Abstract
The RNA helicase Upf1 is a multifaceted eukaryotic enzyme involved in DNA replication, telomere metabolism and several mRNA degradation pathways. Upf1 plays a central role in nonsense-mediated mRNA decay (NMD), a surveillance process in which it links premature translation termination to mRNA degradation with its conserved partners Upf2 and Upf3. In human, both the ATP-dependent RNA helicase activity and the phosphorylation of Upf1 are essential for NMD. Upf1 activation occurs when Upf2 binds its N-terminal domain, switching the enzyme to the active form. Here, we uncovered that the C-terminal domain of Upf1, conserved in higher eukaryotes and containing several essential phosphorylation sites, also inhibits the flanking helicase domain. With different biochemical approaches we show that this domain, named SQ, directly interacts with the helicase domain to impede ATP hydrolysis and RNA unwinding. The phosphorylation sites in the distal half of the SQ domain are not directly involved in this inhibition. Therefore, in the absence of multiple binding partners, Upf1 is securely maintained in an inactive state by two intramolecular inhibition mechanisms. This study underlines the tight and intricate regulation pathways required to activate multifunctional RNA helicases like Upf1.
INTRODUCTION
RNA helicases are highly conserved enzymes that use NTPs to drive the dissociation of RNA duplexes or the remodelling of RNA–protein complexes (1,2). Most RNA helicases are monomeric, non-toroidal enzymes, which fall into two distinct superfamilies (SF1 and SF2) on the basis of sequence conservation (3,4). These enzymes are involved in nearly all aspects of RNA metabolism, acting on many distinct substrates and being associated with a wide range of functions. The basic structural fold of the helicase core is highly conserved in both superfamilies. It is composed of two RecA-like tandem domains facing each other and forming a cleft for nucleotide binding. Despite this structural homology, each RNA helicase has very specific physiological targets and functions (4). Although the exact mechanisms that confer this specificity remain unknown in most cases, they depend presumably on the presence of accessory domain insertions and/or N- and C-terminal domain additions to the helicase core (2,5). Such molecular appendages can allow the regulation of helicase activity by mediating the binding of molecular partners or the formation of multimeric complexes (6). One important challenge is to determine how these helicases are regulated to achieve specific functions.
Upf1 (for up-frameshift) is a SF1 RNA helicase that is essential for several cellular processes including Staufen-1-mediated mRNA decay (SMD) (7), histone mRNA degradation (8), DNA replication (9) and telomere metabolism (10). To date, the best-documented function of Upf1 concerns its role in nonsense-mediated mRNA Decay (NMD). NMD is a quality control process that identifies and degrades mRNAs harbouring a premature termination codon (PTC). This ‘surveillance’ mechanism limits the synthesis of truncated proteins potentially harmful for the cell. PTCs arise from DNA mutations or errors that occur during mRNA synthesis and processing (11). About one-third of human genetic diseases and many types of cancers are associated with mutations that produce a PTC (12,13). NMD also participates in the regulation of gene expression by targeting a significant fraction (5–10%) of physiological mRNAs (14–16). Upf1 is essential for NMD in all eukaryotes even though the signals that define a premature stop codon differ between organisms (17). One pathway that links PTC recognition by the translation machinery to Upf1 action has been well described in mammals (18). Briefly, mammalian mRNAs are associated with exon–junction complexes (EJC) that are deposited as a consequence of pre-mRNA splicing upstream of exon–junctions (19,20). EJCs accompany mRNAs from the nucleus to the cytoplasm where NMD factors Upf3 and Upf2 are recruited. During the first round of translation, the PTC induces the recruitment of the SMG1-Upf1-eRF1-eRF3 (SURF) complex, including translation termination factors eRF1-3, Upf1 and its kinase SMG1 (21–23). If an EJC is present downstream of the stalled ribosome, the SURF complex interacts with EJC to form the decay-inducing complex [DECID; (23)]. The formation of this surveillance complex triggers Upf1 phosphorylation by SMG1 (23–25). Both the phosphorylation state and enzymatic activity of Upf1 are essential to commit definitively the targeted mRNA to degradation (26,27).
Recent biochemical and structural studies have begun to explain how the activity of Upf1 is regulated by its NMD-binding partners. Upf1 possesses a conserved helicase core able to unwind double-stranded nucleic acids in a 5′- to 3′-direction (21,28,29). The helicase core of the human protein contains two internal insertions and is surrounded by two terminal domains. The N-terminal domain is rich in cysteines and histidines (CH domain) and the C-terminal region is rich in serine-glutamine clusters (SQ domain). The role of the CH domain, which exerts a cis inhibitory action on both the ATPase and unwinding activities of the Upf1 helicase core (29), has been recently elucidated. The domain forces Upf1 to bind RNA in a clamping conformation that prevents its function. Inhibition can be relieved by binding of the Upf2 factor to the CH domain. This interaction triggers the displacement of the CH domain from its original position, thereby promoting a conformational change and the activation of the helicase core (30). At present, the molecular function of the SQ domain is much less understood as most biochemical studies have been performed with Upf1 constructs truncated at their C-terminal domain.
Comparison of Upf1 sequences from eukaryotes shows that the SQ domain is much less conserved than the CH domain (31,32). The SQ domain is notably absent in lower eukaryotic organisms such as yeasts. However, the alignment of SQ domain sequences from 12 distantly related vertebrate genomes reveals a high degree of identity over the whole domain (Supplementary Figure S1), suggesting functional importance of the SQ domain in higher eukaryotes. Consistent with this, the SQ domain of human Upf1 contains several phosphorylation sites that are essential for its activity in vivo and its interaction with cofactors (23–25). At present, direct structural information is lacking for the SQ domain. Secondary structure prediction programs identify only small structured regions within a largely unstructured fold (Supplementary Figure S1).
Here, we have used a combination of biochemical assays to investigate the role of the SQ domain in the activity of human Upf1. We found that the SQ domain is able to lock the Upf1 helicase core in a conformation that is not productive for ATP hydrolysis or duplex unwinding. This inhibitory activity is mediated by direct binding of the SQ domain to the helicase core and is not directly affected by SQ phosphorylation. Moreover, SQ-mediated inhibition does not require the CH repressor domain and is not subject to direct regulation by Upf2 or Upf3 cofactors. Altogether our data unveil a new auto-inhibitory mechanism of human Upf1 and reveal a complex regulatory network whereby distinct intramolecular switches independently govern Upf1 activity. This intricate regulation is, to the best of our knowledge, unprecedented for RNA helicases and most likely reflects the necessity for the tight enzymatic control of central, multifunctional enzymes such as human Upf1.
MATERIALS AND METHODS
An exhaustive description of the following methods can be found in Fiorini et al. (33).
Purification of Upf1 proteins
The Upf1 expression vectors were prepared as previously described (29,33). The human proteins were expressed in Escherichia coli BL21 (DE3) Rosetta cells (Novagen) grown in LB medium and induced overnight at 16°C. The cells were lysed in buffer A [1.5× PBS pH 7.5, 225 mM NaCl, 1 mM magnesium acetate, 0.1% (w/v) NP-40, 20 mM imidazole, 10% (w/v) glycerol] supplemented with 100 µg/ml of egg white lysozyme (Sigma-Aldrich) and with 1× protease inhibitor cocktail EDTA-Free (Sigma-Aldrich). Soluble lysate was applied to a pre-packed nickel column (HisTrap FF crude, GE Healthcare) and fractioned on an Äkta Purifier (GE Healthcare) using a linear gradient from buffer A to B (buffer A added of 0.5 M imidazole) over 20 column volumes. The Upf1 proteins fused to a hexahistidine tag were subsequently purified on a heparin column (HiTrap heparin HP, GE Healthcare). Proteins that contained a calmodulin-binding peptide (CBP) were further purified on a calmodulin affinity column. Loading of the samples and column washes were performed with a buffer composed of 1× PBS pH 7.5, 150 mM NaCl, 1 mM magnesium acetate, 0.1% (w/v) NP-40, 1 mM DTT, 4 mM calcium chloride and 10% (w/v) glycerol. Sample elution from the column was performed with the same buffer containing 20 mM EGTA instead of calcium chloride. To purify the Upf1-HD–Upf1-SQ complex, the two proteins (Upf1-HD-His and CBP-Upf1-SQ-His) were co-purified after co-lysis of bacterial pellets obtained from two separate bacterial cultures. Lysates were treated with RNase A (50 µg/ml) for 2 h at 4°C before column purification of the complex. Alternatively, a Upf1-HD–Upf1-SQ complex was also obtained by co-expression of the two proteins (CBP-Upf1-HD and Upf1-SQ-His) expressed from the same plasmid (34) before co-purification onto Nickel and Calmodulin resins. The Upf1-CH/HD K498A and Upf1-HD-SQ S1078E/S1096E mutants were engineered with the QuikChange kit (Stratagene) and purified using the above protocol.
Duplex unwinding assay
For this assay, we used a 21-nt RNA–DNA hybrid containing a 12-nt long 5′-ssRNA overhang (Figure 1B). Transcription of the RNA, radiolabelling of the DNA oligonucleotide, purification of the RNA–DNA duplex and single-run helicase reactions were carried out as in Fiorini et al. (33) with minor modifications. Briefly, the 32P-labelled RNA–DNA hybrid substrate (1 nM, final concentration) was mixed with an excess of Upf1 protein (50 nM) in helicase buffer (20 mM MES, pH 6.0, 100 mM potassium acetate, 1 mM DTT, 0.1 mM EDTA) and incubated for 5 min at 37°C. The reaction was initiated by adding a mixture containing ATP and MgCl2 (1 mM, final concentrations), an excess of cold DNA strand (0.3 µM; to trap released RNA strands) and heparin (1 mg/ml; to trap free/released Upf1 molecules). In preliminary experiments, we observed that lower concentrations of heparin were not sufficient to establish the pseudo-first-order kinetic regimen characteristic of single-run helicase conditions (35). However, heparin interferes with Upf1–Upf2 interaction, so that all the unwinding reactions containing Upf2 were carried out without heparin. Reaction aliquots were withdrawn at various times, quenched with 150 mM sodium acetate, 10 mM EDTA, 0.5% (w/v) SDS, 25% (w/v) Ficoll-400, 0.05% (w/v) xylene cyanol, 0.05% (w/v) bromophenol blue and analysed by polyacrylamide gel electrophoresis and phosphorimaging on a Typhoon-Trio (GE Healthcare), as described (30,33,36).
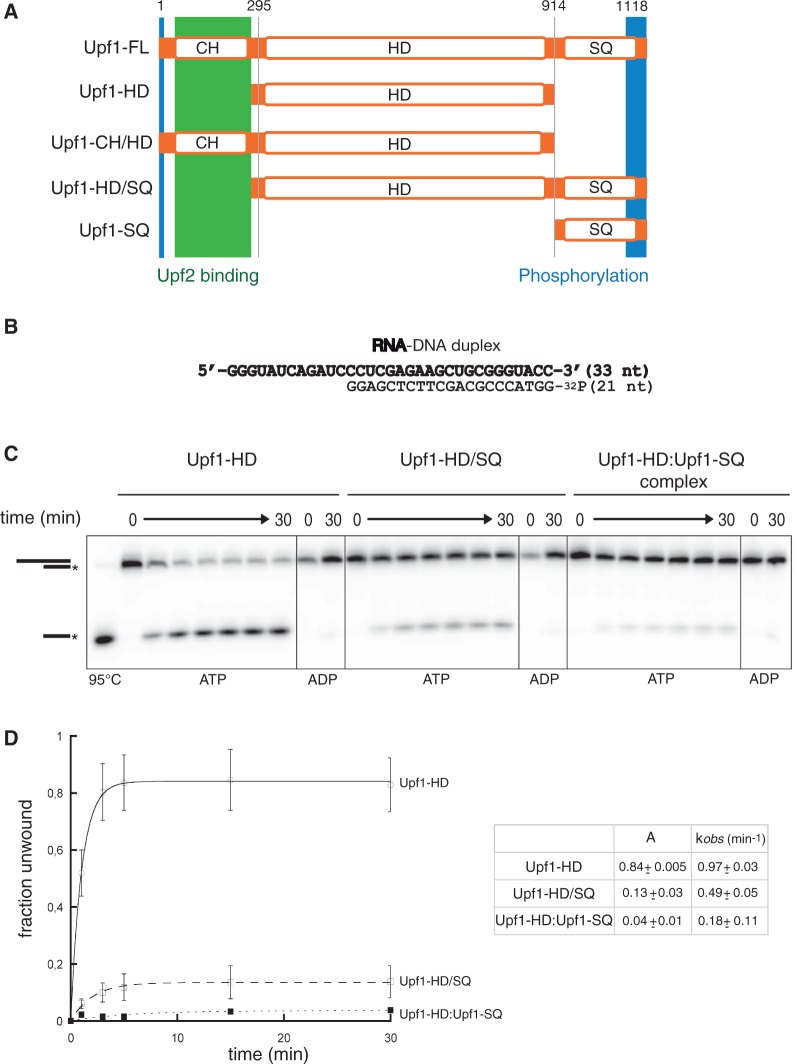
Figure 1.
The SQ domain inhibits Upf1 helicase activity. (A) Schematic diagram showing the Upf1 protein constructs used in this study. Structural domains and regions are represented by open rectangles. The Upf2-binding region and the in vivo phosphorylated regions are indicated by green and blue rectangles, respectively. (B) Diagram and sequence of the RNA–DNA hybrid used in the helicase experiments. (C) Representative gels showing ATP-induced unwinding of the RNA–DNA hybrid depicted in panel B by the Upf1 proteins. In control lanes, ATP was replaced by ADP or samples were heat-denatured (95°C). (D) Graph showing the fractions of oligonucleotide released as a function of time upon incubation of the RNA–DNA hybrid with the Upf1 proteins and ATP. Data points (Mean ± SD) are derived from three independent experiments. Data were fitted with Kaleidagraph (Synergy software) to the pseudo-first order equation y = A[1 − e(−kt)], where A and k represent, respectively, the amplitude and the rate constant of the unwinding reaction (35). The values for the amplitudes (A) and rate constants (k) are tabulated in the panel inset.
RNA and protein co-precipitation
The RNA co-precipitation and CBP pull-down were performed as described previously (29,33,34). Briefly, in RNA co-precipitation assay, proteins or preformed protein complex (2 µg) were mixed with 70 pmol of 3′-end biotinylated ssRNA (30 nt) in binding buffer [20 mM HEPES pH 7.5, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM DTT, 6.3% (v/v) glycerol and 0.1% (w/v) NP-40]. The reactions were supplemented (or not) with 3.3 mM ADPNP or ADP in a final volume of 30 µl and incubated for 20 min at 30°C. Then, 5 µl of pre-coated streptavidin-coupled magnetic beads (Dynabeads, Life technologies) were added before further incubation for 1 h at 4°C. Unless indicated otherwise, the beads were washed with a buffer containing 200 mM potassium acetate. Proteins were eluted by adding 7.5 µl of SDS loading buffer directly to the beads. The various fractions were subsequently analysed by 13.5% SDS–PAGE. For precipitation of protein complexes by CBP-bait protein, the magnetic beads were replaced with 12 µl of pre-coated calmodulin resin (50% slurry, Stratagene). The resin was washed three times with 500 µl BB-200 (200 mM potassium acetate) and eluted with 20 mM EGTA in 20 ml BB-150. Eluates were separated on 10% SDS–PAGE and visualized by comassie staining.
ATP binding
For ATP binding, we essentially followed the protocol described in Cheng et al. (37). Briefly, hUpf1 proteins (2 µg) were spotted on nitrocellulose membrane. The membrane was soaked in blocking buffer [20 mM HEPES pH 7.0, 50 mM K Acetate, 2.5 mM Mg Acetate, 2 mM DTT, 3% BSA (w/v), 10% (v/v) glycerol] and incubated on a rocking platform for 1 h at room temperature. Then, the blocking buffer was replaced by binding buffer [20 mM HEPES pH 7.0, 50 mM K acetate, 2.5 mM Mg acetate, 2 mM DTT, 1.5% (w/v) BSA, 10% (v/v) glycerol] supplemented with 30 µCi of [α32P]-ATP (800 Ci/mmol; Perkin Elmer) or 30 µCi of [γ32P]-ATP (10 mCi/ml, 3000 Ci/mmol; Perkin Elmer) before further incubation for 20 min at room temperature. The membrane was washed twice with blocking buffer before being dried and analysed by phosphorimaging.
ATP hydrolysis
For steady-state experiments, Upf1 (5 pmol) was incubated at 30°C in a 10-μl reaction mixture containing 1× ATPase buffer [20 mM MES pH 6.0, 100 mM potassium acetate, 1 mM DTT, 0.1 mM EDTA, 1 mM magnesium acetate, 1 µM zinc sulphate and 5% (v/v) glycerol], 2 µCi of [α32P]-ATP (800 Ci/mmol, Perkin Elmer), 25 µM ATP and 0.5 mM polyU (concentration in rU residues). The single-cycle ATPase experiments were performed by incubating 7.5 pmol of Upf1 with 5 nM of [α32P]-ATP (800 Ci/mmol) and 50 µM of polyU in 15 µl of 1× ATPase buffer at 30°C. Control experiments were also performed in absence of polyU in the reaction. To account for potential RNA contamination of the protein preparations, samples deprived of polyU were first pre-incubated with 500 U of RNaseT1 (Thermo scientific), 5 µg of RNaseA (Thermo scientific) and 50 U of RNaseI (Ambion) for 10 min at 30°C before addition of [α32P]-ATP. Single-cycle ATPase samples containing polyU were treated under the same conditions but without RNases. Reaction aliquots (2 µl) were withdrawn at various times and quenched with a buffer (5 µl) containing 10 mM EDTA and 0.5% (v/v) SDS. Samples were analysed by phosphorimaging after thin layer chromatography on polyethyleneimine cellulose plates (Merck) with 0.35 M potassium phosphate (pH 7.5) as elution buffer.
Electrophoretic mobility shift assay
Samples were prepared by mixing a radiolabelled 30-mer oligoribonucleotide (1 nM) with Upf1 protein (0, 0.5, 1, 2, 5, 10, 20, 30 or 50 nM) and with or without ADPNP (3.5 mM) in a buffer containing 20 mM MES pH 6.0, 100 mM potassium acetate, 2 mM DTT, 0.2 µg/µl of BSA and 6% (v/v) glycerol. The samples were incubated at 30°C for 20 min before being resolved by native 6.5% polyacrylamide (19:1) gel electrophoresis and analysed by phosphorimaging.
RESULTS
Production of recombinant Upf1 proteins
To determine the function of the SQ domain, we expressed and purified different recombinant versions of human Upf1 (Figure 1A). The boundaries between the N-terminal CH domain, the central helicase domain (HD) and the C-terminal SQ domain were defined according to previous structural studies (30,38–40). We produced the full-length protein [Upf1-FL; amino acids (aa) 1–1118], the helicase core domain (Upf1-HD; aa 295–914), a protein containing both the CH and HD domains (Upf1-CH/HD; aa 115–914), and a protein containing both the HD and the SQ domain (Upf1-HD/SQ; 295–1118 aa). Attempts at purifying the SQ domain alone (Upf1-SQ; 915–1118 aa) were unsuccessful, although the domain could be obtained in co-purification experiments (described later). Each protein was fused to a CBP at the N-terminus and to a hexahistidine tag at the C-terminus and was purified by successive affinity steps with nickel and calmodulin resin columns (33,41).
The helicase activity of Upf1 is inhibited by the SQ domain
Previous observations showed that Upf1 directionally unwinds double-stranded nucleic acids (21,28,29). To determine whether the SQ domain affects Upf1 activity, we first monitored the capacity of the enzyme to unwind an RNA–DNA duplex using a gel shift assay (42). The duplex substrate was formed by annealing a 33-mer RNA strand with a 32P-labelled DNA oligonucleotide and contained a 21-bp RNA–DNA hybrid helix [ΔG°37 = −31.6 kcal mol−1, (43)] downstream of a single-stranded RNA (ssRNA) ‘tail’ of 12 nt (Figure 1B). According to the size of the RNA-binding site deduced from crystal structures of Upf1-HD [9–11 nt; (30)], the length of the ssRNA tail is sufficient to accommodate only one Upf1 molecule upstream from the reporter RNA–DNA helix and short enough to prevent cooperative effects between multiply-bound helicase molecules (44,45). The RNA–DNA hybrid substrate was pre-incubated with an excess of Upf1 protein to favour the formation of an enzyme–substrate complex and to monitor the unwinding reaction under a pre-steady state regimen (35). The unwinding reaction was initiated by adding a mixture containing ATP, MgCl2 and an excess of unlabelled DNA strand to ‘trap’ the RNA strands released over time [see ‘Materials and Methods section and (33)]. The initiation mix also contained heparin in amounts sufficient to trap free Upf1 molecules (i.e. RNA-unbound) so that only the first round of unwinding was monitored during the reaction (single-run conditions). Unwinding efficiencies were deduced from the proportions of 32P-labelled single-stranded DNA accumulated over time.
Since the CH domain exerts an inhibitory effect on the adjacent HD (29,30), we first tested whether the SQ domain has similar effect. We monitored the activities of the Upf1-HD and Upf1-HD/SQ proteins. Remarkably, the unwinding activity of Upf1-HD is strongly affected by the presence of the SQ domain (Figure 1C). Indeed, the amplitude of the reaction with Upf1-HD/SQ is reduced by ∼6-fold when compared with that obtained with Upf1-HD (Figure 1D). The rate of the single-run unwinding reaction, kobs, is also affected by the presence of the SQ domain but to a lesser extent (∼2-fold; see table in Figure 1D). Moreover, when ATP is replaced by ADP in the reaction, the fraction of duplex unwound over time is negligible and comparable for both the Upf1-HD and Upf1-HD/SQ proteins (Figure 1C and data not shown). These data clearly demonstrate that the SQ domain inhibits the helicase activity of Upf1.
We next tested whether the inhibitory effect of the SQ domain is also exerted in the context of the full-length protein (Upf1-FL). Upf2 interacts with the CH domain relieving its inhibitory effect on helicase activity (30). Thus, to discount a potential interference of the CH domain, we first verified that its binding activator Upf2 is able to interact with Upf1-FL. To do this, we used a co-precipitation assay (29,33,34) wherein CBP-tagged Upf2 (761–1227) was used as bait and the protein complexes were purified on calmodulin affinity-resin before analysis by SDS–PAGE (Supplementary Figure S2A). As expected, Upf1-HD did not co-precipitate with Upf2 (lane 4), which confirms that the CH domain is required for the interaction (29). In contrast, Upf1-FL produced in E. coli (Upf1-FLc) as well as a phosphorylated form produced in baculovirus (Upf1-FLb; kindly provided by Elena Conti) co-precipitated with Upf2 (Supplementary Figure S2A, lanes 5 and 6). Controls performed without Upf2 rule out that co-precipitation could stem from non-specific binding of the Upf1-FL proteins to the beads (Supplementary Figure S2A, lanes 1–3). Thus, Upf2 is able to interact specifically with Upf1-FL, which supports that the CH domain remains fully accessible and amenable to Upf2 regulation in the context of full-length protein. Based on these findings, the unwinding activity of Upf1-FL was monitored in the presence of a molar excess of Upf2 to relieve the inhibitory effect of the CH domain (30). We observed that Upf2 is not sufficient to activate Upf1-FL (Supplementary Figure S2B), whereas under identical conditions, Upf2 activates Upf1-CH/HD (30). These results show that the repression exerted by the SQ domain cannot be relieved by the presence of Upf2 bound to the N-terminal CH domain. Taken together, these data strongly suggest that the helicase core domain of Upf1 is independently repressed by both the CH and the SQ flanking domains.
The SQ region physically interacts with the HD
In order to dissect the mechanism(s) used by the SQ domain to inhibit Upf1, we attempted to prepare the domain as a separate polypeptide through expression in E. coli cells. However, the isolated SQ domain, most likely unstructured, cannot be satisfactorily solubilized under native conditions (data not shown). We thus tempted to co-purify the SQ and Upf1-HD proteins, each one individually expressed from independent cellular batches and then co-purified (see ‘Materials and Methods’ section). We made the hypothesis that, if the two domains interact, the HD may help to solubilize the SQ domain. In this particular experiment, both Upf1-HD and Upf1-SQ contained a C-terminal hexahistidine tag while only Upf1-SQ was also fused to a N-terminal CBP tag. Following co-lysis of the cell pellets (see ‘Materials and Methods’ section), sequential purification on Nickel and calmodulin resins yielded a bimolecular complex (Supplementary Figure S3A, lane 8). This bimolecular complex was obtained after RNase A treatment and extensive washes under stringent conditions (0.5 M NaCl) during calmodulin affinity purification, which shows that the SQ domain makes strong and specific contacts with the HD in a RNA-independent manner. We also tested the co-expression of CBP-Upf1-HD and Upf1-SQ-His from the same plasmid but this method yielded too little material for enzymatic assays (Figure 2A). To confirm the pertinence of our purification procedure, we attempted to co-purify the SQ domain with the non-cognate RNA helicase eIF4AIII. This enzyme is an archetypal member of the DEAD-box family (SF2 helicases) and contains the two conserved RecA-like domains characteristic of eukaryotic RNA helicases (4). However, we were unable to co-purify eIF4AIII with CBP-Upf1-SQ-His (Supplementary Figure S3B). This observation supports that only strong and specific complexes such as the one formed between Upf1-SQ and Upf1-HD sustain our harsh purification conditions.
Figure 2.
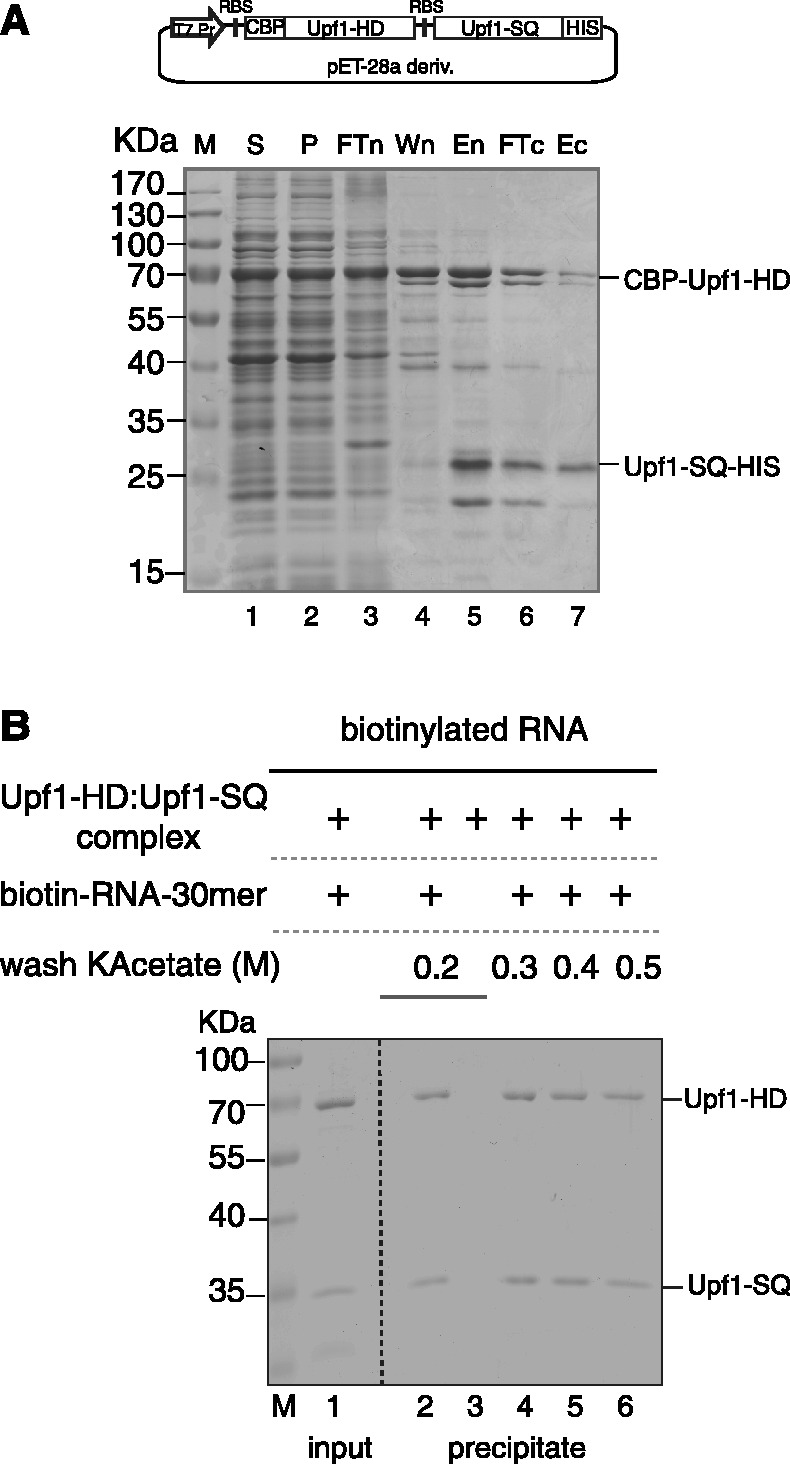
The helicase core of Upf1 interacts with the SQ domain. (A) A SDS–PAGE gel illustrating the co-expression and co-purification of the Upf1-HD and Upf1-SQ proteins. Lane M: marker. Samples from the supernatant (lane S) and pellet (lane P) fractions obtained after centrifugation of the E. coli lysate were loaded together with samples of the flow through (FTn), wash (Wn) and elution (En) fractions obtained after nickel affinity chromatography or flow through (FTc) and elution (Ec) fractions obtained after the final calmodulin affinity purification. (B) A representative 13.5% SDS–PAGE gel showing streptavidin pull-down experiments with a 30-mer biotinylated ssRNA bait and the binary Upf1-HD–Upf1-SQ complex. After incubation with the bait and/or the protein complex (input), the beads were subjected to washes of different stringencies before recovery and analysis of bound materials (precipitate).
The stability of the complex formed between the helicase and SQ domains was further tested using RNA pull-down experiments. The preformed complex was incubated with a 30-mer biotinylated ssRNA and streptavidin beads. Then, the beads were washed with a solution of defined ionic strength (0.2–0.5 M potassium acetate) before analysing the co-precipitated proteins. As shown in Figure 2B, both Upf1-SQ and Upf1-HD proteins were precipitated with the beads in the presence of the biotinylated RNA (lane 2) but not in its absence (lane 3). Moreover, increasing the concentration of potassium acetate from 0.2 to 0.5 M did not markedly affect the amounts of Upf1-SQ and Upf1-HD proteins that were recovered with the beads (lanes 4–6). These results suggest that neither RNA nor the SQ domain dramatically alter each other’s capacity to interact with the Upf1 helicase core in vitro. They also suggest that the inhibitory effect of the SQ domain cannot be ascribed to a dramatic weakening of the Upf1–substrate interaction. Since the SQ domain cannot be tested alone, we, however, cannot rule out an alternative scenario whereby both the HD and SQ domains would interact with RNA without contacting one another.
We next determined if the SQ domain could inhibit Upf1 activity when provided in trans by measuring the unwinding activity of the preformed Upf1-HD–Upf1-SQ complex. We observed that the inhibition of Upf1 activity was even stronger for the bimolecular Upf1-HD–Upf1-SQ complex than for the Upf1-HD/SQ protein (Figure 1C and D; compare middle and right panels and corresponding graphs), supporting that the SQ domain modulates helicase activity by directly interacting with the enzyme core. The difference in inhibitory extents between Upf1-HD/SQ and the binary complex may be due to a greater conformational heterogeneity of the Upf1-HD/SQ protein in which the SQ domain may not always interact adequately with the HD. In contrast, entropic constraints imposed by a physical link between both domains do not exist for the binary Upf1-HD–Upf1-SQ complex wherein the SQ domain must strongly and specifically interact with the helicase core.
The region of the SQ domain containing phosphorylation sites is not required for the inhibition of Upf1 helicase activity
Upf1 phosphorylation is a critical consequence of PTC recognition and EJC interaction during NMD process in human (23,25,26). Several phosphor-acceptor residues have been identified in vitro, but only phosphorylation at Thr28 in the N-terminus and at Ser1078 and Ser1096 in the C-terminus have been detected in vivo so far (24–26). Despite its functional importance, the role of Upf1 phosphorylation remains unclear at the molecular level. One possibility is that phosphorylated Ser1078 and Ser1096 residues modulate the regulatory function of the SQ domain by affecting directly (or not) the binding of the SQ domain to the HD. It has been shown that the replacement of serine by a negatively charged residue such as glutamate could possibly mimic phosphorylation (46,47). To test the potential role of the Ser1078 and Ser1096 residues in the regulation of Upf1 activity, we have thus prepared a phosphomimetic Upf1-HD/SQ variant that contains two glutamate substitutions at positions 1078 and 1096. However, these substitutions had no stimulatory effect on the unwinding activity of Upf1 (data not shown), suggesting that the phosphorylation state of the Ser1078 and Ser1096 residues do not directly interfere with SQ domain action.
In order to test a genuinely phosphorylated form of Upf1, we also used full-length Upf1-FLb produced in Baculovirus. Mass spectrometry analysis confirmed the presence of 1–6 phosphorylated residues without precisely localizing them [E. Conti (personal communication)]. To account for the presence of the CH repressor domain, we have monitored the unwinding activity of Upf1-FLb in the presence of the bound Upf2 activator (Supplementary Figure S2A, lane 6). Again, the Upf1–Upf2 complex showed no unwinding activity (Supplementary Figure S2B, right-most panel), suggesting that the inhibitory effect of the SQ domain is not alleviated by its phosphorylation.
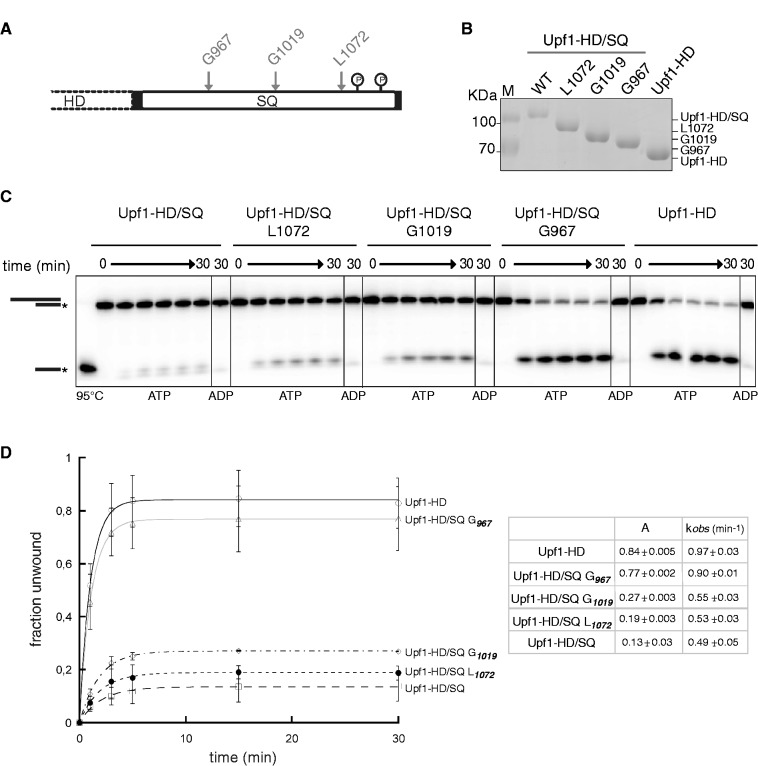
To confirm the above conclusions and determine the portion of the SQ region responsible for Upf1 inhibition, we have generated three C-terminal truncated variants of Upf1-HD/SQ. As depicted in Figure 3A, the corresponding C-terminal deletions start from Leucine 1072 (Upf1-HD/SQL1072 variant; 46 aa deletion), Glycine 1019 (Upf1-HD/SQG1019; 99 aa deletion) or Glycine 967 (Upf1-HD/SQG967; 151 aa deletion), all of which remove the phosphorylation sites identified in vivo. We have purified each truncated Upf1-HD/SQ variant (Figure 3B) in order to probe their respective unwinding activity. We have observed that both Upf1-HD/SQL1072 and Upf1-HD/SQG1019 are about as ineffective as the full-length Upf1-HD/SQ protein, whereas the shortest Upf1-HD/SQG967 variant was almost as efficient as Upf1-HD (Figure 3C and D). These data show that the central region of the SQ domain that is located between residues Gly967 and Gly1019 is important for its inhibitory function. They also show that the C-terminal region containing the major phosphorylation sites (Figure 3A) does not contribute by itself to SQ-mediated inhibition. However, the data do not rule out the possibility that phosphorylation of the C-terminus is important for recognition by accessory factor(s) (see ‘Discussion’ section).
Figure 3.
The region spanning residues 967–1019 of the SQ domain is sufficient to inhibit Upf1 activity. (A) Schematic showing the starting points of the C-terminal deletions in the truncated variants Upf1-HD/SQG967, Upf1-HD/SQG1019 and Upf1-HD/SQL1072 with respect to the phosphorylation sites of the SQ domain. (B) SDS–PAGE gel showing the truncated variants after affinity purification. Gels (C) and graph (D) show unwinding reactions performed with the truncated and control Upf1 variants. Experiments were carried out and analysed as described in Figure 1. Values for the amplitudes (A) and rate constants (k) of the unwinding reactions are tabulated in the inset.
The SQ domain impedes Upf1 ATP hydrolysis
To gain further insight into the mechanism of Upf1 inhibition by the SQ domain, we have first used a dot blot assay to determine if the Upf1-HD and Upf1-HD/SQ proteins and the Upf1-HD–Upf1-SQ complex can bind ATP. In this assay, the amounts of [α32P]-ATP retained on a nitrocellulose membrane by the bound proteins were compared qualitatively. Although differences were observed, neither the Upf1-HD and Upf1-HD/SQ proteins nor the Upf1-HD–Upf1-SQ complex displayed drastic defects in ATP binding (Figure 4A). In contrast, a variant of Upf1-CH/HD carrying a detrimental mutation (R865A) in the nucleotide-binding site (21,28,38) was completely unable to retain ATP (Figure 4A). Thus, the SQ domain does not appear to drastically alter the ability of Upf1 to bind ATP. Similar results were obtained with [γ32P]-ATP (Supplementary Figure S4A), suggesting that the Upf1-HD and Upf1-HD/SQ do not efficiently hydrolyze the bound ATP [which is expected in the absence of RNA cofactor; (29)] or, alternatively, that they do not readily release the Pi produced upon hydrolysis.
Figure 4.
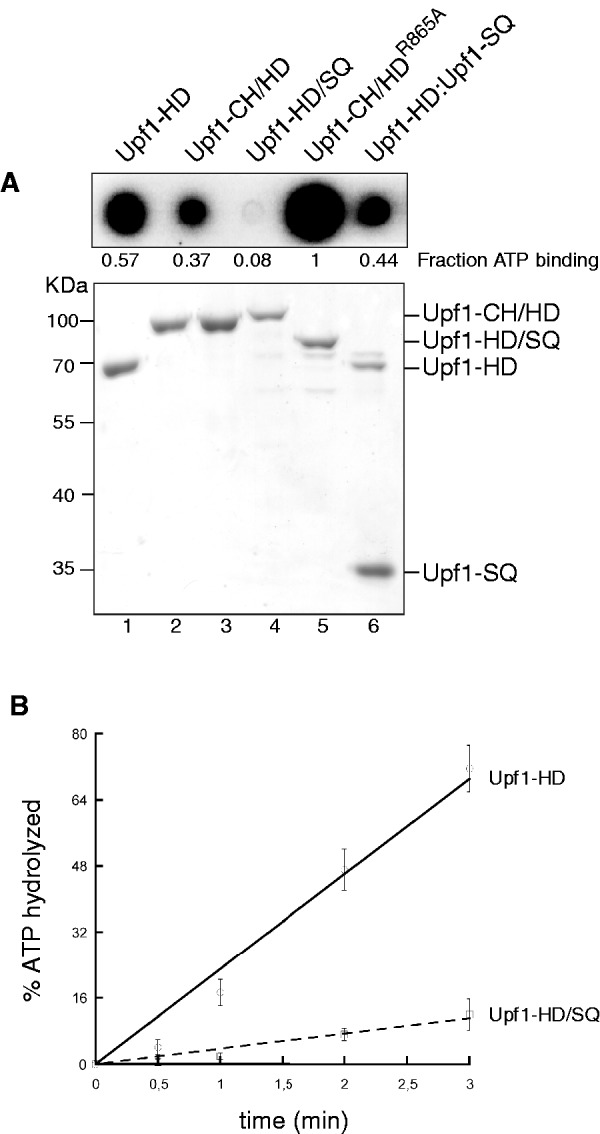
The SQ domain prevents ATP hydrolysis without precluding nucleotide binding. (A) Truncated and mutant Upf1 proteins were spotted on a nitrocellulose membrane and exposed to [α32P]-ATP (top). Corresponding protein samples were analysed by SDS–PAGE (bottom). (B) Graph showing the percentage of ATP hydrolysed as a function of time by the Upf1 variants under conditions of steady-state ATPase turnover (see ‘Materials and Methods’ section).
Next, we have measured the steady-state ATP hydrolysis rates of the Upf1-HD and Upf1-HD/SQ proteins. The purified proteins were incubated with large excesses of ATP (including [α32P]-ATP) and RNA cofactor (polyU), to favour efficient ATPase turnover (Figure 4B). Under these conditions, Upf1-HD/SQ protein showed a weak ATPase activity compared with Upf1-HD (Figure 4B) in agreement with the poor helicase activity of Upf1-HD/SQ (Figure 1D). To ensure that these defects were due to inhibition of ATP hydrolysis by the SQ domain rather than an ATPase turnover defect (e.g. ADP/Pi release defect), we have also performed single-cycle ATPase experiments in the presence of a large excess of Upf1–RNA complex with respect to ATP concentration (100-fold). Reaction mixtures contained saturating concentration of polyU (representing ∼100 equivalents of 10-nt-long binding sites per protein molecule) to offset potential RNA-binding defects triggered by the SQ domain (described later). In this case, the rate of single-cycle ATP hydrolysis was about five times higher for Upf1-HD than for Upf1-HD/SQ (Supplementary Figure S4B and C). In absence of RNA cofactor, single-cycle ATP hydrolysis was much slower (∼15 times) and it was independent of the SQ domain (Supplementary Figure S4B and C). Altogether, these data attest that the SQ domain impedes RNA-stimulated ATP hydrolysis even though it does not physically block access of the nucleotide to the Upf1 active site.
The SQ domain uncouples Upf1 RNA-binding state from the presence of ATP
The affinity of Upf1 for RNA varies during the ATP hydrolysis cycle. Notably, the presence of the nucleotide in the Upf1 active site shifts the helicase core into a ‘closed’ conformation that has a lower affinity for nucleic acids (30). As is the case for other helicases, cycling between ‘weak’ and ‘tight’ RNA-binding states most likely prompts Upf1 activity. Although the SQ domain does not prevent Upf1 from binding RNA (Figure 2B), we wondered whether it could interfere with the NTP-dependent interconversion between the ‘weak’ and ‘tight’ RNA-binding states. Using an electrophoretic mobility shift assay, we have thus assessed the effect of the SQ domain on the affinity of Upf1 for RNA in the presence or absence of the non-hydrolysable nucleotide analogue ADPNP. We have incubated a 30-mer radiolabelled ssRNA with increasing concentrations of purified Upf1-HD or Upf1-HD/SQ and with or without ADPNP. In each case, a band shift was observed upon gel electrophoresis as a result of Upf1–RNA complex formation (Figure 5). Surprisingly, in the absence of ADPNP, the SQ domain markedly reduces the affinity of Upf1 for the ssRNA (compare outermost panels in Figure 5). The presence of ADPNP in the incubation mixture has a similar weakening effect on Upf1-HD affinity for RNA (Figure 5, left panels), confirming previous observations (29,30). In contrast, the presence of ADPNP hardly changes the affinity of Upf1-HD/SQ for the ssRNA (Figure 5, right panels). Similar results were obtained by RNA pull-down assays in which Upf1-HD or Upf1-HD/SQ were co-precipitated by a 30-mer biotinylated ssRNA in the presence/absence of ADPPNP or ADP (Supplementary Figure S5). Thus, the SQ domain appears to constrain Upf1 in a ‘weak’ RNA-binding state uncoupled from the presence of the nucleotide cofactor.
Figure 5.
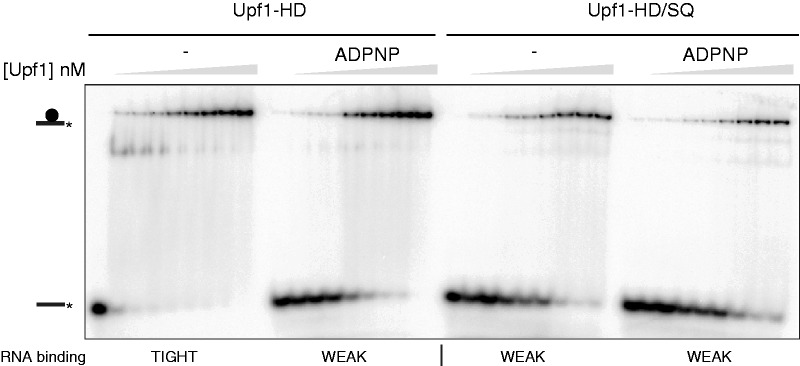
The SQ domain affects nucleotide-dependent RNA binding to Upf1. (A) Representative native 8% polyacrylamide gels illustrating the interaction of Upf1-HD and Upf1-HD/SQ proteins (depicted by a black circle symbol) with a 30-mer oligoribonucleotide substrate (black line) that had been labelled with 32P (black star). The RNA substrate was incubated with increasing concentrations of proteins and with (+) or without (−) ADPNP under the conditions described in ‘Materials and Methods’ section.
DISCUSSION
Helicases are multitasking enzymes. Many of them combine the ability to walk on RNA and/or DNA with unwinding double-stranded nucleic acids. They can also displace proteins blocking their track or remodel nucleoprotein particles. Often the helicase function is quiescent and has to be activated through extra-domains, self-assembly and/or interactions with molecular partners (5,6). Specific activities such as translocation, protein displacement or duplex unwinding may also be engaged selectively and in a timely fashion. Precise regulation reflects the importance and complexity of the physiological tasks implicating helicases, whereby remodelling of supramolecular targets has to occur at the right moment, in the right context, and in the appropriate cellular compartment.
The central role of Upf1 in NMD represents a striking example of a highly regulated RNA helicase. Recent structural and biochemical studies have illuminated the molecular mechanisms governing the regulation of Upf1 during NMD. The Upf1 ATPase and helicase activities are repressed by an intramolecular interaction between the N-terminal CH domain and the central helicase core. Binding of the Upf2 cofactor to the CH domain disengages it from its inhibitory position, thereby prompting the enzymatic activity of Upf1 (29,30,38,39). Although the roles of Upf2 and the CH domain in the control of the helicase core are now well established, a global view of how regulation is orchestrated in the full-length, multi-domain Upf1 enzyme is still missing. Notably, the function of the C-terminal SQ domain (Figure 1A)—which, in higher eukaryotes, is highly conserved (Supplementary Figure S1) and a target for phosphorylation and cofactor interactions has long remained elusive.
Using a combination of biochemical approaches, we assigned a regulatory function to the SQ domain and demonstrated that the CH domain is not the only intramolecular component that represses the activity of Upf1. Inhibition of Upf1 by the SQ domain (Figure 1C and D) is mediated by its stable and specific interaction with the helicase core of the enzyme (Figure 2 and Supplementary Figure S3). The activity of Upf1 is no longer restored by the binding of Upf2 to the CH domain if the SQ domain is present (i.e. in the context of the full-length enzyme; Supplementary Figure S2). This suggests that an optimal enzymatic activity of Upf1 is achieved only if the inhibitory actions of both terminal domains are relieved.
Recently, a fundamental role for ATPase activity of Upf1 has emerged in vivo. This activity is essential for NMD and is required for the disassembly of the NMD factors from mRNA during the late steps of the decay process (27,48). We observed that the binding of ATP to Upf1 is not drastically affected by the presence of the SQ domain (Figure 4A and Supplementary Figure S4A), whereas ATP hydrolysis is strongly inhibited (Figure 4B, Supplementary Figure S4B and C). Thus, the SQ domain prevents the ability of Upf1 to convert the energy stored in the ATP fuel into physical work (mechanochemical transduction) and hence inhibits the helicase activity. Crystal structures show that the helicase core of Upf1 is made of two flexible RecA-like lobes (A1 and A2) that create a cleft for ATP binding at their interface. The two lobes are relatively distant and can move with respect to each other in the absence of ATP. The active site adopts a geometry suitable for catalysis only when the nucleotide is bound (30,38). Moreover, the presence of ATP has an allosteric effect on Upf1 helicase core, diminishing the affinity of the enzyme for RNA (21,28–30,38). The SQ region also reduces the affinity of Upf1 for RNA but in a manner that becomes unresponsive to the presence of a nucleotide cofactor (Figure 5 and Supplementary Figure S5). These results suggest that the SQ domain constrains Upf1 in a conformation wherein RNA-binding components are not adequately positioned for interaction with RNA and for stimulation of ATP hydrolysis. Alternatively, the binding sites for RNA and the SQ domain on the Upf1 helicase core may overlap to the extent that SQ binding prevents the development of a fully productive RNA-bound state of Upf1. The ATP-binding/hydrolysis properties of the SQ-containing protein are reminiscent of the enzymatic phenotypes displayed by the Upf1-HD (Q665A) and Upf1-HD (DE636AA) mutants. These detrimental mutations disrupt the network of active site contacts to the β− and γ-phosphates of the nucleotide cofactor without affecting its binding (38). By analogy, we speculate that the inhibitory HD–SQ interaction affects the (RNA-driven) capacity of phosphate and/or catalytic residues to adopt position(s) suitable for ATP hydrolysis.
PTC-containing mRNA degradation and disassembly of NMD factors occur only after Upf1 ATPase activation (27). This suggests that the protein remains inactive until both CH and SQ inhibitions are relieved to ensure efficient NMD. A key step during NMD is represented by Upf1 phosphorylation at N- and C-terminal ends (23–26). One may envision that C-terminal phosphorylation of Upf1 restores enzyme activity by eliminating the auto-inhibition exerted by the SQ domain. However, phosphorylation of the C-terminal domain (or replacement of phosphorylation target sites by glutamate residues) is not sufficient by itself to relieve the inhibition of Upf1 in vitro (Supplementary Figure S2B and data not shown). In fact, we observed that Upf1 inhibition critically depends on a region of the SQ domain located upstream the in vivo phosphorylation sites (Figure 3). Since this phosphorylation target region is not a direct contributor to SQ-mediated inhibition of Upf1, we envision two distinct scenarios, whereby the phosphorylation target region mediates physiological inactivation of the SQ domain and derepression of Upf1. The first scenario relies solely on the binding of the SMG1 kinase to Upf1 that follows interaction of the SURF complex with Upf2-Upf3-EJC (49). The second scenario invokes a subsequent step whereby additional activating factor(s) specifically recognize and bind phosphorylated Upf1. In this context, it is tempting to speculate that the hyperphosphorylated SQ domain is bound by the SMG5–SMG7 heterodimer, which itself recruits protein phosphatase 2A (PP2A), which in turn catalyses Upf1 dephosphorylation (23,24,26,50,51). Another candidate worth considering is the proline-rich nuclear receptor coregulatory protein 2 (PNRC2) that was also found to interact with hyperphosphorylated Upf1 (52). Interestingly, PNRC2 mediates the interaction of phosphorylated Upf1 with the decapping machinery that precedes mRNA decay events (52).
From all these observations, we propose that during assembly of the SURF complex, Upf1 is completely inactivated by two distinct intramolecular repressor interactions operated respectively by the CH and SQ terminal domains (Figure 6, top diagram). Activation of Upf1 is initiated during formation of the DECID complex whereupon Upf2 binding alleviates the repression exerted by the CH domain (Figure 6, middle diagram). In the resulting intermediate state, Upf1 remains catalytically incompetent due to persisting SQ-mediated repression. Complete activation of Upf1 may then occur only when the bound SMG1 kinase triggers phosphorylation of Upf1 or when additional cofactors, such as the SMG5–SMG7 heterodimer and/or PNRC2, are recruited on the phosphorylated SQ domain (Figure 6, bottom diagram). Analogous to the mechanism of Upf2 derepression, the binding of cofactors to the SQ domain would disrupt SQ inhibitory contacts to the helicase core and achieve Upf1 activation (Figure 6). This sophisticated regulation mechanism cannot occur in lower eukaryotes such as Saccharomyces cerevisiae wherein Upf1 proteins do not contain SQ domains (40). SMG orthologues responsible of Upf1 phosphorylation states are also absent in yeasts (40), suggesting that NMD has evolved towards higher complexity and greater stringency of regulation from lower eukaryotes to metazoans.
Figure 6.
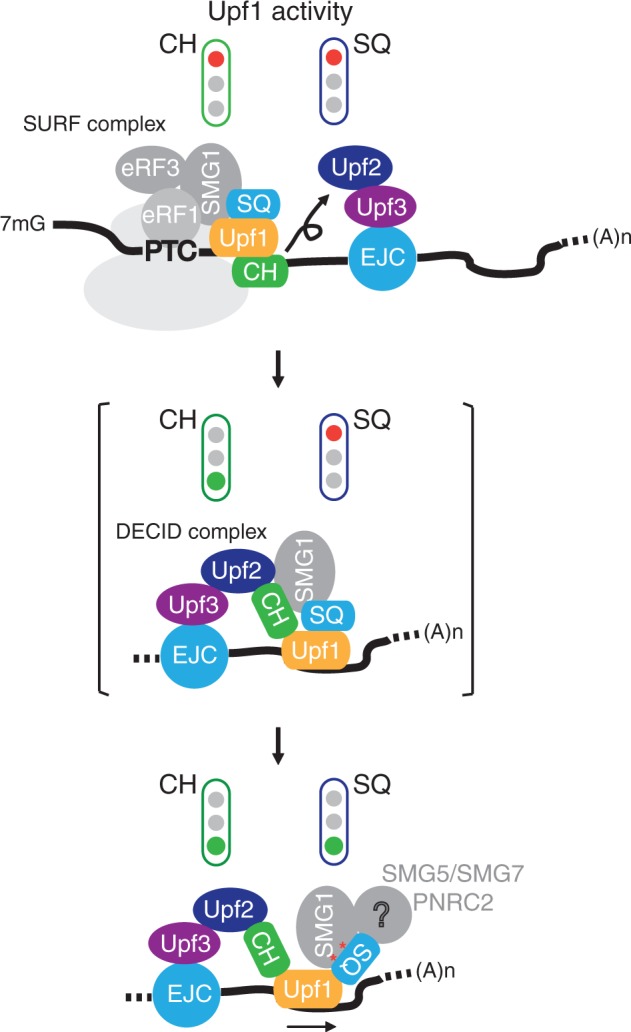
A model of the activation of human Upf1 during NMD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
The Centre National de la Recherche Scientifique (ATIP programme blanc 2008, H.L.H.); Agence Nationale de la Recherche (2008-BLAN-0323, 2011-BLAN-01801, H.L.H.); Foundation Pierre-Gilles de Gennes. Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Elena Conti and Sutapa Chakrabarti (Max Planck Institute, Martinsried) for the gift of materials. They thank Emelie Marquenet (IBENS, Paris), Guillaume Pellerin (IBENS, Paris), Annie Schwartz (CNRS, Orléans) and Hugues Roest Crollius for helpful technical advices. They acknowledge our laboratory for helpful advices, comments and discussions and Catherine Dock-Brégeon for carefully reading the manuscript.
REFERENCES
- 1.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationship. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 4.Jankowsky E, Fairman ME. RNA helicases–one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 7.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 9.Azzalin CM, Lingner J. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle. 2006;5:1496–1498. doi: 10.4161/cc.5.14.3093. [DOI] [PubMed] [Google Scholar]
- 10.Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J. 2011;30:4047–4058. doi: 10.1038/emboj.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat. Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 15.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3' untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 19.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 20.Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 27.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 30.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Culbertson MR, Neeno-Eckwall E. Transcript selection and the recruitment of mRNA decay factors for NMD in Saccharomyces cerevisiae. RNA. 2005;11:1333–1339. doi: 10.1261/rna.2113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imamachi N, Tani H, Akimitsu N. Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. Drug Discover. Therapeut. 6:55–61. [PubMed] [Google Scholar]
- 33.Fiorini F, Bonneau F, Le Hir H. Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay. Methods Enzymol. 2012;511:255–274. doi: 10.1016/B978-0-12-396546-2.00012-7. [DOI] [PubMed] [Google Scholar]
- 34.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 35.Walmacq C, Rahmouni AR, Boudvillain M. Influence of substrate composition on the helicase activity of transcription termination factor Rho: reduced processivity of Rho hexamers during unwinding of RNA-DNA hybrid regions. J. Mol. Biol. 2004;342:403–420. doi: 10.1016/j.jmb.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Boudvillain M, Walmacq C, Schwartz A, Jacquinot F. Simple enzymatic assays for the in vitro motor activity of transcription termination factor Rho from i. Methods Mol. Biol. 2010;587:137–154. doi: 10.1007/978-1-60327-355-8_10. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Z, Morisawa G, Song H. Biochemical characterization of human Upf1 helicase. Methods Mol. Biol. 2010;587:327–338. doi: 10.1007/978-1-60327-355-8_23. [DOI] [PubMed] [Google Scholar]
- 38.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 2003;13:207–214. doi: 10.1016/s0959-437x(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 41.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 42.Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential ‘n-step’ kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 44.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 45.Li N, Henry E, Guiot E, Rigolet P, Brochon JC, Xi XG, Deprez E. Multiple Escherichia coli RecQ helicase monomers cooperate to unwind long DNA substrates: a fluorescence cross-correlation spectroscopy study. J. Biol. Chem. 285:6922–6936. doi: 10.1074/jbc.M109.069286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eidenmuller J, Fath T, Hellwig A, Reed J, Sontag E, Brandt R. Structural and functional implications of tau hyperphosphorylation: information from phosphorylation-mimicking mutated tau proteins. Biochemistry. 2000;39:13166–13175. doi: 10.1021/bi001290z. [DOI] [PubMed] [Google Scholar]
- 47.Leger J, Kempf M, Lee G, Brandt R. Conversion of serine to aspartate imitates phosphorylation-induced changes in the structure and function of microtubule-associated protein tau. J. Biol. Chem. 1997;272:8441–8446. doi: 10.1074/jbc.272.13.8441. [DOI] [PubMed] [Google Scholar]
- 48.Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24:2440–2450. doi: 10.1101/gad.604610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders KR, Grimson A, Anderson P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.