Abstract
Titin, a sarcomeric protein expressed primarily in striated muscles, is responsible for maintaining the structure and biomechanical properties of muscle cells. Cardiac titin undergoes developmental size reduction from 3.7 megadaltons in neonates to primarily 2.97 megadaltons in the adult. This size reduction results from gradually increased exon skipping between exons 50 and 219 of titin mRNA. Our previous study reported that Rbm20 is the splicing factor responsible for this process. In this work, we investigated its molecular mechanism. We demonstrate that Rbm20 mediates exon skipping by binding to titin pre-mRNA to repress the splicing of some regions; the exons/introns in these Rbm20-repressed regions are ultimately skipped. Rbm20 was also found to mediate intron retention and exon shuffling. The two Rbm20 speckles found in nuclei from muscle tissues were identified as aggregates of Rbm20 protein on the partially processed titin pre-mRNAs. Cooperative repression and alternative 3′ splice site selection were found to be used by Rbm20 to skip different subsets of titin exons, and the splicing pathway selected depended on the ratio of Rbm20 to other splicing factors that vary with tissue type and developmental age.
INTRODUCTION
Titin, the gene containing the largest number of exons (363 exons in human), encodes the largest polypeptide in nature [2.97–3.7 megadaltons (MDa)] (1,2). The titin molecule is elastic, with a size ∼1 µm long and 3–4 nm wide (3–5). One molecule spans half of the sarcomere with the amino terminus located in Z-line and the carboxyl terminus in the M-line (2,6–8). The elasticity of titin mainly comes from the folding and extending of polymeric immunoglobulin regions (middle Ig) and the PEVK region [rich in proline (P), glutamate (E), valine (V) and lysine (K)] (2,9). The giant size and the specialized structure enables titin to play a mechanical role in maintaining sarcomere length and structure integrity: it accounts for most of the passive tension of striated muscles in the physiological extension range to restore the sarcomere to normal length after stretch and reposition the thick and thin filaments (5,10–13). Besides its mechanical function, titin also plays important roles in many other physiological processes. Titin acts as a scaffold for myofibrillar assembly during muscle development, and it interacts with many structural proteins (14–17).
Titin undergoes developmental isoform transition from large to small in both cardiac and skeletal muscles (1,2,18–21). Diverse titin isoforms result from the alternative splicing of titin mRNA in the regions corresponding to the middle Ig (exons 50–96) and PEVK regions (exons 115–225) (1,2,9). In heart, the titin-based passive tension determines the stiffness of the myocardial wall during ventricular filling, so it is important to maintain the proper isoform ratios; abnormal titin isoform expression has been associated with heart disease (22–27). Many classes of titin isoforms (such as N2A, N2B and N2BA) and their splicing pathways have been characterized (2,9,18,21), but the mechanism underlying these splicing pathways remains unknown.
We found a mutant rat lacking in titin alternative splicing (19,21,28) and identified the mutation as a nearly complete deletion of the Rbm20 gene (29). Rbm20 protein is a putative RNA-binding protein with one RNA recognition motif and one arginine–serine rich domain. Thus far, mechanistic studies on RBM20 are lacking. Only a few articles have reported mutations in the human RBM20 gene, and these were associated with human dilated cardiomyopathy (DCM), with cell function descriptions lacking (30–34). Our previous study found that the mutant rat with Rbm20 deficiency had a similar pathological phenotype as found in human DCM, and we also found titin splicing was altered in a human DCM subject with an RBM20 mutation. These observations suggest that the deficiency in Rbm20-regulated titin alternative splicing may be an underlying cause for DCM (29). The current work reports investigations on the mechanism of Rbm20 in regulating titin alternative splicing. We demonstrate that Rbm20 mediates intron retention, exon skipping and exon shuffling of titin mRNA, forms microscopically identified aggregates with partially processed titin pre-mRNAs in the nucleus and uses different splicing pathways to skip different subsets of titin exons to form different isoforms. We found muscle cells use a relative simple system, by controlling the Rbm20/splicing factors ratio expression level, to switch splicing pathways and regulate the extremely complex titin alternative splicing process.
MATERIALS AND METHODS
RT–PCR analysis
RNA was purified from the indicated tissues with TRIzol regent (Invitrogen, 15596026) and further treated with RQ1 RNase-free DNase (Promega) to remove genomic DNA contamination. One microgram of total RNA was reverse transcribed with ImProm-II Reverse Transcription System (Promega) using Random primers (Promega). The RT reaction was used as template for PCR to characterize intron retention, exon skipping and exon shuffling in titin mRNA. The primers are listed in Supplementary Table S1. The PCR products were analyzed on ethidium bromide agarose gels. The bands of interest were gel purified and sequenced to confirm their identity.
Antibodies
Polyclonal antibodies directed against full-length Rbm20 were produced from chicken and rabbit and affinity purified (29). For immunohistochemistry, the concentration of chicken anti-Rbm20 was adjusted to 1 mg/ml and used at 1:500 dilution, and rabbit polyclonal anti-PSPC1 (sc-84117, Santa Cruz), mouse monoclonal anti-SC-35 (S4045, Sigma) and mouse monoclonal anti-fibrillarin (ab45661, Abcam) were used at 1:200 dilution. The secondary antibody for chicken anti-Rbm20 was Alexa Fluor 594 goat anti-chicken IgG (Invitrogen), used at 1:200 dilution. The secondary antibodies for PSPC1, SC-35 and fibrillarin were Alexa Fluor 488–conjugated goat anti-rabbit and goat anti-mouse IgG (Invitrogen), used at 1:200 dilution. For western blots, the concentration of rabbit anti-Rbm20 was adjusted to 1 mg/ml and used at 1:5000 dilution, and mouse monoclonal anti-SFRS1 (sc-73026, Santa Cruz), rabbit polyclonal anti-U2AF65 (sc-48804, Santa Cruz) and rabbit polyclonal anti-hnRNP L (sc-28726, Santa Cruz) were used at 1:3000 dilution. The secondary antibodies for western blots were horseradish peroxidase–conjugated ECL donkey anti-rabbit IgG (NA934V, GE Healthcare) and horseradish peroxidase–conjugated ECL sheep anti-mouse IgG (NA931V, GE Healthcare) used at 1:5000 dilution.
Immunofluorescence and mRNA fluorescence in situ hybridization
The procedure for immunochemistry in HL-1 cells is described previously (29). For the immunochemistry in samples from living tissue, homogenized muscle was fixed in fresh 4% formaldehyde in phosphate-buffered saline (PBS) for 15 min, spread on a cover slip and allowed to attach by air drying. The tissues were then permeabilized for 10 min in ice-cold PBS with 0.1% Triton X-100. After three washes in PBS, the homogenized tissues attached on cover slips were immunostained using the same procedures as used for staining HL-1 cell. The procedures for combined immunofluorescence and mRNA fluorescence in situ hybridization (FISH) were described previously (35). Titin exons 1–50 were amplified by four pairs of primers listed in Supplementary Table S1 and cloned into four pGEM-T easy vectors, respectively (Promega). The biotin-labeled cDNA probe against titin exons 1–50 was prepared by nick translation (Roche, 11745824910) using 0.5 µg of each of these plasmids. Chicken anti-Rbm20 and Alexa Fluor 594 goat anti-chicken IgG were used to immunostain Rbm20; fluorescein isothiocyanate–conjugated streptavidin (Sigma, S3762) was used to detect biotin-labeled cDNA probe hybridized with titin mRNA.
RNA immunoprecipitation
The RNA immunoprecipitation (RIP) procedure described previously (36) was followed with some modifications. Nuclei from a 150-day-old wild-type (Wt) rat left ventricle (LV) were enriched first (37); the nuclei were suspended in 1 ml of FA lysis buffer containing 1 mM ethylenediaminetetraacetic acid, 50 mM [(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] (HEPES)–KOH (pH 7.5), 140 nM NaCl, 0.1% sodium dexycholate (w/v), 1% Triton X-100 (v/v) and 1× protease inhibitor cocktail (Sigma, P8340). The lysis buffer is supplemented with 400 U/ml RNAsin inhibitor (Promega, N2511), 10 mM vanadyl ribonucleoside complex (New England Biolabs, S1402S), 10 U/ml RQ1 RNAse-free DNase (Promega, M6101) and 0.5 µg/ml yeast tRNA (Invitrogen, 15401) during RIP. Cross-linking protein with the RNA is not necessary in this study. Sonication was adopted to break down the nuclei and the ribonucleoprotein complex. After centrifugation at 15 000g for 10 min at 4°C, the supernatant was collected. The supernatant was first pre-cleaned by incubation with 200 µl of Dynabeads® Protein G (Invitrogen, 10004D) for 20 min at room temperature and then incubation with antibody-coated Dynabeads® Protein G for 1 h at room temperature. After incubation, the supernatant was discarded, and the beads were washed five times with FA lysis buffer containing 10 mM vanadyl ribonucleoside complex and submitted for protein extraction and RNA extraction. The precipitated protein was extracted with urea–thiourea sodium dodecyl sulphate buffer as described previously (38). The precipitated RNA was extracted with TRIzol reagent. To precipitate Rbm20 protein and the associated RNA, 8 µg of rabbit anti-Rbm20 coated on 200 µl of Dynabeads® Protein G was used. As a parallel control, 8 µg of rabbit anti-spectrin α coated on 200 µl of Dynabeads® Protein G was used.
Plasmid construction
Full-length Rbm20 was in-frame cloned into pcMS-EGFP vector (Clontech) using SalI and NotI restriction sites. MS2–Rbm20 fusion construct was made by inserting MS2 coat sequence before Rbm20 in PCMS-EGFP using the restriction sites NheI and MluI. The sequence coding MS2 coat protein was amplified from the pMAL-MS2-MBP construct (provided by Zhaolan Zhou, University of Pennsylvania). Titin minigene 64–70, which spans from −120 bp upstream of titin exon 64 to 75 bp downstream of titin exon 70, was cloned into PCDNA 3 vector (Invitrogen) using the restriction sites EcorV and XhoI. The fragment containing three MS2 hairpins was amplified from the pCR2.1 MS2 construct (provided by Zhaolan Zhou, University of Pennsylvania). MS2–exons 64–70 construct was made by inserting MS2 stem–loop sequence before exon 64 of the titin minigene in pcDNA 3 using restriction sites BamHI and EcorI; MS2–exons 64–70 construct was made by inserting MS2 stem–loop sequence after exon 70 of the titin minigene in pcDNA 3 using restriction sites XhoI and ApaI. Rbm20 without MS2 coat protein is cloned into PCMS-EGFP, and titin minigene expressing exons 25–27, 35–37, 52–58, 56–62, 60–66 and 64–70 without MS2 stem–loop sequence is constructed into PCDNA 3 vector (for primers, see Supplementary Table S1). All expression plasmid and minigene structures were confirmed by sequencing.
Mammalian cell culture, transfection and transcription inhibition
HL-1 cells were cultured on fibronectin-coated coverslips as previously described (29). Transcription in HL-1 cells was inhibited by adding 1 µg/ml of actinomycin D to the medium for a period of 2.5 h before immunofluorescence staining. HeLa cells were cultured in minimum essential medium (Invitrogen, 11095080) in standard conditions. HeLa cells (105) were plated in each well of 24-well plates and grown for 24 h before transfection. Cells are co-transfected with 1 µg of MS2-Rbm20-pcMS-EGFP and 0.2 µg of each plasmid expressing titin minigene using FuGENE HD transfection reagent (Promega, E2311). An equivalent amount of empty pcMS-EGFP was co-transfected with titin minigenes to serve as a parallel control.
Titin isoform characterization
Titin isoforms were analyzed using the vertical sodium dodecyl sulfate–agarose gel electrophoresis system (38).
RESULTS
Rbm20 represses titin mRNA splicing
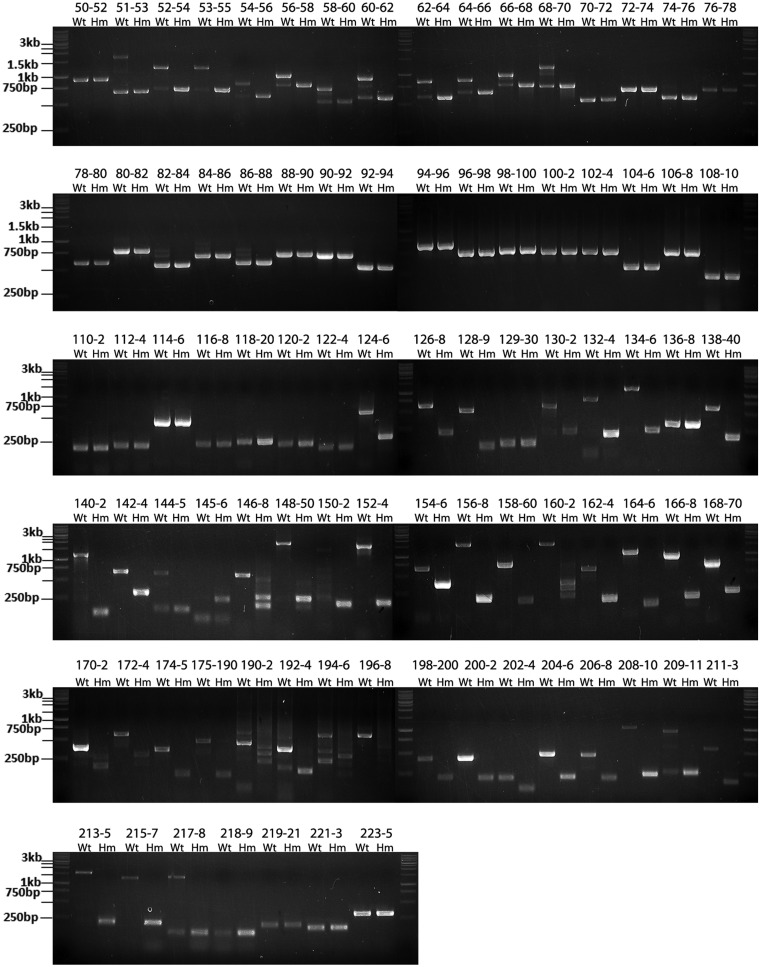
Our previous study showed that Rbm20 is required for the alternative splicing of titin mRNA. In Wt (Rbm20+/+) rats, the cardiac titin mRNA undergoes extensive exon skipping within the regions corresponding to the middle Ig (from exons 50 to 96) and the PEVK (from exons 115 to 225) regions. These exon-skipping events are inhibited in homozygote mutant (Hm, Rbm20−/−) rats where 13 of the 14 coding exons of Rbm20 gene are deleted (29). Theoretically, if RT-PCR is used to amplify the cardiac titin mRNA for the middle Ig and PEVK regions, we should expect smaller PCR products in the Wt than in the Hm. This is true when the primer pairs are designed to span large number of exons (e.g. >10 exons) (21). However, when using primers spanning just two or three exons, we amplified larger PCR products in the Wt than in the Hm for many primer pairs (Figure 1 and Supplementary Figure S1A–E). Sequencing these larger PCR products revealed retained intronic sequences in titin mRNA. Intron retention in Wt titin mRNA occurs between exons 50 and 70, 80 and 88, 117 and 118, 120 and 122, 124 and 129, 130 and 136, 138 and 145 and 146 and 218, but it never occurs in the Hm. A no-RT control was used to exclude the possibility of genomic contamination (Supplementary Figure S1F). The fact that introns cannot be spliced out in the Wt but can be normally removed in the Hm directly suggests that Rbm20 functions to repress the splicing of titin mRNA.
Figure 1.
Splicing of titin mRNA is inhibited in the wild type (Wt, Rbm20+/+) but not in the homozygote mutant (Hm, Rbm20−/−). RT-PCR was used with primer pairs spanning two or three exons to amplify cardiac titin mRNA from Wt and Hm left ventricle (LV). Usually three bands were obtained in the Wt for the middle Ig region between exons 50 and 70 and exons 80 and 88, but only one band in the Hm. As exemplified by exons 68–70 in the top panel, the lower Wt band has the same size as the adjoining Hm band and is the product of constitutive splicing; the upper band contains product where two introns for the primer-spanned region are both retained; the middle band has one of the two spanned introns retained. The splicing in the PEVK region (exons 124–218) is repressed to a greater extent in the Wt. Under most circumstances (as exemplified by exons 124–126), PEVK introns cannot be removed from Wt titin mRNA so that only one band, representing the retained intron pattern with a larger size than the adjoining band in the Hm, can be detected for most primer pairs. No retained introns can be detected in the Hm. The size of marker is indicated on the left, and the expected size of the PCR product for each primer pairs are listed in the Supplementary Table S2. The PCR products match their predicted size for all the primer pairs.
This repression varies in extent along the titin mRNA. For the region between exons 50 and 70, splicing patterns corresponding to retained introns and constitutive splicing co-exist, but the retained intron pattern dominates (Figure 1). This means the splicing between exons 50 and 70 is repressed in the majority of titin mRNAs. The pattern is reversed for the region between exons 80 and 88 so that retained intron and constitutive splicing patterns co-exist but the constitutive splicing pattern dominates (Figure 1), indicating the splicing is repressed only in the minority of titin mRNAs. The retained intron is the only splicing pattern detected from most of the primer pairs for the PEVK region between exons 124 and 218 (Figure 1). This demonstrates the splicing in much of the PEVK region is completely inhibited in all titin mRNAs.
Rbm20 repression is associated with exon skipping and exon shuffling
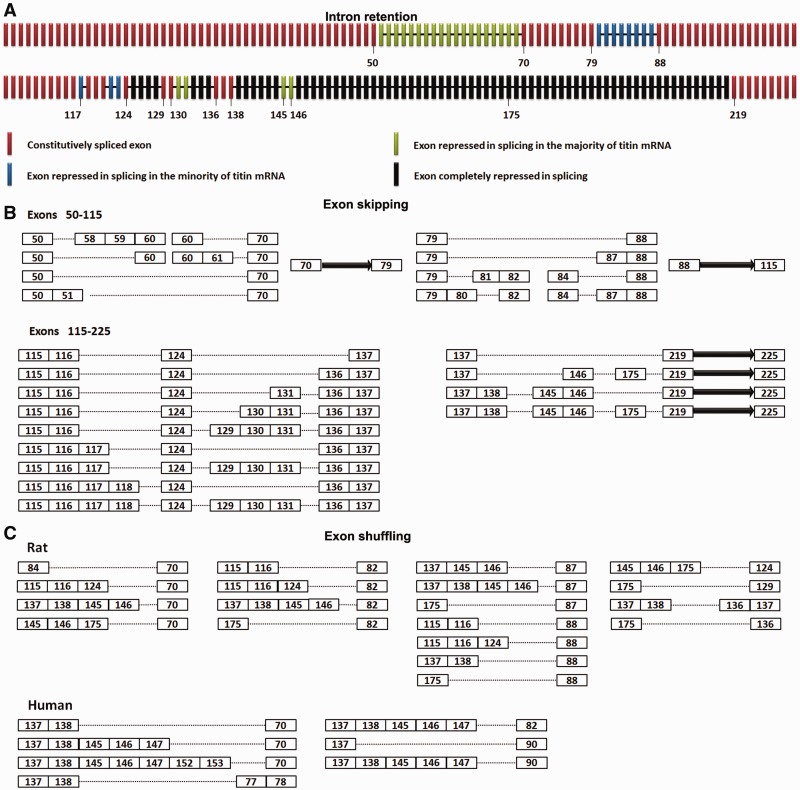
The intron retention pattern along the cardiac titin mRNA in Wt rats is summarized in Figure 2A. The middle Ig and PEVK regions of titin mRNA (exons 50–225) can be further divided into regions repressed by Rbm20 and regions not repressed. We then investigated whether the repression of Rbm20 on splicing is related to titin exon skipping. The exon skipping in Wt is characterized by RT-PCR using primer pairs that span large numbers of middle Ig and PEVK exons (>8 exons). As expected, exon skipping only occurs in the Rbm20-repressed regions: exons 50–70, exons 79–88 and most of the PEVK region (Figure 2B and Supplementary Figure S2A–F). The PEVK exons finally included in the alternative spliced titin mRNAs (such as exons 124, 129, 130, 131, 136, 137, 138, 145 and 146, Figure 2B) are exactly the same exons that are not completely repressed by Rbm20 (Figure 2A). No retained introns can be detected from the regions between exons 70 and 79, between exons 88 and 116 and after exon 219; correspondingly, no exon skipping occurs in these regions (Figure 2B). Our results suggest that the exon skipping of titin mRNA is closely related with the repression of Rbm20 on splicing (Supplementary Figure S2G). No intron retention can be detected where Rbm20 is absent, and correspondingly all titin exons between exons 50 and 225 are constitutively spliced in the Hm (21).
Figure 2.
Rbm20 mediates exon skipping and exon shuffling. (A) Schematic illustration of intron retention along cardiac titin mRNA in Wt rats. The titin exons and retained introns are shown with bars and lines, respectively. Colors denote the extent of Rbm20 repression on different regions. Black: complete repression, splicing repressed in all mRNAs; green: strong repression, splicing repressed in the majority of mRNAs; blue: mild repression, splicing repressed in the minority of mRNAs; red, no repression, normally spliced. The numbers of some exons are indicated. (B) Schematic illustration of exon skipping along cardiac titin mRNA in the Wt rat. The exons are displayed as numbered boxes; the solid arrow represents consecutive exons that result from constitutive splicing between the adjacent numbered exons. The dotted lines denote the exon skipping mediated by direct ligation between adjacent numbered exons. Results show that exon skipping only occurs in the Rbm20-repressed regions: exons 50–70, exons 79–88 and most of the PEVK region.(C) Diagram depicts the exon composition of exon-shuffling isoforms from the cardiac titin mRNA in the Wt rat and human. The exons are shown in numbered boxes, and the dotted lines denote direct attachment between the adjacent numbered exons.
Exon shuffling was found with cardiac titin mRNA in Wt rats. When we amplified titin mRNA using a forward primer set on exon 137 and reverse primer set on multiple upstream exons ranging from exon 24 to exon 96, multiple PCR products were obtained (Supplementary Figure S3A). Sequencing these PCR products revealed exon shuffling in titin mRNA: downstream exons were spliced before upstream exons. We then tested many primer combinations and identified exons 84, 116, 124, 138, 146 and 175 as additional donor exons and exons 70, 82, 87, 88, 124, 129 and 136 as acceptor exons for exon shuffling (Figure 2C). Each donor exon is able to splice with multiple acceptor exons and vice versa, demonstrating that the splicing between downstream donor exons and upstream acceptor exons was somewhat random. Exon shuffling is significantly inhibited in the Hm (Supplementary Figure S3B), indicating this event is also regulated by Rbm20. Exon shuffling can barely be detected from polyadenylated titin mRNA; this means most of them are not from mature titin mRNA (Supplementary Figure S3C). The exon shuffling with titin mRNA is conserved between species. Many exon-shuffling events were detected with human cardiac titin mRNA, and human exons mostly orthologous to those in rat mediated exon shuffling (Figure 2C). Exon shuffling in rat and human titin mRNA is detailed in Supplementary Figure S4.
Nuclear Rbm20 speckles are titin pre-mRNA processing sites
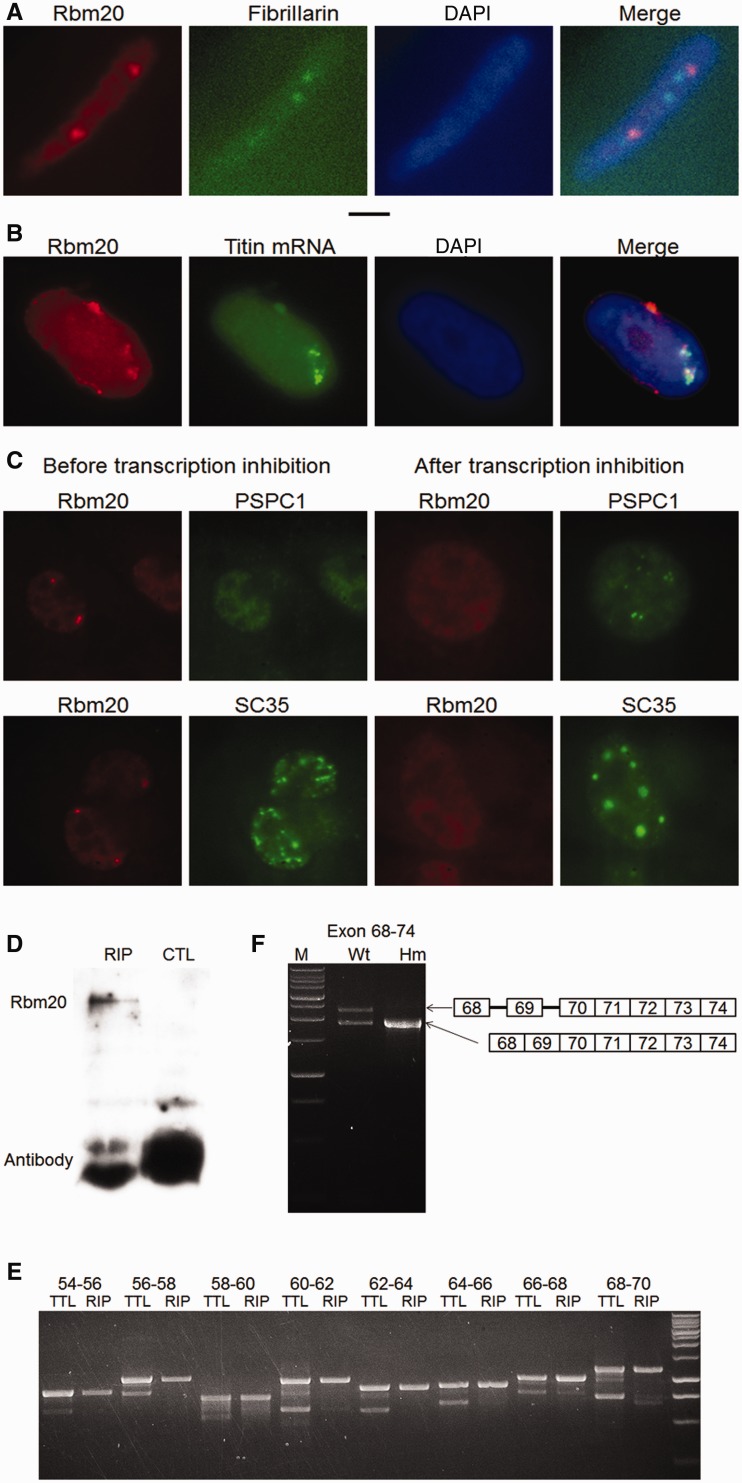
Immunofluorescence staining always revealed two Rbm20 speckles per nucleus with tissue obtained from living muscles (Supplementary Figure S5A). Co-staining of Rbm20 speckles with a nucleolus marker (fibrillarin) showed that Rbm20 speckles have a size similar to nucleoli (0.6 µm in diameter), but the two are not co-localized (Figure 3A). Rbm20 was not co-localized with other nuclear bodies (Supplementary Figure S5B). We suspected that the Rbm20 speckles are aggregates of Rbm20 proteins on titin mRNAs. Immunofluorescence combined with RNA FISH demonstrated that titin mRNAs co-localized with the Rbm20 speckles in nuclei from in vivo muscle (Figure 3B). The two Rbm20/titin mRNA speckles are possibly formed by recruiting Rbm20 protein onto the newly transcribed titin mRNAs near the two titin transcription loci on sister chromosome 3s. Based on this hypothesis, the formation of Rbm20 speckles should depend on transcription. As expected, the Rbm20 speckles dissociated after transcription is inhibited (Figure 3C), indicating that the Rbm20/titin mRNA speckles are formed on newly synthesized titin mRNAs. We isolated Rbm20-bound titin mRNA by RIP. Results showed that Rbm20 protein can be specifically precipitated (Figure 3D), and that the titin mRNAs co-precipitated with Rbm20 are the titin mRNAs with retained introns. In contrast, the constitutively spliced titin mRNAs are barely precipitated (Figure 3E). This result directly shows that Rbm20 binding represses titin splicing. The repression of Rbm20 on titin splicing in the repressed region remains even after the splicing on the rest of the regions is finished: introns 68 and 69 remain retained even after their downstream introns are successfully spliced out (Figure 3F). These results demonstrate that the titin mRNAs bound by Rbm20 are partially processed titin pre-mRNAs; they are not spliced in the Rbm20-repressed regions but are normally spliced in the rest, suggesting that the intron retention we characterized in Figure 1 is from this kind of titin mRNAs. The partially processed titin pre-mRNAs are restricted in the Rbm20 speckle regions in the nucleus instead of being exported.
Figure 3.
Rbm20 aggregated on newly synthesized titin mRNAs forms Rbm20 speckles and represses the splicing of titin mRNA. (A) Immunofluorescence shows two Rbm20 speckles in the nucleus of cardiomyocytes from LV. The size of Rbm20 speckles was similar to nucleoli; Rbm20 is stained red, and nucleoli are stained green with antibody against fibrillarin. 4′,6 diamidino-2-phenylindole (DAPI)-stained nuclei are blue. Black scale bar: 2 microns. (B) Immunostaining of Rbm20 protein combined with titin mRNA FISH shows the co-localization of titin mRNA with Rbm20 speckles. Rbm20 is stained red; titin mRNA is stained green; and the overlap of red and green yields yellow. (C) Rbm20 speckles are transcription dependent. When the transcription of HL-1 cell is inhibited, Rbm20 speckles (red) dissociate. The behavior of paraspeckles (PSPC1) and splicing speckles (SC35) verified that transcription in the specific cell is inhibited. After transcription inhibition, crescent-shaped paraspeckles were formed, and the splicing speckles (green) become concentrated. Rbm20 is stained red; PSPC1 and SC35, marker proteins for paraspeckles and splicing speckles, respectively, are stained green. (D) Rbm20 can be specifically immunoprecipitated by anti-Rbm20 but not by control antibody. RIP: with anti-Rbm20, CTL: with control antibody (anti-spectrin α). (E) When using RT-PCR to amplify the total titin mRNA (TTL) from LV and the titin mRNA co-immunoprecipitated with Rbm20 (RIP), for the total titin mRNA, both retained introns (upper band in each primer pair) and constitutive splicing pattern (lower band) can be detected. However, for the Rbm20-immunoprecipitated titin mRNA, only the retained intron pattern can be clearly detected, meaning the constitutively spliced titin mRNA can barely be precipitated by Rbm20. (F) The co-existence of retained intron and constitutive splicing patterns on the same titin message were found in the Wt. The splicing on introns before exon 70 remains inhibited when the splicing on the downstream introns was finished. This situation does not occur in the Hm.
Rbm20 cooperatively represses the splicing between exons 52 and 68
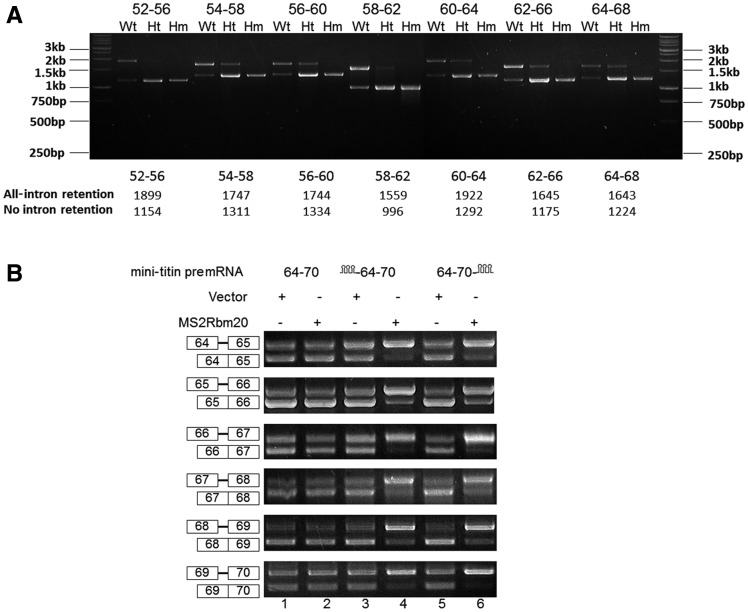
We amplified the Wt cardiac titin mRNA for the region between exons 52 and 68 using primer pairs with partially overlapped spanning regions. We unexpectedly found constitutive splicing and all-intron retention are the two major splicing patterns for all the primer pairs (Figure 4A). This result illustrates that the all-intron–retained patterns are joined together and constitutive splicing patterns are joined together for the entire region between exons 52 and 68 so that the splicing on this region is cooperative: either all repressed or not repressed. In heterozygote mutants (Ht, Rbm20+/−), where the Rbm20 expression level is only half of the Wt, the splicing patterns are the same as in the Wt (Figure 4A). The only difference between Wt and Ht was that the all-intron–retained pattern accounts for the majority in the Wt but only for minority in the Ht. Thus, when the Rbm20 concentration is lowered from Wt to Ht, less titin mRNAs are repressed in splicing, but the repression is still cooperative: all or none. To test whether Rbm20 can repress the splicing of titin mRNA on this region cooperatively in other cell types, we constructed minigene expressing titin pre-mRNA from exons 64 to 70 and transfected this minigene into Hela cells with and without Rbm20. To strengthen the interaction between Rbm20 and titin pre-mRNA in Hela cells, we ligated the MS2 stem–loop sequence on either upstream or downstream of titin minigene and fused MS2 coat protein on the N terminus of Rbm20. The splicing assay on the mini-titin pre-mRNA shows that when Rbm20 is artificially recruited to either upstream or downstream positions of titin mRNA, the splicing of all the exons in the mini-titin pre-mRNA is repressed at a higher level (Figure 4B). Therefore, the presence of Rbm20 at a certain position can repress the splicing of not only its adjacent exons but also the exons >2 kb away. In Hela cells, Rbm20 can repress the splicing of mini-titin pre-mRNAs expressing exons/introns in the repressed regions but not those expressing exons/introns outside the repressed regions (Supplementary Figure S6A–C), indicating there should be some native Rbm20-binding sites within the Rbm20-repressed regions.
Figure 4.
Rbm20 represses the splicing of titin mRNA in a cooperative manner for the region between exons 52 and 68. (A) All-intron retention and constitutive splicing are two major splicing patterns for all the primer pairs across the region from exon 52–68 in the Wt and Ht. Each primer pair used in this study spans five exons and four introns, with three exons and two introns overlapped with the spanned region of the adjacent primer pairs. The ratio of all-intron retention to constitutive splicing decreased from Wt to Ht; no intron retention can be found with Hm. The marker indication and the expected size of the PCR product for each primer pairs are shown. (B) The binding of Rbm20 to certain positions of titin mRNA can strengthen the repression of Rbm20 on distant exons. Lanes 1, 2, 3 and 5 show that neither MS2 structure ligated to titin mRNA nor Rbm20 fused with MS2 coat protein can repress the splicing of titin minigenes by themselves. Lane 4 and 6 show that when Rbm20 is recruited to titin mRNA at either upstream or downstream positions, all the titin exons on the titin mRNA are repressed, even for the exons that are far away from the Rbm20-binding site.
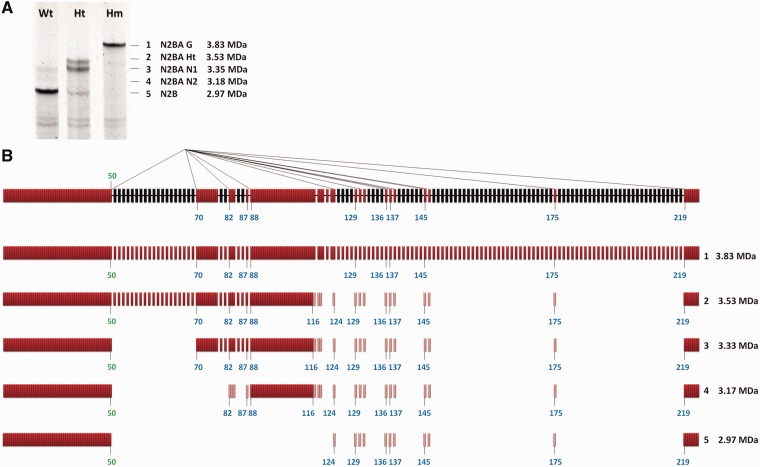
Rbm20-dependent 3′ alternative splice site selection
Thus far, we found the exon skipping in titin mRNA is associated with the repression of Rbm20 on the splicing in the repressed regions (Figure 2A and B). However, some titin-splicing pathways show skipping of exons in regions not repressed by Rbm20. The N2B-splicing pathway is such an example, in which exon 50 directly splices with exon 219 and skips all the exons in between, including the regions between exons 70 and 79 and exons 88 and 116, even though the splicing in these regions is not repressed by Rbm20. The N2B-splicing pathway is predicted to encode the 2.97-MDa N2B titin isoform (9,18), which is the dominating titin isoform in Wt adult heart (Figure 5A). Based on our intron retention data, we found that exon 50 and exon 219 are located at the 5′ and 3′ end of Rbm20-repressed regions, respectively (Figure 2A), so we have reason to hypothesize that other exons located at the border of Rbm20-repressed regions can be involved in similar alternative splicing pathways. As expected, we found alternative 3′ splice site (ss) pathways in which exon 50 can directly splice with many other exons along titin mRNA besides exon 219, and most acceptor exons involved in the alternative 3′ ss pathway are located at the 3′ borders of Rbm20-repressed regions such as exons 70, 82, 88, 124, 136, 145 and 219; exons 130, 137, 138 and 146 are one or two exons downstream of an Rbm20-repressed region but can also be involved in this splicing pathway (Figure 5B). Whether the unrepressed regions between exons 70 and 79 and exons 88 and 116 would be skipped depends on which acceptor exon is spliced with exon 50 (Figure 5B).
Figure 5.
Rbm20-regulated alternative 3′ splice sites (ss). (A) The cardiac titin protein isoforms in the LV of the Wt, Ht and Hm. Based on their size from large to small, the titin isoforms are numbered from 1 to 5. The apparent molecular weights and the name of different titin isoforms are shown on the right. The 2.97-MDa titin isoform is the N2B titin isoform, which is dominant in the Wt rat heart. (B) Schematic illustration of the exon 50–involved alternative 3′ ss pathways in which the 5′ ss of exon 50 can splice with the 3′ ss of many other exons. On top is shown the partially processed titin mRNA in which the splicing in the regions not repressed by Rbm20 (colored in red) is finished but the splicing in the repressed regions (colored in black) remains inhibited. Exons and introns are indicated with bars and thick lines, respectively. The thin lines connecting exon 50 with other exons illustrate different splicing pathways; most of the acceptor exons are located at the 3’′ end of Rbm20-repressed regions. The molecular mass of the titin isoform resulting from each splicing pathway is calculated and shown on the right. The light-colored red bars in splicing isoforms 2, 3, 4 and 5 represent the alternative exons. They do not always exist in the alternatively spliced titin mRNAs because different titin mRNAs contain different combination of these exons (shown in Figure 2B).
Based on the molecular mass of different cardiac titin isoforms and the splicing pathways we have characterized thus far, the splicing pathways corresponding to the five major cardiac titin isoforms in Hm, Ht and Wt rat from large to small should be as follows: (i) constitutive splicing of all the exons between exons 50 and 225, (ii) skipping of the completely repressed PEVK exons, (iii) additional skipping of the strongly repressed middle Ig exons 50–70, (iv) additional skipping of the mildly repressed middle Ig exons 80–87 plus the unrepressed exons between exons 70 and 79, and (v) additional skipping of unrepressed exons between exons 88 and 116. All these splicing isoforms have been verified by sequencing (Figure 5B).
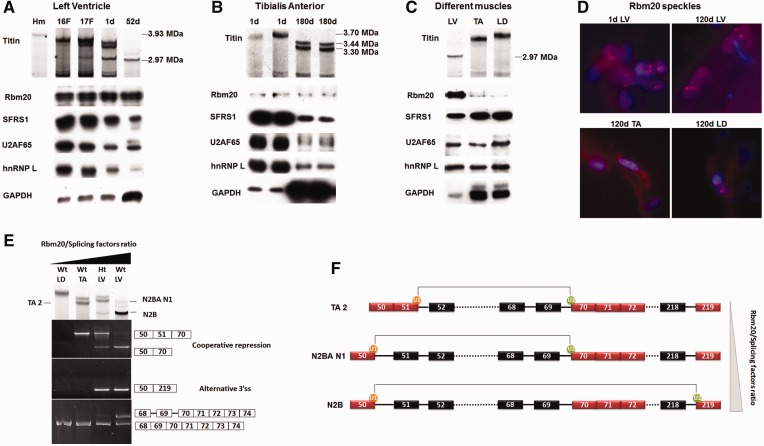
Titin isoform transformation during development as well as between muscles is related to the alteration of Rbm20/splicing factors ratio
Titin undergoes developmental transition from larger neonatal isoforms to smaller adult isoforms, but the development-associated mechanism remains unknown. Because Rbm20 is the protein that regulates titin alternative splicing, we investigated whether developmental changes in Rbm20 lead to the developmental titin isoform transitions. Because Rbm20 is involved in pre-mRNA processing, we tested whether Rbm20 undergoes developmental changes in parallel with other splicing factors: SFRS1 (SF2/ASF), U2AF65 and hnRNP L. Results using LV showed that during development, the ratio between the latter three splicing factors remained essentially constant, but the ratio of Rbm20 to these splicing factors increased with development. This increased ratio coincides with the progressive reduction in titin size (Figure 6A). Similar results were also found in the skeletal muscle tibialis anterior (TA) (Figure 6B). However, if GAPDH was used as a control, we got exactly the opposite conclusion; namely, the ratio of Rbm20 to GAPDH decreased with age (Figure 6A and B). Because titin isoforms vary between muscles, we could test the relationship between Rbm20 expression and titin size between different muscles. LV, TA and longissimus dorsi (LD) with titin size from small to large were selected for this study. Results show that Rbm20/splicing factors ratio in LV, TA and LD is inversely related with the titin size (Figure 6C). The Rbm20 in vivo always forms two Rbm20 speckles in each nucleus from cardiac and skeletal muscles, and the size of the Rbm20 speckles remains relative constant during development as well as between muscles (Figure 6D), indicating the number and size of Rbm20 speckles are determined by newly transcribed titin mRNA instead of the Rbm20 expression level. The fact that the cellular localization of Rbm20 remains the same during development and between different muscles also suggests that the localization of Rbm20 in the muscle cell is not the factor that affects titin alternative splicing.
Figure 6.
Titin size is determined by the relative ratio of Rbm20 to other splicing factors. (A) Upper panel shows titin proteins separated on an sodium dodecyl sulphate–agarose gel. The left lane is from an Hm adult LV, and the rest are Wt from fetal (F) and postnatal heart aged in days (d). Lower panel shows western blots. Sample loads were balanced for Rbm20 content. Results show that during development, the size reduction in titin protein is accompanied by increased ratios of Rbm20 to other splicing factors. (B) Similar developmental changes in the ratios of Rbm20 to other splicing factors occur in the skeletal muscle tibialis anterior (TA). The Rbm20 to splicing factor ratios increase with age, whereas the titin size decreases. (C) Tissue specific Rbm20 and titin isoform expression in LV, TA and longissimus dorsi (LD). Sample loads were balanced for the splicing-related factors. The results demonstrate that the ratio of Rbm20 to other splice factors is inversely related to titin isoform size. (D) Characterization of Rbm20 speckles during development and between muscle tissues. Two speckles per nucleus were observed in all the indicated tissues. d, day(s). (E) Titin isoforms and splicing pathways from different muscle tissues. Titin isoforms from different tissues are shown in the upper panel. TA 2 and N2BA N1 correspond to the cooperative repression between exons 50 and 70. TA2 is smaller than N2BA N1 because only the latter contains protein coded by exon 49, which accounts for an additional 0.1 MDa of protein mass (21). N2B results from the alternative 3′ ss pathway (shown by exons 50–219). The 5′ ss of exon 70 is always active for splicing, but the 3′ ss of exon 70 can be repressed in some titin mRNA in Ht LV and Wt LV (bottom panel). The unspliced 3′ ss of exon 70 cannot be found in Wt LD and Wt TA. (F) A schematic diagram demonstrating the Rbm20/splicing factors ratio–dependent extension of Rbm20-repressed region between exons 50 and 70 and the consequence of each situation. As the Rbm20/splicing factors ratio increases from top to bottom, the Rbm20-repressed region extends from exons 51–70 to exons 50–70. When Rbm20/splicing factors ratio is high enough like in Ht and Wt LV, we found the splicing of the 5′ ss of exon 50 to downstream alternative 3′ ss, for example the 3′ ss of exon 219. The 5′ ss of exon 50 can splice with many other alternative 3′ ss (refer to Figure 5B); they are not shown in this diagram. The dotted lines denote the continuous exons (and introns if repressed by Rbm20) that are not shown.
Diverse titin isoforms expressed in different muscles suggested different splicing pathways, which allows us to do the correlation analysis on these protein and splicing isoforms. With increasing Rbm20/splicing factors ratio in Wt LD, Wt TA, Ht LV and Wt LV, cooperative skipping of internal exons between exons 50 and 70 starts from Wt TA, which leads to the direct splicing of exon 50 or 51 with exon 70 (Figure 6E). In Ht LV and Wt LV, where Rbm20/splicing factors ratio is at still higher levels, the alternative 3′ ss pathway, shown by direct splicing of exon 50 to exon 219, can be detected, and corresponding to it, N2B titin isoform is found in these two tissues (Figure 6E). Interestingly, the intron retention between exons 68 and 70 can only be detected in Ht LV and Wt LV (Figure 6E bottom panel), which indicates that when exons 50–70 is cooperatively repressed, the 3′ ss of exon 70 could either splice with 5′ ss of exon 50 or 51 or remain repressed; the repressed 3′ ss of exon 70 is coincident with the appearance of alternative 3′ ss pathway. In Wt TA where the Rbm20/splicing factors ratio is at a lower level, cooperative skipping of exons 51–70 can be detected, but the repression on the 3′ ss of exon 70 cannot; correspondingly, neither alternative 3′ ss pathway nor the titin isoform similar to cardiac N2B can be detected in this tissue (Figure 6E). The cooperative skipping of internal exons between exons 51 and 70 in Wt TA correlates with titin isoforms with discrete sizes. In Wt LD where the Rbm20/splicing factors ratio is too low to initiate the cooperative repression between exons 50 and 70, titin does not show isoforms with discrete size differences (Figure 6E).
We found that the border of the Rbm20-repressed region is not fixed but somewhat flexible. As the relative Rbm20/splicing factors ratio increases from Wt TA to Wt LV, the 5′ border of the Rbm20-repressed region can be extended from exon 51 to 50 (Figure 6E and F). Flexible 5′ and 3′ borders are common for almost all the Rbm20-repressed regions along the titin mRNA: the variable 5′ border includes exon 50 or 51, exon 116 or 117 or 118 and exon 137 or 138; the variable 3′ border includes exon 87 or 88, exons 129 or 130 or 131, exons 136 or 137 and exons 145 or 146 (Figure 2B). Higher Rbm20/splicing factors ratio favors the extension of Rbm20-repressed regions (Figure 6F).
Full-length titin PEVK exon characterization
Previous studies with human and rat cardiac titin cDNA have resulted in incomplete amplification of the full set of PEVK exons. This is presumably due to the early embryonic appearance and influence of Rbm20 on titin splicing (Figure 6A). The Rbm20-deficient rat model allowed us to amplify the complete PEVK exon set (Supplementary Table S3). Although most of the exons were similar to the predicted ones in humans, a few individual exons were inserted, and a 14-exon group between exons 175 and 190 (old numbering system to align with human) was missing in the rat. These exons do not appear to be expressed in the human sequences reported to date.
DISCUSSION
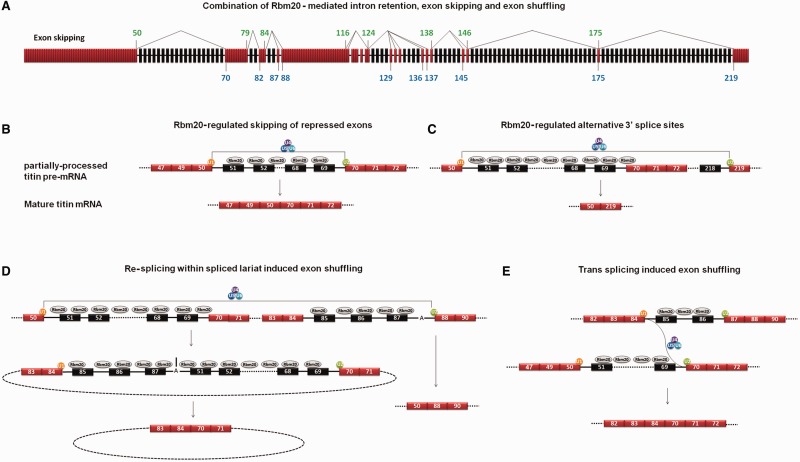
After the donor and acceptor exons involved in exon skipping and exon shuffling were located to the partially processed titin pre-mRNA, we found the donor exons (84, 116, 124, 138, 146 and 175) and acceptor exons (70, 82, 87, 88, 124, 129 and 136) involved in exon shuffling also serve as donor and acceptor exons in exon skipping (Figure 7A). Moreover, all these donor and acceptor exons were located at the border of Rbm20-repressed regions, with a donor exon at the 5′ end and an acceptor exon at the 3′ end (Figure 7A). Considering that the aggregation of Rbm20 protein on titin mRNA repressed splicing, we propose that Rbm20-regulated titin alternative splicing occurs as follows: Rbm20 binds to certain regions of the newly transcribed titin pre-mRNA before the splicing in these regions is initiated, and the binding of Rbm20 on titin pre-mRNA blocks splicing so that the introns in these regions cannot be spliced out. At the same time, the rest of titin pre-mRNA can be normally spliced, leading to partially processed titin pre-mRNAs. The partially processed titin pre-mRNAs are retained in the nucleus, awaiting further processing, instead of being exported. Finally, the 5′ ss and 3′ ss on the exons flanking the Rbm20-repressed regions splice with each other to skip the repressed internal exons/introns (Figure 7B). A similar two-step sequential splicing model was recently reported in which the spliced tumor susceptibility gene 101 (TSG101) and fragile histidine triad (FHIT) mRNA can undergo a second round of splicing to mediate extra exon skipping (39).
Figure 7.
Mechanisms of Rbm20-regulated alternative splicing. (A) Schematic diagram depicts the partially processed titin pre-mRNA and the location of the donor and acceptor exons involved in exon skipping. The same donor and acceptor exons are also involved in exon shuffling. The numbers of donor exons and acceptor exons are colored in green and blue, respectively. The thin lines connecting the donor exons with acceptor exons illustrate different splicing pathways. (B) Hypothesized model showing the process of Rbm20-regulated exon skipping as exemplified by exon skipping between exons 50 and 70. The exons are indicated with numbered boxes, and introns with black lines. The unrepressed exons are colored in red, and the Rbm20-repressed exons in black. SnRNPs are indicated with colored circles. U1 defines the 5′ ss and U2 defines the 3′ ss on titin exons. The association of U4/5/6 to U1 and U2 fulfills the splicing. Splicing between exons 50 and 70 skips the internal exons. (C) Hypothesized model related the upstream 3′ ss repression to the use of downstream alternative 3′ ss. (D) Hypothesized model showing the process of exon shuffling induced by re-splicing within the lariat spliced out during alternative 3′ ss pathway. Here we only show one example, many other alternative 3′ ss pathways and exon-shuffling events have been detected from titin mRNA (refer to Figure 5B and Figure 2C). (E) Hypothesized model for trans splicing–induced exon shuffling. Active 5′ ss on exon 84 splice with active 3′ ss on exon 70 from different titin messages to cause exon shuffling.
The 2.97-MDa N2B titin isoform dominates in Wt LV and results from the exon 50–involved alternative 3′ ss pathway that skips most exons between exons 50 and 219 (9,18), including exons 70–79 and 88–116, which are normally spliced in the partially processed titin pre-mRNA. Our correlation study showed the coincidence of alternative 3′ ss pathway with the repressed 3′ ss on exon 70 (Figure 6E), suggesting the use of downstream alternative 3′ ss may result from the unavailability of upstream 3′ ss in the partially processed titin pre-mRNA (Figure 7C). The repression of Rbm20 on the border exons of different repressed regions should be dynamic: the temporal repression of upstream 3′ ss will lead to the use of downstream 3′ ss to induce large-scale exon skipping and the formation of lariats composed of repressed exons/introns and unrepressed exons (Figure 7D). The temporal relief of Rbm20 on border exons within the lariat will lead to re-splicing within the lariat and cause exon shuffling. The model shown in Figure 7D may be the main mechanisms for exon shuffling in Wt titin mRNA, considering the alternative 3′ ss pathway that produce lariats with different exon/intron combinations is the main splicing pathway in Wt LV (Figure 5A and B). No matter what exon/intron combination the lariat has, the exon shuffling should always occur between the border exons, which are consistent with our result. However, we also found some exon-shuffling events in polyA RNA (Supplementary Figure S3C); we think this may result from trans splicing between independent titin messages (Figure 7E). Thus far, we do not have enough data to rule out this possibility.
Our previous transcriptomic analysis identified 31 Rbm20-dependent spliced genes that were shared between rats and humans (29). However, there were not a corresponding number of Rbm20 speckles in the nucleus; instead, there are only two Rbm20 speckles per nucleus. These speckles were co-localizing with newly transcribed titin mRNA as determined by combined immunofluorescence and in situ hybridization. Several pieces of evidence suggest that titin mRNA is the major target of Rbm20 regulation. First, titin is the third most abundant protein in the muscle after actin and myosin, so the titin mRNA is probably more abundant than the other regulated genes. Second, the number of potential Rbm20-binding sites in titin pre-mRNA is likely much larger than for any of the other Rbm20-regulated genes. The titin pre-mRNA is ∼250 000 nucleotides long, with the Rbm20-repressed region spanning ∼100 kb, so there may be a considerable amount of Rbm20 protein bound on each titin pre-mRNA. The apparent absence of speckles associated with other genes could be due to either lower message abundance or fewer Rbm20-binding sites per message.
An interesting question one might ask is whether the exon skipping events shift the open reading frame (ORF) of titin mRNA. The answer is no. After sequencing all the exons in the middle Ig and PEVK region from the Hm (Supplementary Table S3), we found that the nucleotide number of all the other exons from 50 to 225 is a multiple of 3 except for exons 103, 104 and 106. Thus, no matter which exons or combination thereof are skipped, the ORF of titin mRNA will remain the same. The sum of the nucleotide number for exons 103, 104 and 106 is a multiple of 3, and these exons are located in a constitutively spliced region from exon 88 to 116. Therefore, these three exons will either be co-retained or co-skipped during splicing with the other exons of this region, and the ORF of titin mRNA will remain unchanged in either situation.
This is the first mechanistic description of Rbm20 and also the first regarding the mechanism of titin alternative splicing. Our data suggested that Rbm20 proteins aggregate on titin pre-mRNA to regulate alternative splicing by repressing the splicing of certain regions. Different regions of titin pre-mRNA are differentially sensitive to Rbm20 repression, and higher Rbm20/splicing factors ratio can favor the repression on the regions with less sensitivity. However, we do not know whether these Rbm20-repressed blocks on titin pre-mRNA are repressed independently or synergistically. From the data we have characterized thus far, the only factor that is correlated with titin isoform transition is Rbm20/splicing factors ratio, which is fine tuned during development as well as between tissues by totally unknown mechanisms. The undetermined issues also include the mechanism for the cooperative repression of Rbm20 on the region between exons 50 and 70. These questions deserve further efforts to elucidate the molecular mechanism of Rbm20-regulated titin alternative splicing. Our current work provides a foundation for future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–6.
FUNDING
National Institutes of Health (NIH) [HL77196 to M.L.G., and R01HG005232 to C.N.D.]; College of Agricultural and Life Sciences, University of Wisconsin-Madison with support from the National Institute of Food and Agriculture, United States Department of Agriculture [ID number WIS01556 to M.L.G.]. Funding for open access charge: University funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Zhaolan Zhou (University of Pennsylvania, Philadelphia, PA) for kindly providing the pMAL-MS2-MBP plasmid and pCR2.1 MS 2 construct.
REFERENCES
- 1.Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 2.Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 3.Maruyama K, Kimura S, Yoshidomi H, Sawada H, Kikuchi M. Molecular size and shape of beta-connection, an elastic protein of striated muscle. J. Biochem. 1984;95:1423–1433. doi: 10.1093/oxfordjournals.jbchem.a134750. [DOI] [PubMed] [Google Scholar]
- 4.Trinick J, Knight P, Whiting A. Purification and properties of native titin. J. Mol. Biol. 1984;180:331–356. doi: 10.1016/s0022-2836(84)80007-8. [DOI] [PubMed] [Google Scholar]
- 5.Wang K, McCarter R, Wright J, Beverly J, Ramirez-Mitchell R. Regulation of skeletal muscle stiffness and elasticity by titin isoforms: a test of the segmental extension model of resting tension. Proc. Natl Acad. Sci. USA. 1991;88:7101–7105. doi: 10.1073/pnas.88.16.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fürst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregorio CC, Trombitás K, Centner T, Kolmerer B, Stier G, Kunke K, Suzuki K, Obermayr F, Herrmann B, Granzier H, et al. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell Biol. 1998;143:1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obermann WM, Gautel M, Weber K, Fürst DO. Molecular structure of the sarcomeric M band: mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16:211–220. doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaser ML, Berri M, Warren CM, Mozdziak PE. Species variations in cDNA sequence and exon splicing patterns in the extensible I-band region of cardiac titin: relation to passive tension. J. Muscle Res. Cell Motil. 2002;23:473–482. doi: 10.1023/a:1023410523184. [DOI] [PubMed] [Google Scholar]
- 10.Funatsu T, Higuchi H, Ishiwata S. Elastic filaments in skeletal muscle revealed by selective removal of thin filaments with plasma gelsolin. J. Cell Biol. 1990;110:53–62. doi: 10.1083/jcb.110.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granzier HL, Irving TC. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys. J. 1995;68:1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horowits R, Podolsky RJ. The positional stability of thick filaments in activated skeletal muscle depends on sarcomere length: evidence for the role of titin filaments. J. Cell Biol. 1987;105:2217–2223. doi: 10.1083/jcb.105.5.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat. Rev. Mol. Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsuka H, Yajima H, Maruyama K, Kimura S. Binding of the N-terminal 63 kDa portion of connectin/titin to alpha-actinin as revealed by the yeast two-hybrid system. FEBS Lett. 1997;401:65–67. doi: 10.1016/s0014-5793(96)01432-9. [DOI] [PubMed] [Google Scholar]
- 15.Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, Labeit S. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J. Mol. Biol. 1997;270:688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- 16.Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruger M, Linke W. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J. Biol. Chem. 2011;286:9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freiburg A, Trombitas K, Hell W, Cazorla O, Fougerousse F, Centner T, Kolmerer B, Witt C, Beckmann JS, Gregorio CC, et al. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ. Res. 2000;86:1114–1121. doi: 10.1161/01.res.86.11.1114. [DOI] [PubMed] [Google Scholar]
- 19.Greaser ML, Krzesinski PR, Warren CM, Kirkpatrick B, Campbell KS, Moss RL. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J. Muscle Res. Cell Motil. 2005;26:325–332. doi: 10.1007/s10974-005-9039-0. [DOI] [PubMed] [Google Scholar]
- 20.Ottenheijm CA, Knottnerus AM, Buck D, Luo X, Greer K, Hoying A, Labeit S, Granzier H. Tuning passive mechanics through differential splicing of titin during skeletal muscle development. Biophys. J. 2009;97:2277–2286. doi: 10.1016/j.bpj.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Guo W, Schmitt B, Greaser ML. Comprehensive analysis of titin protein isoform and alternative splicing in normal and mutant rats. J. Cell Biochem. 2012;113:1365–1373. doi: 10.1002/jcb.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitás K, Labeit S, Granzier H. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ. Res. 2000;86:59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- 23.Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, del Monte F, Hajjar RJ, Linke WA. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ. Res. 2004;95:708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- 24.Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 25.Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, Linke WA. Titin isoform switch in ischemic human heart disease. Circulation. 2002;106:1333–1341. doi: 10.1161/01.cir.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- 26.Warren CM, Jordan MC, Roos KP, Krzesinski PR, Greaser ML. Titin isoform expression in normal and hypertensive myocardium. Cardiovasc. Res. 2003;59:86–94. doi: 10.1016/s0008-6363(03)00328-6. [DOI] [PubMed] [Google Scholar]
- 27.Williams L, Howell N, Pagano D, Andreka P, Vertesaljai M, Pecor T, Frenneaux M, Granzier H. Titin isoform expression in aortic stenosis. Clin. Sci. (Lond) 2009;117:237–242. doi: 10.1042/CS20080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greaser ML, Warren CM, Esbona K, Guo W, Duan Y, Parrish AM, Krzesinski PR, Norman HS, Dunning S, Fitzsimons DP, et al. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J. Mol. Cell. Cardiol. 2008;44:983–991. doi: 10.1016/j.yjmcc.2008.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo W, Schafer S, Greaser ML, Radk MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, Hershberger RE. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin. Transl. Sci. 2010;3:90–97. doi: 10.1111/j.1752-8062.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millat G, Bouvagnet P, Chevalier P, Sebbag L, Dulac A, Dauphin C, Jouk PS, Delrue MA, Thambo JB, Le Metayer P, et al. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur. J. Med. Genet. 2011;54:570–575. doi: 10.1016/j.ejmg.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Rampersaud E, Siegfried JD, Norton N, Li D, Martin E, Hershberger RE. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog. Pediatr. Cardiol. 2011;31:39–47. doi: 10.1016/j.ppedcard.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, Gutmann R, Zhang ML, Bloom HL, MacRae CA, et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9:390–396. doi: 10.1016/j.hrthm.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaumeil J, Augui S, Chow JC, Heard E. Combined immunofluorescence, RNA fluorescent in situ hybridization, and DNA fluorescent in situ hybridization to study chromatin changes, transcriptional activity, nuclear organization, and X-chromosome inactivation. Methods Mol. Biol. 2008;463:297–308. doi: 10.1007/978-1-59745-406-3_18. [DOI] [PubMed] [Google Scholar]
- 36.Selth LA, Gilbert C, Svejstrup JQ. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. 2009;6 doi: 10.1101/pdb.prot5234. pdb.prot5234. [DOI] [PubMed] [Google Scholar]
- 37.Grabowski PJ. Splicing-active nuclear extracts from rat brain. Methods. 2005;37:323–330. doi: 10.1016/j.ymeth.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Warren CM, Krzesinski PR, Greaser ML. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis. 2003;24:1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- 39.Kameyama T, Suzuki H, Mayeda A. Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites. Nucleic Acids Res. 2012;40:7896–7906. doi: 10.1093/nar/gks520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.