Abstract
The E3 ubiquitin ligase Rad18 mediates tolerance of replication fork-stalling bulky DNA lesions, but whether Rad18 mediates tolerance of bulky DNA lesions acquired outside S-phase is unclear. Using synchronized cultures of primary human cells, we defined cell cycle stage-specific contributions of Rad18 to genome maintenance in response to ultraviolet C (UVC) and H2O2-induced DNA damage. UVC and H2O2 treatments both induced Rad18-mediated proliferating cell nuclear antigen mono-ubiquitination during G0, G1 and S-phase. Rad18 was important for repressing H2O2-induced (but not ultraviolet-induced) double strand break (DSB) accumulation and ATM S1981 phosphorylation only during G1, indicating a specific role for Rad18 in processing of oxidative DNA lesions outside S-phase. However, H2O2-induced DSB formation in Rad18-depleted G1 cells was not associated with increased genotoxin sensitivity, indicating that back-up DSB repair mechanisms compensate for Rad18 deficiency. Indeed, in DNA LigIV-deficient cells Rad18-depletion conferred H2O2-sensitivity, demonstrating functional redundancy between Rad18 and non-homologous end joining for tolerance of oxidative DNA damage acquired during G1. In contrast with G1-synchronized cultures, S-phase cells were H2O2-sensitive following Rad18-depletion. We conclude that although Rad18 pathway activation by oxidative lesions is not restricted to S-phase, Rad18-mediated trans-lesion synthesis by Polη is dispensable for damage-tolerance in G1 (because of back-up non-homologous end joining-mediated DSB repair), yet Rad18 is necessary for damage tolerance during S-phase.
INTRODUCTION
Trans-lesion synthesis (TLS) is a DNA damage tolerance mechanism that permits DNA synthesis using templates containing bulky DNA lesions such as ultraviolet (UV)-induced cyclobutane pyrimidine dimers (CPD), benzo[a]pyrene dihydrodiol epoxide (BPDE) adducts, 8-oxodG and many others (1). TLS uses specialized DNA polymerases that accommodate damaged DNA templates and perform replication- or repair-coupled DNA synthesis. However, TLS polymerases have reduced fidelity and processivity compared with replicative DNA polymerases and, therefore, replicate damaged templates in an error-prone fashion that leads to mutations. In mammalian cells, the major TLS polymerases are members of the Y-family, namely, DNA polymerase eta (Polη), DNA polymerase kappa (Polκ), DNA polymerase iota (Polι) and Rev1 (2). Each TLS polymerase is specialized to perform DNA synthesis across templates harbouring specific DNA lesions (1). For example, Polη can perform relatively error-free synthesis of CPD, accurately placing AA across thymine dimers (3–5). Alternative TLS polymerases are required to replicate Polη non-cognate lesions. Collectively, the TLS polymerases allow replicative bypass across a variety of DNA lesions. However, owing to its versatile nature, Polη may be the default TLS polymerase for most lesions and, thus, plays a central role in TLS. In ‘xeroderma pigmentosum’-variant (XPV) individuals, Polη is functionally inactivated, conferring reduced tolerance of damage from CPD and other Polη-cognate lesions (4,5). Moreover, in XPV individuals, increased error-prone bypass of Polη-cognate lesions (such as CPD) by alternative inappropriate and more error-prone TLS polymerases leads to elevated mutation frequencies and genome instability. Thus, Polη-mediated TLS defects readily explain the sunlight sensitivity and skin cancer propensity of XPV individuals.
Mono-ubiquitination of the homotrimeric DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) contributes to TLS polymerase recruitment at sites of DNA damage (6). The E3 ubiquitin ligase Rad18 is mobilized to damaged DNA where it mono-ubiquitinates PCNA at the conserved residue K164. TLS polymerases interact with K164 mono-ubiquitinated PCNA via specialized ubiquitin-binding motifs termed ‘UBZ’ (ubiquitin-binding zinc finger, present in Polη and Polκ) and ‘UBM’ (ubiquitin-binding motif, present in Polι and Rev1) domains (7). Therefore, the DNA damage-inducible association of Y-family polymerases with mono-ubiquitinated PCNA contributes to efficient TLS. TLS polymerases also interact with PCNA in an ubiquitination-independent manner via PCNA-interacting peptide (PIP) domains (Polη, Polκ and Polι) and BRCA1 C-terminal region (Rev1).
Several mechanisms for genotoxin-inducible PCNA mono-ubiquitination have been proposed. In some instances, Rad18 recruitment to sites of DNA damage is thought to depend on the generation of RPA-coated ssDNA (8,9). During S-phase, when replicative polymerases are stalled by bulky lesions, RPA-coated ssDNA is generated via the uncoupling of replicative helicase and DNA polymerase activities (10). Similar to redistribution mechanisms proposed for other DNA damage responsive proteins (such as ATRIP and Tipin), Rad18 may be targeted to damaged DNA via direct interactions with RPA (8,9). In addition to its RPA-mediated recruitment, Rad18 may localize to damaged DNA via interaction with p95/Nbs1 (11). Whether RPA and p95 cooperate to recruit Rad18 or mediate different mechanisms of Rad18 recruitment is unclear. Nevertheless, redistribution of Rad18 and TLS polymerases to nuclear foci representing sites of ongoing DNA replication and repair is a hallmark of the DNA damage response to replication fork-stalling lesions during S-phase (12–14). Rad18- or TLS polymerase-deficient cells show more persistent S-phase checkpoint signalling via ATR/Chk1 and reduced damage tolerance after treatment with fork-stalling agents (15,16). Clearly, therefore, TLS plays an important role in the DNA damage response during S-phase.
Interestingly, many recent studies agree that TLS is a post-replication repair process that occurs behind (5′) active replication forks. Thus, replication fork stalling may be accompanied by re-priming events downstream (3′) of the DNA lesion, thereby, allowing continued elongation of existing replicons on damaged templates (17). Strong evidence for a re-priming-based model was provided by the demonstration that TLS-deficient yeast cells maintain normal rates of synthesis on damaged templates, but accumulate single-stranded gaps behind sites of ongoing DNA synthesis (18). Similar results have since been reported for vertebrate cells (19). Thus, during S-phase, TLS may play a major role in gap filling behind a newly re-primed leading strand.
Discontinuities or single-stranded breaks (SSB) in the genome can arise throughout the cell cycle; therefore, the potential exists for TLS to participate in DNA damage processing in a manner that is fully uncoupled from DNA synthesis. For example, using direct visualization and quantification of productive post-replication repair (PRR) tracts in Saccharomyces cerevisiae, Ulrich and colleagues (20) showed that Rad18-dependent TLS is operational after completion of S phase and is, therefore, spatially and temporally dissociable from genome replication. Karras and Jentsch (21) also showed that TLS and the error-free PRR operate in G2/M phase after chromosomal replication. TLS pathway activation also seems to be cell cycle-independent in mammalian cells. For example, Y family DNA polymerases are recruited to sites of DNA damage during growth arrest (22–25) and in G2 (26,27), promoting repair of UV-induced lesions (24) and oxidative DNA damage (25).Although several studies have now reported that TLS can occur outside of S-phase in mammalian cells, analysis of Rad18 and TLS polymerase regulation has typically been performed using cancer cell lines, and TLS has generally not been studied in primary untransformed human cells.
Moreover, although it has been shown that Rad18-mediated TLS pathway activation can occur in growth-arrested cells (25), the relative contribution of TLS to DNA damage tolerance in different cell cycle phases has not been determined. Therefore, the objective of this study was to determine the role of Rad18-mediated TLS pathway activation in DNA damage tolerance during distinct cell cycle stages of normal human cells. To this end, we examined cell cycle stage-specific TLS pathway activation in response to UV and hydrogen peroxide (H2O2). Ultraviolet C (UVC) irradiation induces cross-linking of adjacent pyrimidines, generating templates with helix-distorting CPD that are substrates for DNA synthesis by Polη. Reactive oxygen species (ROS), such as H2O2, lead to oxidized bases including (but not limited to) 8-oxodG that are also bypassed by Polη (28–30). Additionally, the apurinic/apyrimidinic (AP) sites generated during base excision repair (BER) (via glycosylases that remove oxidized bases) or because of spontaneous base loss can be bypassed by Polη (28,29). Thus, CPD, 8-oxodG and AP sites represent potential cognate lesions for Polη, and all have mutagenic potential owing to their error-prone replicative bypass by TLS polymerases. UV- and H2O2-induced lesions represent forms of damage that are generated via exogenous environmental exposures (e.g. solar radiation) and endogenous stresses, such as respiratory metabolism, and it is important to elucidate the mechanisms that allow genome maintenance after exposure to these ubiquitous agents. We show here that both UV and H2O2 elicit TLS pathway activation in a DNA replication-independent manner in normal human cells. Unexpectedly, we define a new role for Rad18 and Polη in preventing double strand break (DSB) after acquisition of H2O2 (but not UV)-induced DNA damage, specifically during G1.
MATERIALS AND METHODS
Adenovirus construction and infection
Adenovirus construction and infections were performed as described previously (16,31). In brief, cDNAs encoding epitope-tagged forms of Rad18 and Polη were subcloned into pAC–CMV. The resulting shuttle vectors were co-transfected into 293T cells with the pJM17 plasmid to generate recombinant adenovirus as described previously (15). Cells were routinely infected with adenovirus at a multiplicity of infection of 20 and 50. To control for adenoviral infection, cells received AdCon (‘empty’ adenovirus vector) or AdGFP.
Cell culture and synchronization
Telomerized human dermal fibroblasts (HDF) and LigIV+/+ and LigIV−/− HCT116 cells (32) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% of fetal bovine serum (FBS) and streptomycin sulphate (100 µg/ml) and penicillin (100 U/ml), as described previously (31,33). For cell synchronization studies, cells were grown to 80–90% confluence, then placed in medium containing reduced FBS (0.1 and 0.5% for HCT116 cells and HDF, respectively) for 48 h to induce a state of growth arrest (G0 or quiescence). To stimulate synchronous cell cycle re-entry, quiescent cells were trypsinized and re-plated at a density of 1:2 in full growth medium then returned to the incubator.
Genotoxin treatments and pharmacological inhibitors
For H2O2 treatments, hydrogen peroxide (Fisher) was diluted to 100 mM in 4°C phosphate buffered saline (PBS). The growth medium was removed from cells and reserved at room temperature. The mono-layers of cells were treated with PBS containing the final indicated concentrations of H2O2 (or with PBS for control samples). After 15 min, the H2O2-containing PBS was removed; the mono-layers were washed twice with PBS and then replenished with the reserved growth medium before being returned to 37°C CO2 incubators. For UVC treatment, the growth medium was removed from the cells, reserved and replaced with PBS. The plates were transferred to a UV cross-linker (Stratagene) and then irradiated. The UVC dose delivered to the cells was confirmed with a UV radiometer (UVP Inc.). The reserved medium from the cells was replaced, and cells were returned to the incubator. NU7441/KU-57788 [8-(4-dibenzothienyl)-2-(4-morpholinyl)-4H-1-benzopyran-4-one, also termed ‘DNA-dependent protein kinase (DNA-PK) inhibitor’] was purchased from TOCRIS Bioscience and dissolved in dimethyl sulfoxide. To inhibit DNA-PK, NU7441 was added directly to the culture medium from a 500× stock solution.
RNA interference
siRNA transfections were performed using 0.5% lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. We routinely used siRNAs at a final concentration of 100 nM. A double-knockdown procedure was necessary to attain efficient depletion of target genes in HDF. The first transfection was performed when the cells grew to 80–90% confluence, and cells were incubated in the transfection reagent for 6 h. Immediately after the first transfection, cells were placed in medium containing 0.5% serum to induce growth arrest. A second 6 h transfection was performed 24 h after the first knockdown. Sequences of siRNAs used in this study are as follows: siApeI: 5′-UCACUUUGAGCCUGGGAAATT-3′; siCon, 5′-UAGCGACUAAACACAUCAAUU-3′; sip95: 5′-GUACGUUGUUGGAAGGAAA-3′ (11); siPCNA: AGAAUAAAGUCCAAAGUCA; siPolη: 5′-GCA GAA AGG CAG AAA GUUTT-3′ (34,35); siRad18, 5′-GAGCAUGGAUUAUCUAUUCAAUU-3′ (34,35); siRnf8: 5′-GAGAAGCUUACAGAUGUUU-3′ (36); siRPA34: 5′-GGCTCCAACCAACATTGTT-3′.
Clonogenic survival assays
For experiments in HDF, cells undergoing log-phase growth (∼80% confluent) were electroporated with non-targeting control siRNA or Rad18 siRNA (100 pmol/1 million cells) using the NHDF nucleofector kit VPD-1001 (Lonzo) and program U-23. Electroporated cells were seeded into flasks at high density and were allowed to recover from electroporation for 15 h in 10% of FBS full-growth medium. Full-growth medium was replaced with 0.5% of FBS starving medium to synchronize cells to G0. After 48 h, quiescent cells were trypsinized and plated into 10-cm dishes with complete growth medium at a density of 500 cells per dish. Owing to the low-plating efficiency of primary HDF, 500 plated cells give rise to ∼150 colonies in control (no genotoxin) cultures. Cells were plated in triplicate for each depletion/H2O2 exposure. Cells were treated with H2O2 at 6 h or at 24 h after plating, times at which cells had progressed to G1 or S-phase after application of full-growth medium, respectively. Replicate plates of G1 and S-phase cells were harvested for immunoblot analysis to confirm Rad18 knockdown. Growth medium was replenished every 3 days, and surviving cells were stained with 0.05% of crystal violet in 40% of methanol 14 days after plating. Colonies containing ≥50 cells were counted.
For experiments in the HCT116 (LigIV WT and LigIV−/−) lines, cells were subject to two rounds of transfection with siRNA, each lasting 6 h (as described under ‘RNA Interference’ section). The first transfection was carried out in complete medium containing 10% of serum. Immediately before the second transfection, cells were placed in starvation medium containing 0.1% of serum. After the second transfection, the cells were provided with fresh 0.1% serum-containing medium.
After 48 h of starvation in 0.1% serum, cells were trypsinized and resuspended at a density of 30 000 cells/ml of pre-chilled PBS containing varying concentrations of H2O2. Cells were incubated in H2O2-containing PBS on ice for 15 min. Cells were then seeded at a density of 300 cells/plate in triplicate six-well dishes. Growth medium was replenished every 3 days. Surviving cells were stained with 0.05% of crystal violet in 40% of methanol, 8 days after plating for the LigIV WT cells and 14 days for the LigIV−/− cells. Colonies containing ≥50 cells were counted.
DNA synthesis assays
HDF cells in 10-cm culture dishes were placed in low-serum medium to induce growth arrest, exactly as described earlier in the text. The resulting quiescent cells were trypsinized, resuspended in complete medium containing 10% of FBS and re-plated in 24-well plates (50 000 cells per well) in the presence of 0.5 µCi/ml [3H]-methyl thymidine (Perkin Elmer). At the indicated time points after re-plating, the amount of radiolabel incorporated into the trichloroacetic acid-insoluble genomic DNA fraction was determined using scintillation counting as described previously (37).
Comet assay
Relative levels of DNA strand breaks under different conditions were measured by single-cell gel electrophoresis assay (38,39) using a commercially-available CometAssay® kit (Trevigen). For comet assays, siRNA-transfected cells (see ‘RNA Interference’ section) were replated in 6-well dishes at a density of 105 cells per well. H2O2 treatments were performed as described under ‘Genotoxin Treatments’ section. At various times after H2O2 treatment, cells were cryopreserved according to the manufacturer’s instructions. Cells were embedded in agarose and subject to electrophoresis exactly as per the manufacturer’s instructions. Cells were visualized using an Olympus IX61 inverted microscope, and images of cells were acquired as tif files. For each experimental condition, ‘tail moments’ (defined as the product of tail length and the fraction of total DNA in the tail) were determined for 50 nuclei using Image J software with the Comet assay plug-in (original macro from Herbert M. Geller, added by Robert Bagnell). To compare tail moments between experimental groups, we performed analysis of variation (ANOVA) followed by Tukey’s test to correct for experiment-wide error rates between multiple comparisons.
Fluorescence microscopy
Asynchronous HDF were infected with AdYFP–Polη then serum-deprived to induce growth arrest. Quiescent YFP–Polη-expressing cells were trypsinized and re-plated 1:2 into glass-bottom dishes (Matek). Twenty-four hours after re-plating (a time point corresponding to S-phase), cells were treated with H2O2 or UV-irradiated. Two hours after genotoxin treatments, cells were rinsed with cold PBS, then immersed briefly (<5 s) in PBS containing 0.1% of Triton X-100, rinsed again in ice-cold PBS and then fixed in PBS containing 4% of formaldehyde for 5 min. Fixed nuclei were 4′,6-diamidino-2-phenylindole-stained and mounted with Vectashield solution (Vector Laboratories). Slides were imaged and analysed using a Zeiss 710 Laser Scanning Microscope at 63× magnification with digital zoom. The 0.5 µM z-sections of representative cells from different experimental conditions were collected and de-convolved to generate the images presented.
Immunoblotting
Cells were separated into cytosolic and nucleosolic or chromatin fractions using cytoskeleton buffer as described previously (15,37). The resulting samples were normalized for protein content, then separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), transferred to nitrocellulose and analysed by immunoblotting with the indicated antibodies. Sources of primary antibodies were as follows: Ape1/Ref-1 (SC-17774), p-ATM S1981 (SC-47739), β-actin (SC-130656), mouse monoclonal PCNA clone PC10 (SC-56), Rb (SC-50), Rnf8 (SC-134492), GFP (SC-9996) were purchased from Santa Cruz Biotechnology Inc.; Cdt1(A300-786A), NBS1/P95 (A300-187A), Polη (A301-231A), Rad18 (A301-340A) were purchased from Bethyl Laboratories Inc.; p-Chk1 S317 (#2344) and p-Chk2 (#2661) antibodies were purchased from Cell Signaling Technology; ATM (GTX70104), GAPDH (GTX627408) were purchased from Gene Tex; mouse monoclonal γH2AX S139 (05-636) was from EMD Millipore; RPA34/RPA32 (NA19L) was from Calbiochem; P21(556430) was from BD Pharmingen; rat monoclonal antibodies against Cdc45 have been described previously (37).
Velocity sedimentation
HDF were pre-labelled with radioactive nucleotide precursors exactly as described previously (40). Cells were treated with H2O2 under standard experimental conditions described elsewhere in this report. Cell lysis and analysis of distribution of labelled ssDNAs based on their differential density sedimentation profiles after centrifugation through alkaline sucrose gradients were performed exactly as described previously (40).
RESULTS
PCNA mono-ubiquitination in synchronized primary human cells
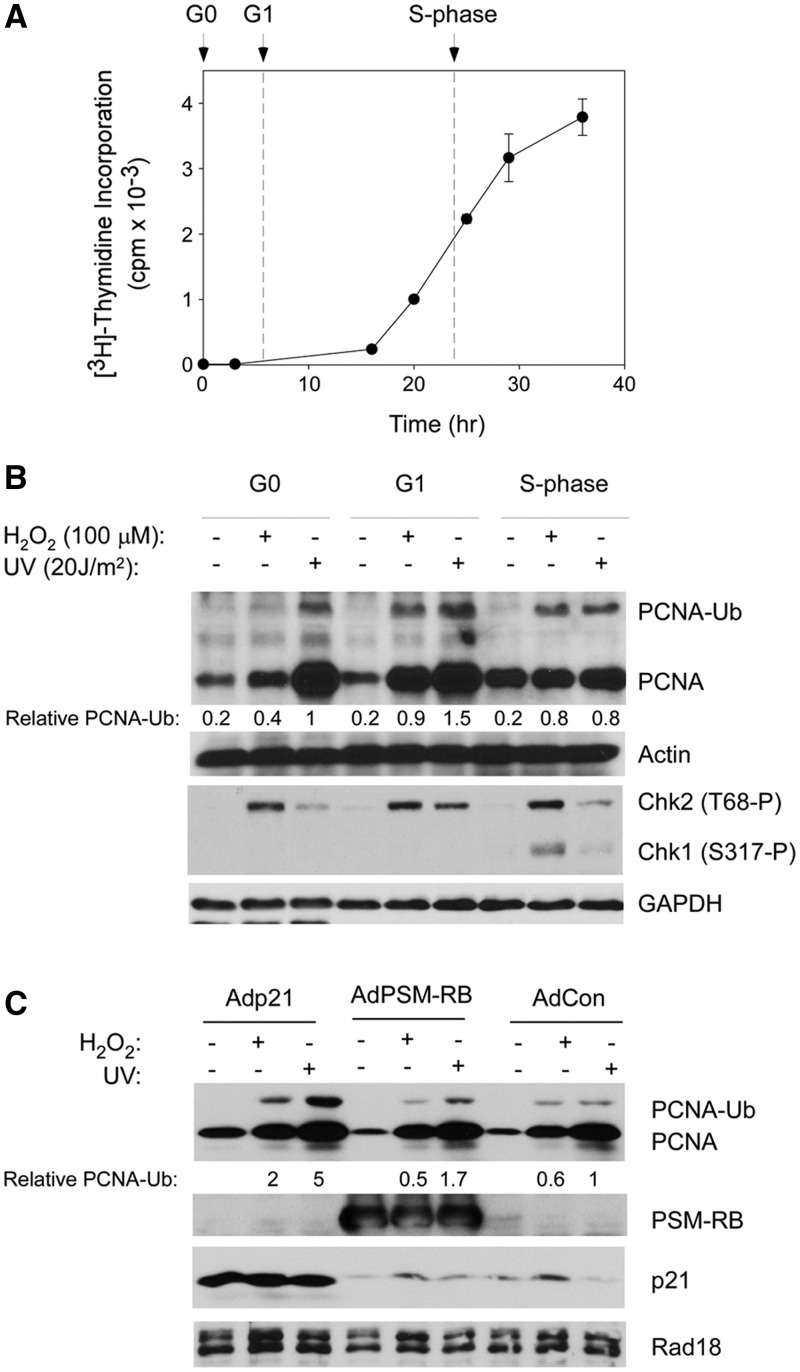
We tested the cell cycle stage-dependency of PCNA mono-ubiquitination in primary untransformed HDF. HDF were synchronized in G0, G1 and S-phase (as described under ‘Materials and Methods’ section). Measurements of [3H]-thymidine incorporation into genomic DNA (Figure 1A) were used to confirm synchrony of the resulting HDF cultures. Cells in G0, mid-G1 and mid-S-phase were treated with UVC (20 J/m2) or H2O2 (100 µM). In normal human fibroblasts, the consensus frequency of CPDs is 1 per ∼60 kb per J/m2 of UVC (41). Therefore, under our experimental conditions, 20 J/m2 of UVC elicits ∼20 CPD per 60 kb. Based on unpublished mass-spectrometry measurements, 100 µM of H2O2 led to approximately two 8-oxodG adducts per 106 dG under our standard experimental conditions. We compared the PCNA mono-ubiquitination and other responses to UVC and H2O2-induced DNA damage in the three cell cycle phases.
Figure 1.
PCNA mono-ubiquitination in different cell cycle phases of synchronized HDF. (A) HDF were synchronized as described under ‘Materials and Methods’ section. At the indicated times after release from quiescence, rates of DNA synthesis were determined using [3H]-thymidine incorporation assays. Each time-point represents a mean of triplicate determinations, and the error bars represent the deviations from the mean. The time points used in all subsequent experiments for cell cycle stage-specific genotoxin treatments in G0, G1 and S-phase cells are indicated by the arrows. (B) G0-, G1- or S-phase-synchronized HDF were treated with 100 µM of H2O2 or 20 J/m2 of UVC (or were left untreated for control samples) as described under ‘Materials and Methods’ sections. Chromatin (PCNA, actin) and soluble (Chk1, Chk2, GAPDH) fractions were isolated and analysed by SDS–PAGE and immunoblotting with antibodies against the indicated proteins. Bands corresponding to mono-ubiquitinated PCNA were quantified by densitometry. The amount of mono-ubiquitinated PCNA in each lane is expressed relative to the amount of mono-ubiquitinated PCNA in UV-treated G0 fibroblasts. In this and subsequent figures, ‘relative PCNA-Ub’ compares total amount of chromatin-associated mono-ubiquitinated PCNA in the cells under different experimental conditions. No attempt was made to normalize the amount of mono-ubiquitinated PCNA-Ub to the level of chromatin-associated unmodified PCNA. (C) Quiescent HDF were infected with adenovirus vectors encoding p21, PSM7-RB or with an ‘empty’ control vector (AdCon). Adenovirus-infected cells were stimulated to enter G1 and subject to genotoxin treatments as indicated. Chromatin extracts were isolated 1 h after genotoxin treatments and analysed by SDS–PAGE and immunoblotting with antibodies against the indicated proteins.
As shown in Figure 1B, UVC and H2O2 treatments induced mono-ubiquitination of chromatin-associated PCNA in G0, G1 and during S-phase. Under our standard experimental conditions, UVC (20 J/m2) induced a slightly higher level of PCNA mono-ubiquitination when compared with H2O2 (100 µM). Both genotoxins typically induced a higher level of PCNA mono-ubiquitination during G1 compared with G0 and S-phase (Figure 1B). Both genotoxic agents also induced chromatin loading of PCNA during G0 and G1 (Figure 1B, lanes 5 and 6), presumably reflecting roles of PCNA in repair synthesis. Genotoxin-induced phosphorylation of cytosolic Chk1 (an S-phase-specific checkpoint kinase) at S317 was only observed during S-phase (Figure 1B), indicating good synchronization of HDF in these experiments. As also expected, activating phosphorylation of cytosolic Chk2 at T68 (an event that is cell cycle-independent) was observed in G0, G1 and S-phase.
It was formally possible that incomplete synchrony and the presence of small numbers of contaminating S-phase cells in the G0- and G1-synchronized populations account for apparent replication fork-independent PCNA mono-ubiquitination. To eliminate this possibility, we examined PCNA mono-ubiquitination in HDF that were G1-arrested because of massive overexpression of the cyclin-dependent kinase inhibitor p21 or a constitutively active retinoblastoma protein (RB) allele harbouring phosphorylation site mutations (termed ‘PSM-RB’). We and others have shown that these cell cycle regulators efficiently induce G1 arrest in HDF and other cells (33,37). As shown in Figure 1C, genotoxin-induced PCNA mono-ubiquitination was actually increased in p21 or PSM-RB-expressing and G1-arrested cells relative to control cultures. Therefore, PCNA mono-ubiquitination in response to H2O2 and UV is not restricted to S-phase and also occurs efficiently in a replication fork-independent manner in primary human cells.
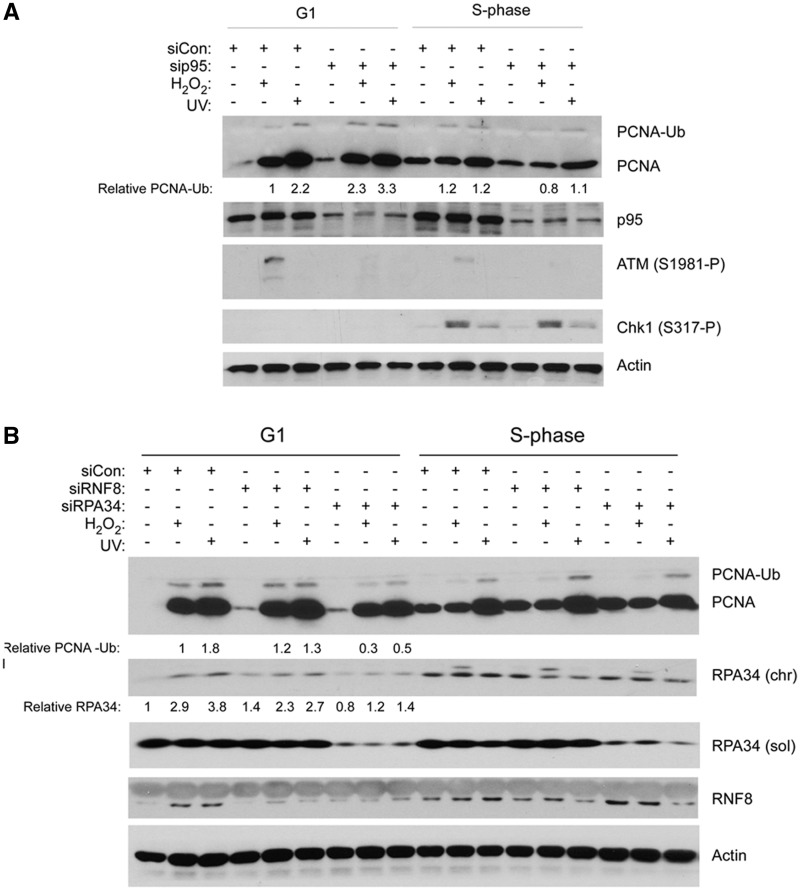
PCNA mono-ubiquitination during G1 is RPA-dependent
Rad18 is the major PCNA-directed E3 ubiquitin ligase. UVC and H2O2-induced PCNA mono-ubiquitination during G0, G1 and S-phases of the cell cycle was attenuated in Rad18-depleted cells (as shown in Figure 2A and several other figures in this study). Three main mechanisms have been proposed for Rad18 recruitment to damaged DNA during S-phase: (i) direct association with RPA-coated ssDNA (8,9); (ii) association with histones that are ubiquitinated by RNF8 at DSB (42); and (iii) direct binding to p95/Nbs1 at sites of UV damage (11). Whether these proposed mechanisms of Rad18 activation show cell cycle or DNA lesion specificity have not been addressed. Therefore, we used siRNA to investigate the participation of p95, Rnf8 and RPA, in H2O2- and UV-induced PCNA ubiquitination in G1-synchronized HDF. As shown in Figure 2A, p95-depletion did not attenuate PCNA mono-ubiquitination during G1, yet fully inhibited genotoxin-induced ATM S1981 phosphorylation. Similarly, Rnf8 depletion did not affect UV- or H2O2-induced PCNA mono-ubiquitination (Figure 2B), yet inhibits Rad18-dependent FA pathway activation by camptothecin in these cells (36). In contrast, partial depletion of RPA34 attenuated PCNA mono-ubiquitination during G1 (Figure 2B). We conclude that RPA-dependent Rad18 activation (but not p95- or Rnf8-based mechanisms) mediates PCNA mono-ubiquitination during G1. We were unable to downregulate chromatin-bound RPA efficiently in S-phase synchronized HDF, most likely explaining why H2O2- and UV-induced PCNA mono-ubiquitination was unaffected by siRPA.
Figure 2.
PCNA mono-ubiquitination during G1 is RPA-dependent and p95-independent. (A) Replicate cultures of HDF were transfected with siRNA targeting p95 or with a scrambled control siRNA. The control and p95-depleted cells were synchronized in G1 and S-phase and treated with genotoxins. Chromatin extracts were isolated 1 h after genotoxin treatments and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. (B) Replicate cultures of HDF were transfected with siRNA targeting RPA34, RNF8 or with a scrambled control siRNA. The resulting cells were synchronized in G1 and S-phase and treated with genotoxins. Chromatin extracts were isolated and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. Bands corresponding to chromatin-bound RPA34 were quantified by densitometry. The amount of RPA34 in each lane is expressed relative to the amount of RPA34 in scrambled control siRNA-treated HDF in G1 that did not receive DNA damage.
A specific role for Rad18 in responses to oxidative DNA damage acquired during G1
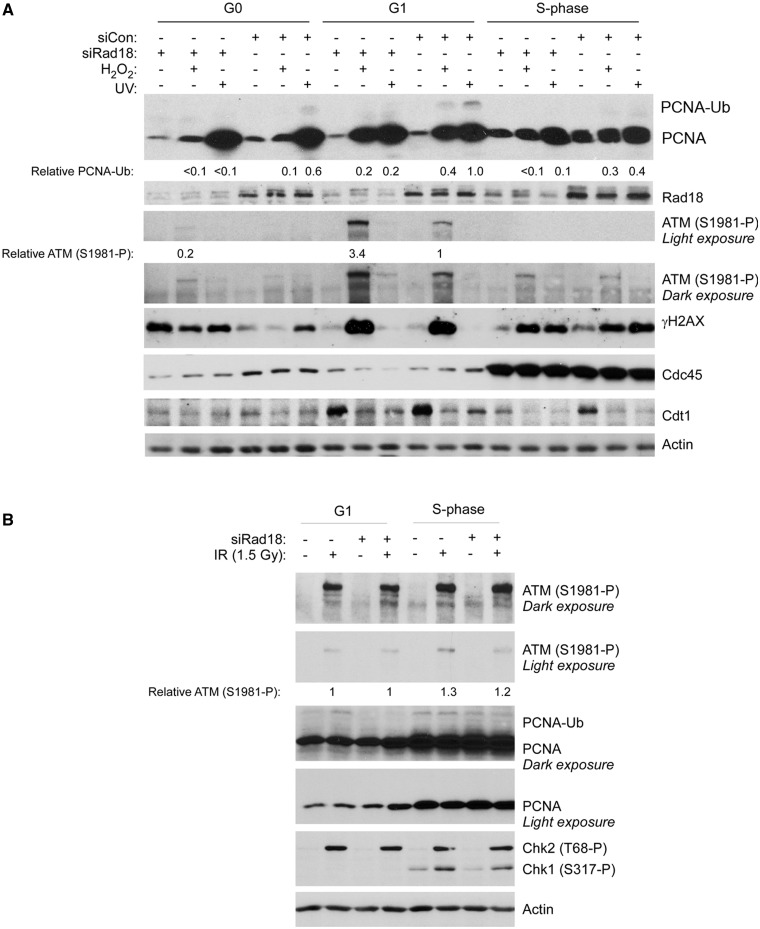
In S-phase cells, loss of Rad18-mediated DNA damage tolerance mechanisms can lead to persistent DNA damage signalling via ATR/Chk1 (15,16). ATR/Chk1 signalling is generally not active outside S-phase. Therefore, to investigate consequences of Rad18-deficiency on DNA damage signalling in G1 cells, we determined the effects of Rad18-depletion on the ATM pathway (whose activation is not restricted to S-phase).
Control and Rad18-depleted HDF were synchronized and subject to genotoxin treatments in G0, G1 and S-phase. We analysed several cell cycle markers to confirm cell cycle synchrony and expected responses to DNA damage: as expected, the licensing factor Cdt1 was present at low levels in chromatin fractions from quiescent cells but was loaded onto chromatin during G1 concomitant with replication licensing (Figure 3A). The initiation factor and S-phase marker Cdc45 was expressed at low levels in G0 and G1 cells but was highly expressed in S-phase, further confirming synchrony of the HDF (Figure 3A). Consistent with previous work (43), Cdt1 was downregulated in response to DNA damage.
Figure 3.
Rad18-deficiency confers H2O2-induced ATM hyper phosphorylation specifically during G1. (A) Replicate cultures of HDF were transfected with siRNA targeting Rad18 or with a scrambled control siRNA. The control and Rad18-depleted cells were synchronized in G0, G1 and S-phase and treated with genotoxins. Cell extracts were isolated 1 h after genotoxin treatments and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. (B) Replicate cultures of HDF were transfected with siRad18, synchronized in G1 and S-phase and irradiated with IR (1.5 Gy). One hour later, cell extracts were isolated and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. Bands corresponding to ATM (S1981-P) in panels A and B were quantified by densitometry. In panel A, the amount of ATM (S1981-P) in each lane is expressed relative to the amount of ATM (S1981-P) in scrambled control siRNA-transfected and H2O2-treated G1 HDF. In panel B, the amount of ATM (S1981-P) in each lane is expressed relative to the amount of ATM (S1981-P) in Rad18 siRNA-transfected and IR-treated G1 HDF.
We observed modest ATM S1981 phosphorylation in response to H2O2, specifically in G1 cells (Figure 3A). Interestingly, Rad18-depletion led to a 3.4-fold increase in H2O2-induced ATM S1981 phosphorylation (Figure 3A). Moreover, the increase in ATM S1981 phosphorylation resulting from Rad18-depletion was evident in G1, but not in G0 or S-phase cells. UV treatment induced a higher level of PCNA mono-ubiquitination when compared with H2O2, yet failed to induce ATM phosphorylation in Rad18-replete or Rad18-depleted cells.
ATM mediates both G1 and S-phase checkpoints in response to DSB. Therefore, the G1-specificity of ATM S1981 phosphorylation after H2O2 treatment (Figure 3A) was unexpected. We considered the formal possibility that failure of H2O2 to activate ATM in S-phase was an idiosyncrasy of primary untransformed HDF. However, as shown in Figure 3B, ionizing radiation (IR) treatment induced ATM phosphorylation efficiently in both G1 and S-phase HDF. We conclude that DSB can potentially activate ATM in a cell cycle-independent manner, but that H2O2 preferentially induces DNA lesions (most likely DSB; see Figure 4) capable of triggering an ATM response during G1-phase. The results of Figure 3 suggest that Rad18-deficiency specifically compromises the repair of H2O2-induced lesions during G1, thereby leading to persistent DNA damage and ATM signalling.
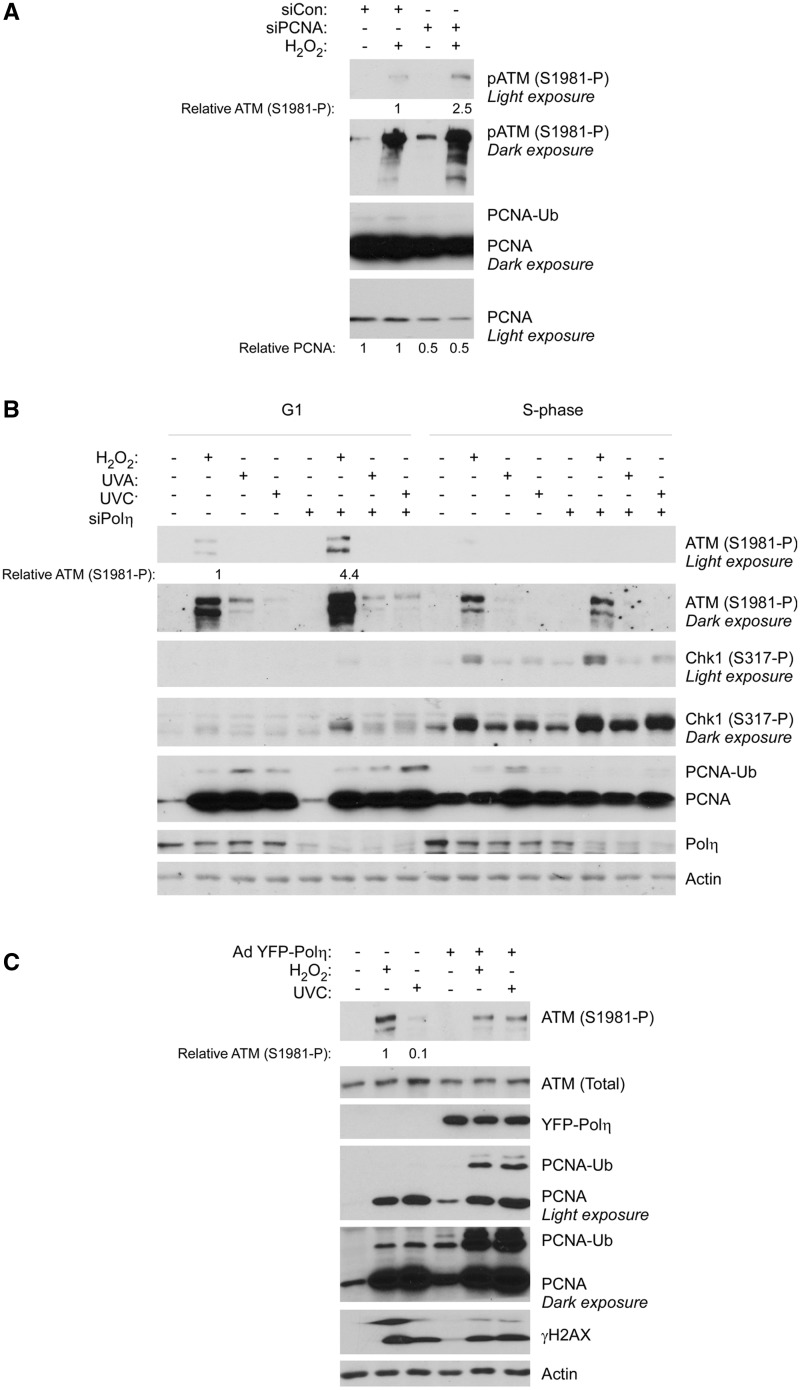
Figure 4.
Depletion of PCNA or Polη recapitulates the H2O2-induced ATM hyper phosphorylation phenotype of Rad18-depleted HDF. (A) Replicate cultures of HDF were transfected with siRNA against PCNA or non-targeting control siRNA, synchronized in G1 and treated with H2O2.One hour later, cell extracts were isolated and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. Bands corresponding to ATM (S1981-P) were quantified by densitometry. The amount of ATM (S1981-P) in each lane is expressed relative to the amount of ATM (S1981-P) in scrambled control siRNA-transfected HDF cells that were treated with H2O2. (B) Replicate cultures of HDF were transfected with siRNA against Polη or non-targeting control siRNA, synchronized in G1 or S-phase and treated with H2O2, UVC or ultraviolet A. After 1 h, cell extracts were isolated and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. (C) Control or YFP–Polη-overexpressing cells were treated with H2O2 or UVC. One hour later, cell extracts were isolated and analysed by SDS–PAGE and immunoblotting with the indicated antibodies. The amount of ATM (S1981-P) in each lane is expressed relative to the amount of ATM (S1981-P) in control H2O2-treated cells.
Because H2O2 induces PCNA ubiquitination and as Polη can replicate DNA templates containing oxidative DNA lesions, such as 8-oxodG and AP sites (28,30,44,45), we hypothesized that Polη-mediated TLS was the likely mechanism by which Rad18 suppressed ATM activation in H2O2-treated cells. Consistent with a role of TLS in repressing ATM, partial downregulation of PCNA using siRNA also exacerbated ATM S1981 phosphorylation after H2O2 treatment (Figure 4A). PCNA is involved in multiple DNA repair pathways including, but not limited to, TLS. To specifically test a role of TLS in preventing H2O2-induced ATM activation, we used siRNA to deplete Polη. As shown in Figure 4B, Polη knockdown fully recapitulated the phenotype of Rad18-depleted cells, increasing ATM S1981 phosphorylation in a manner that was both lesion-specific (H2O2 not UV) and cell cycle-specific (G1 not S-phase). Conversely, we found that ectopic overexpression of YFP–Polη led to repression of ATM S1981 phosphorylation after H2O2 (but not UVC) treatment (Figure 4C). Overexpressed Polκ and Polι (TLS polymerases that do not bypass oxidative DNA lesions efficiently) did not repress ATM S1981 phosphorylation in H2O2-treated cells (data not shown). ATM S1981 phosphorylation in Rad18- and Polη-depleted cells was rapid and transient: in TLS-deficient cells, maximal ATM S1981 phosphorylation levels were attained 30–60 min after H2O2 exposure (Supplementary Figure S1), and ATM S1981 phosphorylation declined to basal levels 3–4 h post-H2O2 treatment (not shown).
As shown in Figure 4B, PCNA mono-ubiquitination was reduced in Polη-depleted cells, particularly during S-phase. Conversely, PCNA mono-ubiquitination was enhanced in Polη overexpressing cells (Figure 4C). We have previously shown that Rad18 and Polη exist as a complex whose formation is enhanced by the S-phase kinase Cdc7 (35). In work that will be described elsewhere, we have found that Polη plays a scaffold role that facilitates recruitment of Rad18 to PCNA. Remarkably, we have also found that endogenous Rad18 is present in excess of endogenous Polη (by ∼70-fold). Thus, Polη levels are limiting for recruitment of Rad18 to PCNA and for Rad18-mediated PCNA mono-ubiquitination (particularly during DNA replication when the S-phase kinase Cdc7 promotes Rad18–Polη complex formation). Consequently, in the Polη depletion experiments, PCNA mono-ubiquitination is reduced, whereas Polη overexpression promotes PCNA mono-ubiquitination.
Taken together, the results of Figure 4 indicate that Rad18/Polη-mediated TLS plays an important role in preventing accumulation of ATM-activating lesions during G1 specifically in response to oxidative DNA damage.
Rad18 and Polη prevent accumulation of H2O2-induced DSB during G1
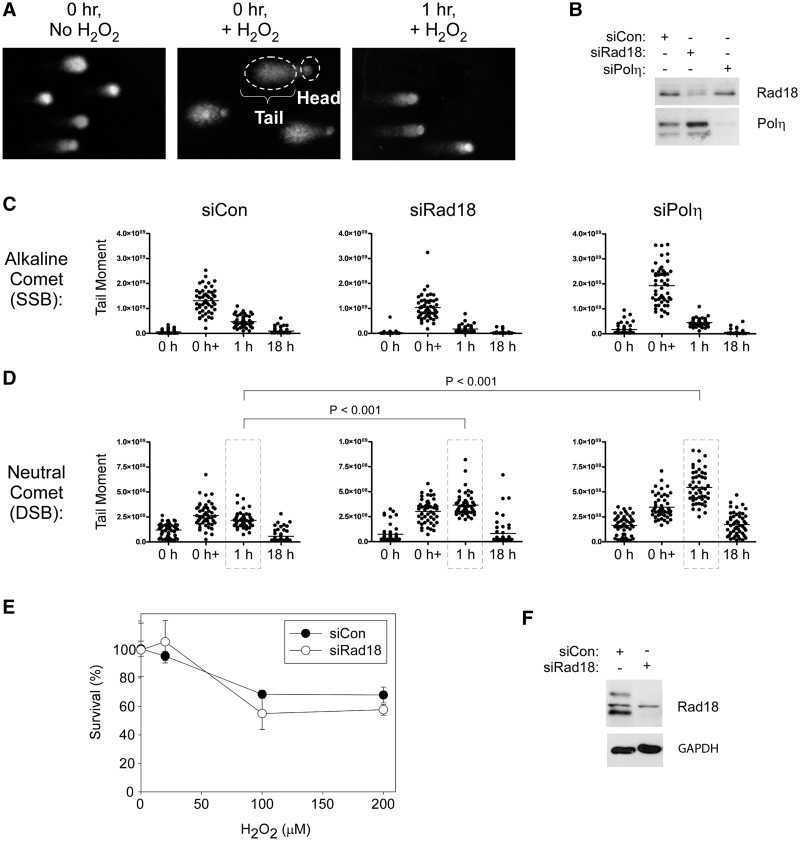
The ssDNA-binding protein RPA was recruited to chromatin in response to genotoxin treatments and RPA-depletion attenuated PCNA mono-ubiquitination during G1 (Figure 2). Moreover, the H2O2 concentrations used in our experiments are reported to induce ssDNA (46,47). Most likely, therefore, H2O2-induced TLS pathway activation during G1 is elicited by ssDNA. However, ATM activation generally correlates with acquisition of DSB not SSB (48). Therefore, we hypothesized that impaired SSB processing and repair in TLS-deficient cells led to DSB formation and ATM activation. To test this hypothesis, we performed alkaline and neutral ‘comet’ assays to test effects of TLS-deficiency on accumulation of SSB and DSB, respectively (Figure 5A–D). Immediately after H2O2 treatment, SSB levels were elevated by 6.9-, 9.1- and 8.6-fold in siCon, siRad18 and siPolη-transfected cultures (Figure 5C). For each time-point, we performed ANOVA between groups followed by a Tukey multiple comparison of means test. One hour after H2O2 treatment, SSB returned to basal levels and was not significantly different between siCon, siRad18 and siPolη-transfected cultures (P > 0.05 for siCon versus siRad18, siCon versus Polη and siRad18 versus siPolη). We conclude that elevated levels of SSB do not account for the increased ATM S1981 phosphorylation in TLS-deficient cells relative to control cultures.
Figure 5.
Effect of TLS-deficiency on DSB and SSB formation and cell survival after H2O2-treatment. (A) Images from a representative alkaline comet assay showing kinetics of SSB formation and repair in H2O2-treated cells. (B) HDF were transfected with siCon, siRad18 or siPolη oligonucleotides, then synchronized and replated in replicate six-well dishes. Lysates from one plate of each replicate were analysed by SDS–PAGE and immunoblotting to validate Rad18 and Polη knockdown. Replicate cultures of siCon-, siRad18- and siPolη-transfected cells in G1 phase were treated with H2O2 (or left untreated for control samples), then harvested immediately or 1 and 18 h after H2O2 treatment for comet assays. Relative levels of SSB and DSB are shown in panels (C) and (D), respectively. (E) HDF were electroporated with siCon or siRad18 oligonucleotides, synchronized in G1 and treated with the indicated concentrations of H2O2. Clonogenic survival was determined by colony formation assays, as described under ‘Materials and Methods’ section. (F) Replicate plates of Rad18- and Polη-depleted G1-phase cells used for the survival assays shown in panel (E) were lysed and analysed by SDS–PAGE and immunoblotting to validate efficiency of Rad18 and Polη knockdowns.
We also observed an increase in DSB levels immediately after H2O2 treatment in siCon, siRad18 and siPolη-transfected cultures (Figure 5D). Interestingly, however, in Rad18-depleted and Polη-depleted cells, DSB levels continued to rise 1 h after H2O2 treatment. In the experiment shown in Figure 5D, 1 h after H2O2 treatment, DSB levels in siRad18 and siPolη-transfected cells were ∼1.68- and 2.5-fold higher than in siCon-transfected cultures (the data points corresponding to these conditions are enclosed by the dashed lines in Figure 5D). To determine the statistical significance of the differences in DSB levels between groups, we performed ANOVA between groups followed by a Tukey multiple comparison of means test. In ANOVA, P < 0.0001 is significant. Results of the Tukey test were as follows: siCon versus siRad18, P < 0.001; siCon versus siPolη, P < 0.001. We conclude that Rad18- or Polη-deficiency leads to increased formation of DSB in H2O2-treated G1 cells.
We hypothesized that the increased formation of DSB in TLS-deficient G1 cells would lead to reduced damage tolerance. Therefore, we performed clonogenic survival assays to compare the H2O2 sensitivities of control and Rad18-deficient G1 cells. Unexpectedly, we did not observe statistically significant differences in survival of control and Rad18-depleted cultures that were treated with H2O2 in G1 (Figure 5E), although in the same experiment, Rad18-deficient cells were more sensitive to H2O2 treatments received during S-phase (see Figure 7). The H2O2-tolerance of Rad18-depleted cells suggested the existence of redundant repair mechanisms for H2O2-induced DNA damage.
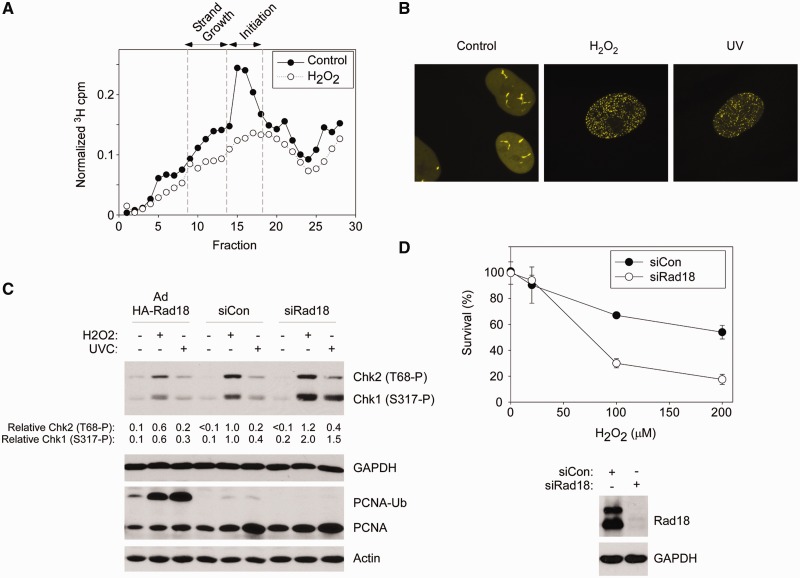
Figure 7.
S-phase-specific roles of Rad18 in tolerance of oxidative DNA damage. (A) Velocity sedimentation profiles showing size distribution of labelled ssDNAs from control and H2O2-treated HDF. (B) YFP–Polη-expressing HDF were synchronized in S-phase and then treated with H2O2 or UVC. Representative nuclei showing the subcellular distribution of YFP–Polη under different conditions are presented. (C) Control (siCon), Rad18-depleted (siRad18) and HA-Rad18-overexpressing HDF were synchronized in S-phase and treated with H2O2 or UVC (or left untreated for control samples). One hour later, cells were lysed, and extracts were analysed by SDS–PAGE and immunoblotting with the indicated antibodies. Bands corresponding to Chk1 (S317-P) and Chk2 (T68-P) were quantified by densitometry. The amount of Chk1 (S317-P) and Chk2 (T68-P) in each lane is expressed relative to the amount of Chk1 (S317-P) and Chk2 (T68-P) in scrambled control siRNA-transfected and H2O2-treated cells. (D) HDF were electroporated with siCon or siRad18 oligonucleotides, synchronized in S-phase and treated with the indicated concentrations of H2O2. Clonogenic survival of H2O2-treated cells was determined by colony formation assays, as described under ‘Materials and Methods’ section. Replicate plates of Rad18-depleted S-phase cells used for the survival assays were lysed and analysed by SDS–PAGE and immunoblotting to validate efficiency of Rad18 and Polη knockdowns (upper panel). The experiments presented here were performed with the same synchronized cultures used for the experiment described in Figure 4E (which showed no sensitivity of Rad18-depleted cells to H2O2 treatments during G1).
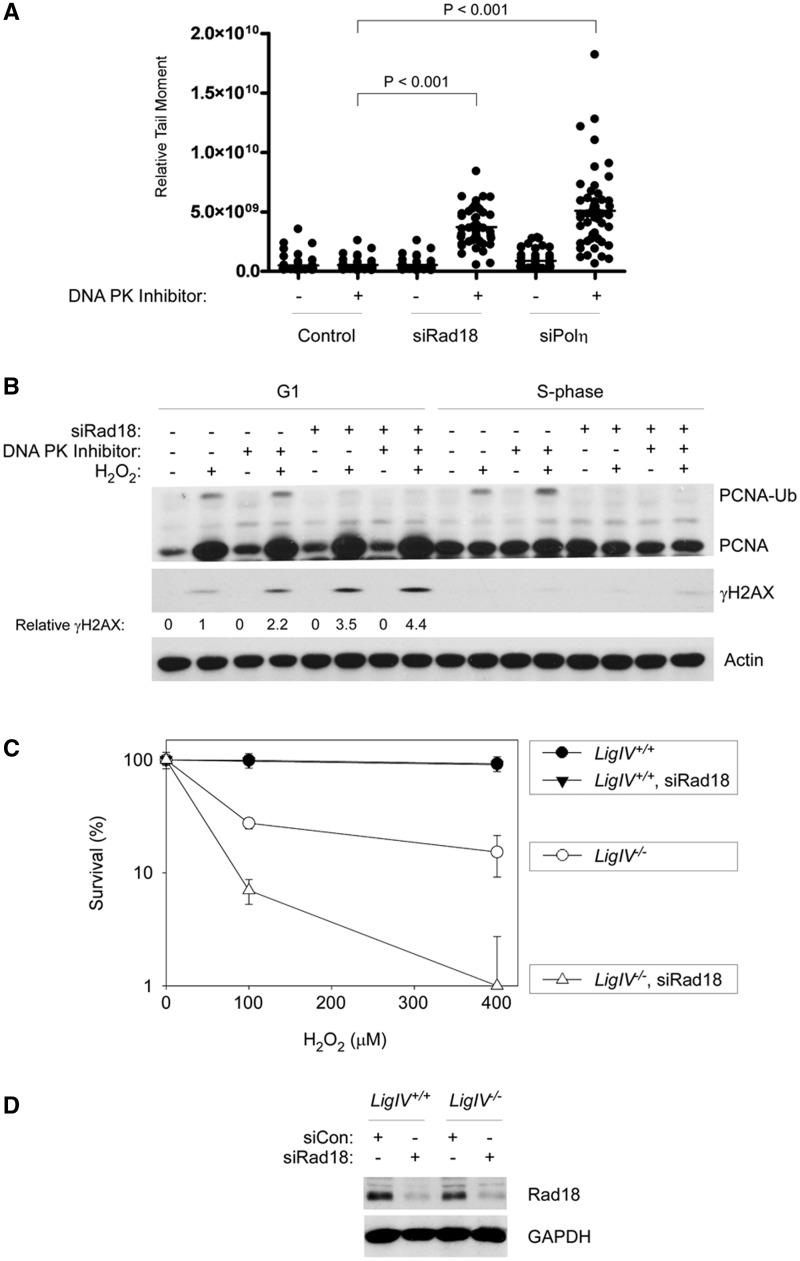
DNA DSB is repaired via a high-capacity non-homologous end joining (NHEJ) pathway throughout the cell cycle. To test whether NHEJ constitutes a back-up mechanism for repair of H2O2-induced DSB, we used NU7441, also termed ‘KU-57788’ (49), to inhibit DNA-PK in TLS-deficient cells during G1. In control cultures (that did not receive NU7441), H2O2-induced DSB levels essentially returned to baseline 4 h post-H2O2 treatment in control as well as in Rad18- and Polη-depleted cells and were indistinguishable between the three groups (Figure 6A). Interestingly, however, in NU7441-treated cells 4 h post-H2O2, levels of DSB were 2.8- and 3.9-fold higher in Rad18 and Polη-depleted cells relative to siCon-transfected cells. In ANOVA, P < 0.0001 indicates significant differences between groups. Results of the Tukey’s multiple comparison test were: siCon versus siRad18, P < 0.001 (indicating significant increase in DSB after Rad18-depletion); siCon versus siPolη, P < 0.001 (indicating significant increase in DSB after Polη-depletion); siRad18 versus siPolη, P < 0.01 (indicating an insignificant difference in DSB levels between Polη and Rad18-depleted cells). An effect of DNA-PK inhibition on DSB repair was also indicated by immunoblot analysis of γH2AX. As shown in Figure 6B, the induction of γH2AX by H2O2 in Rad18-depleted G1 cells was further increased by NU7441.
Figure 6.
In TLS-deficient cells, NHEJ provides a back-up pathway for preventing H2O2-induced DSB. (A) Replicate cultures of siCon-, siRad18- and siPolη-transfected cells in G1 phase were treated with 10 µM of NU7441 to inhibit DNA-PK or were left untreated for control samples. The NU7441-treated cells were then H2O2-treated (or left untreated) 4 h before harvest for neutral comet assays. (B) HDF were transfected with siCon siRad18 oligonucleotides, then synchronized in G1 and treated with NU7441 and H2O2 as described in (A). One hour after H2O2 treatment, cells were harvested, and chromatin fractions were analysed by SDS–PAGE and immunoblotting with the indicated antibodies. Bands corresponding to γH2AX were quantified by densitometry. The amount of γH2AX in each lane is expressed relative to the amount of γH2AX in scrambled control siRNA-transfected cells that were treated with H2O2 in the absence of NU7441. (C) LigIV+/+ and LigIV−/− HCT116 cells were transfected with siRad18 or siCon oligonucleotides, then synchronized and treated with the indicated doses of H2O2. Survival of control and H2O2-treated cells was measured by colony formation assays. Note that the data points for LigIV+/+ and siRad18-transfected LigIV+/+ cells overlap completely. (D) Immunoblot analysis confirming reduced Rad18 expression in siRad18-transfected LigIV+/+ and LigIV−/− HCT116 cells.
The NU7441 treatments used in these experiments only partially inhibited NHEJ, as indicated by the eventual return of DSB levels to basal (not shown). NU7441 concentrations in excess of those used in these experiments were very toxic to HDF, thereby precluding survival assays. As an alternative approach to testing the contribution of NHEJ to survival of Rad18-deficient cells after H2O2 treatment, we depleted Rad18 in DNA LigIV−/− HCT116 cells (32) and in an isogenic parental DNA LigIV+/+ control cell line. Non-replicating HCT116 cultures were treated with 0–400 µM H2O2, and cell survival was determined by colony formation assays. As shown in Figure 5C, Rad18-depleted LigIV+/+ cells showed normal H2O2-tolerance, consistent with the results of Figure 4 showing no change in H2O2-sensitivity in Rad18-depleted HDF. Interestingly, however, Rad18 depletion exacerbated the H2O2 sensitivity of LigIV−/− HCT116 cells (Figure 6C). Taken together, the results of Figure 6 indicate that the DSB resulting from H2O2 treatment of TLS-defective cells are repaired via NHEJ (the major, if not sole, mechanism available to repair DSB in G1 cells). We conclude that NHEJ provides a back-up mechanism for tolerance of H2O2-induced DNA damage when TLS is impaired.
Rad18 confers tolerance of H2O2-induced DNA lesions specifically in S-phase
The results of Figure 5E (showing no H2O2-sensitivity of TLS-deficient cells during G1) seemingly contradict a report by Zlatanou et al. (25) who inferred a role for Polη in tolerance of H2O2-induced lesions during G1. However, those workers performed their viability assays in asynchronous cultures. Under our standard experimental conditions using G1-synchronized HDF, Polη depletion did not confer H2O2-sensitivity (Supplementary Figure S2). We hypothesized, therefore, that the reported H2O2-sensitivity of TLS-deficient cells (25) was primarily because of defective DNA damage tolerance of S-phase populations. Accordingly, we determined the contribution of Rad18 to H2O2-tolerance during S-phase.
First, we used velocity sedimentation of nascent DNA fragments (40) to define the effects of H2O2 treatment on DNA replication dynamics in HDF. As shown by the sedimentation profiles in Figure 7A, H2O2 treatment inhibited elongation (i.e. blocked replication fork progression) and initiation phases of DNA synthesis. The role of Polη in responding to replication blocks in cells harbouring UV-damaged genomes and its contribution to UV-tolerance is well-established (6,13). Similar to UVC treatment, H2O2 induced redistribution of Polη to nuclear foci (representing sites of replication fork stalling), potentially consistent with a role for Polη in tolerance of H2O2-induced damage during S-phase (Figure 7B).
In TLS-deficient cells, genotoxin-induced Chk1 signalling is exacerbated (15,16) because of the persistence of unfilled post-replicative single-stranded gaps in the newly replicated dsDNA (18,50). In Rad18-depleted S-phase cells, H2O2-induced Chk1 phosphorylation was increased relative to control (Rad18-replete) cultures, suggesting a role for Rad18 in PRR at forks encountering ROS-induced DNA damage (Figure 7C). Conversely, Rad18 overexpression led to increased PCNA mono-ubiquitination and repressed Chk1 activation in response to both H2O2 and UVC (Figure 7C). These results suggest that Rad18-mediated TLS enables post-replicative gap filling in response to H2O2-induced replication fork stalling.
The results of Figure 7A–C suggested a possible role for Rad18 in tolerance of H2O2-induced DNA damage acquired during S-phase. Therefore, we determined the effect of Rad18-depletion on clonogenic survival of H2O2-treated S-phase cells. As shown in Figure 7D, siRad18-transfected cells were sensitive to H2O2 when compared with Rad18-expressing (siCon-transfected) cultures. The results shown in Figure 7D were from a synchronization experiment, in which we also measured clonogenic survival of replicate cultures (of siCon- and siRad18-transfected HDF) that were treated with H2O2 in G1-phase (shown in Figure 5E). Taken together, the results of Figures 5E and 7D demonstrate that the Rad18-deficiency in primary human cells specifically results in H2O2-sensitivity during S-phase (and not in G1).
DISCUSSION
The main findings of this study are (i) Rad18 pathway activation occurs during G0, G1 and S-phase in response to both UV- and H2O2-induced DNA lesions in primary human cells; (ii) Rad18 prevents acquisition of DSB specifically after acquisition of oxidative DNA damage during G1; (iii) the role of Rad18 in preventing H2O2-induced DSB during G1 is non-essential owing to backup NHEJ-mediated DSB repair; (iv) in contrast with its redundant role in G1, Rad18 plays an essential role in facilitating completion of DNA replication and conferring cell survival after oxidative injury in S-phase. We conclude that Rad18 plays distinct roles in protecting the genome from oxidative DNA damage in different cell cycle stages.
The synergistic effect of Rad18- and NHEJ-ablation on H2O2-sensitivity in G1 is unexpected. In previous studies (using asynchronous DT40 cells), loss of NHEJ rescued the DNA damage-sensitivity of Rad18-deficient cells (51). The restoration of normal damage tolerance in Rad18−/− DT40 cells after LigIV-deletion was attributed to loss of NHEJ-mediated toxic end-joining events and redirecting of DNA damage processing to error-free homologous recombination (HR) during S-phase. Thus, our analysis of G1-synchronized cells has revealed a new relationship between the Rad18 pathway and NHEJ.
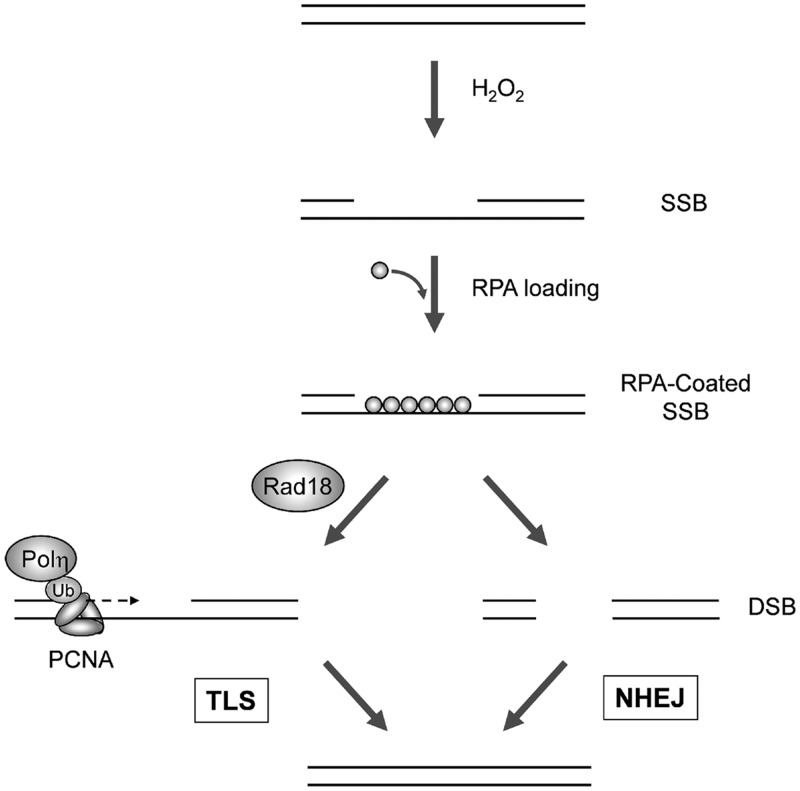
As described in Figure 8, our work indicates that H2O2-induced lesions acquired during G1 lead to Rad18-mediated PCNA mono-ubiquitination and Polη recruitment, facilitating repair synthesis at single-stranded gaps. In the absence of Rad18/Polη, SSB repair synthesis is compromised, leading to breaks in both strands. The resulting DSB activate ATM but are repaired via NHEJ conferring DNA damage tolerance.
Figure 8.
Hypothetical model for functional redundancy of Rad18 and NHEJ in responding to oxidative DNA damage during G1. During G1, H2O2-treatment generates SSB. The exposed ssDNA is RPA-coated leading to Rad18 recruitment and PCNA mono-ubiquitinated by Rad18 at sites of repair synthesis, thereby promoting Polη recruitment. Polη facilitates gap-filling (presumably at templates containing bi-stranded and clustered damage), thereby conferring DNA damage tolerance via TLS (left). In the absence of Rad18, inefficient gap-filling by the TLS pathway leads to accumulation of DSB, which is repaired by NHEJ, also conferring DNA damage tolerance.
The G1-specificity of ATM activation by H2O2 in our experiments is also unexpected, as ATM is well known to mediate DSB-induced S-phase checkpoints (52,53). It is possible that replication-coupled mechanisms for processing of oxidative lesions lead to S-phase-specific DNA structures that fail to activate ATM or are rapidly channelled through the HR pathway. It will be interesting to determine whether preventing HR during S-phase recapitulates the persistence of DSB and ATM hyper-phosphorylation phenotype we observed in H2O2-treated G1 cells.
Our results complement and extend recent reports from Lehmann (24) and Kannouche (25) groups: Lehmann and colleagues (24) demonstrated that nucleotide excision repair (NER) of UV-induced DNA damage in quiescent growth-arrested cells is Polκ-dependent. Kannouche and colleagues (25) described TLS pathway activation in response to H2O2 in both growth-arrested (quiescent) and exponentially growing cells.
Our findings demonstrating DNA synthesis-independent PCNA mono-ubiquitination and Polη recruitment to chromatin are fully consistent with both studies. However, in contrast with the Lehmann and Kannouche groups (who compared G0 and asynchronous cells), we systematically defined responses to both H2O2 and UV in three cell cycle stages: G0, G1, and S-phase. Surprisingly, we show that defective TLS (achieved by Rad18 or Polη depletion) elicits a robust ATM response only outside of S-phase. Moreover, we show that the ATM pathway activation of TLS-defective cells occurs specifically in response to H2O2 (but not UV). Interestingly, however, the increased DSB formation in Rad18-deficient HDF after H2O2 treatment during G1 is not associated with increased lethality because of the high-DSB repair capacity in those cells. On the other hand, we show that Rad18-deficient cells are sensitive to H2O2 treatment during S-phase. Thus, the reported H2O2-sensitivity of asynchronous Polη-deficient cells (25) most likely resulted from post-replication repair defects during S-phase.
The RPA-dependence of PCNA mono-ubiquitination during G1 is fully consistent with the TLS activation mechanism proposed by Ulrich and colleagues (9) involving Rad18 recruitment to RPA-coated ssDNA. It is important, therefore, to consider mechanisms by which RPA-coated DNA is generated outside of S-phase. NER incision events lead to ssDNA patches of ∼30 bp. The minimal length of ssDNA that can bind RPA interaction site is eight nucleotides, and RPA has high affinity for oligonucleotides of 20–30 residues (54). Therefore, NER of bulky lesions has the potential to generate the RPA-coated ssDNA intermediates required for Rad18 recruitment and PCNA mono-ubiquitination. Indeed, Ogi et al. (24) demonstrated that NER-deficient xeroderma pigmentosum cells were defective for recruitment of TLS polymerases to chromatin in non-cycling cultures.
In contrast with the bulky UV lesions (that are processed by NER) H2O2 induces many forms of base damage, including 8-oxodG. Damaged bases, such as 8-oxodG, are repaired by short-patch and long-patch BER pathways that generate ssDNAs of 1 and 2–12 nucleotides, respectively (55,56). Therefore, BER alone is unlikely to generate of sufficient ssDNA to trigger an RPA-mediated TLS response. In our experimental system, BER-deficiency resulting from knockdown of Ape1, or pharmacological inhibition of PARP did not affect PCNA mono-ubiquitination during G1 (Supplementary Figure S3). The elegant study by Zlatanou et al. (25) also found that BER is dispensable for PCNA mono-ubiquitination during G0. Interestingly, however, those workers observed that the mismatch repair (MMR) proteins MSH2 and MSH6 are specifically required for PCNA mono-ubiquitination after H2O2 treatment. Therefore, Kannouche and colleagues suggest that clustered DNA damage resulting from H2O2-induced oxidative lesions is recognized by MSH2–MSH6, promoting loading of Exo1 (or other exonucleases) that generate sufficient ssDNA for Rad18 recruitment, PCNA mono-ubiquitination and ensuing repair synthesis (56). The MMR-mediated mechanism proposed by Kannouche and co-workers likely contributes to TLS-pathway activation in our experimental system.
In our study, H2O2 induced substantial (∼50–70-fold) increases in loading of PCNA onto chromatin after H2O2 treatments during G0 and G1 (but not during S-phase when PCNA is already largely chromatin bound). It will be interesting to identity of the clamp loader(s) involved in responses to H2O2-induced damage. MSH2 is reported to interact with PCNA (57). It is possible that MMR proteins contribute to both PCNA loading (via direct interactions) and PCNA mono-ubiquitination (by generating ssDNA gaps necessary for RPA loading and Rad18 recruitment).
As Rad18 and TLS polymerases facilitate repair of both UV- and H2O2-induced discontinuities in dsDNA (24,25), it is interesting that the ATM ‘hyper-phosphorylation’ phenotype of TLS-deficient G1 cells is highly specific to H2O2-induced lesions.
Paull and colleagues (58,59) have demonstrated that ATM may be activated directly by oxidative stress. However, because depletion of Rad18 or of Polη promotes ATM S1981 phosphorylation by H2O2, we consider it unlikely that the increased ATM phosphorylation associated with TLS-deficiency is related to direct effects of reactive oxygen species on ATM activity. Moreover, Walker and colleagues (60) have shown that the redox-dependent activation of ATM occurs in the cytosol. All the ATM immunoblots presented here were from chromatin fractions. We have never detected S1981 phosphorylation of cytosolic ATM, also arguing against a mechanism involving direct activation of ATM by H2O2 under our experimental conditions.
Another possible explanation for the H2O2-specificity of ATM activation in TLS-deficient cells is that Polη is redundant with other DNA polymerases for repair synthesis at UV-induced discontinuities but not at H2O2-induced SSB. The differential ATM activation in response to H2O2 and UV could also be explained by the nature of the single-stranded breaks induced by the two agents. For example, oxidative damage may be clustered in H2O2-treated cells, whereas it is unlikely that UV fluences used in our experiments result in bi-stranded or tandem clusters of CPD. It is also possible that the putative MMR-dependent exonuclease(s) activated during processing of oxidative lesions (25) generates longer patches of RPA–ssDNA (relative to the ∼30 bp patches generated during NER of UV lesions) that are more prone to breakage if left unrepaired.
Rad18 is implicated in multiple DNA repair pathways, yet Rad18-deficient mice display only mild phenotypes (61). Therefore, redundant mechanisms likely exist to maintain the genome and confer DNA damage tolerance in the absence of Rad18. We show here that H2O2-induced DSB generated in Rad18/Polη-deficient cells can be repaired by NHEJ. Therefore, it is possible that combined defects in Rad18 and other DNA repair genes (including DNA-PK or other NHEJ genes) will reveal more profound DNA damage tolerance phenotypes than are evident in Rad18 mutant mice. The finding that the Rad18 pathway is activated by H2O2 in a DNA replication-independent manner suggests a potential role for Rad18 in tolerance of oxidative stress-induced DNA damage in non-proliferating cells, such as neurons and cardiomyocytes, that can experience considerable ROS exposure in vivo. Further experiments are underway to test roles of Rad18 in tolerance of ROS-induced DNA lesions in non-dividing cells in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Funding for open access charge: National Institutes of Health [ES09558 and ES016280 to C.V.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Dr. Robert Bangnell (Microscopy Services Laboratory, UNC-CH, School Of Medicine) assisted with microscopy and comet assays. We thank Dr. Dale Ramsden and Dr. Satoshi Tateishi for helpful discussions.
REFERENCES
- 1.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 2.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 4.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 6.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 7.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji Y, Watanabe K, Araki K, Shinohara M, Yamagata Y, Tsurimoto T, Hanaoka F, Yamamura K, Yamaizumi M, Tateishi S. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–354. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 9.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol. Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagihara H, Kobayashi J, Tateishi S, Kato A, Matsuura S, Tauchi H, Yamada K, Takezawa J, Sugasawa K, Masutani C, et al. NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol eta-dependent translesion DNA synthesis. Mol. Cell. 43:788–797. doi: 10.1016/j.molcel.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima S, Lan L, Kanno S, Usami N, Kobayashi K, Mori M, Shiomi T, Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J. Biol. Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- 13.Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 16.Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. Rad18 Regulates DNA Polymerase {kappa} and Is Required for Recovery from S-Phase Checkpoint-Mediated Arrest. Mol. Cell. Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan S, Michael WM. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 2009;8:2877–2884. doi: 10.4161/cc.8.18.9485. [DOI] [PubMed] [Google Scholar]
- 18.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 20.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 22.Soria G, Belluscio L, van Cappellen WA, Kanaar R, Essers J, Gottifredi V. DNA damage induced Pol eta recruitment takes place independently of the cell cycle phase. Cell Cycle. 2009;8:3340–3348. doi: 10.4161/cc.8.20.9836. [DOI] [PubMed] [Google Scholar]
- 23.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 24.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, Nakajima S, Yasui A, Ishchenko AA, Kannouche PL. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell. 43:649–662. doi: 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Diamant N, Hendel A, Vered I, Carell T, Reissner T, de Wind N, Geacinov N, Livneh Z. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 40:170–180. doi: 10.1093/nar/gkr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl Acad. Sci. USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase eta. J. Biol. Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 30.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 31.Guo N, Faller DV, Vaziri C. Carcinogen-induced S-phase arrest is Chk1 mediated and caffeine sensitive. Cell Growth Differ. 2002;13:77–86. [PubMed] [Google Scholar]
- 32.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P, Slater DM, Lenburg M, Nevis K, Cook JG, Vaziri C. Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells. Cell Cycle. 2009;8:125–136. doi: 10.4161/cc.8.1.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barkley LR, Palle K, Durando M, Day TA, Gurkar A, Kakusho N, Li J, Masai H, Vaziri C. c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Poleta recruitment to stalled replication forks. Mol. Biol. Cell. 2012;23:1943–1954. doi: 10.1091/mbc.E11-10-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Day TA, Palle K, Barkley LR, Kakusho N, Zou Y, Tateishi S, Verreault A, Masai H, Vaziri C. Phosphorylated Rad18 directs DNA polymerase eta to sites of stalled replication. J. Cell Biol. 2010;191:953–966. doi: 10.1083/jcb.201006043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palle K, Vaziri C. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle. 2011;10:1625–1638. doi: 10.4161/cc.10.10.15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J. Biol. Chem. 2006;281:30631–30644. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 38.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 39.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the "comet" assay. Radiat. Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 40.Day TA, Sproul C, Cordeiro-Stone M, Vaziri C. Analyzing DNA replication dynamics of genotoxin-treated cells using velocity sedimentation. Methods Mol. Biol. 2011;782:159–170. doi: 10.1007/978-1-61779-273-1_11. [DOI] [PubMed] [Google Scholar]
- 41.Boyer JC, Kaufmann WK, Brylawski BP, Cordeiro-Stone M. Defective postreplication repair in xeroderma pigmentosum variant fibroblasts. Cancer Res. 1990;50:2593–2598. [PubMed] [Google Scholar]
- 42.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell. Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- 44.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 45.McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic Acids Res. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradley MO, Kohn KW. X-ray induced DNA double strand break production and repair in mammalian cells as measured by neutral filter elution. Nucleic Acids Res. 1979;7:793–804. doi: 10.1093/nar/7.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismail IH, Nystrom S, Nygren J, Hammarsten O. Activation of ataxia telangiectasia mutated by DNA strand break-inducing agents correlates closely with the number of DNA double strand breaks. J. Biol. Chem. 2005;280:4649–4655. doi: 10.1074/jbc.M411588200. [DOI] [PubMed] [Google Scholar]
- 49.Hardcastle IR, Cockcroft X, Curtin NJ, El-Murr MD, Leahy JJ, Stockley M, Golding BT, Rigoreau L, Richardson C, Smith GC, et al. Discovery of potent chromen-4-one inhibitors of the DNA-dependent protein kinase (DNA-PK) using a small-molecule library approach. J. Med. Chem. 2005;48:7829–7846. doi: 10.1021/jm050444b. [DOI] [PubMed] [Google Scholar]
- 50.Callegari AJ, Clark E, Pneuman A, Kelly TJ. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc. Natl Acad. Sci. USA. 2010;107:8219–8224. doi: 10.1073/pnas.1003449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 53.Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 2002;30:290–294. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 54.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 55.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 56.Caldecott KW. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 57.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 58.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 59.Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle. 2010;9:4805–4811. doi: 10.4161/cc.9.24.14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl Acad. Sci. USA. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J, Yomogida K, Sakao S, Yamamoto H, Yoshida K, Watanabe K, Morita T, Araki K, Yamamura K, Tateishi S. Rad18 is required for long-term maintenance of spermatogenesis in mouse testes. Mech. Dev. 2009;126:173–183. doi: 10.1016/j.mod.2008.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.