Abstract
The transition of mammalian cells from quiescence to proliferation is accompanied by the differential expression of several microRNAs (miRNAs) and transcription factors. However, the interplay between transcription factors and miRNAs in modulating gene regulatory networks involved in human cell proliferation is largely unknown. Here we show that the miRNA miR-22 promotes proliferation in primary human cells, and through a combination of Argonaute-2 immunoprecipitation and reporter assays, we identified multiple novel targets of miR-22, including several cell-cycle arrest genes that mediate the effects of the tumor-suppressor p53. In addition, we found that miR-22 suppresses interferon gene expression by directly targeting high mobility group box-1 and interferon regulatory factor (IRF)-5, preventing activation of IRF3 and NF-κB, which are activators of interferon genes. The expression of interferon genes is elevated in quiescent cells and their expression is inhibitory for cell proliferation. In addition, we find that miR-22 is activated by the transcription factor Myc when quiescent cells enter proliferation and that miR-22 inhibits the Myc transcriptional repressor MXD4, mediating a feed-forward loop to elevate Myc expression levels. Our results implicate miR-22 in downregulating the anti-proliferative p53 and interferon pathways and reveal a new transcription factor–miRNA network that regulates the transition of primary human cells from quiescence to proliferation.
INTRODUCTION
Most cells in eukaryotic organisms are quiescent, that is, they exist in the G0 stage of the cell cycle and are not actively dividing. On appropriate physiological or pathological stimuli, many types of somatic cells may leave the quiescent state, re-enter the cell cycle and begin to proliferate. The ability of cells to remain viable while quiescent, exit quiescence and re-enter into the cell cycle is essential for tissue homeostasis and is the basis for varied physiological processes such as wound healing, lymphocyte activation and hepatocyte regeneration, but it is also a hallmark of cancer (1–5). During wound healing, an intracellular signaling cascade drives global changes in gene expression that result in dermal fibroblasts and epidermal stem cells proliferating rapidly until tissue repair is accomplished and then the cells exit the cell cycle and re-enter quiescence (6).
Primary human dermal fibroblasts are an excellent model for studying the global gene expression programs regulating the transition from quiescence to proliferation. In response to serum containing growth factors, an important mitogenic signal in wound healing and tissue homeostasis, fibroblasts may be induced to proliferate or enter quiescence. Serum activation of fibroblasts induces a transcriptional program activating many aspects of wound healing, and the wound response program is recapitulated in many human cancers (7). Many key cell-cycle regulatory genes that are differentially expressed in primary human fibroblasts exiting quiescence are also deregulated in cancer (6,8). In addition, expression profiles of proliferating fibroblasts are good predictors of cancer progression (7).
Of the many genes activated in fibroblasts during the exit from quiescence, several microRNAs (miRNAs) have been identified (9). miRNAs are endogenous, non-coding small RNAs that repress gene expression post-transcriptionally (10). miRNAs play important regulatory roles in many diverse biological processes, such as development, differentiation, proliferation, apoptosis, the stress response and cancer (11–16). Roughly 50% of all human protein coding genes are predicted to be directly regulated by miRNAs (17,18). miRNAs have emerged as important regulators of cell proliferation driving tumorigenesis, and several studies have functionally linked miRNAs and transcription factors in regulatory networks that govern cell proliferation and cancer (19,20).
The miRNA miR-22 has been reported to act as both a proto-oncogene in various cancer cell lines and a tumor suppressor in other cancer cell lines, as well as being involved in panic disorder, hypoxia signaling, differentiation and cardiomyocyte hypertrophy (21–24). miR-22 was shown to promote cell proliferation, invasion and survival in multiple cancer cell types by regulating PTEN (21,24–26). However, miR-22 has also been demonstrated to repress proliferation by inhibiting the binding partners, MAX and MYCBP, of the oncogene Myc (27–29). In addition, miR-22 was shown to inhibit cancer progression by inducing cellular senescence (30), and to repress cell migration and invasion in ovarian cancer (23). miR-22 has also been linked to the p53 regulatory network, is a direct target of the tumor-suppressor p53 and mediates p53-induced cell-cycle arrest and apoptosis in colon cancer (28). Despite the broad involvement of miR-22 in tumorigenesis in various cancers, the activity of miR-22 in primary cells has not been explored.
In this study, we investigate the regulation of cell proliferation in primary cells by miR-22 and identify a new regulatory network mediating the transition of primary cells from quiescence to proliferation. This network involves the activation by the oncogene Myc of miR-22, which in turn suppresses the interferon and cell-cycle arrest pathways active in quiescent cells, thus facilitating their re-entry into the cell cycle. Our data reveal novel cross-talk between the p53 and Myc regulatory networks that is mediated by miR-22.
MATERIALS AND METHODS
Normal cell culture conditions
HeLa cells and human foreskin fibroblasts (ATCC CRL #2091) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C under 5% CO2. Fibroblasts were made quiescent by first growing them under normal conditions until 40% confluent, then replacing the medium with DMEM supplemented with 0.1% FBS and growing them for a further 48 h.
Fibroblast serum stimulation
Fibroblast cell cultures were grown under normal cell culture conditions until 40% confluent. The medium was removed and cell cultures were washed three times with phosphate buffered saline (PBS). Replacement medium was DMEM supplemented with 0.1% FBS and 100 units penicillin–streptomycin. Cell cultures were grown at 37°C for 48 h. Cell cultures were washed once with PBS. Reference cell cultures were harvested following total RNA isolation. Replacement medium was DMEM supplemented with 10% FBS and 100 units penicillin–streptomycin. Separate cell cultures were allowed to proliferate under serum-rich conditions for time points of 5, 10, 20, 30, 60 and 180 min. At the end of each of these time points, cell cultures were harvested for total RNA isolation using the Trizol reagent (Invitrogen) according to the manufacturer’s protocol.
Myc overexpression
HeLa cell cultures were grown under normal cell culture conditions. Six-well plates were seeded with 1.5 × 105 cells/well. Cell cultures were allowed to grow for 24 h. Cell cultures were lipotransfected with Invitrogen Lipofectamine 2000 according to the manufacturer’s protocol (for DNA plasmid transfection). Myc overexpression plasmid was purchased from Open Biosystems [MHS1010-57504, Human MGC Verified FL complementary DNA (cDNA), CloneID = 298544]. GFP co-transfection plasmid was purchased from Clontech (Vector=pEGFP-N1, Accession=U55762). Cell cultures were grown under normal cell culture conditions for 48 h and then harvested for total RNA.
Myc knockdown
Myc-specific small interfering RNA (siRNA) and negative control siRNA were purchased from Dharmacon. HeLa cell cultures were grown under normal cell culture conditions. Six-well plates were seeded with 1.5 × 105 cells/well. Cell cultures were allowed to grow for 24 h. Cell cultures were lipotransfected with Invitrogen Lipofectamine 2000 according to the manufacturer’s protocol (for siRNA transfection). Cell cultures were grown under normal cell culture conditions for 48 h and then harvested for total RNA. For serum stimulation experiments under conditions of Myc knockdown, fibroblasts were transfected with siRNAs against Myc or negative control siRNA and cells were starved into quiescence. Quiescent fibroblasts were then serum-stimulated for the indicated time points and miR-22 expression was assayed by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR).
miRNA enrichment, labeling and microarrays
Ambion FlashPAGE Fractionator System, Pre-cast Gels, Buffer Kit and Clean-Up Kit were used according to the manufacturer’s protocol. Invitrogen ULYSIS 546 Nucleic Acid Labeling Kit was used for the typical Cy3 sample. Invitrogen ULYSIS 647 Nucleic Acid Labeling Kit was used for the typical Cy5 sample. The kits were used according to the manufacturer’s protocol. Ambion FlashPAGE Clean-Up Kits were used for removal of unincorporated dye according to the manufacturer’s protocol. Labeled small RNAs were hybridized to dual channel miRNA microarrays, quantitated and normalized. The microarrays were printed in house with the v1 Ambion array probe set that includes 281 human miRNAs registered in miRBase.
Real-time miRNA PCR
Quantitative real-time PCR was performed for miR-22 using Applied Biosystems TaqMan miRNA Assays according to the manufacturer’s protocol.
Quantitative RT-PCR for interferon-stimulated genes
RNA from quiescent and proliferating fibroblasts was extracted with the Trizol reagent (Invitrogen) and reverse transcribed using random hexamers and the Superscript II system from Invitrogen. PCR was performed using the SYBR GREEN PCR Master Mix from Applied Biosystems. The target-gene messenger RNA (mRNA) expression was normalized to the expression of GAPDH, and relative mRNA fold changes were calculated by the ΔΔCt method. The primer sequences are shown in Supplementary Table S1.
Myc-binding site detection and motif analysis
Chromatin immunoprecipitation (ChIP) for Myc was performed as previously described (31). ChIP-enriched DNA was sequenced by Illumina sequencing technology. Short reads from the ends of DNA fragments were mapped back to the genome using Maq. Conservation was quantified according to the mammalian conservation track from UCSC, specifically, the 17-species conservation and alignment track (phastCons17way). Conservation was evaluated for each locus through local installation of a portion of UCSC’s Genome Browser and command-line execution of UCSC toolsets (hgWiggle) that retrieved conservation metrics for a specified genomic range. The mean conservation of conserved motifs was 90% (standard deviation is 6.7%), whereas the mean conservation of the non-conserved motifs was 0.6% (standard deviation is 0.3%). The presence of proximal CpG islands was evaluated using the CpG enrichment track (cpgIslandExt). Conserved binding motif coordinates for Myc across the genome were obtained from UCSC’s catalog of conserved binding sites for all mammalian transcription factors (TFBS conserved). Motif search was performed within 20 kb upstream of the miRNA start sites. For the ChIP-PCR assay, fibroblasts were starved into quiescence and then serum-stimulated to proliferate. Cells were harvested 3 h after stimulation and ChIP was performed as described above. Myc binding was confirmed using quantitative PCR performed using the SYBR GREEN PCR Master Mix from Applied Biosystems. Fold enrichment was calculated with respect to the negative control by the ΔΔCt method. Primer sequences used for ChIP-qPCR are shown in Supplementary Table S1.
RNA oligos, transfections and microarray analysis
miR-22 guide and anti-guide mature sequences were obtained from miRBase (http://microRNA.sanger.ac.uk/sequences/), while sequences for siRNA against GFP (control siRNA) were obtained from (32). The corresponding RNA oligos were ordered from Invitrogen or IDT and annealed in RNA annealing buffer (20 mM HEPES, pH 7.3, 50 mM KCl, 2 mM MgCl2). Both the miR-22 and control siRNA oligos contain the same chemical modifications of 5′-phosphate and 3′-OH. The RNA duplexes were transfected at a final concentration of 100 nM using Lipofectamine 2000 according to the manufacturer’s instructions. The miR-22 inhibitor and control were a miRCURY Locked Nucleic Acid (LNA) miRNA Inhibitor and Negative Control Inhibitor obtained from Exiqon and transfected at a final concentration of 10 nM using Lipofectamine 2000. Poly I:C was obtained from Sigma Aldrich and co-transfected at a final concentration of 200 ng/ml with miR-22 or control siRNA duplexes as described above. RNA from transfected cells was extracted using the Trizol reagent (Invitrogen), amplified and hybridized to in-house cDNA expression microarrays as previously described (9). The primary microarray data are available at NCBI’s GEO (Accession GSE42788). Functional analysis for repressed genes was performed using the freely available online software DAVID (33).
Flow cytometry
Fibroblasts were seeded at 50 000 cells per well in six-well plates. Cells were cultured in DMEM supplemented with 10% FBS. Cells were allowed to grow 24 h, and then the medium was replaced with low-serum DMEM 0.1% FBS. Eight h after replacement with DMEM 0.1% FBS, cells were transiently transfected with miR-22 or control siRNA duplexes (100 nM final concentration). Twenty-eight hours after transfection, cells were trypsinized, washed with PBS and fixed for 24 h in 70% ethanol at −20°C. After ethanol fixation, cells were washed with Stain Buffer (BD Pharmingen), incubated 30 min with fluorescein isothiocyanate Mouse Anti-Human Ki67 antibody (BD Pharmingen), washed and resuspended in 500 µl Stain Buffer with Propidium Iodide Staining Solution (5 µg/ml) (BD Pharmingen). Flow cytometry analysis for Ki67 was done using a FACsCalibur flow cytometer and 10 000 events above threshold levels were counted for each sample (BD Biosciences). Data analysis was done using FlowJo.
Luciferase assays
Entire 3′untranslated regions (UTRs), if possible, or at least 0.8–1.2 kb around the predicted miR-22 site in a 3′UTR was cloned into a Renilla vector under a cytomegalovirus (CMV) promoter. For CARF, we included the last exon in addition to the 3′UTR for the luciferase assays. miRNA seed site mutants were made by mutating 3 bp in the 6-mer seed sequence using Agilent’s QuikChange MultiSite-Directed Mutagenesis Kit. Another vector containing the Firefly luciferase under a CMV promoter was used as a normalization control. HEK293 cells were plated in 24-well plates at 105 cells/well and Renilla and Firefly vectors were co-transfected at 25 ng each along with 100 nM final concentration of miR-22. Control siRNA was used as a negative control. Cells were harvested 24 h post-transfection, and luciferase activity was measured using the Promega Dual Luciferase kit according to manufacturer’s instructions. Fold suppression was calculated as the ratio of Renilla to Firefly values for miR-22 normalized by the mean of the Renilla to Firefly ratios for the control siRNA. For the NF-κB reporter assays, HEK293 cells were plated in 24-well plates at 1 × 106 cells/well. Twenty-four hours after plating, cells were transfected with 100 ng/well of NF-κB reporter plasmid (SABiosciences) and co-transfected with poly I:C and miR-22 or the control siRNA. Cells were harvested 16 h post-transfection, and luciferase activity was measured as described above. For Figure 5E and F, we performed luciferase assays using a pGL3 promoter plasmid (Promega) as previously described (34). Around 550 bp of PCR-amplified insert from each of four putative Myc-binding sites was cloned into the vector. All primers used for cloning are listed in Supplementary Table S1.
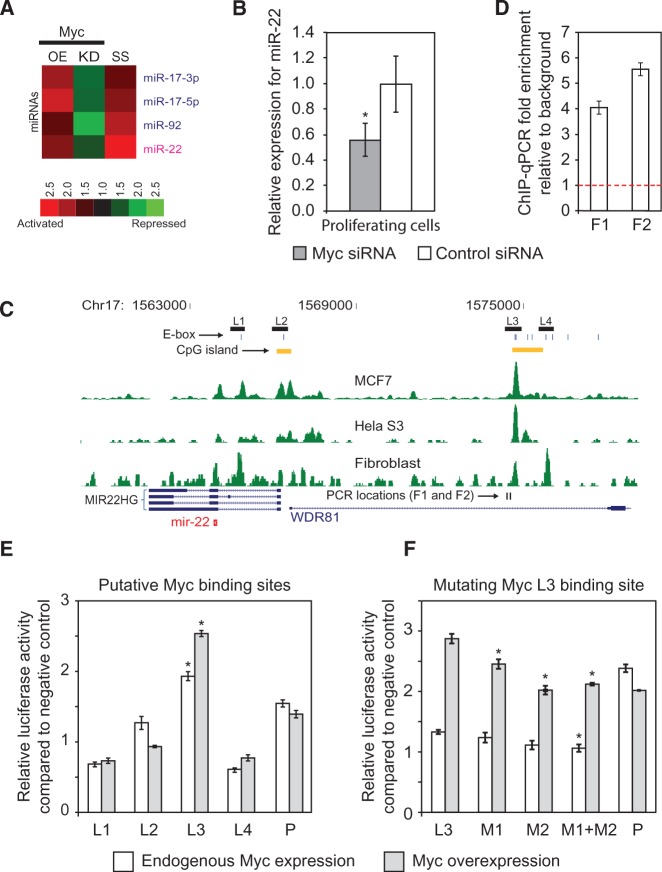
Figure 5.
Myc activates miR-22 during the transition from quiescence to proliferation. (A) Heat map showing the response of miRNAs to Myc overexpression (OE), and Myc knockdown (KO) in HeLa cells, and to serum stimulation of quiescent fibroblasts (SS) measured using microarrays. Ratios displayed under the SS column represent the maximum activation across all measured time points (5, 10, 20, 30, 60 and 180 min) after serum stimulation. miR-17 and miR-92 are previously known downstream targets of Myc. (B) miR-22 expression is inhibited by Myc knockdown in proliferating fibroblasts. miR-22 expression was assayed by qRT-PCR in fibroblasts transfected with an siRNA against Myc or a control siRNA. Relative expression values were normalized to those obtained using the control siRNA (Y-axis). (C) ChIP-seq data for Myc in HeLaS3, MCF7 and serum-stimulated fibroblasts show Myc binding upstream of miR-22. The profiles of overlapping extended ChIP-seq reads are indicated in green. Chromosome coordinates are indicated on top and genes are shown at bottom. MIR22HG refers to the primary miR-22 transcript as annotated in RefSeq, and the mature miR-22 sequence is indicated by a red symbol. Regions (L1–L4) subsequently tested for promoter activity in luciferase assays are shown by black rectangles, phylogenetically conserved E-boxes by vertical blue lines and CpG islands and qPCR primer loci by vertical black lines. (D) ChIP-qPCR verification of Myc binding upstream of miR-22. An independent ChIP for Myc was performed in serum-stimulated fibroblasts. Primers were designed to amplify two loci (F1 and F2) indicated as binding sites by ChIP-seq. Fold enrichment, indicative of Myc binding, was calculated relative to input DNA, normalized to a negative control region (Y-axis). The threshold for ChIP enrichment relative to the control is indicated by the red dashed line. (E) Promoter assays for 4 Myc putative promoter regions upstream of miR-22. L1–L4 on the X-axis indicate cloned candidate promoter regions. P is a positive control. The Y-axis shows expression fold change of a luciferase promoter reporter normalized to a negative control vector. The L3 region shows a significant increase in reporter activity when Myc is overexpressed. (F) Mutation of putative Myc-binding sites M1 and M2 in the L3 promoter region. M1 + M2 indicates both sites were mutated. For E and F, gray bars indicate Myc overexpression and white bars indicate endogenous levels of Myc (Myc was not overexpressed). For B, D, E and F, bars indicate the mean, and error bars denote ±SD, n = 3. For B, E and F, *P < 0.05.
Western blots
For western blots of phosphorylated and native interferon regulatory factor (IRF)-3, HeLa cells were seeded in six-well plates at 8 × 104 cells/well and co-transfected with 200 ng/ml poly I:C and miR-22 or the control siRNA. Twelve hours after transfection, cells were harvested for protein analysis. Cell lysates were separated on 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and proteins were transferred onto polyvinylidene fluoride (PVDF) membranes. Membranes were blocked with 5% milk in Tris-buffered saline and Tween 20 (TBST) (25 mM Tris pH 8.0, 150 mM NaCl, 0.05% Tween-20) and probed with corresponding primary antibodies against specific proteins (phosphorylated and native IRF3; Cell Signaling Technology). horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz Biotechnology) were used to detect primary antibodies, and proteins were visualized by chemiluminescence.
For western blots of putative miR-22 targets, primary human fibroblasts were seeded in six-well plates at 2 × 104 cells/well in DMEM supplemented with 10% FBS. Twenty-four hours after plating, the medium was replaced with low-serum DMEM 0.1% FBS. The medium for proliferating samples was replaced with more DMEM 10% FBS. Forty-eight hours after serum starvation, miR-22 or control siRNA was transfected at a 100 nM concentration, and proliferating and quiescent samples were lysed. Transfected cells were lysed at 12, 24, 48 or 72 h post-transfection. Cell lysates were separated on 4–20% gradient SDS-PAGE gels (Biorad) and proteins were transferred onto PVDF membranes. Membranes were blocked with 5% milk in TBST and probed with corresponding primary antibodies against specific proteins [high mobility group box-1 (HMGB1): Cell Signaling Technology, IRF5: Abcam ab33478, REDD1: Abcam ab106356, TP53INP1: Abcam ab9755, p21: Abcam ab7960, CARF: ab88322, MXD4: Santa Cruz Biotechnology sc-771, MYC: Santa Cruz Biotechnology sc-764X]. Actin was used as a loading control (Actin: Santa Cruz Biotechnology sc-10731). HRP-conjugated secondary antibodies (Santa Cruz Biotechnology sc-2004 and sc-2005) were used to detect primary antibodies, and proteins were visualized by chemiluminescence. Western blots shown are for the time point that showed the strongest repression.
Argonaute-2 immunoprecipitation
We adapted the protocol developed by Hendrickson et al. (35) for immunoprecipitating Argonaute-2 (Ago2)–mRNA complexes. Briefly, HeLa cells were grown in 10 cm2 tissue culture plates and transfected with either miR-22 mature duplexes at a final concentration of 100 nM or mock transfected. After 24 h, 0.5 ml of lysis buffer was added drop-wise onto the cell monolayer followed by incubation at 4°C for 30 min. Cell lysate was collected by scrapping and cleared by centrifugation at 14 000 rpm at 4°C. Cleared lysate was then incubated with 50 µl of protein-G beads (Roche) for 3 h at 4°C (pre-clearing). Before pre-clearing, 50 µl of the cleared lysate was removed for total RNA estimation. Pre-cleared lysate was incubated with 15 µg of Ago2 antibody (ab57113; Abcam) and incubated at 4°C overnight. The next day, 50 µl of protein-G beads were added to the lysate and incubated for 4 h at 4°C. Beads were washed eight times with lysis buffer and Ago2–RNA complexes were extracted by adding 1 ml Trizol reagent (Invitrogen) directly to the beads. RNA extraction was carried out as per the manufacturer’s instructions.
Cell counting assays
For miRNA overexpression experiments, fibroblasts were seeded at 20 000 cells per well in six-well plates. Cells were cultured in DMEM supplemented with 10% FBS. Cells were allowed to grow 24 h, and then the medium was replaced with low-serum DMEM 0.1% FBS. Twenty-four hours after replacement with DMEM 0.1% FBS, cells were transiently transfected with miR-22 or siGFP duplexes (100 nM final concentration); 0, 24, 48, 72 and 96 h after transfection, cells were trypsinized and counted in a hemacytometer. Five fields were averaged for each biological replicate. For miRNA inhibition experiments, fibroblasts were seeded at 20 000 cells per well in six-well plates. Cells were cultured in DMEM supplemented with 10% FBS. Cells were allowed to grow 24 h and then transiently transfected with miR-22 miRCURY LNA miRNA Inhibitor and Negative Control Inhibitor obtained from Exiqon (10 nM final concentration); 0, 24, 48, 72 and 96 h after transfection, cells were trypsinized and counted in a hemacytometer. Five fields were averaged for each biological replicate.
Statistical analysis
Statistical significance was estimated using a one-sided Student’s t-test.
RESULTS
miR-22 is induced during the transition from quiescence to proliferation and promotes proliferation
Microarray expression profiling identified multiple miRNAs that are differentially expressed in primary human fibroblasts induced to transition from quiescence to proliferation by serum stimulation (Figure 1A and Supplementary Table S2). We focused on miR-22 because it was one of the miRNAs most strongly and consistently induced by serum stimulation, and bioinformatic analysis suggested that it could be transcriptionally regulated by immediate-early transcription factors (not shown). qRT-PCR for miR-22 expression after serum stimulation of fibroblasts in an independent biological replicate experiment confirmed its induction (Figure 1B).
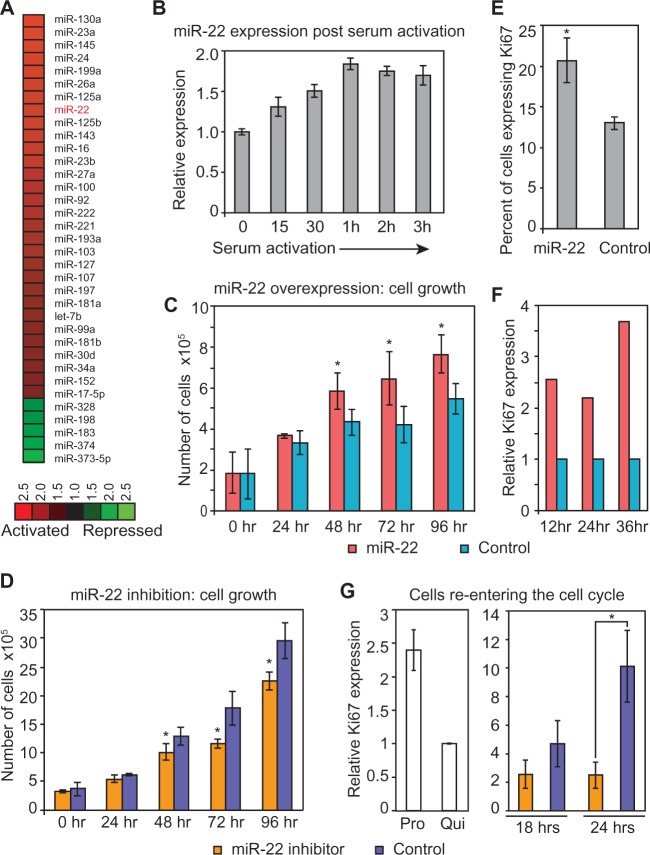
Figure 1.
miR-22 is induced during the transition from quiescence to proliferation and activates proliferation. (A) Heat map showing the response of miRNAs to serum stimulation of quiescent primary human fibroblasts measured using microarrays. Ratios displayed represent the average activation across all measured time points after serum stimulation (5, 10, 20, 30, 60 and 180 min). miRNAs shown were activated or repressed >1.5-fold on average in serum-stimulated compared with serum-starved cells. (B) Independent qRT-PCR verification of miR-22 expression during the transition from quiescence to proliferation. The Y-axis indicates expression of miR-22 after serum stimulation relative to quiescence, at each of the indicated time points on the X-axis. Data shown are for a representative experiment measured in quadruplicate (mean ± SD). (C) Cell growth assay following miR-22 overexpression. Average cell number following miR-22 or control siRNA transfection is shown for each time point indicated. Error bars denote ± SD, n = 3. (D) Cell growth assay following miR-22 inhibition. Average cell number following transfection of a LNA targeting miR-22 or a LNA negative control is shown for each time point indicated. Error bars denote ±SD, n = 3. (E) Flow cytometry analysis for proliferation marker Ki67 of miR-22-transfected quiescent fibroblasts. miR-22-transfected fibroblasts showed a larger population of cells expressing Ki67 compared with control siRNA-transfected fibroblasts. The Y-axis shows the percentage of cells expressing Ki67. Bars are the mean percentage of cells expressing Ki67, and error bars denote the mean ± SD, n = 4. (F) qRT-PCR data for Ki67 expression in quiescent fibroblasts transfected with miR-22 duplexes compared with control siRNA. miR-22-transfected fibroblasts show elevated levels of Ki67 expression as compared with control at the indicated time points. Ki67 expression was normalized using GAPDH mRNA levels, and fold changes were normalized with respect to the control transfection. (G) Inhibition of miR-22 decreases proliferation as measured by Ki67 expression. Quiescent fibroblasts were transfected with a LNA miR-22 inhibitor or LNA control. Twenty-four hours post-transfection, cells were serum-stimulated to proliferate for 18 or 24 h before harvesting for qRT-PCR. Y-axis indicates relative Ki67 expression levels compared with quiescent cells. Bars indicate the normalized mean expression, and error bars denote ±SD, n = 3. Control panel depicts Ki67 expression in normally proliferating fibroblasts and serum-starved quiescent fibroblasts. GAPDH was used as a control for normalizing input RNA levels. For C, D, E and G, P-values were estimated by Student’s t-test. *P < 0.05.
Because miR-22 was activated during the transition of primary human cells from quiescence to proliferation, we hypothesized that miR-22 regulates proliferation in primary human cells. To test this hypothesis, we transiently overexpressed miR-22 by transfecting the mature form of the miR-22 duplex RNA into quiescent fibroblasts and assayed cell growth. Compared with the control, miR-22-transfected cells showed increased growth by cell counting assay (Figure 1C). Conversely, proliferating cells in which we introduced a stable antisense inhibitor of miR-22 showed decreased cell growth (Figure 1D). To further explore the effect of miR-22 on cell growth, we assayed the expression of the proliferation marker Ki67 (26,36). We transfected miR-22 into quiescent fibroblasts and counted cells expressing Ki67 protein using flow cytometry. miR-22 transfection significantly increased the percentage of Ki67-positive cells compared with control, indicating that miR-22 activates proliferation (Figure 1E). We confirmed this result by performing a biologically independent experiment where we transfected miR-22 into both quiescent fibroblasts and proliferating fibroblasts and measured Ki67 expression by qRT-PCR. Compared with the control, miR-22-transfected cells showed elevated levels of Ki67 in both quiescent fibroblasts and proliferating fibroblasts (Figure 1F and Supplementary Figure S1). In addition, inhibition of miR-22 decreased levels of Ki67 compared with the control in cells exiting quiescence (Figure 1G).
Identifying miR-22 targets by microarrays
To identify the downstream targets of miR-22 in proliferating fibroblasts, we overexpressed miR-22 by transfecting miR-22 duplexes and then analyzed gene expression using microarrays, relying on the fact that miRNAs frequently cause downregulation of their mRNA targets (13,37). To investigate whether this approach was able to identify miR-22 targets, we analyzed the 3′UTRs of genes that were downregulated following miR-22 transfection for enrichment of the miR-22 seed match sequence. We also performed the same experiment and analysis in HeLa cells. In both cell lines, 6-mer matches to nucleotides 2–7, the miR-22 seed region, were significantly enriched in genes repressed when miR-22 was overexpressed (P < 10−8 assuming a binomial model, Supplementary Figure S2A and B). Thus, genes whose mRNAs were downregulated in response to high levels of miR-22 contained a significant proportion of direct targets of miR-22.
In addition to conducting gene expression profiling following miR-22 overexpression in proliferating cells, we also transfected miR-22 duplexes into fibroblasts that were rendered quiescent by serum deprivation and analysed the resulting gene expression changes and corresponding seed enrichment as described above. Although there was a significant enrichment of seed matches to the 5′-end of miR-22 in the 3′UTRs of the repressed genes (P < 10−4 assuming a binomial model, Supplementary Figure S2C), the magnitude of this enrichment was much lower than that observed for genes that were repressed by miR-22 during proliferation, suggesting that many mRNAs downregulated by miR-22 in quiescent cells may be indirect targets.
miR-22 inhibits genes involved in cell-cycle arrest
Overexpression of miR-22 repressed several cell-cycle-related genes, suggesting miR-22 may regulate proliferation through repression of cell-cycle arrest genes. From the miR-22 overexpression and gene expression profiling experiments, we selected genes for further analysis that were cell-cycle related, repressed by miR-22 overexpression and contained miR-22 target sites. Of these genes, TP53INP1, REDD1 (also known as DDIT4) and p21 (also known as CDKN1A) are transcriptionally activated by the tumor-suppressor p53 (38–40), while CARF (also known as CDKN2AIP) enhances p53 function and induces premature senescence in primary fibroblasts (41). TP53INP1, p21 and CARF have a functional role in inducing cell-cycle arrest (40–42), while REDD1 is a DNA-damage-inducible protein involved in apoptosis (39). TargetScan and RNA22 predicted REDD1, TP53INP1 and p21 as miR-22 targets. Interestingly, CARF did not show miR-22 seed sequences in its 3′UTR but showed multiple seed matches in its last exon.
To determine whether these genes were direct targets of miR-22, we used a strategy based on Ago2 IPs (Figure 2A) (35,43). Ago2 is an essential component of the RNA-induced silencing complex that is directed to target mRNAs by the miRNA guide strand (10). Hence, changes in Ago2 occupancy on a mRNA that occur in a miR-22-dependent manner can serve to identify direct targets. We measured Ago2 occupancy of candidate target mRNAs by immunoprecipitating Ago2–mRNA complexes and quantifying enrichment of the purified transcripts by qRT-PCR. We performed parallel Ago2 IPs in HeLa cells that were transfected with miR-22 mature duplexes as well as in mock-transfected cells. HeLa cells were used for the Ago2 IPs owing to their greater RNA content and smaller size than human fibroblasts, resulting in much higher mRNA yield per Ago2 IP. Transcripts that showed an increase in Ago2 occupancy in the miR-22-transfected samples as compared with mock transfections were expected to be direct miR-22 targets. We measured Ago2 occupancy by qRT-PCR using GAPDH as a control. Transcript abundance in the IPs was normalized to that in an aliquot of the total RNA before Ago2 IP.
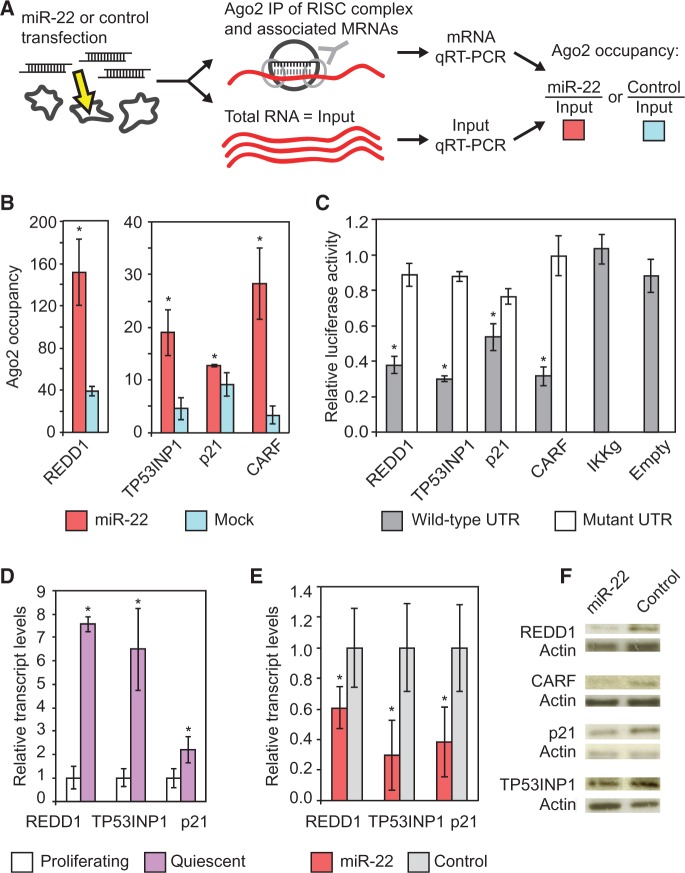
Figure 2.
miR-22 targets genes mediating cell-cycle arrest. (A) Ago2 IP. Ago2 forms a complex with the miRNA and the target mRNA. This complex is immunoprecipitated and the mRNA levels are quantified with qRT-PCR. (B) Ago2 IPs identify genes mediating cell-cycle arrest and apoptosis as direct targets of miR-22. REDD1, TP53INP1, p21 and CARF transcripts show significantly higher Ago2 occupancy in miR-22–transfected HeLa cells compared with mock-transfected cells. Ago2 occupancy of the target genes transcripts was measured using qRT-PCR as described in the text. The Y-axis shows fold change in mRNA levels from Ago2 IP–isolated RNA normalized to input RNA, which served as the control. (C) Luciferase assays show that miR-22 directly targets the 3′UTRs of the genes indicated on the X-axis. IKKg and Empty are negative controls. IKKg was not repressed by miR-22 and Empty is the luciferase vector with no UTR. The Y-axis shows relative luciferase units from miR-22-transfected cells normalized to control siRNA transfection. For the mutant 3′UTRs, 3 bp in each 6-mer miR-22 target site in the 3′UTRs were mutated. (D) qRT-PCR results for REDD1, TP53INP1, p21 and CARF transcripts show increased expression in quiescent fibroblasts relative to proliferating fibroblasts. Y-axis indicates fold enrichment in quiescent versus proliferating fibroblasts. (E) qRT-PCR shows miR-22 suppresses TP53INP1, REDD1 and p21 transcript levels in quiescent fibroblasts. Fold changes are denoted on the Y-axis relative to the control siRNA transfection. (F) REDD1, TP53INP1, p21 and CARF protein expression in fibroblasts was downregulated by transfection with miR-22 compared with control siRNA transfection. For B, D and E, GAPDH was used as a control for normalizing input RNA levels. For B, C, D and E, bars indicate the mean, and error bars denote ±SD, n = 3. P-values were estimated by Student’s t-test. *P < 0.05.
Ago2 IP confirmed REDD1, TP53INP1, p21 and CARF as direct targets of miR-22, as all four genes specifically showed increased Ago2 occupancy in a miR-22-dependent manner (Figure 2B), whereas other cell-cycle regulatory genes did not (Supplementary Figure S3A). We confirmed that all four genes were directly downregulated by miR-22 using a 3′UTR luciferase reporter assay and that miR-22 mediated repression of the 3′UTRs in the assay was dependent on the predicted miR-22 target sites in the 3′UTRs (Figure 2C).
The transcripts of three of these four genes (TP53INP1, p21 and REDD1) were upregulated as fibroblasts entered quiescence (Figure 2D). We therefore assayed whether ectopic miR-22 could inhibit these genes in quiescent cells. We transfected miR-22 into quiescent fibroblasts and assayed the changes of these three genes transcript levels by qRT-PCR and changes in protein expression by western blot. miR-22 suppressed the transcript and protein expression of the three genes in quiescent cells (Figure 2E and F). Protein levels of CARF were also downregulated in quiescent cells transfected with miR-22 compared with control (Figure 2F).
miR-22 targets genes that regulate the interferon response
Remarkably, 25 out of the top 50 genes identified by microarray to be downregulated by miR-22 in quiescent fibroblasts were interferon-inducible genes (Figure 3A and Supplementary Table S3). However, most of the interferon response genes that were repressed by miR-22 under quiescence did not show matches to the miR-22 seed sequence in their 3′UTRs (Figure 3A), suggesting that the downregulation of the majority of these genes in response to miR-22 was occurring through an indirect mechanism. Indeed, the seed match enrichment improved if we excluded the interferon pathway genes before analyzing the seed enrichment (Supplementary Figure S4A).
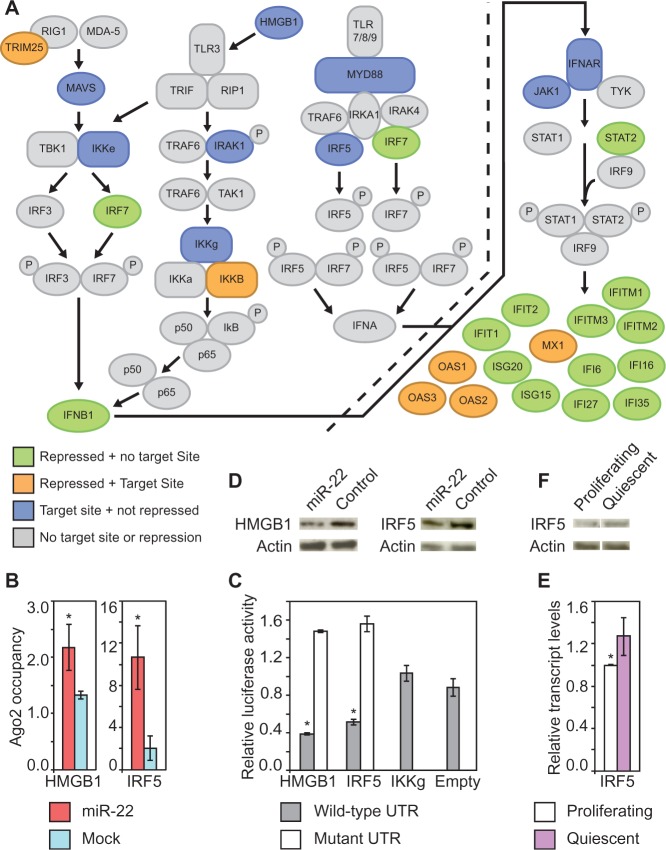
Figure 3.
miR-22 target genes mediating interferon suppression. (A) The interferon response pathway. Green indicates genes in the pathway that were repressed by miR-22 in quiescent fibroblasts but have no predicted miR-22 target sites; orange indicates genes repressed by miR-22 in quiescent fibroblasts and have predicted miR-22 target sites; blue indicates genes that have predicted target sites but were not repressed by miR-22 in quiescent fibroblasts. Genes in gray do not meet any of these criteria. (B) Ago2 IPs identify HMGB1 and IRF5 as direct miR-22 targets. HMGB1 and IRF5 transcripts show significantly higher Ago2 occupancy in miR-22-transfected HeLa cells compared with mock-transfected cells. Ago2 occupancy of the target genes transcripts was measured using qRT-PCR as described in the text. The Y-axis shows fold change in mRNA levels from Ago2 IP–isolated RNA normalized to input RNA. (C) Luciferase assays show that miR-22 directly targets the 3′UTRs of HMGB1 and IRF5. The Y-axis indicates relative luciferase units normalized to the control siRNA transfection. IKKg and Empty are negative controls. IKKg was not repressed by miR-22 and Empty is the luciferase vector with no UTR. For the mutant 3′UTRs, 3 bp in each 6-mer miR-22 target site in the 3′UTRs were mutated. (D) IRF5 protein expression in fibroblasts was downregulated by transfection with miR-22 compared with control siRNA transfection. (E) Increased expression of IRF5 in quiescent fibroblasts relative to proliferating fibroblasts assayed by qRT-PCR. Y-axis indicates fold enrichment in quiescent versus proliferating fibroblasts. (F) IRF5 protein expression was increased in quiescent compared with proliferating fibroblasts. For B and E, GAPDH was used as a control for normalizing input RNA levels. For B, C and E, bars are the mean, and error bars denote ±SD, n = 3. For B, C, and E, P-values were estimated by Student’s t-test. *P < 0.05.
To identify the direct targets of miR-22 that mediate suppression of interferon genes, we used Ago2 IPs. We screened several candidate target genes that had established roles in activating the interferon pathway and were potential miR-22 targets based on the presence of the miR-22 seed sequence in their 3′UTRs. From this screen, we identified two target genes, HMGB1 and IRF5, which showed increased occupancy by Ago2 in miR-22-transfected cells compared with control and had established roles in activating the interferon pathway (Figure 3A and B). Both HMGB1 and IRF5 have multiple miR-22 seed match sequences in their 3′UTRs, and HMGB1 was predicted to be a miR-22 target by RNA22 (44), while IRF5 was predicted to be a target of miR-22 by TargetScan (18). Other candidate genes in the interferon response pathway, which also contained miR-22 seed matches in their 3′UTRs, did not show increased Ago2 occupancy, indicating that this assay identified specific targets (Supplementary Figure S3B). To verify that these targets identified by Ago2 IPs could be repressed by miR-22, we cloned the 3′UTRs of HMGB1 and IRF5 into luciferase reporters to assay repression by miR-22. The 3′UTRs of both genes were indeed repressed by miR-22, and introducing mutations into the predicted miR-22 target sites relieved repression, confirming that miR-22 directly targets HMGB1 and IRF5 (Figure 3C). Additionally, the transcript levels of both of these genes were suppressed by miR-22 overexpression in HeLa cells, and protein expression was repressed by miR-22 overexpression in quiescent primary human fibroblasts (Figure 3D and Supplementary Figure S4B). Evidence for these targets is summarized in Supplementary Table S4. Surprisingly, neither IRF5 nor HMGB1 were among the many interferon genes that showed strongly decreased expression in HeLa cells transfected with miR-22 as determined by microarray profiling (Figure 3A and Supplementary Table S3). We assayed transcript and protein levels for both HMGB1 and IRF5 in proliferating compared with quiescent primary human fibroblasts by qRT-PCR and western blot. Only IRF5 was significantly repressed at both the transcript and protein level in proliferating compared with quiescent cells (Figure 3E and F).
miR-22 suppresses the interferon response
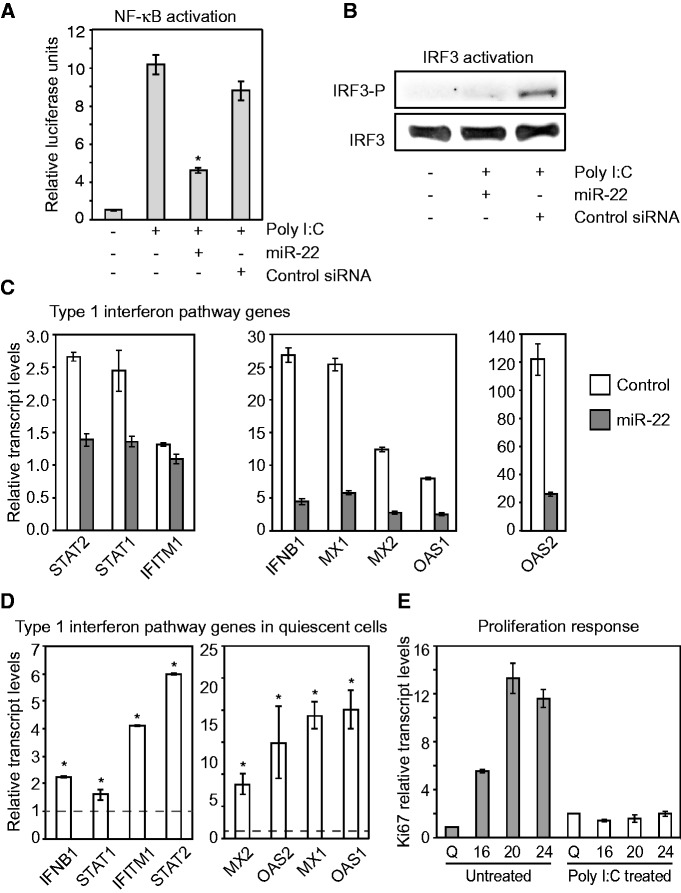
Both IRF5 and HMGB1 mediate interferon signaling pathways. IRF5 is involved in the induction of proinflammatory cytokines in response to several different Toll-like receptor (TLR) ligands and mediates signaling downstream of the TLR7 and TLR8 pathways to activate the interferon-beta promoter (45,46). HMGB proteins are highly represented in the nucleus and are responsible for regulating transcription and chromatin structure (47). However, HMGB1 was recently shown to act as a universal sensor for double-stranded RNA as well as DNA and stimulate the interferon pathway by activating the transcription factors IRF3 and NF-κB, which are essential components of the interferon-beta promoter ‘enhanceosome’. Additionally, HMGB1-deficient fibroblasts (Hmgb1−/−) show significantly impaired type I interferon induction (48).
To establish whether miR-22 was able to functionally suppress HMGB1- and IRF5-activated interferon-signaling pathways, we examined the response of NF-κB and IRF3 to miR-22 after we artificially activated the interferon pathway using poly I:C. Poly I:C is a double-stranded RNA polymer that mimics a viral infection and induces a strong type I interferon response (49). HMGB1 and IRF5 have been shown to mediate the activation of the transcription factors NF-κB and IRF3, both of which are key regulators of the interferon response (48,50). Using a reporter of NF-κB activation, we confirmed that miR-22 inhibits NF-κB activation in response to a trigger of interferon activation (Figure 4A). miR-22 also suppressed IRF3 phosphorylation, a hallmark of IRF3 activation, in response to activation of the interferon response (Figure 4B). To further characterize miR-22-mediated suppression of interferon pathway genes, we asked whether miR-22 could suppress an induced interferon response. We co-transfected miR-22 duplexes and poly I:C into HeLa cells and harvested cells 12 h later. Induction of the type I interferon response was measured by assaying interferon beta-1 (IFNB1) and several other interferon-stimulated gene (ISG) mRNA levels by qRT-PCR. Co-transfection of miR-22 severely impaired normal IFNB1 and subsequent ISG induction by poly I:C (Figure 4C and Supplementary Figure S5). These results strongly indicated that miR-22 was able to suppress the type I interferon response.
Figure 4.
miR-22 suppresses the interferon response. (A) miR-22 inhibits NF-κB activation as measured by reporter activity. Columns correspond to transfections of Poly I:C and miR-22 or control siRNA as indicated under the X-axis. Poly I:C was used to trigger the interferon response. Bars indicate the mean, and error bars denote ±SD, n = 3. *P < 0.05. (B) miR-22 inhibits IRF3 activation. Western blot assay for the phosphorylated form of IRF3 shows that miR-22 prevents IRF3 activation as compared with the control. Transfections of Poly I:C and miR-22 or control siRNA were as indicated. (C) miR-22 suppresses a poly I:C-induced interferon response. Poly I:C was co-transfected in HeLa cells with either miR-22 or the negative control siRNA. miR-22 suppression of the poly I:C-induced interferon pathway was assayed by measuring inhibition of interferon-beta (IFNB1) and several other ISGs, using qRT-PCR. The Y-axis indicates relative transcript levels normalized to control. Data shown are for a representative experiment of two biological replicates measured in triplicate (mean ± SD). Data from an independent replicate are shown in Supplementary Figure S5. (D) The type I interferon pathway is activated in quiescent cells. The column graph shows qRT-PCR data for expression levels of IFNB1 and other ISGs in the type I interferon pathway in quiescent fibroblasts relative to proliferating fibroblasts. The Y-axis indicates relative transcript levels in quiescent fibroblasts normalized to proliferating fibroblasts. Relative transcript levels in proliferating fibroblasts are shown by the dashed line. Bars indicate the mean, and error bars denote ±SD, n = 3. *P < 0.05. (E) Active interferon pathway impairs the transition from quiescence to proliferation. The interferon pathway was activated by transfecting quiescent fibroblasts with poly I:C, and then treated cells were serum-stimulated to proliferate. Cells were harvested at the time points indicated in hours on the X-axis. Q: quiescent fibroblasts. Cell proliferation was assayed by measuring Ki67 transcript levels using qRT-PCR. Relative Ki67 expression as indicated on the Y-axis was calculated with respect to untreated quiescent fibroblasts. Data shown are for a representative experiment measured in triplicate (mean ± SD). For C, D, and E, GAPDH was used as a control for normalizing input RNA levels.
These results combined with the observed downregulation of many interferon response genes with miR-22 overexpression in quiescent cells suggested that interferon response genes were active in quiescent cells and that miR-22 was capable of repressing the interferon response pathway. Previous cDNA profiling studies in quiescent fibroblasts have noted that interferon response genes are activated as fibroblasts enter quiescence (51). To confirm that interferon response genes were activated under quiescence, we measured relative mRNA expression levels of IFNB1, a key marker and inducer of the type I interferon response, as well as several ISGs in proliferating and quiescent fibroblasts. All of the interferon genes that we tested were expressed at higher levels in quiescent cells (Figure 4D). The interferon response is potently anti-proliferative (52), and the elevated expression of interferon genes could potentially be part of intrinsic mechanisms maintaining cellular quiescence.
To verify that an active interferon response would be detrimental to proliferation, we artificially activated the interferon pathway by transfecting poly I:C into quiescent fibroblasts, then stimulated them with serum to induce proliferation and assayed expression of proliferation marker Ki67. Quiescent fibroblasts that were not treated with poly I:C showed efficient exit from quiescence into a proliferative state as measured by Ki67 expression levels (Figure 4E). However, poly I:C-treated quiescent fibroblasts showed no change in Ki67 expression levels, indicating that a heightened interferon response inhibited transition into a proliferative state (Figure 4E).
Myc activates miR-22 during the transition from quiescence to proliferation
Interestingly, miRNA expression profiling of serum-stimulated primary human fibroblasts also showed miRNAs from the miR-17-92 cluster were induced along with miR-22 during the serum-stimulated transition from quiescence to proliferation (Figure 5A). The miR-17-92 cluster has been shown to be directly activated by the transcription factor Myc (53). To investigate whether miR-22 and other serum-stimulated miRNAs were also regulated by Myc, we first overexpressed or knocked down Myc in HeLa cells and assayed the resulting changes in miRNA levels using microarrays. miR-22, and the known targets of Myc, miR-17 and miR-92, were activated by Myc overexpression and were repressed by Myc knockdown (Figure 5A). To examine Myc regulation of miR-22 in primary human fibroblasts, Myc was knocked down by siRNA in proliferating primary human fibroblasts and in primary human fibroblasts exiting quiescence. Proliferating fibroblasts with Myc knocked down showed a significant decrease in miR-22 expression compared with control (Figure 5B and Supplementary Figure S6A). qRT-PCR for Myc expression confirmed Myc activation in response to serum stimulation of fibroblasts, at time points very similar to miR-22 induction (Supplementary Figure S6B). This pattern of miR-22 expression in response to serum stimulation and modulation of Myc activity suggested that miR-22 may be activated by Myc.
To investigate whether Myc directly activates miR-22 by binding to upstream cis-regulatory sequences, we examined genome-wide Myc ChIP-sequencing (ChIP-seq) data generated in our lab from serum-stimulated fibroblasts as well as HeLaS3 and MCF7 cells (54). In all cell types, particularly in the rapidly proliferating HeLaS3 and MCF7 cells, we observed Myc binding at several sites around the predicted transcription start site of miR-22 (55) (Figure 5C). Myc is known to bind its chromosomal target sites in mammalian cells, including human fibroblasts, through a DNA motif known as the E-box (31). Motif analysis of the region upstream of miR-22 revealed phylogenetically conserved E-box elements within the Myc-binding sites that we identified by ChIP-seq, many of which occurred within CpG islands (Figure 5C). Phylogenetic conservation of putative regulatory elements is frequently an indication of functional significance, and the genomic binding of Myc is known to be associated with the proximal presence of CpG islands (56). As an example, we verified Myc binding to one of the sites detected by ChIP-seq in an independent experiment by carrying out ChIP-qPCR in fibroblasts that were induced to proliferate (Figure 5D). In addition, luciferase promoter reporter assays using Myc-binding regions upstream of miR-22 showed that Myc could functionally activate gene expression through one of these upstream promoter regions (L3) (Figure 5E). The L3 promoter region site contains two putative Myc-binding site motifs (M1 and M2), but mutation of the putative Myc-binding site motifs did not impair activation by Myc in the reporter assay in a consistent manner (Figure 5F). These results nevertheless show that miR-22 expression is responsive to Myc and that Myc binds to the promoter of miR-22 and activates its transcription in a manner that is consistent with direct activation, both in HeLa cells as well as fibroblasts during their exit from quiescence.
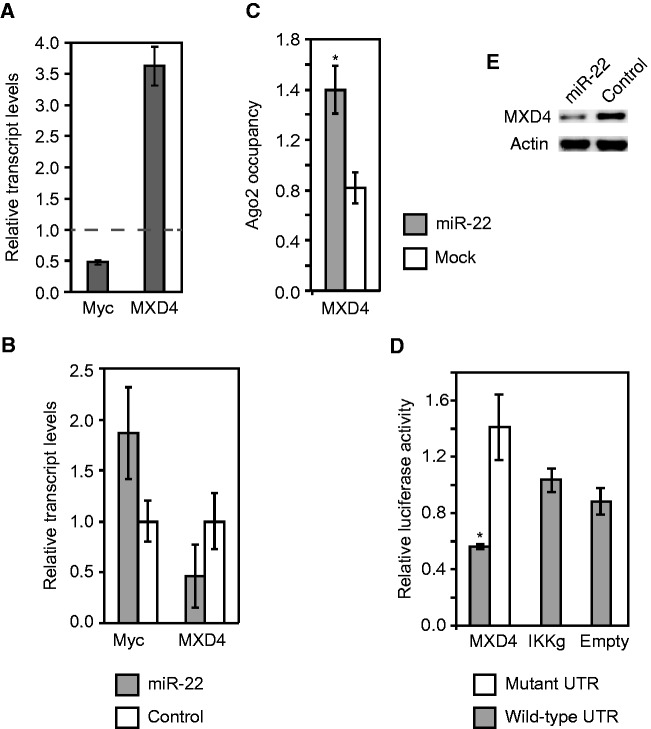
miR-22 inhibits the Myc repressor MXD4
Another target of miR-22 that we identified was MXD4, a transcriptional repressor of Myc, which in turn is repressed by Myc (57,58). We found that MXD4 transcript levels were activated and Myc levels were suppressed when cells entered quiescence (Figure 6A and Supplementary Figure S7). miR-22 transfection impaired MXD4 activation and Myc suppression under quiescence (Figure 6B). We verified using Ago2 IP and 3′UTR luciferase reporter assays that MXD4 was indeed a direct target of miR-22 (Figure 6C and D). Western blot confirmed miR-22 downregulated MXD4 expression at the protein level in primary human fibroblasts (6E). These results suggest the existence of a feedback loop in which Myc activates miR-22 to suppress MXD4, which causes the upregulation of Myc expression.
Figure 6.
miR-22 inhibits the Myc repressor MXD4. (A) qRT-PCR data showing MXD4 transcripts are activated while Myc transcript levels are inhibited in quiescent cells. qRT-PCR measurements of transcript levels in quiescent cells were made with respect to proliferating fibroblasts, as indicated on the Y-axis. Relative transcript levels in proliferating fibroblasts are shown by the dashed line. Data shown are for a representative experiment of two biological replicates measured in triplicate. Data from an independent replicate are shown in Supplementary Figure S7. (B) miR-22 transfection impairs MXD4 activation and indirectly reduces Myc suppression in quiescent fibroblasts compared with control siRNA transfection. (C) Ago2 IPs identify MXD4 as a direct target of miR-22. MXD4 transcripts showed higher Ago2 occupancy in miR-22-transfected HeLa cells compared with mock-transfected cells. Ago2 occupancy of the target genes transcripts was measured using qRT-PCR as described in the text. The Y-axis shows fold change in mRNA levels from Ago2 IP–isolated RNA normalized to input RNA. (D) Luciferase assays confirm that miR-22 directly targets the 3′UTR of MXD4. The Y-axis indicates relative luciferase units from miR-22-transfected cells normalized to control siRNA-transfected cells. IKKg and Empty are negative controls, IKKg was not repressed by miR-22 and Empty is the luciferase vector with no UTR. For the mutant 3′UTRs, 3 bp in each 6-mer miR-22 target site in the 3′UTRs were mutated. (E) MXD4 protein expression in fibroblasts was downregulated by transfection with miR-22 compared with control siRNA transfection. For A, B and C, GAPDH was used as a control to normalize input RNA levels. For B, C and D, bars indicate the mean, and error bars denote ±SD, n = 3. P-values were estimated by Student’s t-test. *P < 0.05.
DISCUSSION
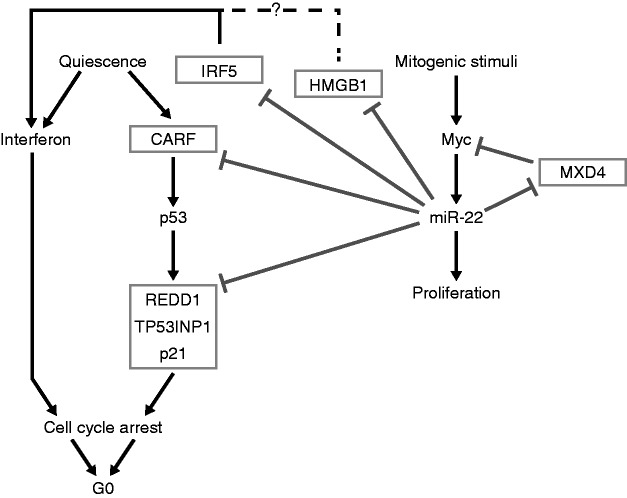
Our results show a TF–miRNA network that is activated as primary fibroblasts exit quiescence and re-enter the cell cycle. The oncogenic transcription factor Myc binds upstream of miR-22 and activates miR-22 expression when quiescent fibroblasts are stimulated to proliferate. In addition, we show that miR-22 promotes proliferation in primary human fibroblasts, and we identified targets of miR-22 that include several cell-cycle arrest genes, REDD1, TP53INP1, p21 and CARF, as well as MXD4, which represses Myc. Although the effect of miR-22 on the protein levels of cell-cycle arrest genes was modest, moderate regulation of multiple cell-cycle arrest genes may have large phenotypic effects in concert. We also found that miR-22 targets genes that mediate the interferon response, HMGB1 and IRF5, and that miR-22 repressed the interferon response.
The early activation of miR-22 we observed as cells exit quiescence and re-enter the cell cycle may be rationalized in terms of its effect on the interferon response and cell-cycle arrest genes. Although the interferon response is known to be antagonistic to cell proliferation, the exact role of the interferon response genes during quiescence is unclear (52). It has been suggested that signaling pathways actively maintain cells in a viable, reversibly arrested stage (59). It is possible that high levels of interferon response genes are required for maintaining or inducing a state of cell-cycle arrest in quiescent cells. Indeed, upregulation of interferon response genes in cells entering quiescence has been observed, and interferon has long been known to repress proliferation in fibroblasts (51,60). Consequently, to exit quiescence and begin to proliferate, cells must overcome this inhibitory influence on proliferation. When we artificially upregulated the interferon pathway under quiescence, entry into proliferation in response to serum-stimulation was severely impaired. One of the means by which cells may overcome this inhibitory effect is to use an immediate-early transcription factor like Myc to activate a suppressor of the interferon pathway, namely, the miRNA miR-22 (Figure 7). We found that miR-22 functionally inhibits NF-κB and IRF3 activation by directly targeting and downregulating HMGB1 and IRF5, which are known activators of the interferon activation pathway, thus identifying the molecular basis of how miR-22 mediates interferon suppression. In addition, HMGB1 has recently been shown to regulate proliferation, and it influences both wound healing and cancer progression (58,59). Surprisingly, HMGB1 was not downregulated in quiescent primary human fibroblasts. However, IRF5 was downregulated in quiescent primary human fibroblasts, indicating that the interferon-mediated maintenance of quiescence may only be regulated by IRF5 in primary human fibroblasts.
Figure 7.
The anti-quiescence network mediated by the Myc-miR-22 pathway.
We also present evidence that reveals novel cross-talk between the p53 and Myc regulatory networks that is mediated by miR-22. We show that miR-22 targets cell-cycle arrest pathways that are mediated by the tumor-suppressor p53. IRF5 is known to be a direct target of p53 and promotes cell-cycle arrest and cell death (61,62). IRF5 also induces senescence when overexpressed in immortalized Li–Fraumeni fibroblasts (63). Additionally, we find that miR-22 directly inhibits other pro-apoptotic and cell-cycle arrest genes such as REDD1, TP53INP1, p21 and CARF that are also direct targets of or regulate p53 (Figure 7). Interestingly, miR-22 itself is known to be a direct target of p53, and miR-22 determines p53-dependent cellular fate through direct regulation of p21 (28). In addition, previous studies have shown a p53-dependent suppression of Myc at the transcriptional and post-transcriptional level (64,65). Furthermore, our data show upstream binding and activation of miR-22 by Myc. A group previously observed similar results and concluded that this indicated direct activation of miR-22 by Myc, but our further analysis of Myc-binding sites does not conclusively support direct activation (29). Taken together, our data demonstrate novel interaction between the p53 and Myc regulatory networks that is mediated by miR-22 (Figure 7). We postulate that the combined effect of inhibiting interferon and cell-cycle arrest pathways downstream of p53 enables miR-22 to induce quiescent fibroblasts to proliferate.
Recently, other groups have reported that miR-22 acts like a proto-oncogene promoting proliferation and invasion of cancer cells by inhibiting the tumor-suppressor PTEN (21,24,25). On the other hand, other recent studies have reported that miR-22 inhibits proliferation of cancer cell lines (not including the ones we have tested) by inhibiting MAX (27) and MYCBP (28,29). However, we observed no inhibitory effect of miR-22 in primary cells or HeLa cells (data not shown), and in fact observed a modest activation of proliferation marker Ki67 by miR-22 in proliferating fibroblasts, consistent with our other results (Supplementary Figure S1). We found that miR-22 indirectly upregulates Myc expression levels by inhibiting the Myc transcriptional repressor MXD4, thus participating in a regulatory feedback loop (Figure 7). Other groups observed inhibition of Myc by miR-22 repression of Myc-binding partners MAX or MYCBP in cancer cell lines, suggesting miR-22 may activate or repress Myc activity depending on cell type (27,29). Because haploinsufficiency of Myc leads to impairment of proliferation, a small change in Myc expression can be expected to have significant outcomes on cell-cycle progression. The above results taken together raise the intriguing possibility that miR-22 may act as a switch inducing or inhibiting cellular proliferation in a context-dependent manner. This study provides evidence that miRNAs upregulated during the transition of quiescent cells into a proliferative state have a defined functional role in reprogramming gene expression to enable the transition of G0 arrested cells into the cycling G1 stage. Furthermore, our results provide evidence of the complex interplay between transcription factors and miRNAs to transduce extracellular signals into physiological responses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–7.
FUNDING
National Institutes of Health (NIH) [CA130075]; Cancer Prevention and Research Institute of Texas [RP120194 to V.R.I.]. Funding for open access charge: NIH [CA130075], Cancer Prevention and Research Institute of Texas [RP120194].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Chris Sullivan for the Luciferase reporters and for discussion, Klause Linse for help with the Luciferase assay, Anna Battenhouse for help with the ChIP-Seq analysis and Richard Salinas for help with flow cytometry.
REFERENCES
- 1.Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends Immunol. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 2.White P, Brestelli JE, Kaestner KH, Greenbaum LE. Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 2005;280:3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- 3.Tzachanis D, Lafuente EM, Li L, Boussiotis VA. Intrinsic and extrinsic regulation of T lymphocyte quiescence. Leuk. Lymphoma. 2004;45:1959–1967. doi: 10.1080/1042819042000219494. [DOI] [PubMed] [Google Scholar]
- 4.Martin P. Wound healing–aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 6.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang HY, Nuyten DSA, Sneddon JB, Hastie T, Tibshirani R, Sørlie T, Dai H, He YD, Van’t Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl Acad. Sci. USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 9.Gu J, Iyer VR. PI3K signaling and miRNA expression during the response of quiescent human fibroblasts to distinct proliferative stimuli. Genome Biol. 2006;7:R42. doi: 10.1186/gb-2006-7-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 15.Yekta S, Shih I-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 17.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 20.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 21.Bar N, Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS One. 2010;5:e10859. doi: 10.1371/journal.pone.0010859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, Wang S, Bian C, Yang Z, Zhou H, Zeng Y, Li H, Han Q, Zhao RC. Up-regulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 Protein Expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Liang S, Yu H, Zhang J, Ma D, Lu X. An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecol. Oncol. 2010;119:543–548. doi: 10.1016/j.ygyno.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Jiang Y, Zhang H, Greenlee AR, Yu R, Yang Q. miR-22 functions as a micro-oncogene in transformed human bronchial epithelial cells induced by anti-benzo[a]pyrene-7,8-diol-9,10-epoxide. Toxicol. In Vitro. 2010;24:1168–1175. doi: 10.1016/j.tiv.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan P-H, Bay B-H, Yip G, Selvarajan S, Tan P, Wu J, Lee C-H, Li K-B. Immunohistochemical detection of Ki67 in breast cancer correlates with transcriptional regulation of genes related to apoptosis and cell death. Mod. Pathol. 2005;18:374–381. doi: 10.1038/modpathol.3800254. [DOI] [PubMed] [Google Scholar]
- 27.Ting Y, Medina DJ, Strair RK, Schaar DG. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem. Biophys. Res. Commun. 2010;394:606–611. doi: 10.1016/j.bbrc.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchiya N, Izumiya M, Ogata-Kawata H, Okamoto K, Fujiwara Y, Nakai M, Okabe A, Schetter AJ, Bowman ED, Midorikawa Y, et al. Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res. 2011;71:4628–4639. doi: 10.1158/0008-5472.CAN-10-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong J, Du Q, Liang Z. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene. 2010;29:4980–4988. doi: 10.1038/onc.2010.241. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi R-U, Takata T, Shimamoto A, et al. miR-22 represses cancer progression by inducing cellular senescence. J. Cell Biol. 2011;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katome T, Obata T, Matsushima R, Masuyama N, Cantley LC, Gotoh Y, Kishi K, Shiota H, Ebina Y. Use of RNA interference-mediated gene silencing and adenoviral overexpression to elucidate the roles of AKT/protein kinase B isoforms in insulin actions. J. Biol. Chem. 2003;278:28312–28323. doi: 10.1074/jbc.M302094200. [DOI] [PubMed] [Google Scholar]
- 33.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 34.Lee B-K, Bhinge AA, Iyer VR. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011;39:3558–3573. doi: 10.1093/nar/gkq1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brizova H, Kalinova M, Krskova L, Mrhalova M, Kodet R. A novel quantitative PCR of proliferation markers (Ki-67, topoisomerase IIalpha, and TPX2): an immunohistochemical correlation, testing, and optimizing for mantle cell lymphoma. Virchows Arch. 2010;456:671–679. doi: 10.1007/s00428-010-0922-8. [DOI] [PubMed] [Google Scholar]
- 37.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 39.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 40.Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 41.Hasan K, Cheung C, Kaul Z, Shah N, Sakaushi S, Sugimoto K, Oka S, Kaul SC, Wadhwa R. CARF Is a vital dual regulator of cellular senescence and apoptosis. J. Biol. Chem. 2009;284:1664–1672. doi: 10.1074/jbc.M805778200. [DOI] [PubMed] [Google Scholar]
- 42.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc. Natl Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 45.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 46.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 47.Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol. Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 48.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 49.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 50.Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, Sassetti CM, Kelliher MA. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Adler AS, Segal E, Chang HY. A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 2007;3:e91. doi: 10.1371/journal.pgen.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fensterl V, Sen GC. Interferons and viral infections. Biofactors. 2009;35:14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 53.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 54.Lee B-K, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012;22:9–24. doi: 10.1101/gr.127597.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, Farnham PJ, Huang TH, Penn LZ. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 57.Kime L, Wright SC. Mad4 is regulated by a transcriptional repressor complex that contains Miz-1 and c-Myc. Biochem. J. 2003;370:291–298. doi: 10.1042/BJ20021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcotte R, Chen JM, Huard S, Wang E. c-Myc creates an activation loop by transcriptionally repressing its own functional inhibitor, hMad4, in young fibroblasts, a loop lost in replicatively senescent fibroblasts. J. Cell. Biochem. 2005;96:1071–1085. doi: 10.1002/jcb.20503. [DOI] [PubMed] [Google Scholar]
- 59.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor-Papadimitriou J, Balkwill F, Ebsworth N, Rozengurt E. Antiviral and antiproliferative effects of interferons in quiescent fibroblasts are dissociable. Virology. 1985;147:405–412. doi: 10.1016/0042-6822(85)90142-4. [DOI] [PubMed] [Google Scholar]
- 61.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 2003;63:6424–6431. [PubMed] [Google Scholar]
- 62.Mori T, Anazawa Y, Iiizumi M, Fukuda S, Nakamura Y, Arakawa H. Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target for p53. Oncogene. 2002;21:2914–2918. doi: 10.1038/sj.onc.1205459. [DOI] [PubMed] [Google Scholar]
- 63.Li Q, Tang L, Roberts PC, Kraniak JM, Fridman AL, Kulaeva OI, Tehrani OS, Tainsky MA. Interferon regulatory factors IRF5 and IRF7 inhibit growth and induce senescence in immortal Li-Fraumeni fibroblasts. Mol. Cancer Res. 2008;6:770–784. doi: 10.1158/1541-7786.MCR-07-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho A, Dowdy SF. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr. Opin. Genet. Dev. 2002;12:47–52. doi: 10.1016/s0959-437x(01)00263-5. [DOI] [PubMed] [Google Scholar]
- 65.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl Acad. Sci. USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.