Abstract
Human DNA polymerase mu (Polμ), a family X member involved in DNA repair, has both template-directed and terminal transferase (template-independent) activities. In addition to their ability to incorporate untemplated nucleotides, another similarity between Polµ and terminal deoxynucleotidyl transferase (TdT) is their promiscuity in using ribonucleotides (NTPs), whose physiological significance is presently unknown. As shown here, Polµ can use NTPs instead of deoxynucleotides (dNTPs) during non-homologous end joining (NHEJ) of non-complementary ends, a Polµ-specific task. Moreover, a physiological concentration of Mn2+ ions did benefit Polµ-mediated NHEJ by improving the efficiency and accuracy of nucleotide insertion. Analysis of different mutations in the ‘steric gate’ of the active site indicated that Polµ is taking advantage of an open active site, valid for selecting alternative activating metal ions and nucleotides as substrates. This versatility would allow ad hoc selection of the most appropriate nucleotide/metal ion combination for individual NHEJ events to gain efficiency without a cost in terms of fidelity, thus widening the spectrum of available solutions to position a discontinuous template strand in proper register for connection.
INTRODUCTION
The non-homologous end-joining (NHEJ) pathway is the principal mechanism used to repair double-strand breaks (DSBs) in higher eukaryotes. The NHEJ pathway is considered to be error-prone but, nevertheless, it can often restore the original sequence at DSB sites, even when containing damaged ends and fragmented nucleotides (1). DNA ends may contain regions with a certain level of microhomology but, in the most extreme case, the two ends would be non-complementary. The template-directed polymerases from the human X family (Polβ, Polλ and Polµ) select their optimal DNA substrates on the basis of a gradient of template-dependence [maximal in Polβ, moderate in Polλ and much lower in the case of Polµ (2)]. In agreement with these differences, Polβ is not a player of the NHEJ pathway, while Polλ and Polµ are the main polymerases involved (3–7); however, they are non-redundant, as in the presence of NHEJ accessory factors such as Ku70/80 and XRCC4/LigaseIV, both of them can use complementary ends, but only Polµ is able to bridge and polymerize on non-complementary ends (2).
Regarding the metal cofactor, the use of manganese ions increases polymerization efficiency of X family DNA polymerases (8), but also reduces fidelity for most of the polymerases tested to date (8–11), a behaviour that has been also true for Polµ (12). However, manganese could be the natural metal activator of Polλ in vivo, as this polymerase is active over a wide range of Mn2+ concentrations while inhibited by high levels (>2.5 mM) of Mg2+ (8). The same observations have been made for Polι, that is most strongly activated at physiological (as low as 0.1 mM) MnCl2 concentrations but is inhibited by physiological MgCl2 concentrations; moreover, activation of Polι by low concentration of manganese ions improves its nucleotide insertion fidelity opposite T (13).
Are deoxynucleotides the only valid substrates for DNA repair? Ribonucleotides are highly abundant in human cells, their levels being 40- to 350-fold (depending on the dNTP/NTP pair) higher that those of dNTPs in cycling cells, and 160- to 2000-fold in confluent cells (14,15). In most instances, DNA polymerases discriminate against NTP insertion by binding them weakly and inserting them more slowly than dNTPs. Crystallographic studies of DNA polymerases from most families have suggested that a discrete side chain, referred to as a ‘steric gate’, could sterically interfere with binding an NTP (16,17). In contrast, the X family polymerases Polβ and Polλ have been suggested to deter insertion of a ribonucleotide using the protein backbone near the carboxyl terminus of α-helix M with a tyrosine residue that acts more as a ‘steric fence’ than a ‘gate’ (18,19).
As previously reported (20,21), NTPs are valid substrates for human Polµ during gap filling, with relatively low efficiency (about 10-fold lower) compared with dNTP insertion, but still in sharp contrast with the >1000-fold discrimination exhibited by most DNA-dependent DNA polymerases. This ability critically resides on a single residue, Gly433, shown to play a permissive role in sugar selection of the incoming nucleotide: its mutation to the consensus ‘steric fence’ tyrosine present in Polβ or Polλ produced a strong increase in the discrimination against ribonucleotides (20). Another X family polymerase, S. pombe Pol4, has much lower discrimination values, with almost equal insertion of NTPs and dNTPs in gap-filling reactions (22). There is also recent evidence of the frequent usage of ribonucleotides even by replicative polymerases (23). Moreover, a subfamily of RNA polymerases [related to Archaeo Eukaryotic Primases (AEPs)] has been involved in the NHEJ repair of DSBs in bacteria (24). Given all these evidences, it was important to study the impact of the Mn2+ and/or NTP usage by Polµ, especially during the most delicate end-joining reactions involving non-complementary ends. Our findings indicate that Mn2+ ions at physiological concentrations benefit NHEJ by improving the polymerization efficiency without a cost in terms of fidelity, and that NTPs are not only accepted by the active site as valid physiological substrates, but they also help to improve the fidelity of certain NHEJ reactions.
MATERIALS AND METHODS
DNA and proteins
Synthetic DNA oligonucleotides were obtained from Isogen (Ijsselstein, Holland). PAGE-purified oligonucleotides were labelled at their 5′ ends with [γ-32P]ATP. The oligonucleotides used to generate the DNA substrates were the following: for 1-nt gapped substrates, Sp1C (5′GATCACAGTGAGTAC), T13C (5′AGAAGTGTATCTCGTACTCACTGTGATC) and DG1P (5′AGATACACTTCT); for a gapped substrate containing 8oxoG as the templating base, pBER (5′CTGCAGCTGATGCGCC), tBER (5′GTACCCGGGGATCCGTAC8GGCGCATCAGCTGCAG) and dBER (5′-GTACGGATCCCCGGGTAC); for 2-nt gapped substrates, P15 (5′TCTGTGCAGGTTCTT), T17 (5′TGAAGTCCCTCTCGACGAAGAACCTGCACAGA) and DG2P (5′GTCGAGAGGGACTTCA). For NHEJ assays, three sets of oligos were used. A first set of primers sharing the same common part (5′CCCTCCCTCCC…) and bearing different 3′ protrusions (1AC […CA3′], 1TG […GT3′], 1A […A3′], 1C […C3′], 1G […G3′], 1T […T3′]) were hybridized to 1D-NHEJ (5′-GGGAGGGAGGG). A second set of primers sharing the common part (5′GCACTCACGTCCC…) and bearing different 3′ overhangs (2AC […CA3′], 2TG […GT3′], 2A […A3′], 2C […C3′], 2G […G3′], 2T […T3′]) were hybridized to oligonucleotide 2D-NHEJ (5′-GGGACGTGAGTGC). Also, oligonucleotide D3C (5′-CCCTCCCTCCGCGGC) was used as primer hybridized to D1 (5′-CGGAGGGAGGG), whereas oligonucleotide D4C (5′-CGCGCACTCACGTCCCCGCC) was hybridized to D2 (5′-GGGACGTGAGTGCGCG). Oligonucleotides DG1P, DG2P, dBER, 1D-NHEJ, 2D-NHEJ, D1 and D2 contain a 5′-P group when indicated. Ultrapure dNTPs, ddNTPs, [α-32P] dNTPs (3000 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were purchased from GE Healthcare (USA). T4 polynucleotide kinase was obtained from New England Biolabs (Beverly, MA, USA). Pfu DNA polymerase and DNAse I were purchased from Promega Corporation (Madison, WI, USA). Purified human Polλ was obtained as described (25).
Construction and purification of human Polµ mutant proteins
Wild-type and mutant Polµ variants were over-expressed and purified in an Äkta Purifier FPLC system (GE Healthcare) with the following protocol: the cleared bacterial lysate was loaded on a heparin column followed by an S sepharose column. The selected fractions were then loaded on a HiPrep Sephacryl 26/60 to eliminate contaminant nucleases. The eluted fractions containing highly purified protein were concentrated and stored at −80°C.
DNA polymerization and NHEJ assays
Several DNA substrates containing 5′P-labelled primers were incubated with different proteins, at the concentration indicated in each case. The reaction mixture, in 20 µl, contained 50 mM Tris–HCl (pH 7.5), 1 mM DTT, 4% glycerol and 0.1 mg/ml BSA, in the presence of 5 nM labelled DNA substrate and 5-fold (25 nM) cold substrate, and the indicated concentrations of dNTP and activating metal ions. After incubation for 30 min, reactions were stopped by adding gel loading buffer [95% (v/v) formamide, 10 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromophenol blue] and analysed by 8 M urea/20% PAGE and autoradiography. When indicated, we used dideoxynucleotides (ddNTPs) instead of dNTPs to limit incorporation to a single nucleotide on the 3′-end of the labelled oligonucleotide.
Cell culture and preparation of nuclear cell extracts
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (GIBCO BRL) and penicillin/streptomycin (Sigma). Nuclear cell extracts were prepared as previously described (6).
Yeast culture, protein over-expression and preparation of whole cell extracts
Strains of Schizosaccharomyces Pombe were grown in Yeast Extract with Supplements (YES)-rich medium, or on Edinburgh Minimal Media (EMM) for selection. To repress the nmt1 promoter of pREP41 and pDS473a vectors, 15 mM thiamine was added to the medium. For growth on solid medium, 2% agar was added. The yeast strains were made competent by the lithium acetate method and transformed by heat shock as described (26). Whole cell extracts were obtained as previously described (27).
Western blot
Cells were lysed, and extracted proteins were resolved in 13% SDS–polyacrylamide gels and transferred to nitrocellulose membranes. Endogenous Polµ, Polλ, Ku70, XRCC4 and Ligase IV were detected with specific rabbit polyclonal (α-Polµ and α-Polλ, own source) and mouse monoclonal antibodies (sc9033, sc5606 and sc11750, Santa Cruz Biotechnologies). Over-expressed Pol4, hPolµ and hPolλ were detected with anti-GST monoclonal antibody (G1160, Sigma). Proteins were visualized by enhanced chemiluminescence detection (Amersham Biosciences) using goat anti-mouse or anti-rabbit IgGs coupled to horseradish peroxidase as the secondary antibody (Amersham Biosciences).
RESULTS
Physiological concentration of manganese ions increases Polµ efficiency at no fidelity cost during NHEJ
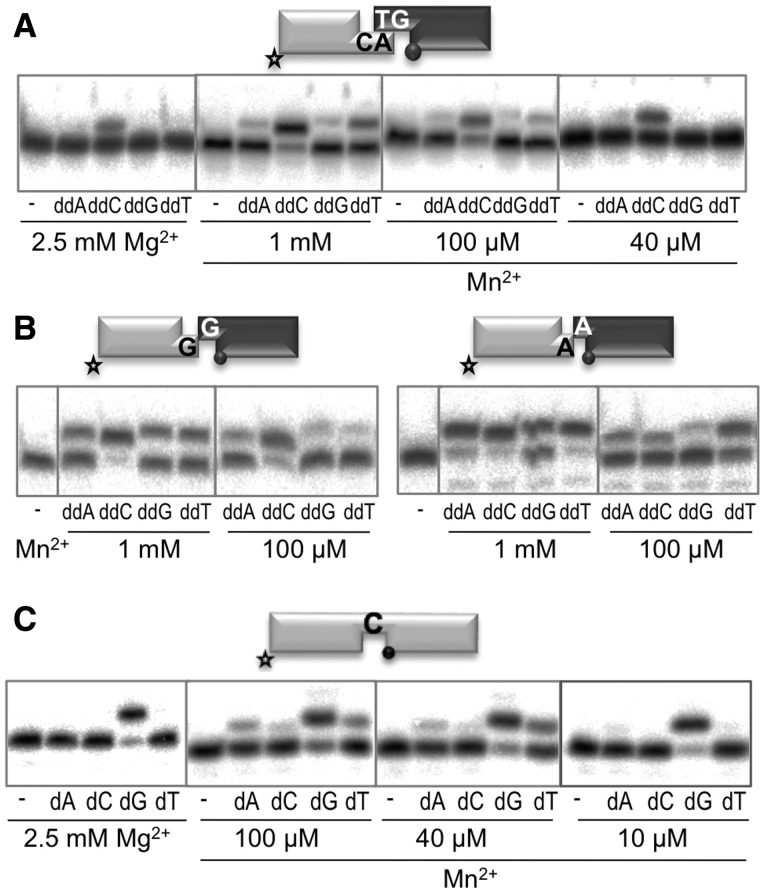
Even in the absence of NHEJ core factors, Polµ can catalyse Mg2+-driven nucleotide incorporation during NHEJ of non-complementary ends (28). However, when confronted with different substrates, there was a gradation of both efficiency and accuracy: some DNA ends allowed a very efficient and error-free reaction (G and C as template or primer), while the levels of polymerization and fidelity achieved on others (T and A as template or primer) were very low (Supplementary Figure S1). Because manganese is a well-known hyperactivator of Polµ during gap filling (12), we tested whether this cofactor could also increase the efficiency of Polµ-mediated NHEJ. We first determined the optimal concentration of Mg2+ or Mn2+ required to promote maximum activity of Polµ on NHEJ substrates (Supplementary Figure S2). In the case of Mg2+ activation, the optimum for the two DNA substrates tested was achieved at a concentration of 2 mM, while 10 mM was inhibitory. For Mn2+ activation, the highest level of activity was obtained at much lower concentrations, reaching the maximum at 125 µM. The efficiency of the reaction was extremely boosted when using Mn2+ instead of Mg2+ (see graph in Supplementary Figure S2B). We next evaluated the effect of Mn2+ activation on the fidelity of Polµ-mediated NHEJ. Strikingly, low concentrations of manganese ions (40, 100 µM) allowed efficient insertion of the correct nucleotide, while insertion of the other three was significantly lower. This was valid for both complementary (Figure 1A) and non-complementary NHEJ substrates (Figure 1B). As shown for the complementary substrates, the efficiency of the incorporation was again strongly increased when using MnCl2, given that the concentration of nucleotide needed was 100-fold lower. Fidelity of NHEJ was also increased, up to 3-fold, when activated by a low (100 µM) versus a high (1 mM) concentration of manganese ions (Figure 2B and Supplementary Figure S3). Considering this, we assayed a range of MnCl2 concentrations to test whether a low concentration of manganese ions could also modulate the fidelity and/or efficiency of Polµ-mediated gap-filling reactions. As expected, at relatively high MnCl2 concentration, nucleotide insertion fidelity was very poor compared with activation with 2.5 mM MgCl2, but progressively improved by reducing manganese concentration (Figure 1C): insertion of the correct nucleotide (dG) was as efficient as that obtained with 2.5 mM MgCl2 and insertion of the other three incorrect nucleotides was barely detectable. Taken together, these results indicate that Polµ takes advantage of the use of manganese ions, at physiological concentrations, to achieve the best efficiency and fidelity when dealing with NHEJ substrates.
Figure 1.
Mn2+ is the preferred activator in terms of both efficiency and fidelity during Polµ-mediated NHEJ reactions. (A) NHEJ reactions were performed with 200 nM Polµ and using one set of complementary substrates: the labelled substrate contained 1AC and 1D-NHEJ, and the cold substrate, 2TG and 2D-NHEJ. When indicated, ddNTPs were added separately at 10 µM in the presence of the indicated amounts of MgCl2, or at 0.1 µM when MnCl2 was used. From now on, the black spheres indicate the presence of a 5′-P group in the downstream strand of the substrate, and the star indicates the labelled substrate. (B) NHEJ reactions were performed with 200 nM Polµ and using two sets of non-complementary substrates: the labelled substrates contained 1C or 1A and 1D-NHEJ, and the cold substrates, 2C or 2A and 2D-NHEJ. When indicated, ddNTPs were added separately at 0.1 µM in the presence of the indicated amounts of MnCl2. (C) Gap-filling reactions (right panels) were performed with Polµ (25 nM) using a 1-nt gapped substrate. When indicated, dNTPs were added separately at 10 nM in the presence of the indicated amounts of MgCl2 or MnCl2.
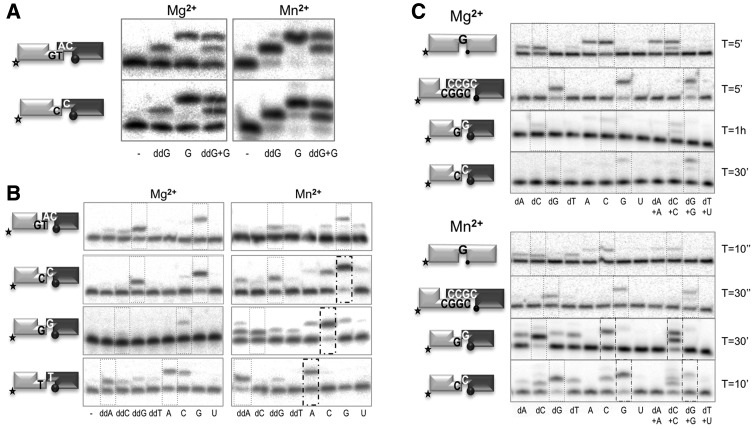
Figure 2.
Ribonucleotides as the ultimate substrates for Polµ during NHEJ. (A) NHEJ reactions were performed with 200 nM Polµ. The labelled substrates contained 1TG or 1C and 1D-NHEJ, and the cold substrates 2AC or 2C and 2D-NHEJ. When indicated, the correct ddNTP (ddG) or/and the correct NTP (rG) were added in the presence of 2.5 mM MgCl2 (1 µM ddNTP/10 µM NTP) or 0.1 mM MnCl2 (10 nM ddNTP/100 nM NTP). (B) NHEJ reactions were performed with 200 nM Polµ and using four sets of substrates: the labelled substrates contained 1TG, 1G, 1T or 1C and 1D-NHEJ, and the cold substrates 2AC, 2G, 2T or 2C and 2D-NHEJ. When indicated, each of the four ddNTPs or NTPs was added in the presence of 2.5 mM MgCl2 (1 µM ddNTP/10 µM NTP) or 0.1 mM MnCl2 (10 nM ddNTP/100 nM NTP). Dotted boxes indicate the complementary nucleotide insertion. Thick-lined boxes indicate a substantial change in efficiency of fidelity of the reaction when using NTPs and Mn2+. (C) Gap-filling and NHEJ reactions were performed with 25 nM and 200 nM Polµ, respectively, in the presence of 2.5 mM MgCl2 or 0.1 mM MnCl2, as indicated. Nucleotides were added at the following concentrations: dATP at 2.4 µM, ATP at 2.1 mM; dCTP at 4.5 µM, CTP at 91 µM; dGTP at 2.7 µM, GTP at 305 µM; dTTP at 17 µM, UTP at 253 µM. Reaction times are indicated in each panel.
Ribonucleotides as ultimate substrates: Insertion of ribonucleotides increases the fidelity of Polµ during NHEJ
The low selectivity of Polµ regarding the use of NTPs as alternative substrates prompted us to reconsider their importance not only during gap filling, but also mainly for both the efficiency and accuracy of NHEJ reactions. For comparison, ddNTPs were used as reference substrates (X family DNA polymerases do not discriminate between dNTPs and ddNTPs) and also to limit primer extension to a single nucleotide. ddNTPs were provided at 10-fold lower concentration than NTPs, as derived from previous studies (20). Figure 2A shows that Polµ can efficiently use NTPs on NHEJ substrates having either a minimal microhomology of one base pair (upper panels), or none (lower panels). According to their similar insertion efficiency (observed either when ddGTP and GTP were independently or simultaneously added), it can be concluded that Polµ displays low (about 10-fold) ddGTP/GTP discrimination during NHEJ reactions.
We then tested the fidelity (base discrimination) of Polµ in these NHEJ contexts by comparing the insertion of the eight possible nucleotides (ribo and deoxy; ribos at 10-fold higher concentration). In this case, we used one set of complementary and three different sets of non-complementary NHEJ substrates as shown in Figure 2B. On complementary ends, preferential insertion of the correct nucleotides (ddGTP and GTP) and a low level of misinsertion were observed when activated by Mg2+ (2.5 mM). By using Mn2+ (100 µM), a similar insertion efficiency of the correct nucleotide was observed, when provided at a 100-fold lower concentration. The minimum level of misinsertion was comparable when using NTPs or ddNTPs in the presence of Mn2+. Therefore, template-directed insertion of complementary ribonucleotides occurs during Polµ-driven NHEJ of complementary ends. When using non-complementary ends, the outcome varied as a function of the sequence of the 3′-protrusions. In all the contexts tested, efficient and predominant insertion of the correct nucleotide was observed with both metals. In most of the cases, the manganese-driven incorporation of the complementary NTP was the most efficient and accurate solution (thick lined, dotted boxes in Figure 2B).
To further perfect our study, we repeated the assays in the presence of physiological concentrations of deoxynucleotides, ribonucleotides or both (14) either with magnesium of manganese ions as cofactors. To set up the correct reaction conditions for measuring fidelity, we performed time-course experiments for each substrate (1-nt gap, complementary and non-complementary DNA ends) with the correct deoxy- or ribonucleotide only (Supplementary Figure S4). After quantification of primer extension, we confirmed the strong boosting effect of manganese, and we could conclude that NTP insertion is in general more efficient than dNTP insertion, although slightly. When both substrates were added at the same time, NTPs were always preferentially incorporated while dNTP insertion was diminished. For the following experiments, we selected the time at which around 50% of primer extension was achieved in each case. For the gapped and complementary NHEJ substrates (Figure 2C, top panels of each part), fidelity levels were similar with NTPs or dNTPs, despite the large concentration of the three incorrect ribonucleotides (Figure 2C). It is especially notable the discrimination against ATP, present at millimolar amounts. For the two non-complementary NHEJ substrates tested, fidelity was improved when NTPs were used in the presence of Mn2+ ions, thus confirming our previous results (Figure 2C, lower panels). When both substrates were added simultaneously, NTPs were preferentially incorporated in every case, regardless of the metal cofactor used. Strikingly, the presence of ribonucleotides led to a decrease in the mutagenic incorporation of deoxynucleotides (for example, see reaction on non-complementary ends with dG as templating base, in the presence of Mn2+ ions).
Therefore, ribonucleotides or deoxynucleotides, and magnesium or manganese, can be used in alternative combinations of substrate and metal activator to achieve the optimal solution for the variety of protrusions to be handled by Polµ during NHEJ.
Ribonucleotide insertion: the consequence of a versatile active site?
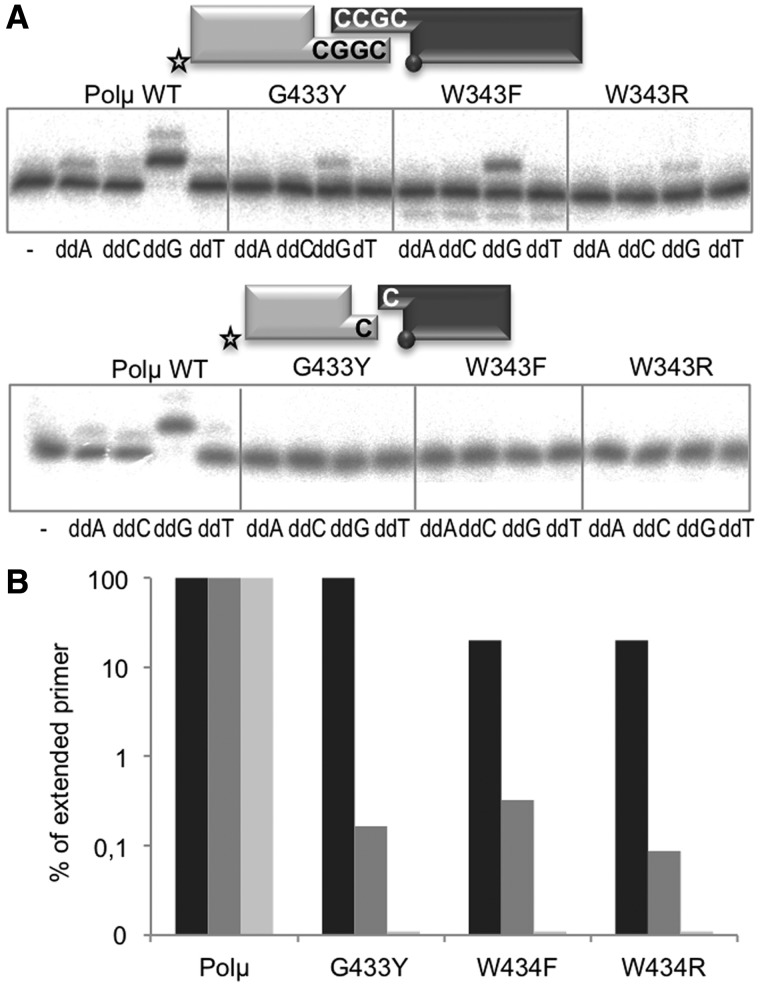
Three Polβ residues make Van der Waals contacts with the sugar moiety of the incoming nucleotide: Phe272, Gly274 and Tyr217, the ‘steric fence’ in this polymerase (29). A counterpart of Polβ Gly274 is invariantly present in all members of the PolX family. Interestingly, Gly and Trp substitute Tyr271 and Phe272, respectively, in TdT and Polµ from different species and in Pol4 from S. pombe, all of them polymerases able to incorporate ribonucleotides. According to this, it seemed very likely that residues Gly433 and Trp434 of Polµ could be responsible for the lack of sugar discrimination between NTPs and dNTPs. In a previous study from our laboratory, Gly433 and Trp434 were mutated to restore individual consensus residues of Polβ-like enzymes (20). Mutant W434F had a polymerization activity similar to that of the wild-type protein, but the insertion of NTPs occurred at lower rate. The deleterious effect of the other mutations in general polymerization, specially the change of the glycine for a larger residue, points to a unique requirement of an open and spacious active site that leads not only to the insertion of NTPs and other nucleotide analogues, but also to higher misincorporation rates and, most specially, to the ability to bind and connect two distinct DNA ends. To test this hypothesis, we studied whether the different competence of these mutants in polymerization would specifically affect the efficiency and fidelity of Polµ-driven NHEJ reactions with ddNTPs. As shown in Figure 3A, the polymerization levels of the mutants were greatly affected (complementary ends) or undetectable (non-complementary ends). Quantification of the +1 incorporation of the correct dNTP/ddNTP on a DNA gap (black) versus complementary (dark grey) or non-complementary (light grey) NHEJ substrates (Figure 3B) demonstrates that the effect of these mutations is sensibly larger when confronting the most difficult reaction of bridging two DNA ends and performing trans-directed polymerization.
Figure 3.
Mutations in the steric gate affect insertion of ddNTPs during Polµ-mediated NHEJ reactions. (A) NHEJ reactions were performed with 200 nM wild-type or mutant versions of Polµ and two sets of substrates: the labelled substrates contained D3C and D1 or 1C and 1D-NHEJ, and the cold substrates D4C and D2 or 2C and 2D-NHEJ. When indicated, each of the four ddNTPs (10 µM) was added in the presence of 2.5 mM MgCl2. (B) Quantification of the percentage of extended primer by the indicated enzymes on gap filling (black), NHEJ of complementary ends (dark grey) and of non-complementary ends (light grey), in a logarithmic scale.
Nucleotides used for NHEJ in nuclear cell extracts
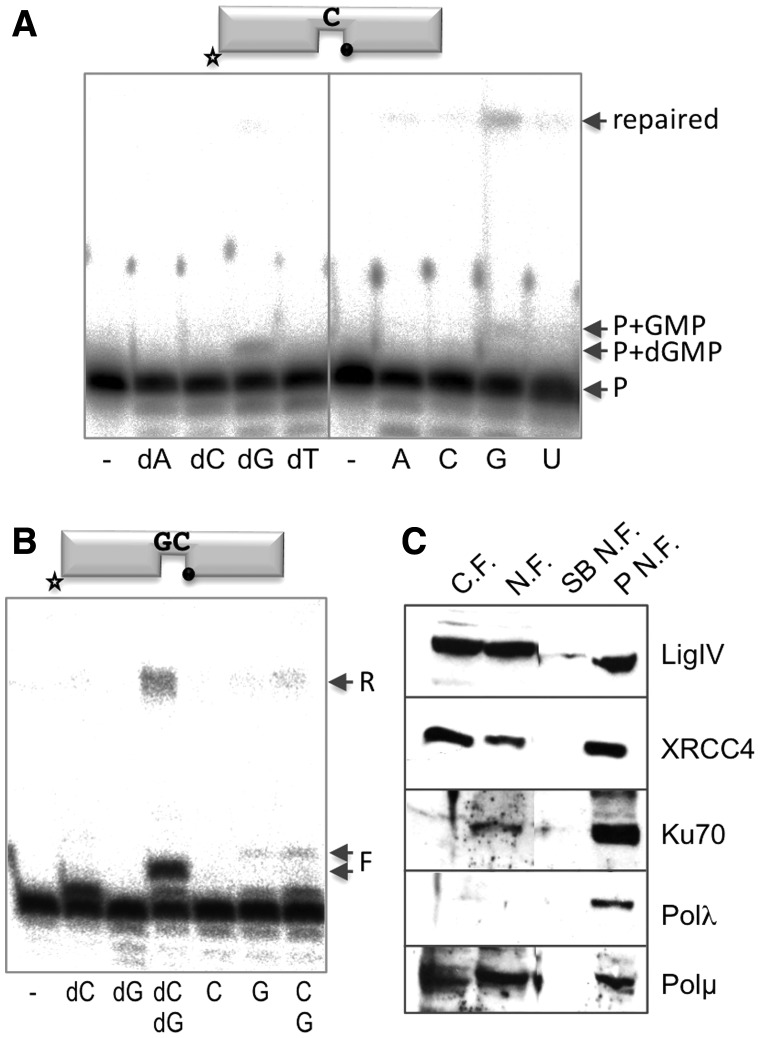
To further understand the role of Polµ and its use of ribonucleotides/manganese in the NHEJ repair pathway in a situation that more closely resembles the in vivo physiological conditions, we used HeLa nuclear cell extracts. The extracts were firstly tested for the presence of NHEJ core factors needed for the reaction. Western blotting showed that XRCC4, Ligase IV and Ku70 were enriched in the nuclear fractions obtained, and Polµ and Polλ were also present (Figure 4C). By using a 1-nt gapped substrate, and after incubation for 6 h at 37°C, the extracts catalysed the insertion of only the correct dNTP (dG) dictated by the templating base (Figure 4A). Ligation (after gap filling/dG incorporation) was also evidenced by the formation of a repaired product of 34 nts. Strikingly, the extracts also catalysed the preferential insertion of GTP and its subsequent ligation even more efficiently than with dNTPs (Figure 4A, right panel). This reaction, compatible with Polµ activity, could also account for the known preference of LigIV of ligating substrates containing a 3′-terminal NTP (21). Looking for an enzymatic activity that could be more specifically attributed to Polµ, we next used a 2-nt gap in which Polµ, but not Polβ or Polλ, will perform ‘template dislocation’: instead of using the templating base closest to the 3′ terminus (position +1), the polymerase will choose another one located further downstream in the template strand (position +2 in the 2-nt gap, closest to the 5′-P group). For this, the previous bases (only one in a 2-nt gap but more in a longer gap) have to be ‘dislocated’ (a ‘bubble’ will be formed in the template strand) and will not be copied, resulting in frameshifts (30). In the presence of dNTPs, the polymerase activity present in the extracts preferentially inserted the first templated nucleotide, dCTP (and not dGTP, the nucleotide complementary to the second templating base), and efficiently filled the 2-nt gap when both dCTP and dGTP were provided simultaneously (Figure 4B). The formation of a 34-mer, product of a very efficient ligation, could be observed when the gap was correctly filled, but some level of ligation was also observed when the incorporation of only the first nucleotide was achieved. When NTPs were provided as substrates, no reaction was observed with CTP (complementary to the first templating base) but incorporation of GTP (complementary to the second templating base of the gap) could be clearly observed; this indicates that the first templating base is being dislocated, thus hallmarking Polµ as the main author of this incorporation. When the two ribonucleotides CTP and GTP were present, the reaction pattern was the same as that with only GTP, again a typical Polµ behaviour. In the cases in which incorporation of GTP was observed, repaired products were also formed (R), implying the formation of a ligated product lacking one nucleotide.
Figure 4.
Use of ribonucleotides during repair reactions by HeLa nuclear extracts. (A) Gap-filling reactions were performed with HeLa nuclear extracts (30 µg) using a 1-nt gapped substrate. When indicated, dNTPs or NTPs were added separately at 100 µM in the presence of 1 mM MnCl2. After 2 h of incubation, reactions were stopped and loaded on 20% PAGE/8 M urea gels. +1 incorporation and ligation (arrows) products could be detected. ‘P’ stands for ‘primer’. (B) Gap-filling reactions were carried out as in (A), but using a 2-nt gapped substrate. ‘R’ stands for ‘repaired’, ‘F’ for ‘gap-filled’. (C) Western blot analysis showing the presence of the different NHEJ factors in the different steps of the purification of the HeLa nuclear extracts: C.F.: cytosolic fraction; N.F.: nuclear fraction; SB N.F.: supernatant of the nuclear fraction; P N.F.: pellet of the nuclear fraction.
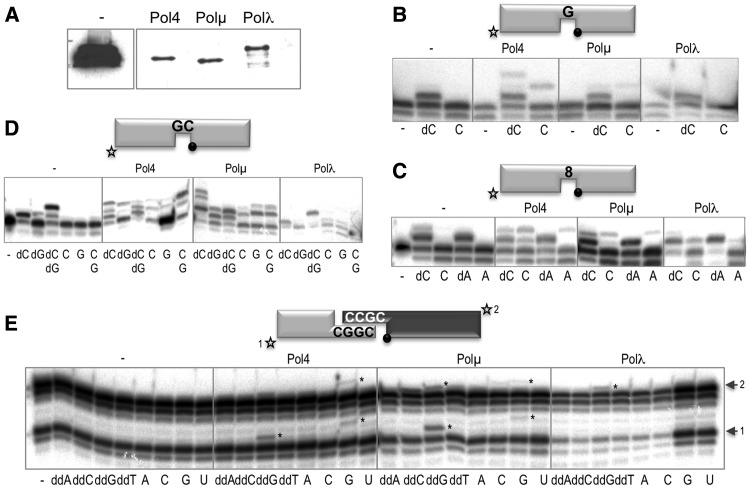
NHEJ activity in S. pombe extracts over-expressing SpPol4, Polλ or Polµ
The only DNA PolX from the fission yeast S. pombe, SpPol4, is more closely related to Polµ, but its DNA polymerization properties combine those of Polβ, Polµ and Polλ (22). Like Polµ, SpPol4 can incorporate an NTP into a primer DNA. Also, mammalian family X polymerases have been shown to differentially complement a Δpol4 mutation in Saccharomyces cerevisiae, depending on the joint structure, demonstrating that these polymerases can participate in yeast NHEJ but with distinct specificities (31). In the light of all this evidence, we decided to use S. pombe as a model organism, as it provides us with a simplified system to test the usage of ribonucleotide substrates by SpPol4, Pol µ and Polλ (over-expressed in this system) with a more physiological approach: using whole cell extracts instead of purified recombinant proteins. To eliminate the background of an endogenous SpPol4 activity, we used an S. pombe mutant strain lacking the pol4 gene, ΔPol4 (22). The level of over-expression of the three polymerases in the whole cell extracts was very similar, as assessed by WB (Figure 5A). Polymerase activity of the extracts was first assayed in the presence of a 1-nt gapped DNA substrate (Figure 5B). The Δpol4 extract displayed a very efficient gap-filling activity when Mn2+ and the correct dNTP were supplied, while the endogenous polymerase activity responsible for this reaction was not able to use the correct NTP as a substrate (Figure 5B, compare lanes 2 and 3). This feature is important since, as it has been already shown (20,22), Polµ and SpPol4 can use NTPs, thus making it a specific activity ascribable only to these polymerases in this context. When either of them was over-expressed, incorporation of the correct NTP was observed (Figure 5B), but not in the case of Polλ over-expression. We next used a 1-nt gapped DNA substrate in which the templating base is 8oxoG, an oxidized damaged base (Figure 5C). Similar levels of the non-mutagenic and mutagenic incorporation of dC and dA were obtained in the case of the four extracts, but the use of NTPs was again restricted to the SpPol4 and Polµ extracts (the faint +1 bands in the Polλ extract correspond to the usage of the corresponding deoxynucleotide, present in the purified ribonucleotide aliquot owing to the spontaneous degradation of the ribose 2′-OH group. Polλ can use this very small amount of deoxynucleotide owing to its very low Km per nucleotide). When using a 2-nt gapped substrate, the Δpol4 and Polλ extracts were not able to efficiently dislocate the template and thus incorporated preferentially the dNTP complementary to the first templating base (Figure 5D); when the two deoxynucleotides were supplied, complete filling of the gap was achieved. In the case of the SpPol4 and Polµ extracts, on the other hand, dislocation was the preferential outcome, both with deoxy and ribonucleotides, indicating that these DNA polymerases compete out the endogenous polymerase activity on this substrate (Figure 5D, second and third panels). Finally, to check the NHEJ activity displayed by the extracts, we used a set of complementary NHEJ substrates that allow detection of nucleotide incorporation in the two sides of the break at the same time. In this case, the Δpol4 extract displayed no activity at all (Figure 5E, first panel), showing that the endogenous polymerase activity, although capable of filling a gap, is not able to bridge two DNA molecules. The Polλ extract showed a limited ability to bridge these DNA ends and inserted the correctly templated ddNTP only on one side of the synapsis (Figure 5E, fourth panel). The SpPol4 and Polµ extracts, on the other hand, were able to efficiently and faithfully polymerize on both sides of the synapsis, and with both ddNTPs and NTPs (Figure 5E, second and third panels).
Figure 5.
Use of ribonucleotides during repair reactions by S.pombe extracts over-expressing Pol4, Polλ or Polµ. (A) Western blot analysis showing the presence of the over-expressed GST-tagged proteins in the yeast extracts. (B) Gap-filling reactions with S.pombe extracts (30 µg) using a 1-nt gapped substrate. When indicated, dNTPs or NTPs were added separately at 100 µM in the presence of 1 mM MnCl2. After 2 h of incubation, reactions were stopped and loaded on 20% PAGE–8 M urea gels. (C) Gap-filling reactions were carried out as in (B), but using a gapped substrate with 8oxoG as the templating base. (D) Gap-filling reactions were carried out as in (B), but using a 2-nt gapped substrate. (E) NHEJ reactions performed with 30 µg of S. pombe extracts. The short substrate (light grey) contained D3C and D1, the long substrate (dark grey) D4C and D2. When indicated, each of the four ddNTPs or NTPs (100 µM) was added in the presence of 1 mM MnCl2.
DISCUSSION
The degree of variability at the DNA substrate level during NHEJ compels Polµ to have a spacious and versatile active site. In fact, our results indicate that this versatility is such that Polµ can take advantage of the widest range of solutions when polymerizing: not only it can use deoxynucleotides, but also ribonucleotides (20,21). This lack of sugar discrimination is due to the change of the YF motif, the ‘steric gate’ conserved in Polβ and Polλ active sites, for a GW motif. Specifically, the change of the tyrosine for a glycine enables Polµ to use NTPs during catalysis (20). Recent results of site-directed mutagenesis in Polλ (18) and Polβ (29) confirmed the initial results in Polµ, consolidating the idea of a common mechanism of sugar selection in this family of polymerases. In the case of Polµ, this selectivity is lost in the way to find a more open architecture for the active site that not only promotes the insertion of NTPs, but surely contributes also to the low fidelity levels of the enzyme. Thus, Polµ has to cope with both ‘collateral damages’ in order to obtain the profit of being able to deal with non-complementary DNA end substrates. But the ability to insert NTPs brought its own benefits: NTPs are a much more abundant resource than dNTPs during almost all the phases of the cell cycle (14), and thus they qualify as favourable substrates in the equation of DNA repair. X family DNA polymerases that exhibit low NTP discrimination would be predicted to insert more NTPs than dNTPs when the in vivo dNTP/NTP imbalance is taken into consideration. Also, the use of NTPs during NHEJ in vivo is supported by the finding that a 3′-terminal ribonucleotide stimulates the activity of eukaryotic ligase IV (21). Moreover, the ability of Polµ to polymerize with NTPs is dramatically reduced with each successive ribonucleotide added, effectively limiting synthesis to few more than one nucleotide (20,21), compatible with ligase IV preference for only one ribonucleotide at the 3′ end. This may be also helpful in curtailing template-independent activity, because long non-complementary tails would be difficult to align and ligate. Thus, insertion of ribonucleotides by Polµ would not jeopardize the NHEJ system, but even make it more efficient from the point of view of both polymerization and ligation.
Our results show that during NHEJ the error proneness of Polµ was lowered when using NTPs. This was valid even in the presence of manganese, an optional activating metal ion that appears to be physiological during NHEJ. Mn2+ was for a long time considered mutagenic, and consequently not physiological, but renewed efforts in the study of polymerase function in vitro have demonstrated that some specialized polymerases, as Polι, clearly prefer to use Mn2+ even when Mg2+ is in large molar excess (13). Our results with Polµ point to a similar conclusion: although able to use other cofactors such as zinc or cobalt, the peak of polymerase activity with NHEJ substrates was reached at the physiological concentration of manganese ions (32–34). The intracellular magnesium concentration is 0.21–0.24 mM (35,36), also in the range for Polµ activation. When NHEJ reactions were performed at low concentration of manganese, the well-known burst in efficiency was achieved, but without the negative effect on fidelity that can be observed at the exceedingly high concentrations of manganese used before in the literature. As already mentioned, these positive effects on both efficiency and fidelity were observed not only with dNTPs, but more interestingly, even better results were obtained with NTPs. Whether a similar situation occurs in vivo is an intriguing possibility certainly worth considering. By using wild-type HeLa nuclear extracts, and yeast extracts over-expressing the NHEJ-related polymerases SpPol4, Polµ and Polλ, we tested ribonucleotide usage by these enzymes during the first step of a NHEJ reaction and also during the second step (gap filling), and evidence shows that under these conditions, SpPol4 and Polµ were able to use NTPs as competent substrates.
Taken together, all these evidences are pointing to the ambidexterity of Polµ active site, prepared for almost any kind of DNA and nucleotide substrates as well as cofactors that need to be used, and speak of Polµ as a highly specialized polymerase, singularly suited for its function: engagement of two different DNA chains and trans-directed polymerization, even in the case of inexistent complementarity, with the only objective of fulfilling the most efficient reaction possible with the minimal loss of sequence during NHEJ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Ministerio de Ciencia y Tecnología [BFU2009-10085]; CONSOLIDER [CSD2007-00015]; Institutional grant to Centro de Biología Molecular ‘Severo Ochoa’ from Fundación Ramón Areces; CAM fellowship (to M.J.M.). Funding for open access charge: MEC [BFU2009–10085].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 2.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Davis BJ, Havener JM, Ramsden DA. End-bridging is required for pol mu to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–3094. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 7.Ramsden DA. Polymerases in nonhomologous end joining: building a bridge over broken chromosomes. Antioxid. Redox. Signal. 2011;14:2509–2519. doi: 10.1089/ars.2010.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanca G, Shevelev I, Ramadan K, Villani G, Spadari S, Hubscher U, Maga G. Human DNA polymerase lambda diverged in evolution from DNA polymerase beta toward specific Mn(++) dependence: a kinetic and thermodynamic study. Biochemistry. 2003;42:7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 9.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 10.Loeb LA, Kunkel TA. Fidelity of DNA synthesis. Annu. Rev. Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 11.Sirover MA, Loeb LA. Metal-induced infidelity during DNA synthesis. Proc. Natl Acad. Sci. USA. 1976;73:2331–2335. doi: 10.1073/pnas.73.7.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez AC, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank EG, Woodgate R. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J. Biol. Chem. 2007;282:24689–24696. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- 14.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 15.Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl Acad. Sci. USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 20.Ruiz JF, Juarez R, Garcia-Diaz M, Terrados G, Picher AJ, Gonzalez-Barrera S, Fernandez de Henestrosa AR, Blanco L. Lack of sugar discrimination by human Pol mu requires a single glycine residue. Nucleic Acids Res. 2003;31:4441–4449. doi: 10.1093/nar/gkg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Barrera S, Sanchez A, Ruiz JF, Juarez R, Picher AJ, Terrados G, Andrade P, Blanco L. Characterization of SpPol4, a unique X-family DNA polymerase in Schizosaccharomyces pombe. Nucleic Acids Res. 2005;33:4762–4774. doi: 10.1093/nar/gki780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Diaz M, Bebenek K, Sabariegos R, Dominguez O, Rodriguez J, Kirchhoff T, Garcia-Palomero E, Picher AJ, Juarez R, Ruiz JF, et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- 26.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 27.Cueille N, Salimova E, Esteban V, Blanco M, Moreno S, Bueno A, Simanis V. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 2001;114:2649–2664. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- 28.Andrade P, Martin MJ, Juarez R, Lopez de Saro F, Blanco L. Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ. Proc. Natl Acad. Sci. USA. 2009;106:16203–16208. doi: 10.1073/pnas.0908492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavanaugh NA, Beard WA, Batra VK, Perera L, Pedersen LG, Wilson SH. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J. Biol. Chem. 2011;286:31650–31660. doi: 10.1074/jbc.M111.253401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz JF, Lucas D, Garcia-Palomero E, Saez AI, Gonzalez MA, Piris MA, Bernad A, Blanco L. Overexpression of human DNA polymerase mu (Pol mu) in a Burkitt's lymphoma cell line affects the somatic hypermutation rate. Nucleic Acids Res. 2004;32:5861–5873. doi: 10.1093/nar/gkh929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 32.Ash DE, Schramm VL. Determination of free and bound manganese(II) in hepatocytes from fed and fasted rats. J. Biol. Chem. 1982;257:9261–9264. [PubMed] [Google Scholar]
- 33.Markesbery WR, Ehmann WD, Alauddin M, Hossain TI. Brain trace element concentrations in aging. Neurobiol. Aging. 1984;5:19–28. doi: 10.1016/0197-4580(84)90081-2. [DOI] [PubMed] [Google Scholar]
- 34.Versieck J. Trace elements in human body fluids and tissues. Crit. Rev. Clin. Lab. Sci. 1985;22:97–184. doi: 10.3109/10408368509165788. [DOI] [PubMed] [Google Scholar]
- 35.Goldschmidt V, Didierjean J, Ehresmann B, Ehresmann C, Isel C, Marquet R. Mg2+ dependency of HIV-1 reverse transcription, inhibition by nucleoside analogues and resistance. Nucleic Acids Res. 2006;34:42–52. doi: 10.1093/nar/gkj411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gee JB, II, Corbett RJ, Perlman JM, Laptook AR. Hypermagnesemia does not increase brain intracellular magnesium in newborn swine. Pediatr. Neurol. 2001;25:304–308. doi: 10.1016/s0887-8994(01)00317-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.