates based on high-throughput transcriptome sequencing indicate that up to 95% of multi-exon human genes are subject to alternative splicing that involves ∼100 000 major alternative events (136). Exceptionally wide spread of alternative initiation and alternative termination of transcription in the genome (6,137,138), coupled with independent alternative splicing events in different regions of the same gene locus, can yield dozens of different transcript variants. Such combinatorial use of alternative exons represents a major source of transcriptome diversity in higher eukaryotes, especially in humans and other mammals, where it allows generation of hundreds of thousands isoforms from 30 000–40 000 protein-coding genes.
Traditionally, pre-mRNA has been viewed as a passive molecule that is kept by hnRNPs in the unfolded and unstructured form to allow snRNPs and other proteins to scan the pre-mRNA for regulatory sequences and process it into mature transcript. However, this view has been largely reconsidered in light of transcriptome studies demonstrating that pre-mRNA itself is actively regulating its own processing (24,139). It is well established that RNA structural elements can directly inhibit or activate splicing. Taking into account current data demonstrating that pre-mRNA can be actively spliced as it is being transcribed (140), it is obvious that not only local but also distant mRNA structural elements might be important for efficient splicing. In many cases, distant and local signals (5′ and 3′ exonic splice sites or branch points) within coding regions have been found involved in mRNA structure formation. U1, U2, U4, U5 and U6 snRNAs participate in excising the major class of introns from pre-mRNAs (24). The secondary structures of these snRNAs are highly conserved from yeast to human, as are their nucleotide sites that are involved in intermolecular interactions. These conserved regions specify the roles of snRNPs and participate in the intricate RNA–RNA interaction network during spliceosome assembly and function (141). Taking into account that this complicated machinery contains many active players with short interacting sites, efficient cross-talk between RNA and protein molecules requires high accessibility of pre-mRNA. Many individual cases of such interactions have been described in the literature with examples of pre-mRNA structures that inhibit or accelerate splicing (9,24,139,142).
Sequence analysis and prediction of RNA secondary structures are useful tools in experimental design aimed at determination of pre-mRNA regulatory sites. Interspecies conservation of local RNA structures identified with co-variation base-pairing models that consider exchanges between paired dinucleotides in the structure (e.g. G-U change to A-U or G-C) may correspond to the functional signals of pre-mRNA processing. Exonic splicing enhancers and silencers, usually located near intron–exon boundaries and represented by oligomeric motifs, are responsible for a cross-talk between RNAs and spliceosomal proteins to facilitate splice-site recognition (143,144). Selection pressure on such elements manifests itself with a high level of interspecies similarity of their conserved mRNA secondary structures (145), with a low frequency of polymorphisms in the paired regions and low density of SNPs at the ends of exons (143,144). Synonymous changes in exonic splicing enhancers and silencers could affect exclusion or retention of exons in mature transcripts. Recent reports have identified proteins and small molecules that can affect splicing by modulating RNA structures, thereby expanding our knowledge of the mechanisms of splicing regulation (24,139).
RNA editing and protein recording
RNA editing is a phenomenon that provides a mechanism for the alteration of particular nucleotides in RNA sequences relative to their genomic templates, resulting in diversification of RNA sequences that consequently change their function (146,147). RNA editing has been found across all kingdoms of life, including viruses (148,149). A surprisingly large number of instances of RNA editing has been identified in humans using bioinformatics screens and high-throughput experimental investigations utilizing next-generation sequencing technologies (150). Analysis of RNA editing events in the human ENCODE RNA-seq data identified frequent editing of housekeeping genes involved in cell division, translation and viral defense across multiple cell types (151).
RNA editing plays a variety of functional roles in regulation of gene expression. Editing of a nucleotide within the protein-coding region may change the identity of a particular encoded amino acid or prematurely terminate the protein, create or deplete entire exons through changes in a splicing site, cause retention of mRNA in the nucleus or miRNA modification, affect RNA stability, efficiency of protection against viral RNA, and heterochromatin formation (151). For example, RNA editing can lead to exonization of the Alu repeat in the nuclear prelamin A recognition factor. Exon 8 in this gene is derived from the recently exonized sequence of the Alu repeat, where a non-valid (AA) 3′ splice site is edited to a valid AG and alternatively spliced in a tissue-dependent manner, leading to a higher transcript abundance in brain tissue than in skeletal muscle (152). The sequence of the new exon contains the in-frame TAG stop codon that is efficiently edited to TGG that code for tryptophane to keep the reading frame (152,153). When editing is needed within a protein-coding sequence, a region of the coding sequence usually forms base-pairing with intronic regions of the same gene. In this example, another Alu element 25 bp upstream to the exonized Alu is crucial for creation of Alu-Alu duplex that is required for RNA editing (153). The selection force acting to maintain these interactions reduces evolutionary rates at synonymous positions of the sites important for the duplex formation. RNA editing, occurring within intron–exon boundaries, can affect splicing, effectively resulting in the generation of alternatively spliced products (154). It also can change functioning of RNA structural elements related to the translation efficiency (155,156). Combinatorial editing is a significant contributor to the transcriptome repertoire, suggesting that editing of synonymous positions, together with alternative exonization, adapted by natural selection, may serve as important mechanisms of transcriptome diversification in primates (157).
RNA secondary structures are also involved in recording of protein sequences that change the meaning of particular codons. One of the best studied examples is the case of selenocystein insertion, which is driven by mRNA secondary structures known as SECIS (158). As in other cases, selective pressure acting on these structures slows down evolutionary rates for synonymous substitutions. Similar stable and conserved RNA structures are often required for frameshifting and stop codon readthrough that are common in viruses (159,160). The most prominent example of frameshifting is antizyme in eukaryotes (161,162). Frameshifting is triggered by mRNA–rRNA interactions and evolutionarily protected by selection pressure on silent sites.
mRNA stability and decay
Mutations, errors in transcription and splicing may create mRNA variants encoding abnormal proteins. mRNA can serve as a quality control template by ensuring that defective proteins, containing aberrant sequences that would result in premature functional truncations and/or other major abnormalities, do not get synthesized at all [reviewed in (163)]. Three major mechanisms of mRNA surveillance and decay function in the nucleus and cytoplasm (164). Nonsense-mediated mRNA decay that exists in all eukaryotes detects and degrades transcripts that contain premature stop codons (165,166). Non-stop mRNA decay targets mRNA that lack a stop codon (167). No-go mRNA decay detects abnormally stalled ribosomes and cleaves transcripts with low translation efficiency near such stalled sites by endonucleases (168). Overall, transcripts with a range of abnormalities resulting in low translation efficiency (defined by low ribosome density, slow ribosome translocation and abnormal initiation rates) are specifically targeted by various mRNA decay complexes in vivo, extending this regulatory mechanism to the translation level (169). Such low translational efficiency arises through multiple mRNA features acting in concert and can result from low translation initiation rate, mediated by stable secondary structure and/or weak initiation sites, as well as low translation elongation speed, mediated by codon usage (169).
mRNA turnover is a highly controlled process. In addition to a nonsense codon, specific downstream sequence elements are required for mRNA destabilization and degradation of abnormal nonsense transcripts. Sequence motifs enriched by pyrimidine can predict potential regions in mRNAs that together with the upstream nonsense codon promote rapid decay of its mRNA. It was also suggested that other sequence elements modulate the activity of the downstream element by forming RNA secondary structures (170).
Several sequence elements can regulate the rate of turnover of a transcript by promoting or by inhibiting decay through stabilizer or destabilizer elements, respectively. Most of these elements, such as the AU-rich sites, are located in 3′-UTRs, but also found in 5′-UTRs and coding regions (171). For example, both AU-rich site in 3′-UTRs and a destabilizing sequence known as the major protein-coding region determinant (mCRD) within c-fos mRNA coding region work together (172). The mCRD usually locates at least 450 nucleotides proximal to the poly(A) tail and requires continuing translation for the destabilizing function. Transit of ribosomes through the mCRD element disrupts the complex and triggers the mRNA decay (173). In addition to the nonsense codon, specific downstream sequence elements enriched by pyrimidine are required for mRNA destabilization and, likely, modulate the activity of the downstream element by forming RNA secondary structures. Another example of translation-dependent instability element within the protein-coding region was localized in yeast MATa1 mRNA (174). Notably, this element corresponds precisely to an mRNA sequence previously shown to be complementary to 18S rRNA. These results suggest a model where the triggering of MATa1 mRNA destabilization results from establishment of an interaction between translating ribosomes and a downstream sequence element.
mRNA decay mechanisms not only serve as important quality control checkpoints in the functioning of a normal cell, but also play a role as major disease barriers in organisms carrying recessive mutations that would cause protein truncations and/or major structural abnormalities that may result in dominant negative or gain-of-function effects at the organismal level. mRNA decay mechanisms also assist in degrading defective physiological transcripts and preventing the effects of routine inaccuracies that occur during transcription initiation, pre-mRNA splicing or transcriptional errors (166,175). Finally, it has been suggested that failure of mRNA decay mechanisms constitutes a strong drive for molecular evolution aimed to increase the overall robustness of genes to errors (176,177).
As mRNA decay constitutes a key mechanism of the overall regulation of mRNA availability and protein synthesis, it is not surprising that a large role in disease and its prevention belongs to mRNA decay mechanisms. When combined with frameshift or nonsense mutations, they can result in premature termination of translation, often leading to deficiencies in critical proteins. Recent genome-wide association studies revealed a substantial fraction of synonymous substitutions linked to human disease risk and other genetic traits by mechanisms that are believed to be largely associated with alterations in translation rates and mRNA decay. Cases of β-thalassemia (178), cystic fibrosis (179), Duchenne muscular dystrophy (180) and a number of cancers have been found to be linked to RNA decay. Other examples include somatic-cell rearrangement and hypermutation of immunoglobulin or T-cell receptor genes that generate immune diversity (181).
mRNA localization and interactions with mRNA-associated proteins
Another level of protein regulation by mRNA arises through mRNA-associated proteins that package mRNA into an mRNP complex and participate in multiple aspects of mRNA functions, including regulation of its stability and turnover, translation initiation and translation rates, as well as its distribution throughout the cell that ensures preferential translation of specific proteins at key functional sites. Much of this interaction has been characterized at the untranslated regions, either upstream (5′-UTR), where binding or release of specific proteins can mediate translation initiation, or downstream (3′-UTR), where RNA-protein binding can regulate various aspects of RNA folding and targeting to different complexes; however, some prominent examples of such elements within the mRNA coding regions have also been found. A particular type of such regulation involves ‘localizer’ or ‘zipcode’ sequences, which are present in a highly specific subclass of mRNAs (182) and target them to key cellular destinations (183). Zipcode-mediated targeting requires specific zipcode-binding proteins (184) that associate with mRNA during transcription (185) and induce RNA looping in the process of recognition and binding (186). Such targeting has proven to be highly physiologically important and has been implicated in a wide range of biological processes, including leading-edge activity in motile cells (182), axon guidance and growth cone activity (187,188), brain development (189), G protein signaling (190), cell polarity and chemotaxis (191) and many others. Moreover, it has been found that zipcode-binding proteins regulate mRNA stability during stress and prevent their premature removal (192).
An important but less explored aspect of RNA and DNA protein binding arises through the likely impact of synonymous nucleotide substitutions on affinity and recognition by regulatory protein factors. A correlation between coding sequence and protein binding has been found at the nucleosome level, where nucleosome positioning apparently defines the rates of coding sequence evolution (193). Other studies found a correlation between nucleosome positioning and evolution of tandem repeats (194). It appears likely that protein–nucleic acid binding should in turn be regulated by nucleotide sequence and represent an interconnected hierarchical chain driving protein expression and mRNA function.
EVOLUTION: RNA-LEVEL SELECTION PRESSURE ON PROTEIN-CODING SEQUENCES
RNA selection pressure and the Ka/Ks metric of amino acid selection pressure
As discussed previously, evolutionary selection pressure is acting at both the protein-coding level and RNA or nucleotide level (32,195–199). Patterns of the RNA-level selective constraint are manifested by the elevated sequence similarity and base-pairing (or hybridization affinity), which is crucial for RNA secondary structure, stability and intermolecular interactions. These patterns are specific for transcripts of different functional groups. The functional importance of the RNA-level selection pressure has been exemplified by the evidence of non-neutral evolution at synonymous sites and by the finding that alternatively spliced exons in mammals are more conserved at their silent sites than constitutive exons (28). RNA selection pressure affects genome architecture at different levels of organization, as identified by conservation of local and global RNA secondary structures in mammalian pre-mRNAs and mRNAs (10,89,145). Two distinct biological manifestations of RNA selection pressure, related to RNA hybridization affinity, are seen in the coding regions; one associated with mRNA folding/stability and the other with mRNA intermolecular interactions.
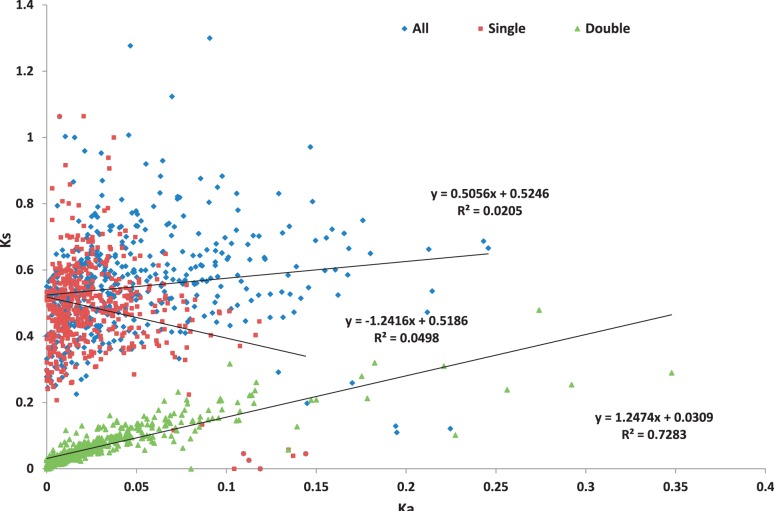
An important question is how one can accurately estimate RNA selection pressure. Evaluations of evolutionary selection are based on the frequencies of substitutions at the non-synonymous and synonymous sites, termed Ka and Ks, respectively. The Ka/Ks ratio is generally accepted as a measure of evolutionary selection on protein-coding sequences, where the frequencies of mutations observed at the non-synonymous and synonymous sites are: Ka = ωρµ, Ks = ρµ and Ka/Ks = ω, which is not dependent on ρ or µ (where ω is amino acid selection pressure, ρ is RNA selection pressure and µ is mutation rate) (200,201).
At first approximation, the RNA selection pressure could be described by a simplified model that considers all potential driving forces of the RNA-level selection as one independent variable, ρ (28). Ks, the key parameter for the estimation of the RNA-level selection pressure, could be measured accurately and independently of Ka, and specific classifications of different events at synonymous positions could be considered for the accurate estimation of ρ (200). A sensitive bioinformatics approach was recently suggested for identifying alternatively spliced exons with evidence of strong RNA selection pressure, where evolutionary selection against mutations changes only the mRNA sequence leaving the protein sequence unchanged (202).
Best studied examples of the RNA-level selection are associated with translation selection on codon usage and selection pressure related to the regulation of pre-mRNA splicing. Selection on codon usage reduces the synonymous divergence rate, and may introduce a bias into the Ka/Ks estimations. In some cases, when purifying selection (selection that eliminates a new mutation from the population, removing deleterious alleles from the population; also known as negative selection) on synonymous sites is strong, a very low Ks might be due to the presence of splicing regulatory signals (9). Evaluation of synonymous evolution in the regions with high Ka/Ks ratio and accurate estimation of Ks and Ka values are helpful to identify Ka peak zones or Ks dips to detect positive selection (natural selection that promotes the spread of a new mutation through the population, resulting in a fixed difference between species; also known as Darwinian selection) in specific genome regions or sites (9,203). Detailed individual analysis of Ka, Ks and Ka/Ks values and classification according to their ranges for regional and/or site-specific applications are essential for the development of accurate models of the specific RNA-level selection. However, the task to classify and model all different aspects of the RNA-level selection pressure is a challenge. Statistical test was developed for identification of purifying and positive selection at synonymous sites in the protein-coding genes (69). To measure selection on synonymous sites, the authors used the substitution rate in intronic sequences (Ki) as a proxy for neutral evolution. The method is based on the difference in the statistical features of the CDS and intron sequences and uses shuffling of the intron sequence alignments such that their statistical properties mimic those of the coding sequences.
Potential driving forces of selection at synonymous sites
The majority of recent studies of the RNA-level selection in mammals were performed by comparing evolutionary rates at synonymous sites with those for flanking introns or within the introns of neighboring chromosome regions. This approach avoids complications resulting from regional variations in mutation rates and transcription-related bias. As discussed previously, GC enrichment at the synonymous sites, as compared with intronic sequences, could indicate selection acting at these sites (34,35,204). Estimations of Ks for synonymous sites and Ki for intronic sequences performed by different authors significantly vary for different gene sequences and mammalian species (9), which may be due to the methodological difficulties in the determination of the RNA-level selection pressure (28).
Notably, although the overall rates of nucleotide substitutions at synonymous sites and for intronic sequences are quite similar, their patterns are dramatically different (53,205). For example, C residues are more common at the four-fold degenerate sites than in introns, and also are relatively less likely to be associated with substitutions (10,53). This is dictated by the structure of the genetic code, where all codons with C at the second position are four-fold degenerate, which could be responsible not only for the strand asymmetry (206), but also for the more stable and ordered mRNA folding (10). Taking into account that C-rich sites in mRNA have a potential to interact with rRNA clingers in both prokaryotes and eukaryotes, the strand asymmetry may also indicate the RNA-level selection pressure to optimize translation levels of differently expressed proteins (14,107).
Results of transcriptome-wide analysis of the human and mouse mRNA folding suggest that selection in favor of G and C may be operating on synonymous codons to maintain a more stable and ordered mRNA, which is likely important for transcript stability and translation (10). These data are in good agreement with theoretical predictions of the average coefficient of selection in favor of nucleotides G and C at human synonymous sites, which shows limited variation across individual sites (34). A plausible explanation for these results is synergistic epistasis (34,207,208), expected, for example, if synonymous sites are involved in maintaining the mRNA secondary structures (10,17,209) or are responsible for mRNA hybridization affinities and RNA interactions (107). Evolutionary rates at synonymous sites are dependent on the mutable CpG content. Rate of evolution at non-CpG synonymous sites is 10% below that of similar intron sites, whereas at postCpreG sites, it is 30% above that of similar intron sites (34). From these data, a reasonable estimation of neutral divergence between two mammalian genomes (expressed as the mutation rates outside CpG context multiplied by the number of generations of their independent evolution) can be approximated as ∼1.1 times the Ks at non-CpG four-fold degenerate synonymous sites (34). Current estimations suggest that ∼40–50% of synonymous positions in mammals have been opposed by selection (10,34,210), whereas at least half of them are under selection in favor of more stable mRNA secondary structure.
The nature of the driving forces of selection at mammalian synonymous sites largely remains an open question. Different factors might contribute to negative and positive selection at synonymous sites, including gene function and expression patterns, codon bias, mRNA folding and stability (69). Analysis of associations between selection on synonymous positions, mRNA stability and expression revealed that the genes with positive selection at synonymous sites showed no correlation between Ks and Ka, indicating that evolution of synonymous sites in such genes is uncoupled from protein evolution. As discussed previously, significant negative correlation between Ks and expression in the group of genes under purifying selection indicates that highly expressed genes evolve slowly. Contrary, synonymous sites in the genes under positive selection show, on average, higher Ks in highly expressed genes, and a significantly lower mRNA stability, compared with the genes under negative selection. Notably, positive selection at synonymous sites of mammalian genes is substantially more common than positive selection on the protein sequences, and might act through mRNA destabilization affecting mRNA level and translation (69). However, purifying-negative selection on synonymous sites is linked to elevated mRNA stability (10,89,102).
RNA selection pressure affects the structure of functional domains and regulatory signals
The RNA-level selection pressure in the protein-coding regions would have to be periodic, with periodicity of three nucleotides, and would not interfere with protein functional requirements (10,200). Codon usage bias could influence this periodicity, but other sources of bias are also important in mammals, such as mRNA stable secondary structure elements and sites interacting with rRNA clingers. Estimation of selection pressure associated with the maintenance of mRNA secondary structure or mRNA intermolecular interactions could be more accurate if analysis of stable hairpins, stem-loop structures and sites responsible for the RNA–RNA interactions would be conducted separately. All these classifications should be based on the experimental results produced by the new SHAPE approach (selective 2′-hydroxyl acylation analysed by primer extension) or similar techniques (92,211) for the reliable estimation of the specific RNA-level selection. Although theoretic predictions of RNA secondary structures and sites of intermolecular interactions are in good agreement with experimentally produced classifications (10,92,107,110,211,212), it is more desirable to measure the RNA-level selection pressure based on reliable experimental data.
What protein-coding regions could be under the strong RNA-level selection pressure? Comparison of 29 placental mammalian species revealed ∼10 000 highly conserved regions with extremely low rates of synonymous substitutions corresponding to overlapping functional signals, such as splicing regulatory elements and miRNA target sites, RNA secondary structure elements and dual-coding genes and enhancers (213). Numerous studies demonstrated that alternatively spliced regions slowly evolve in the flanking intronic regions and synonymous positions near exon boundaries (28). These observations might be indicative of the differences between constitutive and alternative exons, which could be related to the variability in density and composition of the splicing regulatory signals that tend to reside near exon–intron boundaries (28). The lower GC content of alternative exons has been proposed as a support for translation selection (214). Analysis of evolutionary rates (Ks and Ki) at the exon–intron boundaries in the human OPRM1 gene locus (215) showed that both alternative/constitutive status of exon–intron boundaries and exon location (at the termini or core parts of a coding region) might affect the rate of evolution. The usage of certain codons is more biased near exon junctions, owing to significantly more common occurrence of the codon GAA in exonic splicing enhancers (145). Functional importance of such signals is exemplified by disease-related synonymous mutations that disrupt the splicing patterns and impair splicing regulation (9).
Another protein-coding region under the strong RNA-level selection is a leader peptide, where RNA secondary structure is relaxed with specific local elements, compared with the downstream CDS (95,99). Several studies show that selection forces act almost uniformly to reduce the stability of mRNA at the beginning of protein-coding regions in different organisms (10,94,95,216). Relaxed mRNA secondary structures are characteristic for the start and stop codon regions, where they may facilitate initiation and termination of translation (10,97). Thus, we can conclude that certain conserved protein-coding regions are under the strong RNA-level selection pressure.
The evolutionary tradeoff between selective pressure acting at the RNA and protein levels
How the RNA-level selection pressure affects non-synonymous positions is still an open question. Some evidence of the evolutionary tradeoff between selection pressure acting at the RNA and protein levels was found in viral genomes (211,212,217) and provided a better understanding of their evolution and variability. One interesting example is the HIV-1 RNA genome, the secondary structure of which has been experimentally determined (211). A correspondence was found between RNA and protein primary sequences as well as a correlation between high levels of RNA structure and sequences that encode inter-domain loops in HIV proteins. Analysis of this information led authors (212) to the conclusion that mRNA and protein structures do not evolve independently. A negative correlation exists between the extent of base-pairing in the RNA and amino acid variability. Relaxed mRNA secondary structures in the coding regions may favor the accumulation of genetic variation in proteins and, conversely, sequence changes driven by selection at the protein level may disrupt existing RNA structures.
Another evidence of co-evolution of mRNA and protein structures emerged from the analysis of Ka/Ks and Ks values in mammals, where the positive correlation between these values is due to runs of adjacent substitutions (218). Strong positive correlation between Ka and Ks was found for the double mutations in the same codons in mammalian protein kinases genes (114), where in the majority of cases, one of the mutations is synonymous and the second is not (Figure 5). These substitutions may reflect selection acting at both the nucleotide and protein levels. Obviously, such correlation may arise if synonymous and non-synonymous sites are parts of the same structure or same regulatory signal involved in the intra- or intermolecular interactions. Although a definite explanation of the reason for the positive correlation between Ks and Ka is still open (218), there is evidence to suggest that the evolutionary tradeoff between selection forces acting at the RNA and protein levels in mammals exists.
Figure 5.
Correlation between Ka and Ks for codons with single and double substitutions. Ka and Ks were estimated from sequence alignments of complete kinome (∼600 orthologous human and mouse protein kinase mRNAs, analysed in 46) by PAML program (Yang et al.). Strong correlation between Ka and Ks was found for codons with double substitutions, where synonymous and non-synonymous substitutions occur (green dots). Image courtesy of Dr Aleksey Ogurtsov.
CONCLUSIONS
Synonymous nucleotide positions are essential for the maintenance and function of diverse regulatory signals located in the protein coding regions. There are several levels of punctuation complexity and biological signals encoded by mRNAs. A prominent punctuation signal is periodic pattern of RNA secondary structure, which provides for a more ordered and stable structure of transcripts in the protein-coding regions and may also support maintenance of the reading frame during translation (10,219). This basic pattern is overlaid by stable conserved RNA secondary structure elements (29,100,101) that may cause translation pausing or stalling. The functional significance of synonymous positions for the maintenance of local stable RNA structures, which are crucial for protein regulation of expression, is well recognized, especially at the initiation of translation (10,89,92). These stable conserved folding elements, the second class of mRNA punctuation elements, could affect translation and, ultimately, the protein structure and function (10,11,29,103,106), whereas higher-order RNA structures may directly define protein folding, especially at domain junctions (63,64,211). Often this type of signals is located in the sequences encoding protein inter-domain loops, such as in the HIV genome (211). The third class of RNA punctuation signals are sites of intermolecular interactions providing, for example, regulation of translation (Shine-Dalgarno elements and sites interacting with rRNA clingers), splicing sites, and miRNA target sites (9,14,107,109,143).
Ribosome pausing or stalling, caused by the secondary structures of messenger RNA or mRNA hybridization to rRNA, can affect a variety of co-translational processes, including protein folding and targeting (109). Direct base-pairing of mRNAs to rRNA clinger sites within ribosomes may function as upregulating and downregulating elements (13), providing an additional mechanism of translational control. Most of these diverse RNA punctuation signals exist in both prokaryotes and eukaryotes, and enrich regulation of the translation efficiency and protein folding.
The extraordinary complexity of transcriptomes that underpins the structural and functional diversity of mammalian proteomes is created by alternative splicing and transcription with the use of distinct types of RNA splicing and regulatory control elements (5,6). Synonymous codon positions allow further diversification of intra- and intermolecular mRNA hybridization affinity (128,129,131), creating previously unrecognized patterns of RNA punctuation and hidden language of mRNA–miRNA cross-talk, characteristic for the higher eukaryotes and responsible for the regulation of the biological complexity, tissue-specific and condition-specific expression (15).
In the past, transcriptomes have been mostly characterized by transcript sequences and expression levels. The recent progress in experimental techniques (SHAPE, PARS), together with improved computational prediction methods, has enabled genome-wide measurements of RNA structure and has provided the first picture of the structural organization of prokaryotic and eukaryotic transcriptomes (92,211,220,221). With further progress in method refinement and interpretation, structural views of the transcriptome should provide new approaches for the estimation of the RNA level of selection pressure, identification and validation of regulatory RNA patterns and new punctuation signals that are involved in diverse cellular processes, and thereby increase understanding of RNA function.
FUNDING
DHHS (NIH, National Library of Medicine) intramural funds; NIH [R01HL084419 to A.K.]. Funding for open access charge: DHHS (NIH, National Library of Medicine) intramural funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Fangliang Zhang and Aleksey Ogurtsov for providing the data for Figures 2 and 5, Pavel Baranov and Olga Matveeva for stimulating discussions. The number of published materials on synonymous sites in protein-coding genes has grown exponentially over the past decade, and only a small portion of the relevant studies are cited in this review. We sincerely apologize to all colleagues whose important publications are not cited here owing to space constraints.
REFERENCES
- 1.Claverie JM. Gene number. What if there are only 30,000 human genes? Science. 2001;291:1255–1257. doi: 10.1126/science.1058969. [DOI] [PubMed] [Google Scholar]
- 2.Pennisi E. Human genome. A low number wins the GeneSweep Pool. Science. 2003;300:1484. doi: 10.1126/science.300.5625.1484b. [DOI] [PubMed] [Google Scholar]
- 3.Stein LD. Human genome: end of the beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 4.Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 6.Shabalina SA, Spiridonov AN, Spiridonov NA, Koonin EV. Connections between alternative transcription and alternative splicing in mammals. Genome Biol. Evol. 2010;2:791–799. doi: 10.1093/gbe/evq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 10.Shabalina SA, Ogurtsov AY, Spiridonov NA. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;34:2428–2437. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parmley JL, Hurst LD. How do synonymous mutations affect fitness? Bioessays. 2007;29:515–519. doi: 10.1002/bies.20592. [DOI] [PubMed] [Google Scholar]
- 12.Itzkovitz S, Hodis E, Segal E. Overlapping codes within protein-coding sequences. Genome Res. 2010;20:1582–1589. doi: 10.1101/gr.105072.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tranque P, Hu MC, Edelman GM, Mauro VP. rRNA complementarity within mRNAs: a possible basis for mRNA-ribosome interactions and translational control. Proc. Natl Acad. Sci. USA. 1998;95:12238–12243. doi: 10.1073/pnas.95.21.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matveeva OV, Shabalina SA. Intermolecular mRNA-rRNA hybridization and the distribution of potential interaction regions in murine 18S rRNA. Nucleic Acids Res. 1993;21:1007–1011. doi: 10.1093/nar/21.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl Acad. Sci. USA. 2004;101:3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz L, Burge CB. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlini DB. Context-dependent codon bias and messenger RNA longevity in the yeast transcriptome. Mol. Biol. Evol. 2005;22:1403–1411. doi: 10.1093/molbev/msi135. [DOI] [PubMed] [Google Scholar]
- 19.Jia M, Luo L, Liu C. Statistical correlation between protein secondary structure and messenger RNA stem-loop structure. Biopolymers. 2004;73:16–26. doi: 10.1002/bip.10496. [DOI] [PubMed] [Google Scholar]
- 20.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 21.Francino MP, Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol. 2001;18:1147–1150. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- 22.Hershberg R, Petrov DA. Selection on codon bias. Annu. Rev. Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 23.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 24.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 25.Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 28.Xing Y, Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl Acad. Sci. USA. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 30.Akashi H, Kliman RM, Eyre-Walker A. Mutation pressure, natural selection, and the evolution of base composition in Drosophila. Genetica. 1998;102–103:49–60. [PubMed] [Google Scholar]
- 31.Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duret L. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 33.Hershberg R, Petrov DA. General rules for optimal codon choice. PLoS Genet. 2009;5:e1000556. doi: 10.1371/journal.pgen.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondrashov FA, Ogurtsov AY, Kondrashov AS. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. 2006;240:616–626. doi: 10.1016/j.jtbi.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 35.Eyre-Walker A. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152:675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith NG, Eyre-Walker A. Synonymous codon bias is not caused by mutation bias in G+C-rich genes in humans. Mol. Biol. Evol. 2001;18:982–986. doi: 10.1093/oxfordjournals.molbev.a003899. [DOI] [PubMed] [Google Scholar]
- 37.Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Najafabadi HS, Goodarzi H, Salavati R. Universal function-specificity of codon usage. Nucleic Acids Res. 2009;37:7014–7023. doi: 10.1093/nar/gkp792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha EP. The quest for the universals of protein evolution. Trends Genet. 2006;22:412–416. doi: 10.1016/j.tig.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 2010;27:1745–1749. doi: 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 42.Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 43.Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 44.Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3:2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 49.Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- 50.Sorensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 51.Whittle CA, Sun Y, Johannesson H. Genome-wide selection on codon usage at the population level in the fungal model organism Neurospora crassa. Mol. Biol. Evol. 2012;29:1975–1986. doi: 10.1093/molbev/mss065. [DOI] [PubMed] [Google Scholar]
- 52.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chamary JV, Hurst LD. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol. Biol. Evol. 2004;21:1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- 54.Smith DW, McNamara AL. Specialization of rabbit reticulocyte transfer RNA content for hemoglobin synthesis. Science. 1971;171:577–579. doi: 10.1126/science.171.3971.577. [DOI] [PubMed] [Google Scholar]
- 55.Smith DW, Meltzer VN, McNamara AL. A comparison of rabbit liver and reticulocyte transfer RNA: evidence of unique species in reticulocytes. Biochim. Biophys. Acta. 1974;349:366–375. doi: 10.1016/0005-2787(74)90123-3. [DOI] [PubMed] [Google Scholar]
- 56.Comeron JM. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urrutia AO, Hurst LD. Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics. 2001;159:1191–1199. doi: 10.1093/genetics/159.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 2001;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 60.Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 61.Misawa K, Kikuno RF. Relationship between amino acid composition and gene expression in the mouse genome. BMC Res. Notes. 2011;4:20. doi: 10.1186/1756-0500-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh ND, Bauer DuMont VL, Hubisz MJ, Nielsen R, Aquadro CF. Patterns of mutation and selection at synonymous sites in Drosophila. Mol. Biol. Evol. 2007;24:2687–2697. doi: 10.1093/molbev/msm196. [DOI] [PubMed] [Google Scholar]
- 63.Thanaraj TA, Argos P. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996;5:1973–1983. doi: 10.1002/pro.5560051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 65.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- 67.Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 2009;26:1571–1580. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Resch AM, Carmel L, Marino-Ramirez L, Ogurtsov AY, Shabalina SA, Rogozin IB, Koonin EV. Widespread positive selection in synonymous sites of mammalian genes. Mol. Biol. Evol. 2007;24:1821–1831. doi: 10.1093/molbev/msm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shields DC, Sharp PM, Higgins DG, Wright F. “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol. Biol. Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 71.Botzman M, Margalit H. Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol. 2011;12:R109. doi: 10.1186/gb-2011-12-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Withers M, Wernisch L, dos Reis M. Archaeology and evolution of transfer RNA genes in the Escherichia coli genome. RNA. 2006;12:933–942. doi: 10.1261/rna.2272306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonos ES, Goddard JP. Human tRNAGlu genes: their copy number and organisation. FEBS Lett. 1990;276:138–142. doi: 10.1016/0014-5793(90)80527-p. [DOI] [PubMed] [Google Scholar]
- 74.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 75.Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- 76.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 77.Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 78.Moura GR, Pinheiro M, Freitas A, Oliveira JL, Frommlet JC, Carreto L, Soares AR, Bezerra AR, Santos MA. Species-specific codon context rules unveil non-neutrality effects of synonymous mutations. PLoS One. 2011;6:e26817. doi: 10.1371/journal.pone.0026817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lipman DJ, Wilbur WJ. Interaction of silent and replacement changes in eukaryotic coding sequences. J. Mol. Evol. 1984;21:161–167. doi: 10.1007/BF02100090. [DOI] [PubMed] [Google Scholar]
- 80.Trotta E. The 3-base periodicity and codon usage of coding sequences are correlated with gene expression at the level of transcription elongation. PLoS One. 2011;6:e21590. doi: 10.1371/journal.pone.0021590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White HB, 3rd, Laux BE, Dennis D. Messenger RNA structure: compatibility of hairpin loops with protein sequence. Science. 1972;175:1264–1266. doi: 10.1126/science.175.4027.1264. [DOI] [PubMed] [Google Scholar]
- 82.Fitch WM. The large extent of putative secondary nucleic acid structure in random nucleotide sequences or amino acid derived messenger-RNA. J. Mol. Evol. 1974;3:279–291. doi: 10.1007/BF01796043. [DOI] [PubMed] [Google Scholar]
- 83.Meyer IM, Miklos I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution: the sounds of silence. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- 85.Mita K, Ichimura S, Zama M, James TC. Specific codon usage pattern and its implications on the secondary structure of silk fibroin mRNA. J. Mol. Biol. 1988;203:917–925. doi: 10.1016/0022-2836(88)90117-9. [DOI] [PubMed] [Google Scholar]
- 86.Zama M. Correlation between mRNA structure of the coding region and translational pauses. Nucleic Acids Symp. Ser. 1999:81–82. doi: 10.1093/nass/42.1.81. [DOI] [PubMed] [Google Scholar]
- 87.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seffens W, Digby D. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 1999;27:1578–1584. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shabalina SA, Ogurtsov AY, Spiridonov NA. Periodic pattern of secondary structures in prokaryotic and eukaryotic mRNAs. FEBS J. 2007;274:366–366. [Google Scholar]
- 91.Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. Genome-wide Measurement of RNA Folding Energies. Mol. Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 2004;32:1774–1782. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina SA, Shagin D, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum. Mol. Genet. 2005;14:135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 97.Tsao D, Shabalina SA, Gauthier J, Dokholyan NV, Diatchenko L. Disruptive mRNA folding increases translational efficiency of catechol-O-methyltransferase variant. Nucleic Acids Res. 2011;39:6201–6212. doi: 10.1093/nar/gkr165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bonnefoy N, Bsat N, Fox TD. Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol. Cell. Biol. 2001;21:2359–2372. doi: 10.1128/MCB.21.7.2359-2372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Williams EH, Fox TD. Antagonistic signals within the COX2 mRNA coding sequence control its translation in Saccharomyces cerevisiae mitochondria. RNA. 2003;9:419–431. doi: 10.1261/rna.2182903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 101.Shen LX, Basilion JP, Stanton VP., Jr Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc. Natl Acad. Sci. USA. 1999;96:7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duan J, Antezana MA. Mammalian mutation pressure, synonymous codon choice, and mRNA degradation. J. Mol. Evol. 2003;57:694–701. doi: 10.1007/s00239-003-2519-1. [DOI] [PubMed] [Google Scholar]
- 103.Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, 3rd, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- 105.Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, Nussinov R. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 2008;383:281–291. doi: 10.1016/j.jmb.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 107.Shabalina SA. [Region of intermolecular complementarity in Escherichia coli 16S rRNA, mRNA, and tRNA molecules] Mol. Biol. (Mosk) 2002;36:460–465. [PubMed] [Google Scholar]
- 108.Nechipurenko Iu D, Popov NV, Shabalina SA, Isaev MA, Matveeva OV. [A model of multiple contacts, describing the interaction of rRNA sites with mRNA] Biofizika. 1995;40:1208–1213. [PubMed] [Google Scholar]
- 109.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barendt PA, Shah NA, Barendt GA, Sarkar CA. Broad-specificity mRNA-rRNA complementarity in efficient protein translation. PLoS Genet. 2012;8:e1002598. doi: 10.1371/journal.pgen.1002598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kozak M. The scanning model for translation: an update. J. Cell. Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sarge KD, Maxwell ES. Intermolecular hybridization of 5S rRNA with 18S rRNA: identification of a 5′-terminally-located nucleotide sequence in mouse 5S rRNA which base-pairs with two specific complementary sequences in 18S rRNA. Biochim. Biophys. Acta. 1991;1088:57–70. doi: 10.1016/0167-4781(91)90153-d. [DOI] [PubMed] [Google Scholar]
- 113.Mauro VP, Edelman GM. rRNA-like sequences occur in diverse primary transcripts: implications for the control of gene expression. Proc. Natl Acad. Sci. USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ogurtsov AY, Marino-Ramirez L, Johnson GR, Landsman D, Shabalina SA, Spiridonov NA. Expression patterns of protein kinases correlate with gene architecture and evolutionary rates. PLoS One. 2008;3:e3599. doi: 10.1371/journal.pone.0003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Panopoulos P, Mauro VP. Antisense masking reveals contributions of mRNA-rRNA base pairing to translation of Gtx and FGF2 mRNAs. J. Biol. Chem. 2008;283:33087–33093. doi: 10.1074/jbc.M804904200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 117.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 118.Gu W, Wang X, Zhai C, Xie X, Zhou T. Selection on synonymous sites for increased accessibility around mirna binding sites in plants. Mol. Biol. Evol. 2012;29:3037–3044. doi: 10.1093/molbev/mss109. [DOI] [PubMed] [Google Scholar]
- 119.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 120.Nakamoto M, Jin P, O'Donnell WT, Warren ST. Physiological identification of human transcripts translationally regulated by a specific microRNA. Hum. Mol. Genet. 2005;14:3813–3821. doi: 10.1093/hmg/ddi397. [DOI] [PubMed] [Google Scholar]
- 121.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl Acad. Sci. USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 126.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schnall-Levin M, Rissland OS, Johnston WK, Perrimon N, Bartel DP, Berger B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21:1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 129.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 132.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang S, Wu S, Ding J, Lin J, Wei L, Gu J, He X. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 2010;38:7211–7218. doi: 10.1093/nar/gkq564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc. Natl Acad. Sci. USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat. Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 136.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 137.Yamashita R, Wakaguri H, Sugano S, Suzuki Y, Nakai K. DBTSS provides a tissue specific dynamic view of Transcription Start Sites. Nucleic Acids Res. 2010;38:D98–D104. doi: 10.1093/nar/gkp1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baek D, Davis C, Ewing B, Gordon D, Green P. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 2007;17:145–155. doi: 10.1101/gr.5872707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 141.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 142.Wang P, Lyman RF, Shabalina SA, Mackay TF, Anholt RR. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics. 2007;177:1655–1665. doi: 10.1534/genetics.107.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 144.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 145.Chamary JV, Hurst LD. Biased codon usage near intron-exon junctions: selection on splicing enhancers, splice-site recognition or something else? Trends Genet. 2005;21:256–259. doi: 10.1016/j.tig.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 146.Gray MW. Evolutionary origin of RNA editing. Biochemistry. 2012;51:5235–5242. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- 147.Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Casey JL. RNA editing in hepatitis delta virus. Curr. Top. Microbiol. Immunol. 2006;307:67–89. doi: 10.1007/3-540-29802-9_4. [DOI] [PubMed] [Google Scholar]
- 149.Kolakofsky D, Roux L, Garcin D, Ruigrok RW. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J. Gen. Virol. 2005;86:1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- 150.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 151.Park E, Williams B, Wold BJ, Mortazavi A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012;22:1626–1633. doi: 10.1101/gr.134957.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maas S, Gommans WM. Novel exon of mammalian ADAR2 extends open reading frame. PLoS One. 2009;4:e4225. doi: 10.1371/journal.pone.0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 156.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 157.Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc. Natl Acad. Sci. USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011;39:6679–6691. doi: 10.1093/nar/gkr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bekaert M, Ivanov IP, Atkins JF, Baranov PV. Ornithine decarboxylase antizyme finder (OAF): fast and reliable detection of antizymes with frameshifts in mRNAs. BMC Bioinformatics. 2008;9:178. doi: 10.1186/1471-2105-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mazzoni C, Falcone C. mRNA stability and control of cell proliferation. Biochem. Soc. Trans. 2011;39:1461–1465. doi: 10.1042/BST0391461. [DOI] [PubMed] [Google Scholar]
- 164.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 165.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 168.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang Z, Zhou L, Hu L, Zhu Y, Xu H, Liu Y, Chen X, Yi X, Kong X, Hurst LD. Nonsense-mediated decay targets have multiple sequence-related features that can inhibit translation. Mol. Syst. Biol. 2010;6:442. doi: 10.1038/msb.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang S, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ruiz-Echevarria MJ, Munshi R, Tomback J, Kinzy TG, Peltz SW. Characterization of a general stabilizer element that blocks deadenylation-dependent mRNA decay. J. Biol. Chem. 2001;276:30995–31003. doi: 10.1074/jbc.M010833200. [DOI] [PubMed] [Google Scholar]
- 172.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 173.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 174.Hennigan AN, Jacobson A. Functional mapping of the translation-dependent instability element of yeast MATalpha1 mRNA. Mol. Cell. Biol. 1996;16:3833–3843. doi: 10.1128/mcb.16.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang Z, Xin D, Wang P, Zhou L, Hu L, Kong X, Hurst LD. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cusack BP, Arndt PF, Duret L, Roest Crollius H. Preventing dangerous nonsense: selection for robustness to transcriptional error in human genes. PLoS Genet. 2011;7:e1002276. doi: 10.1371/journal.pgen.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.de Lima Morais DA, Harrison PM. Large-scale evidence for conservation of NMD candidature across mammals. PLoS One. 2010;5:e11695. doi: 10.1371/journal.pone.0011695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Galanello R, Cao A. Relationship between genotype and phenotype. Thalassemia intermedia. Ann. N. Y. Acad. Sci. 1998;850:325–333. doi: 10.1111/j.1749-6632.1998.tb10489.x. [DOI] [PubMed] [Google Scholar]
- 179.Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, Collawn JF, Bebok Z. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 2010;285:28741–28748. doi: 10.1074/jbc.M110.154575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Day JW, Ranum LP. RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord. 2005;15:5–16. doi: 10.1016/j.nmd.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 181.Sakaguchi N, Maeda K, Kuwahara K. Molecular mechanism of immunoglobulin V-region diversification regulated by transcription and RNA metabolism in antigen-driven B cells. Scand. J. Immunol. 2011;73:520–526. doi: 10.1111/j.1365-3083.2011.02557.x. [DOI] [PubMed] [Google Scholar]
- 182.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell. Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jansen RP, Niessing D. Assembly of mRNA-protein complexes for directional mRNA transport in eukaryotes—an overview. Curr. Protein. Pept. Sci. 2012;13:284–293. doi: 10.2174/138920312801619493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell. Biol. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J. Neurosci. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. J. Neurosci. 2010;30:9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Perycz M, Urbanska AS, Krawczyk PS, Parobczak K, Jaworski J. Zipcode binding protein 1 regulates the development of dendritic arbors in hippocampal neurons. J. Neurosci. 2011;31:5271–5285. doi: 10.1523/JNEUROSCI.2387-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liao G, Ma X, Liu G. An RNA-zipcode-independent mechanism that localizes Dia1 mRNA to the perinuclear ER through interactions between Dia1 nascent peptide and Rho-GTP. J. Cell Sci. 2011;124:589–599. doi: 10.1242/jcs.072421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH. ZBP1 enhances cell polarity and reduces chemotaxis. J. Cell Sci. 2007;120:3173–3178. doi: 10.1242/jcs.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Warnecke T, Batada NN, Hurst LD. The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genet. 2008;4:e1000250. doi: 10.1371/journal.pgen.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Trifonov EN, Volkovich Z, Frenkel ZM. Multiple levels of meaning in DNA sequences, and one more. Ann. N. Y. Acad. Sci. 2012;1267:35–38. doi: 10.1111/j.1749-6632.2012.06589.x. [DOI] [PubMed] [Google Scholar]
- 195.Akashi H, Eyre-Walker A. Translational selection and molecular evolution. Curr. Opin. Genet. Dev. 1998;8:688–693. doi: 10.1016/s0959-437x(98)80038-5. [DOI] [PubMed] [Google Scholar]
- 196.Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164:1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shabalina SA, Ogurtsov AY, Kondrashov VA, Kondrashov AS. Selective constraint in intergenic regions of human and mouse genomes. Trends Genet. 2001;17:373–376. doi: 10.1016/s0168-9525(01)02344-7. [DOI] [PubMed] [Google Scholar]
- 198.Kondrashov AS, Shabalina SA. Classification of common conserved sequences in mammalian intergenic regions. Hum. Mol. Genet. 2002;11:669–674. doi: 10.1093/hmg/11.6.669. [DOI] [PubMed] [Google Scholar]
- 199.Shabalina SA, Kondrashov AS. Pattern of selective constraint in C. elegans and C. briggsae genomes. Genet Res. 1999;74:23–30. doi: 10.1017/s0016672399003821. [DOI] [PubMed] [Google Scholar]
- 200.Xing Y, Lee C. Can RNA selection pressure distort the measurement of Ka/Ks? Gene. 2006;370:1–5. doi: 10.1016/j.gene.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 201.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lu H, Lin L, Sato S, Xing Y, Lee CJ. Predicting functional alternative splicing by measuring RNA selection pressure from multigenome alignments. PLoS Comput. Biol. 2009;5:e1000608. doi: 10.1371/journal.pcbi.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bazykin GA, Kondrashov FA, Ogurtsov AY, Sunyaev S, Kondrashov AS. Positive selection at sites of multiple amino acid replacements since rat-mouse divergence. Nature. 2004;429:558–562. doi: 10.1038/nature02601. [DOI] [PubMed] [Google Scholar]
- 204.Hughes AL, Yeager M. Comparative evolutionary rates of introns and exons in murine rodents. J. Mol. Evol. 1997;45:125–130. doi: 10.1007/pl00006211. [DOI] [PubMed] [Google Scholar]
- 205.Keightley PD, Gaffney DJ. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proc. Natl Acad. Sci. USA. 2003;100:13402–13406. doi: 10.1073/pnas.2233252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Webster MT, Smith NG. Fixation biases affecting human SNPs. Trends Genet. 2004;20:122–126. doi: 10.1016/j.tig.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 207.Li WH. Models of nearly neutral mutations with particular implications for nonrandom usage of synonymous codons. J. Mol. Evol. 1987;24:337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- 208.Akashi H. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics. 1996;144:1297–1307. doi: 10.1093/genetics/144.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Innan H, Stephan W. Selection intensity against deleterious mutations in RNA secondary structures and rate of compensatory nucleotide substitutions. Genetics. 2001;159:389–399. doi: 10.1093/genetics/159.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hellmann I, Zollner S, Enard W, Ebersberger I, Nickel B, Paabo S. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 2003;13:831–837. doi: 10.1101/gr.944903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sanjuan R, Borderia AV. Interplay between RNA structure and protein evolution in HIV-1. Mol Biol Evol. 2011;28:1333–1338. doi: 10.1093/molbev/msq329. [DOI] [PubMed] [Google Scholar]
- 213.Lin MF, Kheradpour P, Washietl S, Parker BJ, Pedersen JS, Kellis M. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res. 2011;21:1916–1928. doi: 10.1101/gr.108753.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Iida K, Akashi H. A test of translational selection at ‘silent’ sites in the human genome: base composition comparisons in alternatively spliced genes. Gene. 2000;261:93–105. doi: 10.1016/s0378-1119(00)00482-0. [DOI] [PubMed] [Google Scholar]
- 215.Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, Tchivileva IE, Belfer I, Mishra B, Kiselycznyk C, et al. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum. Mol. Genet. 2009;18:1037–1051. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl Acad. Sci. USA. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Moss WN, Priore SF, Turner DH. Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA. 2011;17:991–1011. doi: 10.1261/rna.2619511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stoletzki N, Eyre-Walker A. The positive correlation between dN/dS and dS in mammals is due to runs of adjacent substitutions. Mol. Biol. Evol. 2011;28:1371–1380. doi: 10.1093/molbev/msq320. [DOI] [PubMed] [Google Scholar]
- 219.Lagunez-Otero J, Trifonov EN. mRNA periodical infrastructure complementary to the proof-reading site in the ribosome. J. Biomol. Struct. Dyn. 1992;10:455–464. doi: 10.1080/07391102.1992.10508662. [DOI] [PubMed] [Google Scholar]
- 220.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bindewald E, Wendeler M, Legiewicz M, Bona MK, Wang Y, Pritt MJ, Le Grice SF, Shapiro BA. Correlating SHAPE signatures with three-dimensional RNA structures. RNA. 2011;17:1688–1696. doi: 10.1261/rna.2640111. [DOI] [PMC free article] [PubMed] [Google Scholar]