Abstract
Paternal duplications of chromosome 6q24, a region that contains the imprinted PLAGL1 and HYMAI transcripts, are associated with transient neonatal diabetes mellitus. A common feature of imprinted genes is that they tend to cluster together, presumably as a result of sharing common cis-acting regulatory elements. To determine the extent of this imprinted cluster in human and mouse, we have undertaken a systematic analysis of allelic expression and DNA methylation of the genes mapping within an ∼1.4-Mb region flanking PLAGL1/Plagl1. We confirm that all nine neighbouring genes are biallelically expressed in both species. In human we identify two novel paternally expressed PLAGL1 coding transcripts that originate from unique promoter regions. Chromatin immunoprecipitation for CTCF and the cohesin subunits RAD21 and SMC3 reveals evolutionarily conserved binding sites within unmethylated regions ∼5 kb downstream of the PLAGL1 differentially methylated region and within the PLAGL1 3′ untranslated region (UTR). Higher-order chromatin looping occurs between these regions in both expressing and non-expressing tissues, forming a non-allelic chromatin loop around the PLAGL1/Plagl1 gene. In placenta and brain tissues, we identify an additional interaction between the PLAGL1 P3/P4 promoters and the unmethylated element downstream of the PLAGL1 differentially methylated region that we propose facilitates imprinted expression of these alternative isoforms.
INTRODUCTION
Genomic imprinting is defined by genotype-independent expression from either the maternal or paternal allele. This parent-of-origin expression is mediated by epigenetic modifications that differ between the two parental chromosomes (1). There are currently more than 60 imprinted genes in humans, which are largely conserved in mouse (2; http://igc.otago.ac.nz/home.html). Human diseases associated with cytogenetic abnormalities or (epi)mutations, such as transient neonatal diabetes mellitus (TNDM) and Beckwith–Wiedemann, Silver–Russell, Angelman and Prader–Willi syndromes, and targeted mouse experiments have revealed that imprinted genes are essential for foetal and postnatal growth and behaviour, with aberrant expression in adults associated with multigenic diseases and cancer (3–8).
Imprinted genes are regulated by cis-acting imprinting control regions (ICRs), which manifest as regions of allelic DNA methylation that are set during gametogenesis and are inherited throughout somatic development by the action of the DNMT1 complex (9–11). These differentially methylated regions (DMRs) are associated with a canonical chromatin signature, which comprises H3 lysine 9 trimethylation and H4 lysine 20 trimethylation on the DNA-methylated allele, whereas H3 lysine 4 methylation is associated with the unmethylated allele and may confer protection from de novo DNA methylation (12).
Imprinted genes rarely occur in isolation, as monoallelic expression is achieved by shared cis-regulatory elements that act in distinct ways at different loci (reviewed in 1). Despite information from a few well-studied loci, still little is known how ICRs silence neighbouring genes, and there seems to be two prevailing models of regulation. In general, paternally methylated intergenic ICRs act as methylation-sensitive insulators recruiting CTCF (13), while the majority of maternally methylated ICRs act as promoters. In some cases, these differentially methylated promoters are associated with long non-coding RNAs that confer silencing of neighbouring genes in cis through recruitment of histone-remodelling complexes (14,15). However, for the majority of imprinted domains, the intricacies of ICR action are unknown, and novel mechanisms, such as allele-specific alternative polyadenylation choice, are continuously updating our insight on imprinted regulation (16). Imprinted domains are especially attractive to researchers as they permit the study of inherited epigenetic transcriptional regulation, as both the active and silent alleles are genetically identical and are present within the same nucleus, exposed to the same trans-acting chromatin regulators.
To date, the imprinted domain on human chromosome 6 associated with TNDM contains two paternally expressed transcripts, PLAGL1 (previously known as ZAC1) and the non-coding RNA HYMAI, both initiating from within the maternally methylated PLAGL1-DMR (17,18). To determine whether these transcripts are part of a larger imprinting cluster, we performed allelic expression analysis of the flanking genes in both human and mouse. This revealed that the region is a micro-imprinted domain, as all surrounding transcripts are biallelically expressed in both species, including PHACTR2, which we show exhibits allelic bias in human placenta associated with genotype and not parental origin (19). Interrogation of publicly available CTCF ChIP-seq data shows that PLAGL1 is flanked by CTCF-binding sites conserved between species in both expressing and non-expressing cells. These two CTCF/cohesin regions interact physically despite being separated by more than 70 kb, and we propose this restricts imprinting to only those transcripts contained within the chromatin loop.
MATERIALS AND METHODS
Human tissues
A cohort comprising 65 foetal tissue sets (8–18 weeks) with corresponding maternal blood samples and 96 term placental samples from the Moore Tissue bank are described elsewhere (20). An additional 96 human placenta samples were collected at Hospital St Joan De Deu (Barcelona, Spain). DNA and RNA extraction and cDNA synthesis were carried out as previously described (21). A selection of normal adult brain samples was obtained from BrainNet Europe/Barcelona Brain Bank. The human RNA panel was purchased from Clontech (Human Total RNA master Panel II). Ethical approval for adult blood and foetal tissue collection was granted by the Hammersmith, Queen Charlotte’s & Chelsea and Acton Hospital Research Ethics Committee (Project Registration 2001/6029 and 2001/6028); collection of the HSJD placental cohort was granted by the ethical committee of Hospital St Joan De Deu (Study Number 35/07) and IDIBELL (PR006/08). Written informed consent was obtained from all participants.
Mouse crosses and cell lines
To determine allelic expression in mouse, wild-type embryos and placentas were produced by crossing C57BL/6 (B) with Mus musculus castaneus (C) mice. RNA and DNA were isolated and extracted as previously described (10). Control lymphoblastoid cell lines were established by Epstein–Barr virus transformation of peripheral blood cells and propagated as previously described (22). Before chromatin immunoprecipitation (ChIP), the lymphoblastoid methylation signature throughout the PLAGL1 domain was compared with leucocytes to ensure that the transformation process had not altered the epigenetic profile. The human TCL1 placental trophoblast cell line was grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and antibiotics.
Allelic expression analysis
Genotypes were obtained in the foetal and placental tissues for expressed genes using PCR and direct sequencing. Sequences were interrogated using Sequencher v4.6 (Gene Codes Corporation, MI) to distinguish heterozygous and homozygous samples. Heterozygous sample sets were analysed for allelic expression using RT-PCR that incorporated the polymorphisms in the final PCR product (Supplementary Table S1). The resulting RT-PCR amplicons were sequenced in both directions. The amplification cycle number for each transcript was determined to be within the exponential phase of the PCR, which varied for each gene, but was between 32 and 40 cycles.
Pyrosequencing analysis for allelic expression quantification
Pyrosequencing was used as an accurate method of quantifying allelic expression of PHACTR2 in heterozygous placenta samples. Standard RT-PCR was used to amplify across two independent single nucleotide polymorphisms (SNPs) mapping to exon 6 (an alternative spliced exon) and the 3′ UTR, with the exception that the reverse primers were biotinylated. The entire biotinylated RT-PCR product (diluted to 40 µl) was mixed with 38 µl of binding buffer and 2 µl (10 mg/ml) of streptavidin-coated polystyrene beads. Bead–amplicon complexes were captured on a vacuum prep tool (Qiagen), and the PCR products denatured using 0.2 M NaOH. The denatured DNA was resuspended in 40 pmol of sequencing primer dissolved in 12 µl of water, and primer annealing was achieved by heating the sample to 80°C for 2 min before cooling to room temperature. For sequencing, forward primers were designed to the complementary strand (Supplementary Table S1). The pyrosequencing reaction was carried out on a PyroMark Q96 instrument. The peak heights were determined using the pyrosequencing commercial software.
Real-time RT-PCR
All PCR amplifications were run in triplicate on either an Applied Biosystems 7500 or 7900 Fast real-time PCR machines (Applied Biosystems) following the manufacturers’ protocol. All primers were optimized using SYBR Green (Supplementary Table S1 for PLAGL1 primers, all other genes were amplified using KiCqStart primers from Sigma-Aldrich), and melt curve analysis was performed to ensure that amplicons were specific and free of primer-dimer products. Thermal cycle parameters included Taq polymerase activation at 95°C for 10 min for one cycle, repetitive denaturation at 95°C for 15 s and annealing at 60°C for 1 min for 40 cycles. All resulting triplicate cycle threshold (Ct) values had to be within 1 Ct of each other. The quantitative values for each triplicate were determined as a ratio with the level of RPL19 expression, which was measured in the same sample.
Analysis of allelic DNA methylation
The DNA methylation profile throughout the PLAGL1 domain was determined using published data from our previous Illumina Infinium HumanMethylation27 BeadChIP array analysis (GEO accession number: GSE 28525) (23). Promoter regions not included on this array platform were assessed by conventional bisulphite PCR. Approximately 1 µg of DNA was subjected to sodium bisulphite treatment and purified using the EZ GOLD methylation kit (ZYMO, Orange, CA) and was used for all bisulphite PCR analysis. Bisulphite PCR primers for each region were used with Hotstar Taq polymerase (Qiagen, West Sussex, UK) at 45 cycles, and the resulting PCR product was cloned into pGEM-T easy vector (Promega) for subsequent sequencing (see Supplementary Table S1).
Chromatin immunoprecipitation
We used previously published CTCF ChIP-seq data sets for leucocytes and CD34 cells (GEO accession number: GSM651541) and chromatin interaction analysis with paired-end tags (ChIA-PET; GEO accession number: GSM970213 and GSM970216) (http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.aspx) to identify potential binding sites within the 6q24-imprinted domain. For ChIP, chromatin from ∼80 million cells was aliquoted into 100-µg batches and used for each immunoprecipitation reaction with Protein A Agarose/Salmon Sperm DNA (Millipore, 16-157) and specific antibody. The antibody against CTCF (07-729) was obtained from Millipore, and the antibodies against RAD21 (AB992) and SMC3 (AB9263) were obtained from ABCAM. For each ChIP, a fraction of the input chromatin (1%) was also processed for DNA purification, and a mock immunoprecipitation with a neutral unrelated IgG antiserum was carried out in parallel.
Levels of immunoprecipitated chromatin at specific region were determined by qPCR using an Applied Biosystems 7900 Fast real-time PCR machine, using SYBR Green (Applied Biosystems). Each PCR was run in triplicate, and protein binding was quantified as a percentage of total input material. Normalization of ChIP levels was performed by comparing precipitation levels obtained for each specific region of interest with the level obtained for a known CTCF/cohesin-positive control region mapping to chromosome 6 (24) (see Supplementary Table S1 for primer sequences).
Chromatin conformation capture analysis
The chromatin conformation capture (3C) protocol was performed as previously described (25), with minor amendments. Briefly, HindIII was used to digest 1 × 107 formaldehyde–cross-linked nuclei from leucocytes, placental cell line TCL1, mouse embryonic fibroblasts (MEFs) (for which EcoR1 was used) or 100 mg of archived tissue (overnight digestion, 1200 U, NEB). The efficiency of the restricted enzyme digestion was assessed by qPCR across each restriction site, comparing digested and undigested chromatin fractions. Only chromatin with digestion efficiency more than 70% was used. Subsequently, the DNA was ligated overnight in a 500-ul reaction volume using 1950 units of T4 ligase (Fermentas). DNA was de–cross-linked by incubating overnight at 65°C and purified using phenol/chloroform extraction. This DNA was used for both nested PCR for analysis of allelic interactions, and real-time PCR (LightCycler, Roche Applied Science) to determine the frequency of interactions, using constant primers either in the unmethylated regions ∼5 kb upstream from the DMR, within the last PLAGL1 exon, or between the P3/P4 promoters (see Supplementary Table S1). Primer efficiency and basal interaction frequencies were determined using digested and ligated bacterial artificial chromosomes (BAC) DNA (human RPII RP11 947A22; mouse RP24 399D18) as described by Braem and co-workers (26). All 3C experiments were performed in at least 3 biological samples per tissue in technical triplicates.
RESULTS
PLAGL1 and /HYMAI are the only imprinted genes mapping to 6q24
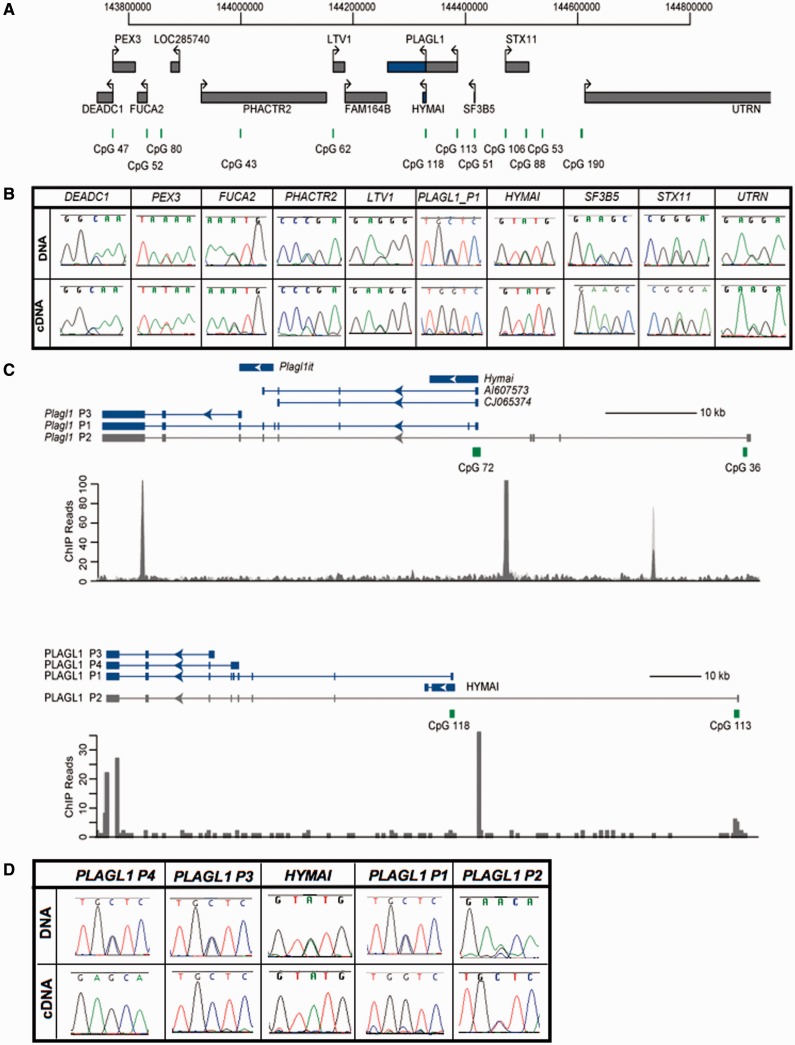
To characterize the boundaries of the imprinted region on human chromosome 6, we determined the abundance and allele-specific expression of nine genes flanking PLAGL1 (DEADC1, PEX3, FUCA2, PHACTR2, LTV1, SF3B5, STX11, BC033369/LOC285740 and UTRN) using RT-PCR amplification across transcribed SNPs in first trimester foetal tissues and term placenta. No SNPs were identified within FAM164B. We confirm that human PLAGL1 transcripts from the P1 promoter and HYMAI were paternally expressed in all tissues analysed at all gestational ages. The PLAGL1 expression from the P2 promoter was biallelic when detectable. Biallelic expression of the remaining transcripts was observed in all tissues analysed, and their associated promoters were unmethylated (Figure 1A and B; Supplementary Figures S1 and S2; Supplementary Table S2).
Figure 1.
Schematic representation of the PLAGL1 domain on human chromosome 6p24. (A) The relative organization of the human genes and CpG islands. The orthologous mouse domain has a similar organization, except the lack of the non-coding RNA LOC285740. (B) The allelic expression for all genes in the human cluster. The sequence traces for heterozygous DNA samples from term placentas are shown for all genes, as well as the resulting RT-PCR. Black boxes depict genes that are expressed biallelically in all tissues throughout gestation, whereas blue boxes represent ubiquitously imprinted, paternally expressed transcripts. (C) A comparison of the structure and position of the imprinted transcripts within the Plagl1/PLAGL1 transcriptional unit, including the location of the CTCF sites. (D) DNA sequences across rs2076684 SNP in the penultimate PLAGL1 exon and the RT-PCR sequence traces for each isoform in term placenta.
To assess the imprinting profile within the mouse orthologous region on chromosome 10, we used various embryonic tissues and placenta (at embryonic stage E14.5) from reciprocal crosses between C57BL/6 (B) and Mus musculus castaneus (C). We confirm the ubiquitous imprinting previously described for the paternally expressed Plagl1 transcript, and observed equal expression from both parental alleles for all other genes (Supplementary Figure S3).
Conserved paternally expressed PLAGL1 isoforms
We recently identified and characterized several paternally expressed Plagl1 alternative transcripts in mouse (27) (Figure 1C). One of these transcripts, originating from the P3 promoter region, results in a transcript containing the protein-coding exons. As a result of expressed sequence tag (EST) and mRNA alignments, we identified an orthologous transcript originating from the equivalent P3 promoter region in the human. This transcript (reference EST AJ006354) originates from a unique promoter region 5′ to the exon 6 acceptor site (tctcacag/GTTTGAAT or tgtacaag/GTCTCTTC) of the P1-PLAGL1 transcript, with a 5′ UTR that extends at least 800 bp into the upstream intron. In addition, we identified another transcript originating from a novel promoter region ∼3.5 kb upstream of the P3 promoter, located 5′ to the exon 5 acceptor site (ctttctag/GTATTTGC) of P1-PLAGL1 transcript, which we named P4-PLAGL1 (Figure 1C). The resulting transcript (reference EST AK091707) also includes the full open reading frame. Both of these promoter regions can produce transcripts with alternative splicing of the penultimate exon, which results in protein lacking the first two zinc finger domains. These transcripts are evident on the northern blot analysis performed by Kas et al. (28) using a probe encompassing last exon of PLAGL1. Using strategically positioned RT-PCR primers, we were unable to link these unique 5′ UTRs with exons 1–3 of P1-PLAGL1, confirming that these are independent transcripts. Consistent with this, we observe that P1-PLAGL1 expression was higher than all other isoforms in a panel of foetal and adult tissues (Supplementary Figure S4), with all transcripts being most plentiful in placenta and absent in leucocytes. Using an SNP located within exon 7, we show that these novel isoforms are paternally expressed in placenta (n = 11) and monoallelically expressed in brain (n = 2) (Figure 1C and D).
Human PHACTR2 exhibits genotype-associated allelic expression in placenta
Partial imprinting of PHACTR2 has been described in some term placentas, with maternal bias observed for the SNP rs1082 within the 3′ UTR of the gene. The expression levels of PHACTR2 were reported to be lower in heterozygous placentas compared with homozygous samples, suggesting that the genotype could play a role in the allelic bias (19). Using both standard Sanger DNA sequencing and quantitative pyrosequencing, we also observed a range of allelic profiles for rs1082, consistent with previous reports. However, on assessing a second SNP (rs2073214) within exon 6 of PHACTR2, three placental samples showed equal expression from both parental alleles, whereas 17 had strong preferential/monoallelic expression of the T allele. Subsequent genotyping of maternal DNAs corresponding to these samples identified two placentas with expression from the paternal allele, revealing that the monoallelic expression observed is not due to imprinting but presumably SNP-associated regulation (Supplementary Figure S5).
CTCF/cohesin form non-allelic boundaries flanking PLAGL1
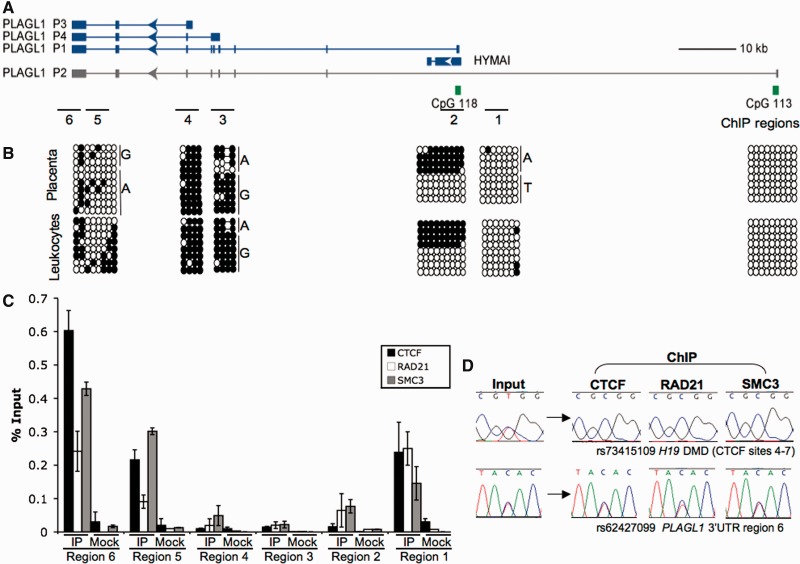
In an attempt to understand the mechanism restricting imprinting to HYMAI and the PLAGL1 transcripts, we interrogated genome-wide CTCF data sets to identify potential CTCF boundaries. Using two published data sets (29,30), we identified two regions of strong CTCF binding, one ∼5 kb downstream of the PLAGL1-DMR and between this and the non-imprinted P2 promoter, and the other within the last exon of PLAGL1. This latter region showed two independent CTCF peaks ∼2 kb apart (Figures 1C and 2). Using PCR on bisulphite-converted DNA, we were able to show that the CpG dinucleotides surrounding these CTCF-binding sites are unmethylated in leucocyte- and placenta-derived DNA (Figure 2B). Comparison of the sequence surrounding these binding sites identified CTCF consensus sites that contain both motifs (M1 and M2) recently described by Schmidt and co-workers (31). The M1 motifs of both regions contain CpG dinucleotides, which ensure methylation-sensitive binding. Interestingly, the second binding site within the last PLAGL1 exon (region 6, Figure 2) lacks the M2 motif and CpG dinucleotides (data not shown).
Figure 2.
DNA methylation and ChIP of CTCF, RAD21 and SMC3 analysis throughout the PLAGL1 locus. (A) The localization of the bisulphite PCR and ChIP regions analysed. (B) Bisulphite sequence in placenta- and leucocyte-derived DNA. Each circle represents a single CpG dinucleotide on the strand: (filled circle) a methylated cytosine, (open circle) unmethylated cytosines. (C) qPCR performed on CTCF, RAD21 and SMC3 ChIP material in normal lymphoblastoid cells at various regions across the PLAGL1 domain. (D) Sequence traces showing monoallelic precipitation of CTCF and cohesion subunits at the control H19-ICR domain and biallelic precipitation at region 6 mapping within the PLAGL1 3′ UTR.
Subsequent ChIP using antisera to CTCF was performed to confirm binding in lymphoblastoid cells and the placental cell line TCL1. We observed strong CTCF enrichment within the regions downstream of the PLAGL1-DMR and in the last exon, but not within the PLAGL1-DMR itself or in the regions overlapping the P3 and P4 promoters (Figure 2C; data not shown). Recently, cohesin has been shown to play a critical role in maintaining CTCF higher-order chromatin conformation at the H19-IGF2 loci (32). To determine whether CTCF and cohesin co-localize within the PLAGL1 regions, we performed ChIP using antisera against the cohesin subunits RAD21 and SMC3 (Figure 2C). In lymphoblastoiod cell lines, the cohesin subunits clearly precipitated with CTCF on one allele at the H19-ICR control region, and on both chromosomes throughout the PLAGL1 domain (Figure 2D).
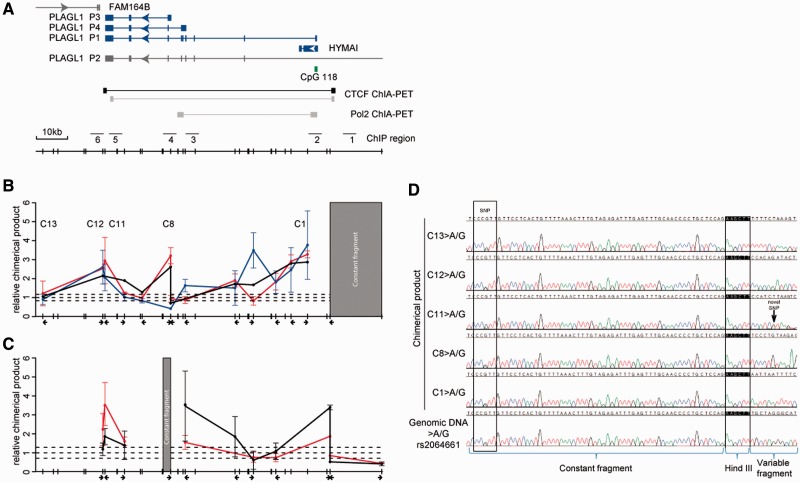
The biallelic CTCF regions adjacent to PLAGL1 physically interact to form chromatin loops
Recent studies have revealed that CTCF is a key regulator of chromatin fibre structure and responsible for organizing chromosomal territories within the cell nucleus (33,34). To determine whether the CTCF/cohesin sites physically interact, we performed 3C experiments to identify potential chromatin folding. 3C-qPCR assays were performed on adult leucocytes and brain samples, term placenta material and the TCL1cell line, and interaction frequencies were determined between a constant HindIII site located ∼500 bp upstream to the unmethylated CTCF/cohesin binding site 5′ to the PLAGL1-DMR and other HindIII sites throughout the locus. We observed high interaction frequencies of sites separated by <20 kb from the constant fragment, which included the PLAGL1-DMR, reflecting non-specific random collision due to their physical proximity. We subsequently identified strong interactions between the CTCF/cohesin constant fragment and the CTCF/cohesin sites in the last PLAGL1 exon, in different tissues irrespective of expression (Figure 3). Direct sequencing of the chimerical products from placenta and brain revealed that the interactions occurred on both chromosomes, and that the appropriate chimeric products result from the 3C ligations (Figure 3D and Supplementary Figure S6). Interrogation of publicly available ChIA-PET (35) data revealed that CTCF mediates an abundant loop identical to the one we identified (Figure 3). In addition, we also identified non-allelic interactions between the constant fragment and the HindIII sites associated with the P3/P4 promoter regions in placenta and brain (Figure 3B). We did not observe these interactions in leucocytes, which suggests this specific contact occurs only in tissues exhibiting transcription from the P3/P4 promoters and appears independent of CTCF binding. Interestingly, we observed a leucocyte-specific chimeric product between the CTCF/cohesin binding site 5′ to the PLAGL1-DMR and a region in intron 1, which may represent a repressive interaction. These interactions were confirmed in two additional 3C experiments using constant primers in the last PLAGL1 exon (Supplementary Figure S6) as well as the intervening region between the P3 and P4 promoter (Figure 3C).
Figure 3.
Chromatin interactions with the CTCF sites upstream of the PLAGL1-DMR. (A) The position of the HindIII sites used for 3C analysis, and annotated ChIA-PET data showing CTCF and POL2 interactions. (B) The looping profile in brain (black), placental TCL1 cell line (red) and normal leucocytes (blue) using a constant primer 5 kb upstream of the PLAGL1-DMR, showing strong interactions with the 3′ UTR and alternative promoters of PLAGL1. The x-axis shows the position of the primers used. (C) The interaction profile in expressing tissues, brain (black) and placental TCL1 cell line (red), using a constant primer between the P3 and P4 promoters. (D) Heterozygous SNP rs2064661 in constant fragment 5 kb upstream of the PLAGL1-DMR allowed for allele-specific chromatin interactions to be assessed in adult brain. Sequence traces reveal biallelic higher-order chromatin interactions between the constant fragment and various contact points throughout the PLAGL1 domain.
CTCF sites at conservation locations physically interact in the mouse
Because the allelic expression patterns were similar between human and mouse for genes surrounding PLAGL1/Plagl1, we predicted that the higher-order chromatin structure will be comparable. Interrogation of the mouse transcription factor binding site (TFBS) data set from ENCODE (data available on the UCSC browser, NCBI37/mm9 assembly) identified two regions of strong CTCF binding at similar locations as in human (Figure 1). To confirm that these sites interact to form a chromatin loop, we performed allelic 3C on mouse embryonic fibroblast cells. This semi-quantitative analysis revealed a biallelic contact between an EcoRI constant fragment containing the CTCF site located ∼4 kb upstream of the Plagl1-DMR and the binding site in the last exon ∼40 kb downstream (Supplementary Figure S7).
DISCUSSION
By analysing the allelic expression of transcripts flanking PLAGL1/HYMAI, we show that this locus is one of only a few true conserved micro-imprinted domains. Previously reported micro-imprinted domains such as Grb10/GRB10 and imprinted transcripts that result from X-derived retrotransposition into host gene introns have subsequently been shown to influence either neighbouring or host genes (36–38).
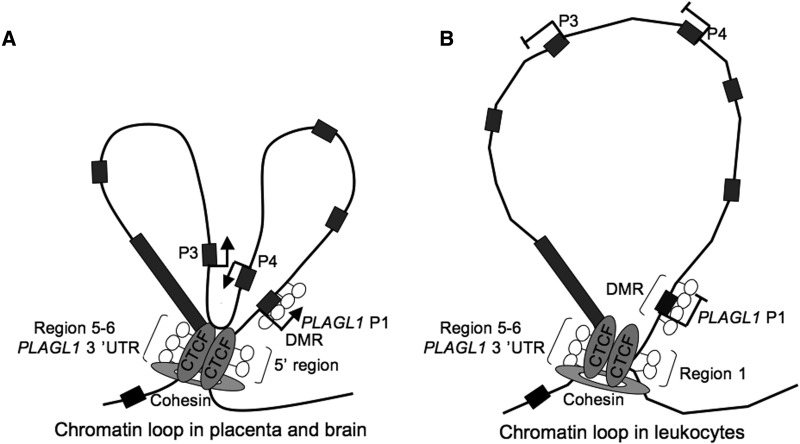
We demonstrate that the imprinted transcriptional unit in both humans and mouse is restricted within CTCF/cohesin boundaries, suggesting that they are associated with a conserved regulatory function. Interestingly, this 70-kb loop is very similar in size to the minimal TNDM critical region identified by micro-duplications (39). By using allelic 3C, we show that the flanking CTCF/cohesin-binding sites physically interact in both species in a biallelic manner. In humans, this confirmation is maintained in both expressing and non-expression tissues, suggesting that the chromatin architecture is maintained by CTCF/cohesin and that additional transcription factors are required to promote expression. This is reminiscent of the CTCF/cohesin long-range interactions observed at the IGF2-H19 domain in lymphoblastoid cells, a cell type that does not express IGF2 despite maintained looping (22,32). In tissues that abundantly express the predominant P1-PLAGL1 transcript, we identified novel imprinted isoforms originating 45 kb downstream. The promoter regions of the P3- and P4-PLAGL1 transcripts are embedded within CpG-poor regions (the upstream 1 kb of P3 and P4 have 12 and 10 CpG dinucleotides, respectively). We hypothesize that the level of transcription from these isoforms is regulated by a combination of the endogenous promoter sequences and shared enhancers with the P1-PLAGL1 promoter, the latter conferring the allelic specificity. This is supported by the observed CTCF-independent chromatin loop in expressed tissues that juxtaposes the P3/P4 promoters and the PLAGL1-DMR so that any methylation-sensitive transcription factor binding at the PLAGL1-DMR could influence P3/P4 (Figure 4). Analysis of ChIA-PET using CTCF and POL2 revealed that this second chromatin interaction is solely present in the POL2 data set, confirming our observations (35).
Figure 4.
Proposed model of the chromatin scaffold of the paternal allele in (A) expressing and (B) non-expressing tissues. In tissues co-expressing transcripts from the P1 and P3/P4 promoters, we speculate that a second chromatin loop unites these regions to share transcription factors.
A meta-analysis of microarray data recently revealed that Plagl1 is a member of a co-regulated imprinted gene network (IGN) important for embryonic growth (40). The mechanism regulating this IGN remains to be elucidated, and may be directly linked to the Plagl1 protein because it encodes a zinc finger transcription factor (41). Alternatively, it may be speculated that CTCF is a common denominator mediating expression of numerous imprinted loci via intra- and interchromosomal interactions (42). Our results suggest that the TNDM phenotype associated with paternal duplications of 6q24 likely results from the overexpression of PLAGL1, the only imprinted protein-coding gene in the region, and it remains to be determined whether PLAGL1 is central to an IGN important for non-syndromic human growth as in mouse.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–7.
FUNDING
Ayuda Merck Serono- Fundación Salud 2000 de Investigación en Endocrinología 2009 (to D.M. and I.I.-P.); Spanish Ministerio de Educacion y Ciencia [SAF2008-1578 to D.M.]; Fundació La Marató de TV3 [101130 to D.M.]; Telethon-Italia N. GGP11122 (to A.R.); Wellcome Trust (to G.E.M.); Medical Research Council (to G.E.M.); Wellbeing of Women (to G.E.M.). D.M. is a Ramon y Cajal research fellow, and A.G.-A. is funded by a FPU studentship. Research in the Neonatal Unit in HSJD is partially funded by an unrestricted grant from BBDue Spain. Funding for open access charge: Internal funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Isidro Ferrer of the Barcelona Brain Bank for supplying human brain specimens, Deborah Bourc’his for B × C tissues and Robert Feil for MEF cells. The authors also thank Juan Sandoval for helpful advice with the ChIP optimization and Thierry Forne for supplying the reagents for the 3C experiments. The authors are especially grateful to all the families and staff members at Hospital Sant Joan de Déu for participating and co-ordinating the placenta tissue collection.
REFERENCES
- 1.Abramowitz LK, Bartolomei MS. Genomic imprinting: recognition and marking of imprinted loci. Curr. Opin. Genet. Dev. 2012;2:72–78. doi: 10.1016/j.gde.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29:275–276. doi: 10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay DJ, Temple IK. Transient neonatal diabetes mellitus type 1. Am. J. Med. Genet. C. Semin. Med. Genet. 2010;154C:335–342. doi: 10.1002/ajmg.c.30272. [DOI] [PubMed] [Google Scholar]
- 4.Eggermann T. Silver-Russell and Beckwith-Wiedemann syndromes: opposite (epi)mutations in 11p15 result in opposite clinical pictures. Horm. Res. 2009;71(Suppl. 2):30–35. doi: 10.1159/000192433. [DOI] [PubMed] [Google Scholar]
- 5.Buiting K. Prader-Willi syndrome and Angelman syndrome (2010) Am. J. Med. Genet. C. Semin. Med. Genet. 2010;15:365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- 6.Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monk D. Deciphering the cancer Imprintome. Brief. Funct. Genomics. 2010;9:329–339. doi: 10.1093/bfgp/elq013. [DOI] [PubMed] [Google Scholar]
- 9.Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;5:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2003;21:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 11.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 12.Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum. Mol. Genet. 2009;18:3375–3383. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- 13.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 14.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Airn noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 15.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, Takada S, Takagi N, Arima T, Wake N, Kamimura K, et al. The cell cycle control gene ZAC/PLAGL1 is imprinted-a strong candidate gene for transient neonatal diabetes. Hum. Mol. Genet. 2000;9:453–460. doi: 10.1093/hmg/9.3.453. [DOI] [PubMed] [Google Scholar]
- 18.Arima T, Drewell RA, Oshimura M, Wake N, Surani MA. A novel imprinted gene, HYMAI, is located within an imprinted domain on human chromosome 6 containing ZAC. Genomics. 2000;67:248–255. doi: 10.1006/geno.2000.6266. [DOI] [PubMed] [Google Scholar]
- 19.Daelemans C, Ritchie ME, Smits G, Abu-Amero S, Sudbery IM, Forrest MS, Campino S, Clark TG, Stanier P, Kwiatkowski D, et al. High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta. BMC Genetics. 2010;11:25. doi: 10.1186/1471-2156-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apostolidou S, Abu-Amero S, O'Donoghue K, Frost J, Olafsdottir O, Chevele KM, Whittaker JC, Loughna P, Stanier P, Moore GE. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J. Mol. Med. 2007;85:379–387. doi: 10.1007/s00109-006-0131-8. [DOI] [PubMed] [Google Scholar]
- 21.Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl Acad. Sci. USA. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nativio R, Sparago A, Ito Y, Weksberg R, Riccio A, Murrell A. Disruption of genomic neighbourhood at the imprinted IGF2-H19 locus in Beckwith-Wiedemann syndrome and Silver-Russell syndrome. Hum. Mol. Genet. 2011;20:1363–1374. doi: 10.1093/hmg/ddr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakabayashi K, Trujillo AM, Tayama C, Camprubi C, Yoshida W, Lapunzina P, Sanchez A, Soejima H, Aburatani H, Nagae G, et al. Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes. Hum. Mol. Genet. 2011;20:3188–97. doi: 10.1093/hmg/ddr224. [DOI] [PubMed] [Google Scholar]
- 24.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 25.Court F, Baniol M, Hagege H, Petit JS, Lelay-Taha MN, Carbonell F, Weber M, Cathala G, Forne T. Long-range chromatin interactions at the mouse Igf2\H19 locus reveal a novel paternally expressed long non-coding RNA. Nucleic Acid Res. 2011;39:1–14. doi: 10.1093/nar/gkr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braem C, Recolin B, Rancourt RC, Angiolini C, Barthes P, Branchu P, Court F, Cathala G, Ferguson-Smith AC, Forne T. Genome matrix attachment region and chromosome confirmation capture quantitative real time PCR assay identify novel putative regulatory elements at the imprinted Dlk1/Gtl2 locus. J. Biol. Chem. 2008;283:18612–18620. doi: 10.1074/jbc.M801883200. [DOI] [PubMed] [Google Scholar]
- 27.Iglesias-Platas I, Martin-Trujillo A, Cirillo D, Court F, Guillaumet-Adkins A, Camprubi C, Bourc’his D, Hata K, Feil R, Tartaglia G, et al. Characterization of novel paternal ncRNAs at the Plagl1 locus, including Hymai, predicted to interact with regulators of active chromatin. PLoS One. 2012;7:e38907. doi: 10.1371/journal.pone.0038907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kas K, Voz ML, Hensen K, Meyen E, Van de Ven WJ. Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J. Biol. Chem. 1998;273:23026–23032. doi: 10.1074/jbc.273.36.23026. [DOI] [PubMed] [Google Scholar]
- 29.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Browm GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin confirmation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol. Syst. Biol. 2010;6:1–6. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;285:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequences. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menheniott TR, Woodfine K, Monk D, Schulz R, Baldwin HS, Moore GE, Oakey RJ. Genomic imprinting of DOPA decarboxylase in heart and reciprocal allelic expression with neighbouring Grb10. Mol. Cell. Biol. 2008;28:386–396. doi: 10.1128/MCB.00862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz R, McCole RB, Woodfine K, Wood AJ, Chahal M, Monk D, Moore GE, Oakey RJ. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum. Mol. Genet. 2009;18:118–127. doi: 10.1093/hmg/ddn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Docherty LE, Poole RL, Mattocks CJ, Lehmann A, Temple IK, Mackey DJ. Further refinement of the critical minimal genetic region for the imprinting disorder 6q24 tranisent neonatal diabetes. Diabetolgia. 2010;53:2347–2351. doi: 10.1007/s00125-010-1853-2. [DOI] [PubMed] [Google Scholar]
- 40.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates and imprinted gene network critically involved in the control of embryonic growth. Dev. Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Huang SM, Stallcup MR. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol. Cell. Biol. 2000;20:1855–1867. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nature Genet. 2011;43:630–640. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.