Abstract
In the mammalian mitochondrial translation apparatus, the proteins and their partner RNAs are coded by two genomes. The proteins are nuclear-encoded and resemble their homologs, whereas the RNAs coming from the rapidly evolving mitochondrial genome have lost critical structural information. This raises the question of molecular adaptation of these proteins to their peculiar partner RNAs. The crystal structure of the homodimeric bacterial-type human mitochondrial aspartyl-tRNA synthetase (DRS) confirmed a 3D architecture close to that of Escherichia coli DRS. However, the mitochondrial enzyme distinguishes by an enlarged catalytic groove, a more electropositive surface potential and an alternate interaction network at the subunits interface. It also presented a thermal stability reduced by as much as 12°C. Isothermal titration calorimetry analyses revealed that the affinity of the mitochondrial enzyme for cognate and non-cognate tRNAs is one order of magnitude higher, but with different enthalpy and entropy contributions. They further indicated that both enzymes bind an adenylate analog by a cooperative allosteric mechanism with different thermodynamic contributions. The larger flexibility of the mitochondrial synthetase with respect to the bacterial enzyme, in combination with a preserved architecture, may represent an evolutionary process, allowing nuclear-encoded proteins to cooperate with degenerated organelle RNAs.
INTRODUCTION
Mitochondria are of endosymbiotic origin and have undergone massive gene transfer to the host nucleus during evolution (1). On sharp reduction (∼70-fold) to a 16.6 kb circular mammalian mitochondrial (mt) DNA, only 37 genes remain coding for 13 respiratory chain subunits, 22 tRNAs and 2 rRNAs (2). The faster evolutionary rate of mt-DNA than the nuclear genome (3,4) leads to abnormal RNAs, shrunken in size and often lacking important signals. mRNAs are deprived of 3′- and 5′-untranslated regions, rRNAs are smaller than their bacterial or eukaryotic homologs, and an RNA moiety for RNase P is absent (5–7). Most mt-tRNAs have shortened sequences, miss crucial folding and recognition nucleotides as compared with ‘classical’ tRNAs and are more flexible (6,8–10). Hence, preservation of mitochondrial translation relies on a continued adaptation of proteins to the peculiarities of partner RNAs. One compensatory mechanism consists in an increase in the number and/or size of partner proteins (encoded in the nucleus and imported) that take over the role of missing RNA domains. For instance, bovine mt-ribosomes contain 30 proteins in the small subunits and 48 in the large subunits, instead of 21 and 33, respectively, in prokaryotes, with N- or C-terminal extensions (11,12). Human RNase P is fully proteinaceous with three protein subunits (7).
Mechanisms to compensate for degeneration of mammalian mt-tRNAs remain mainly unsolved, especially for mt aminoacyl-tRNA synthetases (mt-aaRSs; specificity indicated by the amino acid abbreviated in a one-letter code, the origin of the aaRS is indicated by a three-letter abbreviation of the organism’s name with the mitochondrial origin given by the digit 2 e.g. HsaDRS2 for Homo sapiens mitochondrial aspartyl-tRNA synthetase), the enzymes which activate and bind a specific amino acid to their cognate tRNA (13). These enzymes are nuclear encoded and, with only few exceptions, are of about the same size, have the same structural 2D organization and contain the expected specificity signals and synthetase class signatures as bacterial and cytosolic aaRSs (14–17). The first 3D structures of mammalian mt-aaRSs confirmed the absence of additional domains (18–21).
Properties of mammalian mt-aaRSs enabling them to deal with mt-tRNAs remain mainly unknown (9,22,23). On one side, their catalytic efficiency is usually one to two orders of magnitude below that of bacterial aaRSs, aminoacylation identity rules rely either on minimalist sets of determinants in the tRNA or even are disobeyed (22,23). On the other hand, a long standing remarkable feature of mt-aaRSs is their high capacity in tRNA accommodation: mt-aaRSs charge both mt- and bacterial tRNAs, whereas bacterial aaRSs cannot cross-aminoacylate mt-tRNAs (24). Non-aminoacylation of an mt-tRNA by a bacterial enzyme is linked to the absence of essential identity signals and to the large flexibility of the mt-tRNA (25). These data suggest the existence of peculiar dynamic properties for the mt-aaRSs, which are not shared with a bacterial homolog.
Here, we compared several biophysical properties of human mitochondrial aspartyl-tRNA synthetase, HsaDRS2, with them to those of a bacterial (E. coli) homolog, EcoDRS. The crystal structure of HsaDRS2 was solved, thermal stabilities were quantified, and thermodynamic parameters of substrate binding were determined by isothermal titration calorimetry. As an outcome, both proteins share a common 3D architecture but the mammalian mt-aaRS gained in plasticity, enabling it to handle any tRNAAsp and especially the non-canonical mt-tRNAAsp. We propose that enlarged plasticity represents a co-evolution mechanism of the nuclear and mt genomes in eukaryotic cells subjected to highly different evolutionary pressures. This plasticity will be an important parameter to consider for the molecular understanding of a rapidly increasing number of human disorders linked to mutations in mt-aaRSs.
MATERIALS AND METHODS
Protein and tRNA purification
EcoDRS was overproduced in E. coli JM83, purified to homogeneity and concentrated to 40 mg ml−1 (25). HsaDRS2 [mt-AspRS(I41) in (26); Supplementary Figure S1] was cloned into pDEST14 using the Gateway technology (Invitrogen) and overproduced in E. coli BL21 (DE3) Rosetta 2 strain as described (26). It was purified by affinity chromatography on an Ni-NTA column followed by gel filtration and concentrated to 20 mg ml−1 (concentration of monomers) in 50 mM of HEPES–Na pH 7.5, 150 mM of NaCl, 1 mM of DTT, 0.1 mM of ethylenediaminetetraacetic acid and 10% (v/v) of glycerol. This buffer, called herein ‘isothermal titration calorimetry (ITC) buffer’, prevents denaturation of the mt-aaRS for several days as verified by dynamic light scattering (DLS) (Supplementary Figure S2). Magnesium had to be kept out, as even low concentrations led to precipitation of HsaDRS2. It was also used for the preparation of EcoDRS and of the tRNAs. tRNAs (sequences are indicated in Supplementary Figure S1) were produced by in vitro transcription and were purified to single nucleotide resolution on denaturing 12% polyacrylamide gel electrophoresis. They were solubilized in ITC buffer and denatured/renatured before use as described (16).
Aminoacylation
Assays were performed at 25°C on transcripts as described previously (16,27). Specific activities were measured by incubating 0.2–3 nM EcoDRS or 77–930 nM HsaDRS2, with an excess of 4 µM cognate or non-cognate transcript. Kinetic parameters kcat and KM were derived from assays containing 2 nM of EcoDRS or 50 nM of HsaDRS2 with 0.2–4 µM of Eco tRNAAsp or mt-tRNAAsp transcript.
Crystallography
Crystallization conditions were searched at 20°C using commercial kits as described (26). HsaDRS2 was crystallized by vapor diffusion with 10% (m/v) PEG-3350 and 0.5 M of ammonium sulfate as crystallants in 25 mM of Bis–Tris pH 5.5 (26). Resulting needle-like crystals (∼300 × 20 × 20 µm3) were cryocooled in their mother liquor. They were extremely radiation sensitive and diffracted weakly. Complete data sets were collected with a fast PILATUS 6 M pixel detector (DECTRIS) at the X06DA beamline, Swiss Light Source (Villigen). Diffraction data at 3.7Å resolution in P2 space group were processed using XDS and XSCALE (28) and were converted into structure factors without any sigma cut-off using TRUNCATE from the CCP4 package (29). Molecular replacement solutions were sought with data truncated at 4.5 Å using a homology model derived from the structure of EcoDRS (30) and the PHASER program (31) as implemented in the PHENIX package (32). Two HsaDRS2 dimers were identified in the monoclinic asymmetric unit. Refinement using a dynamic elastic network (DEN) as implemented in CNS 1.3 (33) led to a reorientation of the insertions and to a dramatic improvement of the electron density map. All residues of HsaDRS2 were built except the 26 C-terminal amino acids (including the 6-His tag), which are disordered. The final coordinates (residues 42–630 for each of the four monomers) and the experimental data have been deposited with the PDB (ID: 4AH6). Figures were prepared with Pymol (www.pymol.org), electrostatic surface potentials were calculated using its APBS plug-in and are depicted at the same color scale in all figures.
Dynamic light scattering
Particle size analysis was performed at 25°C using a Malvern Zetasizer NanoS instrument and a 40 µl quartz cell containing 20 µl of protein solution at 0.5–8 mg ml−1 in ITC buffer. Five consecutive measurements were collected in automatic mode, and data were processed with the manufacturer’s software. The hydrodynamic diameter dh was derived from diffusion coefficient via the Stokes–Einstein relation and was corrected for solvent viscosity and refractive index. Thermal stability was estimated by subjecting protein at 3 mg ml−1 to temperature gradients from 20°C to 80°C. The temperature at which particle size was increased by a factor 2 was taken as the melting point Tm. Additives were prepared in water.
Differential scanning fluorimetry
Samples made of 20 µl protein solution at 3 mg ml−1 and 0.5 µl of SYPRO Orange™ 5000 × stock diluted 20-fold in dimethlsulfoxyde (DMSO) were transferred in 0.2 ml tubes. Additives prepared as described earlier in the text were brought in 1 µl samples, so that their final concentration was as indicated. The variation of fluorescence intensity during a temperature gradient from 25°C to 95°C was monitored in a Stratagene Mx3005P quantitative polymerase chain reaction instrument. On unfolding of the protein, hydrophobic residues become accessible to the dye, which becomes fluorescent (34). Melting point Tm was deduced from maximum of the derivative plot. They are ∼5°C higher as compared with data obtained by the DLS method.
Synchrotron radiation circular dichroism
The experimental set-up at the DISCO beamline, SOLEIL synchrotron (Saint-Aubin, France) was calibrated for magnitude and polarization with a 6.1 mg/ml D-10-camphorsulfonic acid solution. Ten micrograms per milliliter of DRS solutions in 100 mM of potassium phosphate, 50 mM of KCl, 10% (v/v) of glycerol and 1 mM of Tris(2-carboxyethyl)phosphine hydrochloride, TCEP, were placed in a CaF2 cuvette, with an optical path of 8 µm. Three spectra between 170 and 280 nm were recorded at temperatures ranging from 20°C to 80°C by 4°C steps to assess the thermal stability of EcoDRS and HsaDRS2. Data were processed (spectrum averaging and solvent baseline subtraction) using CDtool (35).
Isothermal titration calorimetry
Before ITC measurements, conditions were determined in which the four investigated macromolecules (HsaDRS2, EcoDRS, Hsa mt-tRNAAsp and Eco tRNAAsp) were stable during the time of the experiment. This was found to be the case in the so-called ‘ITC buffer’ (50 mM of HEPES–Na pH 7.5, 150 mM of NaCl, 1 mM of DTT, 0.1 mM of ethylenediaminetetraacetic acid and 10% of glycerol). Magnesium had to be omitted, as it leads to precipitation of HsaDRS. To eliminate possible errors to ITC experiments, all samples (proteins and tRNAs) were extensively and extemporaneously dialyzed against a same stock solution of ‘ITC buffer’, degassed with argon and ultracentrifuged at 4°C. ITC experiments were performed at 25°C in a MicroCal iTC200 instrument (GE Healthcare). In a typical experiment, 2 µl samples of 300 µM EcoDRS or of 100 µM HsaDRS2 were injected into 200 µl of 36 µM or 11 µM, respectively, cognate tRNA transcript solution. Injections lasted 4 s at 120 s intervals. Cross-bindings were also performed with protein/RNA concentration in a ∼10:1 molar ratio. Retrieved ΔH and ΔG values were used to calculate the binding entropy ΔS (expressed as −TΔS, with ΔG = ΔH − TΔS). The adenylate analog 5′-O-[N-(l-aspartyl)sulfamoyl] adenosine (Asp–AMS) was titrated against 200 µl of enzyme with 0.7 µl injections to HsaDRS2 at 32 µM and 1.5 µl to EcoDRS at 45 µM. Data were analyzed with the Origin software.
RESULTS
Solution properties of HsaDRS2
HsaDRS2 and EcoDRS are homologous dimeric enzymes, both of bacterial evolutionary type, sharing a same size and 43% sequence identity, including all typical signature motifs of DRSs and the same secondary structure organization (16,36) (Supplementary Figure S1). Recombinant proteins were overexpressed in E. coli and the final enzymatic preparations brought into the same buffer (‘ITC buffer’, see ‘Materials and Methods’ section) suitable for ultimate comparative isothermal titration calorimetry experiments. These conditions were a compromise for non-aggregation of HsaDRS2, the more sensitive of the two enzymes (27), preservation of activity for both enzymes and stability of the tRNAs. HsaDRS2 studied herein is deprived of the 40 first amino acids likely encompassing the mitochondrial targeting sequence (26). Both proteins had the same mean hydrodynamic diameter (dh) of 10 ± 1 nm in DLS, in agreement with a homodimeric structure (Supplementary Figure S2). Kinetic parameters and catalytic efficiencies of both enzymes tested under identical conditions confirmed the ∼60-fold higher aminoacylation rate and efficiency in the EcoDRS cognate system compared with HsaDRS2 (Table 1). However, EcoDRS was unable to aminoacylate the mt-tRNA, whereas the mt-aaRS charged both cognate and non-cognate tRNAs with similar efficiency.
Table 1.
Comparative kinetic properties of HsaDRS2 and EcoDRS in tRNA aspartylation
| DRSa | tRNAAsp (transcript) | kcat (10−3 sec−1) | KM (µM) | kcat/KM (10−3 sec−1 µM−1) | L | ΔΔGb (kcal mol−1) |
|---|---|---|---|---|---|---|
| HsaDRS2 | mt | 4.0 ± 0.4 | 1.1 ± 0.2 | 3.7 ± 0.7 | 72 | −2.45 |
| HsaDRS2 | Eco | 10.3 ± 2.0 | 1.2 ± 0.1 | 8.3 ± 0.8 | 35 | −2.04 |
| EcoDRS | Eco | 230 ± 6.0 | 0.8 ± 0.1 | 293 ± 14.4 | 1 | – |
| EcoDRS | mt | nd | nd | nd | >106 |
All data were obtained at 25°C (T = 298°K) and correspond to mean values from at least duplicated experiments with independent and freshly prepared enzyme and tRNA lots.
akcat and KM values were derived from assays containing 2 nM of EcoDRS or 50 nM of HsaDRS2 and 0.2–4 µM of Eco or Hsa mt-tRNA transcript. L stands for the relative loss in aminoacylation efficiency referred to the tRNA aminoacylation efficiency of EcoDRS for Eco tRNAAsp [L = (kcat/KM)Eco system/(kcat/KM)other]; bΔΔG is the variation of the free energy change at the transition state when comparing aminoacylation in the homologous Eco system with the reactions in the heterologous or Hsa mt-systems [ΔΔG = −RT lnL] (37); nd for not detectable.
Crystal structure of HsaDRS2
Single needle-like crystals of HsaDRS2 were obtained in the presence of PEG-3550 and ammonium sulfate (26). They yielded a complete diffraction data set (26). In spite of their modest resolution (3.7 Å), these data were suitable to determine the architecture of HsaDRS2 (26). The structure, solved herein by molecular replacement using a homology model derived from the structure of the EcoDRS (30), confirmed the dimeric status of the enzyme. It clearly showed the anticodon binding and catalytic domains of the monomers but did not enable visualization of the bacterial insertion domain (residues 337–470, Supplementary Figure S1). This suggested either a structural rearrangement or a local flexibility of this domain. The ambiguity was solved by using a DEN-assisted refinement procedure (33) that allowed repositioning of the insertion in well-defined electron density (Supplementary Figure S3 and Table 2). This domain reorientation, different from EcoDRS, was not an effect of crystal packing because the four independent monomers in the asymetric unit adopted the same conformation.
Table 2.
Crystallographic analysis of HsaDRS2
| Data collection statistics | |
| Beamline | SLS/X10A |
| Space group (number) | P2 (3) |
| Unit cell lengths a, b, c (Å) | 142.4, 82.6, 146.3 |
| Unit cell angles α, β, γ (°) | 90, 100.4, 90 |
| Resolution range (Å) | 82–3.7 |
| Highest resolution shell (Å) | 3.8–3.7 |
| Number of observations0 | 255 882 (9662) |
| Number of unique reflections | 35 768 (2403) |
| Completeness (%) | 98.8 (91.2) |
| Multiplicity | 7.2 (4.0) |
| Rmerge/Rmeas (%)a | 18.0 (63.9)/19.4 (72.5) |
| <I/σ(I)> | 9.4 (2.1) |
| Crystal mosaicity (°) | 0.25 |
| Asymmetric unit content | 2 dimers |
| Solvent content (%) | 62 |
| Refined atomic structure | |
| Resolution range (Å) | 30–3.7 |
| R-factor/R-free (%) | 22.1/28.0 |
| Number of protein residues/atoms | 4 × 589/18 840 |
| Rmsd and bonds (Å) and angles (°)° | 0.005/1.08 |
| Ramachandran plotb residues in most favored unfavored regions (%) | 89.2/0.2 |
| PDBid | 4AH6 |
Values in parentheses are for the highest resolution shell.
aRmerge = Σhkl Σi |Ii(hkl) − <I(hkl)>|/Σhkl Σi Ii(hkl) and redundancy-independent; Rmeas = Σhkl (n/n-1)1/2 Σi |Ii(hkl) − <I(hkl)>|/Σhkl Σi Ii(hkl).
bAnalysis carried out with Molprobity in Phenix package.
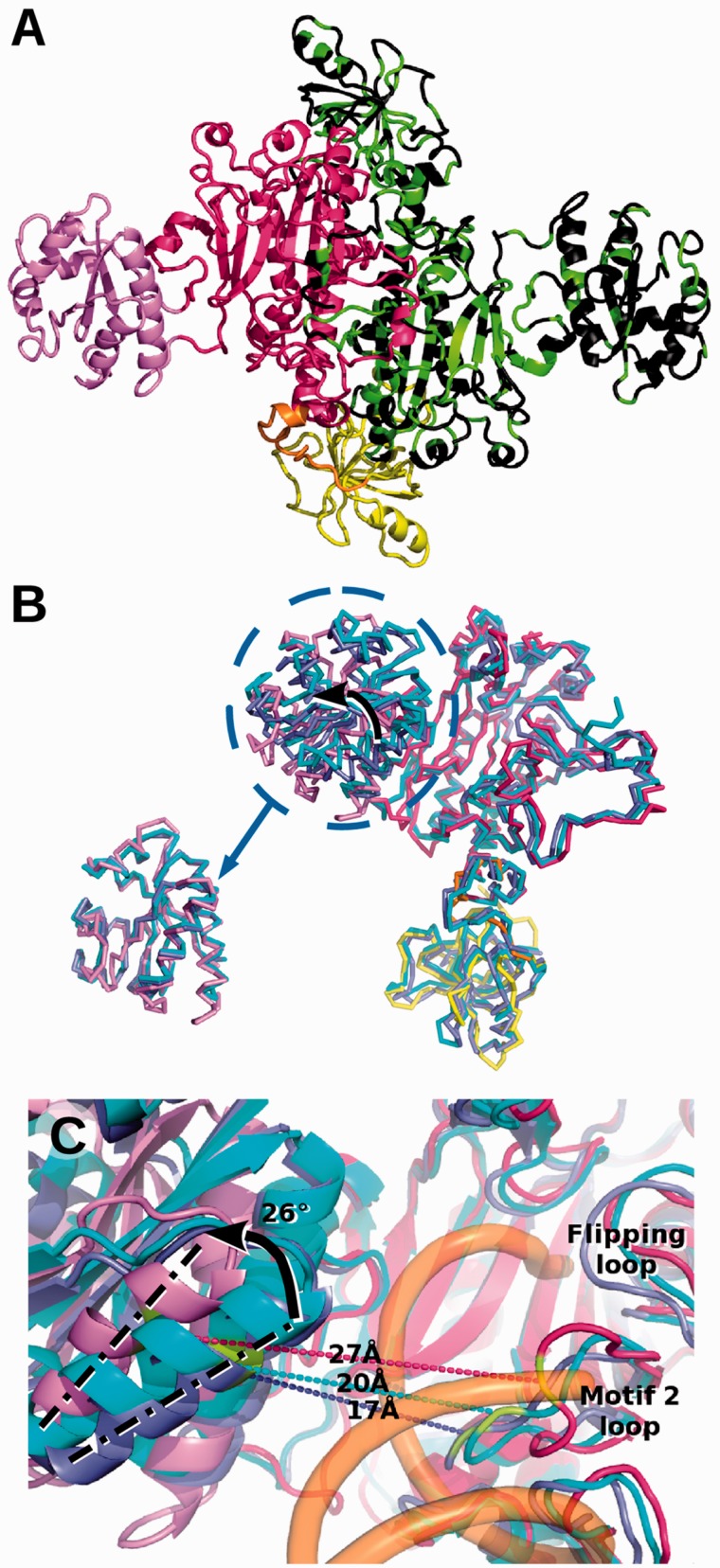
Altogether, 96% of the sequence of the crystallized homodimeric HsaDRS2 is seen (Figure 1A), with a slight asymmetry between its two subunits because of variations in the bacterial insertion domain (Supplementary Figure S4 and Supplementary Table S5). Overall, the architecture of HsaDRS2 is similar to that of bacterial DRSs, in agreement with sequence conservation (16,36). Comparison with EcoDRS crystal structure (30) highlighted a strong conservation. Respective DRS cores, including the anticodon-binding domain, which adopts the so-called OB fold (for oligonucleotide-binding fold), the hinge region and the catalytic domain with its characteristic antiparallel β-sheet (30,38) displayed an overall rmsd (root mean square distance) of ∼2 Å (Supplementary Table S5). These three domains shared a similar position with respect to the dimer interface. The main difference between the two aaRSs resulted from the bacterial insertion that underwent a rigid-body counter-clockwise rotation of ∼26° and clearly made the mitochondrial active site more open for the binding of tRNA (Figure 1B and C). The overall fold of the insertion remained unchanged. This reorientation explains why the insertion was barely visible on the initial maps. Minor conformational diversity occurred in protein segments that diverge in sequence, located in the periphery of the catalytic domain, the flipping loop and the motif 2 loop (short insertions/deletions, see Supplementary Figure S1), which are extremely mobile in DRSs (39,40).
Figure 1.
Newly solved 3D structure of HsaDRS2 in comparison with EcoDRS. (A) Cartoon representation of HsaDRS2 dimer. Characteristic DRS domains are highlighted in the left monomer: anticodon binding domain in yellow, hinge region in orange, catalytic domain in red and bacterial insertion in violet. The right monomer is displayed in black with position of residues conserved in HsaDRS2, and EcoDRS is colored in green. Sequence identity is 40% overall, 47% in the catalytic domain, 41% for the anticodon-binding domain and drops to 25% in the insertion. (B) Superposition of DRS monomers: HsaDRS2 chain colored as in (A), E. coli free (PDBid: 1EQR) and tRNA-bound (PDBid: 1C0A) monomers in light and dark blue, respectively. The bacterial insertion in HsaDRS2 is rotated by 26° compared with its position in EcoDRS (see arrow). Bottom left: zoom on superposed insertions illustrating the overall fold conservation. (C) Zoom on the active site groove with same color code for DRSs as in (B), and Eco tRNA is represented as a semi-transparent orange backbone. The distance between two conserved residues (in green) on each side of the groove is displayed: Phe390 in HsaDRS2 (Phe340 in EcoDRS) in the external helix of the insertion and Arg271 (Arg222) in the motif 2 loop of class II synthetases. The 26° reorientation visualized by the movement of the axis of the external helix opens up the groove for the tRNA acceptor arm by 7–10 Å in HsaDRS2 when compared with the tRNA bound and free states of EcoDRS.
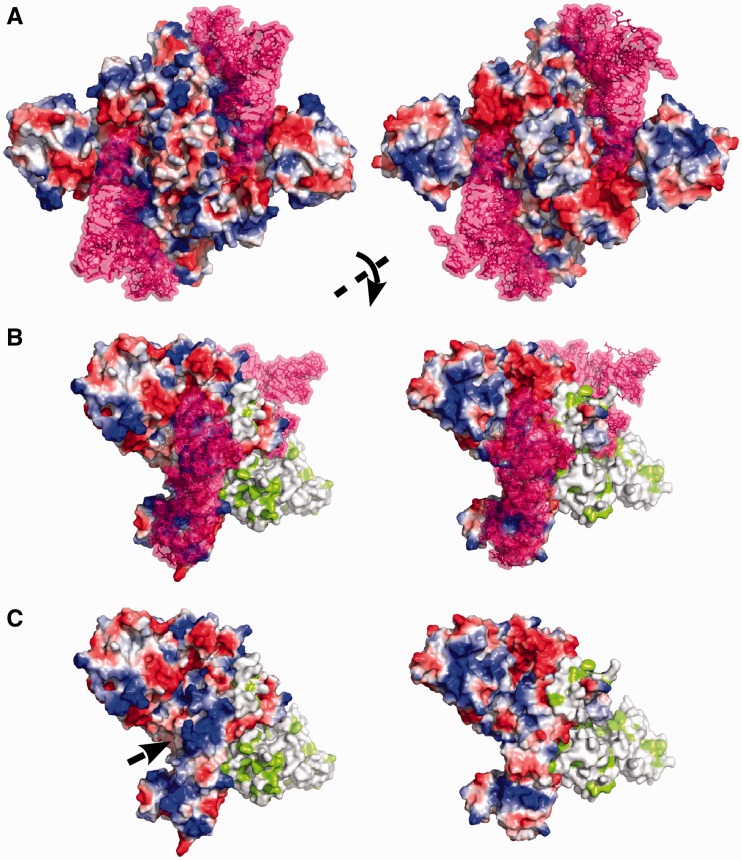
Based on the structural knowledge on human mt-tRNAAsp (41) a model of the complex with HsaDRS2 could be built (Figure 2) and allowed for comparison with the bacterial complex. Notable alterations in the charge distribution within the tRNA interaction surface of the proteins were observed. Electropositive patches render HsaDRS2 more attractive for its RNA partner (Figure 2), similar to what was observed in the three other mt-aaRS crystal structures solved so far (18–21) (Figure 5). Ten additional basic residues (nine Lys and an Arg highlighted in blue in Supplementary Figure S1) are inserted in the close neighborhood of HsaDRS2 catalytic cleft. They may contribute to make the surface more attractive or directly contact the tRNA backbone, especially in the hinge region where such kind of aspecific interactions with the D-stem was described in the E. coli complex (30). Otherwise, almost all residues making direct and specific interactions with bases at both ends of the Eco tRNA are conserved in the human enzyme (indicated in green in Supplementary Figure S1). This is true for E270/R271/R274 or R599, which may contact C74 or nucleotides 69–70, respectively, as well as for Q129/E140, R75/Q91/R125, R125, which are adequately positioned to interact with the anticodon bases C34, U35 and G36, respectively. A few residues (indicated in orange in Supplementary Figure S1) differ without major implication for the conservation of the interaction network. Thus, the main modification is the presence of a Gly at position 269, instead of the usually conserved Asp, which accounts for the insensitivity of HsaDRS2 to the nature of the discriminator base 73 (42).
Figure 2.
Comparison of HsaDRS2 (left) and EcoDRS (right) surface potentials. Surface potentials are depicted from positive to negative in blue to red with the same color scale. (A and B), tRNA-bound HsaDRS2 (model) and EcoDRS (PDBid: 1C0A) with tRNAAsp (shown in purple). (C) The enzymes without tRNA. In (A), the complexes are observed along their two-fold symmetry axis. In (B and C), they are rotated as indicated to better visualize the interaction zone on the surface of the left monomer. The right monomer is colored with hydrophobic patches in green.
Figure 5.
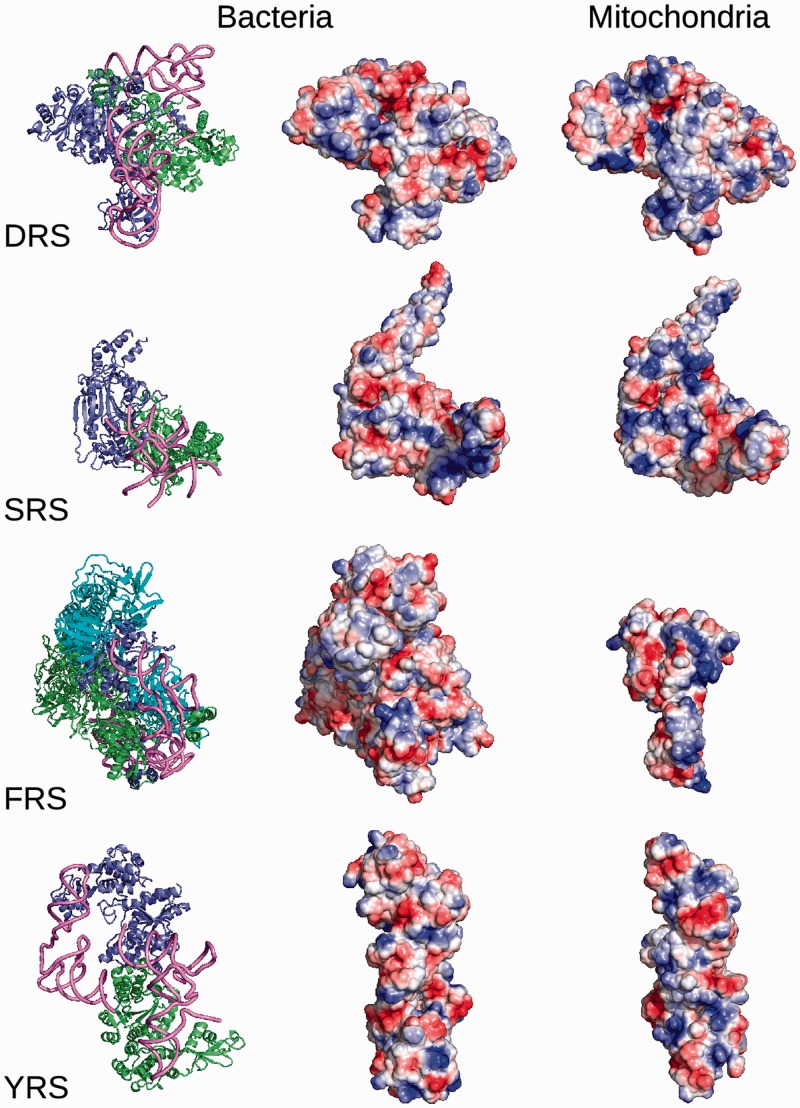
Molecular surface potential of bacterial (E. coli or Thermus thermophilus) and mammalian (bovine or human) mitochondrial aaRSs. On the left, bacterial complexes (PDBids: 1C0A for EcoDRS/tRNA, 1SRS for TthSRS/tRNA, 2IY5 for TthFRS/tRNA and 1H3E for TthYRS/tRNA) are shown on the left with monomers A in blue, monomers B in green and tRNAs in pink, indicating the binding site of the cognate substrates. In the case of tetrameric bacterial FRS, monomers C and D are depicted in cyan. The electrostatic surface potential of free aaRS forms is represented in the same orientation as the complexes for bacterial (1EQR for EcoDRS, 1SRY for TthSRS, 1B7Y for TthFRS, 1H3F for TthYRS without its C-terminal domain) and mitochondrial enzymes (this study for HsaDRS2, 1WLE for BtaSRS2, 3TUP for HsaFRS2, 2PID for HsaYRS2). Overall, the tRNA binding surface seems to be more electropositive in mitochondrial forms, even for FRS, which has a different structural organization (monomer instead of heterotetramer).
The dimeric interfaces of both DRSs differ notably by the distribution and nature of subunit/subunit contacts (Supplementary Figure S6). In total, 70 hydrogen bonds and 28 salt bridges stabilize the EcoDRS interface, whereas HsaDRS2 only contains 60 hydrogen bonds and 20 salt bridges. Considering a 10% larger interface area (5500 versus 5100 Å2), the contribution of specific interactions is 25% lower per Å2, whereas the contribution of aspecific van der Waals interactions is increased, which may on average lead to a less cohesive HsaDRS2 dimer.
Altogether, the crystal structure of HsaDRS2 reveals that it shares the same global architecture with EcoDRS but differs by an enlarged catalytic groove, by a more electropositive surface potential and by a modified network of interactions in the dimer interface.
Temperature stability of HsaDRS2 versus EcoDRS
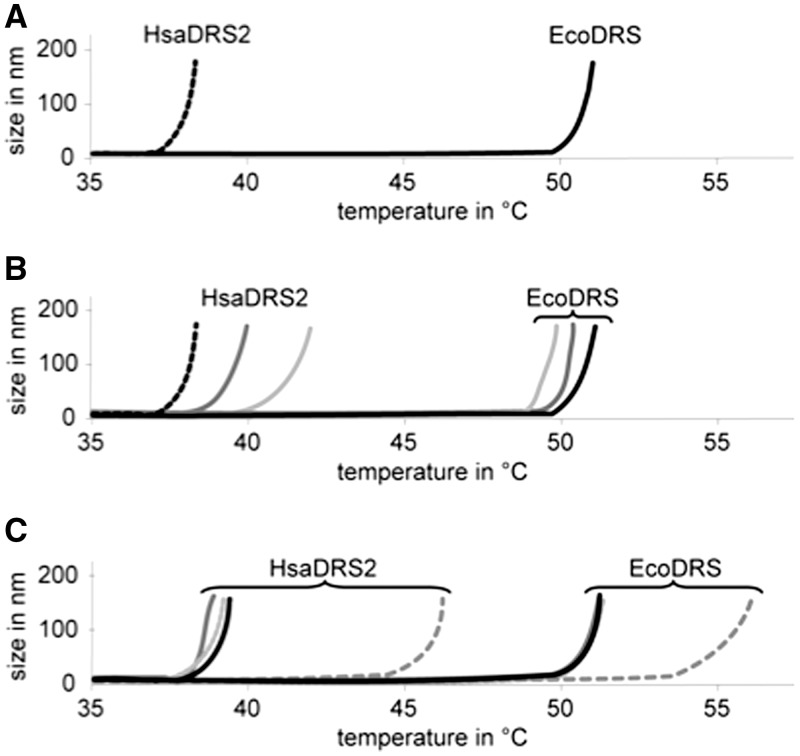
The sensitivity of both proteins to temperature has been compared using DLS and differential scanning fluorimetry (Figure 3 and Supplementary Figure S7). HsaDRS2 unfolded at a temperature of >12°C below that of EcoDRS, in the 37°C–45°C range, under various biochemical conditions and in the presence of various additives. This result is supported by sets of synchrotron radiation circular dichroism spectra recorded at temperatures from 20°C to 80°C (Supplementary Figure S8). For both proteins, a transition from an ordered to a disordered state occurs at temperatures similar to those observed in DLS and differential scanning fluorimetry. This low melting temperature was also valid for an HsaDRS2 variant bearing different N- and C-terminal amino acids but not for HsaYRS2, which melted at ∼55°C (Supplementary Figure S7). Significant stabilization was observed in the presence of Asp–AMS [(5′-O-[N-(L-aspartyl)sulfamoyl]adenosine; Glu-AMS, Gln-AMS, Tyr-AMS for other sulfamoyl-adenylates; AspOH, GluOH, TyrOH for amino alcohol-adenylates], a high affinity analog of the aspartylation reaction transition state (43), but neither in the presence of tRNA, adenosine triphosphate or aspartic acid did (Figure 3 and Supplementary Figure S7). Asp–AMS had a specific effect at a 1:1 stoichiometry per monomer (not shown), suggesting that it is binding to the catalytic site and that this binding protects the protein from melting. Stabilization of aaRSs by binding of adenylate analogs has been taken advantage for successful crystallization (44). Herein, we observe stabilization against temperature induced melting. Along initial attempts to stabilize HsaDRS2 by site-directed mutagenesis, the replacement of amino acids of the hinge domain by those of EcoDRS led to a gain of 5°C in Tm (Supplementary Figure S7).
Figure 3.
Thermostability of HsaDRS2 and EcoDRS. Melting temperatures as measured by DLS. (A) Thermostability of HsaDRS2 (dashed black line) compared with EcoDRS (black line). (B) The presence of in vitro transcribed Hsa mt-tRNAAsp (light grey) or Eco tRNAAsp (dark grey) (in 1.2-fold molar excess) did not significantly influence the thermostability of either DRS. (C) Among the small substrates, only Asp–AMS (dashed grey line) influences the thermostability of the two enzymes positively [aspartic acid: black line, adenosine triphosphate (ATP): grey line, AspOH: light grey line]. Denaturation temperatures of duplicates performed with different enzyme preparations differed by ≤2°C.
Thermodynamics of tRNA binding to HsaDRS2 versus EcoDRS
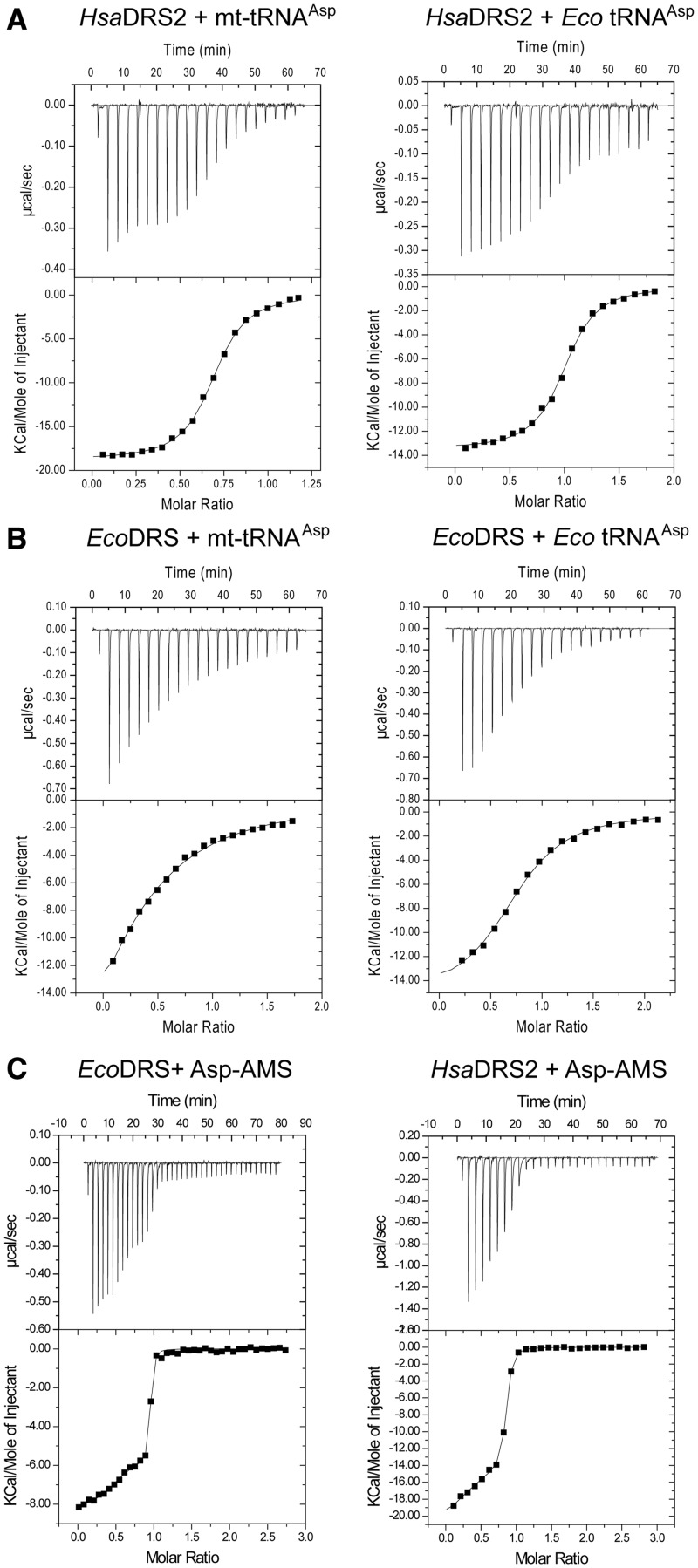
ITC (45) was used to gain insights into the thermodynamics of tRNAAsp binding to DRSs. In vitro transcribed tRNAs were used, as access to milligram amounts of native human mt-tRNAs remains out of scope (9). Mt-tRNAAsp distinguishes drastically from bacterial tRNAAsp by primary, secondary and tertiary structural elements (Supplementary Figure S1), leading to the absence of major aspartate identity elements and to a larger structural flexibility (25). Typical thermodynamic data are presented in Figure 4 and summarized in Table 3. Under the tested conditions, the affinities of HsaDRS2 for either mt- or Eco tRNAAsp were in the same range (Kd ∼250 nM), whereas binding of EcoDRS to its cognate tRNAAsp was weaker by one order of magnitude (Kd of 3 µM) and binding to the non-cognate mt-tRNAAsp dropped the affinity by two orders of magnitude (Kd of 22 µM). Enthalpy and entropy contributions distinguished mitochondrial and bacterial cognate complex formation. HsaDRS2/mt-tRNAAsp was accompanied by about twice as large contributions of both parameters than EcoDRS/Eco tRNAAsp formation. However, both values compensated each other to similar extents, leading to similar variations in free energy namely ΔG = −9.1 kcal/mol and −7.5 kcal/mol, respectively. The difference between these free energies (ΔΔG = −1.6 kcal/mol) is in agreement with a ΔΔG of −2.45 kcal mol−1 measured by comparative kinetic properties of both aaRSs at the transition state (Table 1, 3, and 4).
Figure 4.
Isothermal titration calorimetry of HsaDRS2 and EcoDRS to tRNAs and to the adenyate analog Asp–AMS. (A) Titration of in vitro transcribed tRNAs by HsaDRS2. (B) Titration of in vitro transcribed tRNAs by EcoDRS. (C) Titration of EcoDRS and HsaDRS2 by Asp–AMS.
Table 3.
Comparative thermodynamic properties of HsaDRS2 and EcoDRS: thermodynamic parameters of DRS interaction with tRNA, as measured by ITC
| DRS + tRNA(transcripts) | na | Kd (µM) | ΔH (kcal mol−1) | −TΔS (kcal mol−1) | ΔG (kcal mol−1) |
|---|---|---|---|---|---|
| HsaDRS2 + Hsa mt-tRNAAsp | 0.48 ± 0.26 | 0.26 ± 0.18 | −20.3 ± 0.7 | +11.2 ± 0.4 | −9.1 ± 0.5 |
| HsaDRS2 + Eco tRNAAsp | 0.87 ± 0.16 | 0.24 ± 0.01 | −14.0 ± 0.7 | +5.0 ± 0.7 | −9.0 ± 0.5 |
| EcoDRS + Eco tRNAAsp | 0.76 ± 0.01 | 3.1 ± 0.1 | −13.0 ± 3.1 | +5.5 ± 3.2 | −7.5 ± 0.5 |
| EcoDRS + Hsa mt-tRNAAspb | 0.39 ± 0.02 | 22.0 | −30.8 | +24.4 | −6.4 |
All data were obtained at 25°C (T = 298°K) and correspond to mean values from at least duplicated experiments with independent and freshly prepared enzyme and tRNA lots.
an stands for the number of moles of Asp–AMS bound per mole of DRS monomer.
bCross-binding between EcoDRS and Hsa mt-tRNAAsp performed only once.
Table 4.
Comparative thermodynamic properties of HsaDRS2 and EcoDRS: thermodynamic parameters of DRS interaction with Asp–AMS, as measured by ITC
| DRS + Asp-AMS | na | Kd (nM) | ΔH (kcal mol−1) | −TΔS (kcal Mol−1) | ΔG (kcal mol−1) |
|---|---|---|---|---|---|
| HsaDRS2–monomer 1 | 0.48 ± 0.02 | 129 ± 38.0 | −13.18 ± 0.39 | +3.9 ± 0.78 | −9.38 ± 0.18 |
| HsaDRS2–monomer 2 | 0.29 ± 0.04 | 17.5 ± 12.0 | −21.8 ± 0.31 | +10.57 ± 0.71 | −10.63 ± 0.44 |
| EcoDRS–monomer 1 | 0.49 ± 0.02 | 29.5 ± 5.0 | −5.50 ± 0.01 | −4.75 ± 0.13 | −10.24 ± 0.10 |
| EcoDRS–monomer 2 | 0.44 ± 0.01 | 2.95 ± 1.34 | −8.16 ± 0.35 | −3.52 ± 0.61 | −11.63 ± 0.28 |
All data were obtained at 25°C (T = 298°K) and correspond to mean values from at least duplicated experiments with independent and freshly prepared enzyme and tRNA lots.
an stands for the number of moles of Asp–AMS bound per mole of DRS monomer.
Thermodynamic parameters for non-cognate complex formation (cross-complexes) revealed a pre-eminent role of the tRNA partner as compared with the synthetases (Table 4 and Figure 4A and B). Similarly, low binding enthalpies and entropies drive complexes involving the structurally stable Eco tRNAAsp regardless the enzyme. At opposite, binding of the unstable and highly flexible mt-tRNAAsp required far larger enthalpic and entropic compensations, regardless the enzyme. These large values are indicative of the adaptation of mt-tRNAAsp to both enzymes’ surfaces involving likely conformational changes of the tRNA, releasing of water molecules along the binding surface and reciprocal adaptation of the nucleic acid and the protein.
Thermodynamics of adenylate analog binding to HsaDRS2 versus EcoDRS
Binding of the adenylate analog Asp–AMS to the two dimeric DRSs revealed two different binding sites within each DRS dimer (Figure 4 and Table 4). For both enzymes, a first adenylate bound to one monomer (n ∼0.5, i.e. 0.5 mol of adenylate per mol of monomer) with low affinity (Kd of 129 nM for HsaDRS2 and 29.5 nM for EcoDRS). The second adenylate bound then to the second site with a higher affinity (Kd of 17.5 nM for HsaDRS2 and of 2.95 nM for EcoDRS), in favor of a positive-allosteric event. The gain in affinity for the second binding site was of similar amplitude (5- to 10-fold) in both enzymes, and affinities (in the nM range) were in agreement with the previously measured Ki values for HsaDRS2 (43). Binding of Asp–AMS to each monomer, however, followed a different thermodynamic route depending on the considered enzyme. For EcoDRS, binding was both enthalpy- and entropy-driven. In contrast, binding to HsaDRS2 was strongly enthalpy-driven. This different thermodynamic signature supports different binding mechanisms of Asp–AMS to HsaDRS2 and to EcoDRS.
DISCUSSION
‘Same–same but different’
The crystal structure of HsaDRS2 revealed a typical overall bacterial-type DRS architecture (30, 38–40) that superimposes with the structure of EcoDRS with a global rmsd <2 Å (Figure 1). The two functionally important domains, catalytic site and anticodon-binding domain overlap with an rmsd of <1.5 Å. The important differences between the two structures are (i) the angle formed by the prokaryal-specific insertion domain and the catalytic domain, which is more opened by 26° in the mt-aaRS; (ii) the overall electropositive surface potential (as observed for other mammalian mt-aaRSs); and (iii) the dimer interface, which is weaker in terms of salt-bridges and hydrogen bonds.
A distinguishing property between the two proteins is the unprecedented observation for a human mt-aaRS to melt at near body temperature (37°C) and ∼12°C lower than EcoDRS, in line with a restricted thermo-tolerance of the translation and replication processes in mitochondria (46). It is likely that the weakened dimer interface is one of the contributors to this fragility. Fine prediction of the molecular reasons for the melting temperature of a protein is difficult and may be dependent on as little as a single amino acid (47). Interestingly, a thermosensitive mutant of EcoDRS in which proline 555 was affected, has been reported (48). The equivalent position in HsaDRS2 has kept an equivalent proline residue (Pro 605), so that the contribution of this position to thermosensitivity is ruled out. We favor the contribution of the hinge domain, as partial stabilization by mutation in HsaDRS2 was successful.
ITC was used here for the first time on tRNA/synthetase couples to examine thermodynamics of cognate mt-tRNAAsp and non-cognate Eco tRNAAsp interactions. When tested under strictly the same conditions, HsaDRS2 binds both its cognate and non-cognate tRNAAsp with a same sub-micromolar Kd, whereas EcoDRS binds cognate Eco tRNAAsp in the low micromolar range and non-cognate mt-tRNAAsp in the high micromolar range. It is anticipated that the parameters would be closer to, but still distinct, from those for the bacterial cognate pair when measured with native post-transcriptionally modified tRNAs. Indeed, modifications are known to stabilize structures (49), but the modification pattern of mt-tRNAAsp is far beneath that of Eco tRNAAsp (see Supplementary Figure S1). Binding between cognate mitochondrial partners requires large enthalpic and entropic compensations at opposite to binding between the cognate bacterial partners where compensations were small. Cross-binding revealed that this difference is mostly linked to the tRNA. Binding of the floppy mt-tRNA involved large positive enthalpic and large negative entropic contributions, whereas binding of the well-folded and stable Eco tRNA involved low enthalpy/entropy compensations. These results are in line with thermodynamic parameters determined for cognate and non-congate tRNA/synthetase couples measured earlier by fluorescence titrations and fast kinetic techniques (50,51).
Altogether, comparative investigation of biophysical properties of HsaDRS2 and EcoDRS shed new light on the long-term open question as to non-reciprocal aminoacylation of bacterial and mitochondrial tRNA/synthetase pairs (24). They revealed that the human enzyme has gained structural adaptability to tRNA substrates by combining several properties. A more electropositive tRNA-binding surface [see also (52)] and an opening of the cleft between the insertion domain and the catalytic domain by an angle of as much as 26° improve accessibility and binding of the tRNA end to the catalytic site. Strong binding likely reflects an evolutionary adaptation of the mt-aaRS to compensate for high flexibility of the mt-tRNA and for the absence of otherwise important identity elements. Whether the binding step is accompanied by a concerted movement of the insertion domain towards the catalytic site to further lock the tRNA, is unknown. The larger structural plasticity of the mt-aaRS (enabling it to deal with the highly flexible and degenerated mt-tRNAAsp) does not harm its interaction with classical and less flexible Eco tRNAAsp. At opposite, the bacterial aaRS is unable to bind properly the mt-tRNA. Binding to its cognate more rigid tRNAAsp is closer to a lock-and-key type process.
Gain of plasticity as reflected here by unusual thermodynamic properties, may be a more general property of mt-aaRSs that have to deal with degenerated mt-tRNAs. An extreme case of plasticity has recently been described with the human mitochondrial FRS that undergoes dramatic conformation change on tRNA binding (21). The near-body thermal unstability of HsaDRS2 as well as its electropositive surface potential suggest that different parameters may stabilize the protein in vivo. These may be of biochemical nature or correspond to co-factors, partner proteins or peculiar sub-mitochondrial localization, as is the case for mt-ribosomes and mt-elongation factor that bind to the inner membrane (9).
Cooperative allostery in class II dimeric DRSs
A long-standing question for dimeric aaRSs concerns the communication mechanisms between the two catalytic sites (13). Data obtained herein by ITC demonstrate unambiguously that both EcoDRS and HsaDRS2 undergo an allosteric event on binding to the adenylate analog. The first monomer binds the analog with a weaker affinity than the second monomer. Thermodynamic signatures strongly suggest a different binding mechanism for each enzyme: interaction with HsaDRS2 is a strongly exothermic process, characterized by a large negative enthalpic term, whereas binding to EcoDRS is associated with moderate enthalpic and entropic contributions. These differences likely reflect large obligatory rearrangements in HsaDRS2 but not in EcoDRS.
Allostery has been reported for several aaRSs (53–56). Half of the site activity was mainly observed. Herein, we report direct cooperative allostery for small substrate binding, as detected by ITC. Previous ITC investigations performed on other aaRSs had not detected cooperative allostery (53–56).
CONCLUSION AND PERSPECTIVES
Resolution of the crystal structure and exploration of biophysical properties of a human mitochondrial aaRS, revealed similarities but also distinctive features from a prokaryotic homolog: a conserved architecture and a conserved cooperative allosteric communication within the dimers but also increased structural plasticity. We propose that the modification of intrinsic biophysical properties and new surface properties, along with conservation of size and structural framework, represents an evolutionary mechanism for adaptation of nuclear-encoded proteins to degenerated mitochondrial-encoded RNAs. This mechanism is alternative to domain extension and sustains adaptation of two genomes subjected to different evolutionary constraints. The possibility that mt-aaRSs also undergo functional expansion by structural inventions and have partner proteins as described for cytosolic aaRSs (57,58) is not excluded but remains to be tackled. The properties of an mt-aaRS reported herein provide new insights towards the understanding of the growing number of human disorders linked to mutations in the corresponding genes (9,59,60). Deciphering the fine molecular triggers leading to both specific (i.e. plasticity) and shared (i.e. allostery) properties of human mt-aaRSs, and their evolutionary related homologs requires delineation of discriminating and non-discriminating amino acids within the aaRS architecture. Bioinformatic tools enabling such an analysis are under development.
ACCESSION NUMBERS
The coordinates and structure factors are deposited in the Protein Data Bank under the accession code 4AH6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4 and 6–8, Supplementary Table 5 and Supplementary References [61–63].
FUNDING
Association Française contre les Myopathies (AFM); Agence Nationale pour la Recherche [ANR-09-BLAN-0091-01/03, ANR-PCV07-187047]; French National Program ‘Investissements d’Avenir’ (Labex MitoCross); Centre National de la Recherche Scientifique (CNRS); Université de Strasbourg; France Diplomatie [200930]. Funding for open access charge: Centre National pour la Recherche Scientifique (CNRS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank R. Giegé and P. Dumas for fruitful discussions, G. Eriani for gift of material, R. David and the team of the high-performance computing facility for allocating CPU time to the project to carry out extensive DEN refinement on HsaDRS2 structure (HPC méso-centre de calcul, Université de Strasbourg), V. Olieric and the PX team at the Swiss Light Source (Villigen, Switzerland), as well as Frank Wien and the staff of DISCO at SOLEIL synchrotron (Saint-Aubin, France) for beamtime allocation and assistance during data collection.
REFERENCES
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrel BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon JC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellana S, Vicario S, Saccone C. Evolutionary patterns of the mitochondrial genome in Metazoa: exploring the role of mutation and selection in mitochondrial protein coding genes. Genome Biol. Evol. 2011;3:1067–1079. doi: 10.1093/gbe/evr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol. 2010;2010:1–24. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe K. Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:11–36. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Helm M, Brulé H, Friede D, Giegé R, Pütz J, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 10.Giegé R, Jühling F, Pütz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip. Rev. RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 11.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 12.Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibba M, Francklyn C, Cusack S. The Aminoacyl-tRNA Synthetases. Georgetown, TX: Landes Biosciences; 2005. [Google Scholar]
- 14.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sissler M, Pütz J, Fasiolo F, Florentz C. Mitochondrial aminoacyl-tRNA synthetases. In: Ibba M, Francklyn C, Cusack S, editors. Aminoacyl-tRNA Synthetases. 2005. Landes Biosciences, Georgetown, TX, Vol. Chap 24, pp. 271–284. [Google Scholar]
- 16.Bonnefond L, Fender A, Rudinger-Thirion J, Giegé R, Florentz C, Sissler M. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry. 2005;44:4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
- 17.Brindefalk B, Viklund J, Larsson D, Thollesson M, Andersson SG. Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol. Biol. Evol. 2007;24:743–756. doi: 10.1093/molbev/msl202. [DOI] [PubMed] [Google Scholar]
- 18.Chimnaronk S, Gravers Jeppesen M, Suzuki T, Nyborg J, Watanabe K. Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005;24:3369–3379. doi: 10.1038/sj.emboj.7600811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnefond L, Frugier M, Touzé E, Lorber B, Florentz C, Giegé R, Sauter C, Rudinger-Thirion J. Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features. Structure. 2007;15:1505–1516. doi: 10.1016/j.str.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Klipcan L, Levin I, Kessler N, Moor N, Finarov I, Safro M. The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase. Structure. 2008;16:1095–1104. doi: 10.1016/j.str.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Klipcan L, Moor N, Finarov I, Kessler N, Sukhanova M, Safro MG. Crystal structure of human mitochondrial PheRS complexed with tRNA(Phe) in the active “open” state. J. Mol. Biol. 2012;415:527–537. doi: 10.1016/j.jmb.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 22.Florentz C, Sohm B, Tryoen-Tóth P, Pütz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnefond L, Frugier M, Giegé R, Rudinger-Thirion J. Human mitochondrial TyrRS disobeys the tyrosine idenity rules. RNA. 2005;11:558–562. doi: 10.1261/rna.7246805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumazawa Y, Himeno H, Miura K-I, Watanabe K. Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J. Biochem. 1991;109:421–427. doi: 10.1093/oxfordjournals.jbchem.a123397. [DOI] [PubMed] [Google Scholar]
- 25.Fender A, Gaudry A, Jühling F, Sissler M, Florentz C. Adaptation of aminoacylation rules to mammalian mitochondria. Biochimie. 2012;94:1090–1097. doi: 10.1016/j.biochi.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 26.Gaudry A, Lorber B, Messmer M, Neuenfeldt A, Sauter C, Florentz C, Sissler M. Redesigned N-terminus enhances expression, solubility, and crystallisability of mitochondrial enzyme. Protein Eng. Des. Sel. 2012;25:473–481. doi: 10.1093/protein/gzs046. [DOI] [PubMed] [Google Scholar]
- 27.Sissler M, Lorber B, Messmer M, Schaller A, Pütz J, Florentz C. Handling mammalian mitochondrial tRNAs and aminoacyl-tRNA synthetases for functional and structural characterization. Methods. 2008;44:176–189. doi: 10.1016/j.ymeth.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project,N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Eiler S, Dock-Bregeon AC, Moulinier L, Thierry J-C, Moras D. Synthesis of aspartyl-tRNAAsp in Escherichia coli-a snapshot of the second step. EMBO J. 1999;18:6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral G, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummings MD, Farnum MA, Nelen MI. Universal Screening Methods and Applications of ThermoFluor®. J. Biomol. Screen. 2006;11:854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 35.Lees JG, Smith BR, Wien F, Miles AJ, Wallace BA. CDtool-an integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal. Biochem. 2004;332:285–289. doi: 10.1016/j.ab.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Giegé R, Rees B. Aspartyl-tRNA synthetases. In: Ibba M, Francklyn C, Cusack S, editors. Aminoacyl-tRNA Synthetases. 2005. Landes Bioscience, Georgetown, TX, Vol. Chap 19, pp. 210–226. [Google Scholar]
- 37.Pütz J, Puglisi JD, Florentz C, Giegé R. Additive, cooperative and anti-cooperative effects between identity nucleotides of a tRNA. EMBO J. 1993;12:2949–2957. doi: 10.1002/j.1460-2075.1993.tb05957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delarue M, Poterszman A, Nikonov S, Garber M, Moras D, Thierry J-C. Crystal structure of a prokaryotic aspartyl-tRNA synthetase. EMBO J. 1994;13:3219–3229. doi: 10.1002/j.1460-2075.1994.tb06623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt E, Moulinier L, Fujiwara S, Imanaka T, Thierry J-C, Moras D. Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archeon specificity and catalytic mechanism of adenylate formation. EMBO J. 1998;17:5227–5237. doi: 10.1093/emboj/17.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moulinier L, Eiler S, Eriani G, Gangloff J, Thierry J-C, Gabriel K, McClain WH, Moras D. The structure of an AspRS-tRNAAsp complex reveals a tRNA-dependant control mechanism. EMBO J. 2001;20:5290–5301. doi: 10.1093/emboj/20.18.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Messmer M, Pütz J, Suzuki T, Suzuki T, Sauter C, Sissler M, Florentz C. Tertiary network in mammalian mitochondrial tRNAAsp revealed by solution probing and phylogeny. Nucleic Acids Res. 2009;37:6881–6895. doi: 10.1093/nar/gkp697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fender A, Sauter C, Messmer M, Pütz J, Giegé R, Florentz C, Sissler M. Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J. Biol. Chem. 2006;281:15980–15986. doi: 10.1074/jbc.M511633200. [DOI] [PubMed] [Google Scholar]
- 43.Messmer M, Blais SP, Balg C, Chênevert R, Grenier L, Lagüe P, Sauter C, Sissler M, Giegé R, Lapointe J, et al. Peculiar inhibition of human mitochondrial aspartyl-tRNA synthetase by adenylate analogs. Biochimie. 2009;91:596–603. doi: 10.1016/j.biochi.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Giegé R, Touzé E, Lorber B, Théobald-Dietrich A, Sauter C. Crystallogenesis trends of free and liganded aminoacyl-tRNA synthetases. Cryst. Growth Des. 2008;8:4297–4306. [Google Scholar]
- 45.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr. Opin. Struct. Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 46.Germaniuk A, Liberek K, Marszalek J. A bichaperone (Hsp70-Hsp78) system restores mitochondrial DNA synthesis following thermal inactivation of Mip1p polymerase. J. Biol. Chem. 2002;277:27801–27808. doi: 10.1074/jbc.M201756200. [DOI] [PubMed] [Google Scholar]
- 47.Perl D, Mueller U, Heinemann U, Schmid FX. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 2000;7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- 48.Martin F, Sharpless GJ, Lloyd RG, Eiler S, Moras D, Gangloff J, Eriani G. Characterization of a thermosensitive E. coli aspartyl-tRNA synthetase mutant. J. Bacteriol. 1997;179:3691–3696. doi: 10.1128/jb.179.11.3691-3696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 50.Lam SSM, Schimmel PR. Equilibrium measurements of cognate and noncognate interactions between aminoacyl transfer RNA synthetases and transfer RNA. Biochemistry. 1975;14:2775–2780. doi: 10.1021/bi00683a034. [DOI] [PubMed] [Google Scholar]
- 51.Krauss G, Riesner D, Maass G. Mechanism of discrimination between cognate and non-cognate tRNAs by phenylalanyl-tRNA synthetase from yeast. Eur. J. Biochem. 1976;68:81–93. doi: 10.1111/j.1432-1033.1976.tb10766.x. [DOI] [PubMed] [Google Scholar]
- 52.Tworowski D, Feldman AV, Safro MG. Electrostatic potential of aminoacyl-tRNA synthetase navigates tRNA on its pathway to the binding site. J. Mol. Biol. 2005;350:866–882. doi: 10.1016/j.jmb.2005.05.051. [DOI] [PubMed] [Google Scholar]
- 53.Buddha MR, Crane BR. Structures of tryptophanyl-tRNA synthetase II from Deinococcus radiodurans bound to ATP and tryptophan. Insight into subunit cooperativity and domain motions linked to catalysis. J. Biol. Chem. 2005;280:31965–31973. doi: 10.1074/jbc.M501568200. [DOI] [PubMed] [Google Scholar]
- 54.Retailleau P, Weinreb V, Hu M, Carter CW. Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5' tetraphosphate: evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases. J. Mol. Biol. 2007;369:108–128. doi: 10.1016/j.jmb.2007.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain T, Kamarthapu V, Kruparani SP, Deshmukh MV, Sankaranarayanan R. Mechanistic insights into cognate substrate discrimination during proofreading in translation. Proc. Natl Acad. Sci. USA. 2010;107:22117–22121. doi: 10.1073/pnas.1014299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dignam JD, Guo J, Griffith WP, Garbett NC, Holloway A, Mueser T. Allosteric interaction of nucleotides and tRNAAla with E coli alanyl-tRNA synthetase. Biochemistry. 2011;50:9886–9900. doi: 10.1021/bi2012004. [DOI] [PubMed] [Google Scholar]
- 57.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem. Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Guo M, Schimmel P, Yang X-L. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584:434–442. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheper GC, van der Klok T, van Andel RJ, van Berkel CG, Sissler M, Smet J, Muravina TI, Serkov SV, Uziel G, Bugiani M, et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat. Genet. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 60.Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl Acad. Sci. USA. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fender A, Sauter C, Messmer M, Pütz J, Giegé R, Florentz C, Sissler M. Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J. Biol. Chem. 2006;281:15980–15986. doi: 10.1074/jbc.M511633200. [DOI] [PubMed] [Google Scholar]
- 62.Sekiya T, Mori M, Takahashi N, Nishimura S. Sequence of the distal tRNAAsp1 gene and the transcription termination signal in the Escherichia coli ribosomal RNA operon rrnF (or G) Nucleic Acids Res. 1980;8:3809–3827. doi: 10.1093/nar/8.17.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonnefond L, Frugier M, Touzé E, Lorber B, Forentz C, Giegé R, Rudinger-Thirion J, Sauter C. Tyrosyl-tRNA synthetase: the first crystallization of a human mitochondrial aminoacyl-tRNA synthetase. Acta Crystallogr. Sect. F. 2007;F63:338–341. doi: 10.1107/S1744309107012481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.