Abstract
Synthetic RNA formulations and viral vectors are the two main approaches for delivering small therapeutic RNA to human cells. Here we report findings supporting an alternative strategy in which an endogenous human RNA polymerase (RNAP) is harnessed to make RNA hairpin-containing small RNA from synthetic single-stranded DNA oligonucleotides. We report that circularizing a DNA template strand encoding a pre-microRNA hairpin mimic can trigger its circumtranscription by human RNAP III in vitro and in human cells. Sequence and secondary structure preferences that appear to promote productive transcription are described. The circular topology of the template is required for productive transcription, at least in part, to stabilize the template against exonucleases. In contrast to bacteriophage and Escherichia coli RNAPs, human RNAPs do not carry out rolling circle transcription on circularized templates. While transfected DNA circles distribute between the nucleus and cytosol, their transcripts are found mainly in the cytosol. Circularized oligonucleotides are synthetic, free of the hazards of viral vectors and maintain small RNA information in a stable form that RNAP III can access in a cellular context with, in some cases, near promoter-like precision and biologically relevant efficiency.
INTRODUCTION
Small RNAs (sRNA) can have big effects on gene expression and other biochemical processes. Whether encoded genomically or originating through design or laboratory selection, sRNA from classes such as microRNA (miRNA), small interfering RNA (siRNA), ribozymes, short hairpin RNA and RNA aptamers have demonstrated biological activity in vitro and in cells (1–4). Biologically active RNA from these and related sRNA classes hold promise for translation to clinical applications if a general means to deliver them safely to human tissues can be found (5).
Most sRNA intracellular delivery approaches fall within two general categories. One is the direct chemical synthesis of the sRNA (6) with modifications (7,8) and nanoparticle (2,9) or liposomal (10) packaging to enhance serum stability, tissue targeting and cellular uptake. The other is based on gene therapy viral vectors that carry the genetic information to make the sRNA, or a pre-processed form of it (11–13). Viral vectors are attractive because the otherwise labile and comparatively difficult to synthesize RNA sequence information is usually held in the more stable form of DNA, and comes packaged in its own delivery vehicle. However, viral vectors carry many risks, including severe immune reactions (14) and random integration of the DNA into chromosomes, which can lead to cancer (15). In addition, like other biologics, gene therapy vectors cannot be characterized to the same extent as synthetic compounds, and can therefore bring with them unnoticed bio-contamination. They also suffer from poor nucleotide (nt) economy, i.e. a high ratio of vector nt to mature RNA nt, increasing the chances of off-target effects. Given the cost and difficulties of using RNA directly, and the risks associated with virus-based delivery vectors, alternative approaches for producing sRNA in human cells are needed.
Chemical DNA synthesis is simpler and more economical than RNA synthesis (16), with DNA chains over 100 nt now routinely made (17). We are investigating the possibility that the cell’s own RNA polymerases (RNAPs) can be harnessed to transcribe synthetic single-stranded (ss) DNA into sRNA. To initiate transcription at specific sites, RNAPs are widely understood to have a general requirement for double-stranded (ds) DNA promoter sequences. It is, however, known that most RNAPs can initiate ‘non-specifically’ on ss DNA regions, a property that was often exploited to study RNAPs before promoters were discovered (18). The demonstrated ability of human RNAPs to initiate promoterless transcription raises the possibility that there exist certain sequences, secondary structures or topological forms that might trigger, in the absence of a canonical promoter, significant levels of precise transcription on oligonucleotide templates.
Here we report that circularizing synthetic ss DNA oligonucleotides can convert them into efficient templates for RNAP III. We show that circular oligonucleotides, or coligos (19), encoding RNA hairpin structures resembling pre-miRNA, exhibit site-specific transcription initiation, circumtranscription and predictable termination, leading to the synthesis of discrete sRNA transcripts. In contrast to previous reports on the promoterless transcription of coligos by phage and bacterial RNAPs, in which the RNAP was observed to transcribe many times around the circle (20,21), human RNAP III mainly carries out single circumtranscription events.
MATERIALS AND METHODS
Coligo synthesis
Synthetic DNA Ultramer® oligonucleotides with 5′ phosphorylation were purchased from Integrated DNA Technologies (Coralville, IA, USA). Enzymatic DNA cyclization using the TS2126 RNA ligase, coligo purification, RNA marker, denaturing polyacrylamide gel electrophoresis (DPAGE) were done as previously described (19). In some cases, when the crude Ultramers® were sufficiently free of failure sequences to cyclize without prior gel purification, the DPAGE step was done after cyclization, where it removed any unreacted linear form and the (circular or linear) failure sequences, obviating the need for a final exonuclease treatment. RNA secondary structures were predicted using the online version of the mfold program (22). Escherichia coli RNAP was purchased from USB. Yeast RNAP II was a gift from D. Bushnell and R. Kornberg (23). General molecular biology methods were done following standard procedures (24).
Cell culture and preparation of HEK293T whole cell extracts
In a typical procedure, four 10-cm dishes of confluent HEK293T cells grown in 1× Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum and Pen./Strep. were rinsed three times with cold 1× phosphate buffered saline on ice. Then, 350–700 µl lysis buffer [50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM Ethylenediaminetetraacetic acid, 1% Triton X-100 and Protease Inhibitor Cocktail (Roche, cat. #04693159001)] was added per dish and incubated on ice for 5 min. Lysed cells were scraped to one side of the dish and transferred with a 1-ml pipette tip to pre-chilled 1.5-ml microfuge tubes. Cell lysis was continued for 5–10 more min on ice until the DNA was visibly precipitating. The extract was centrifuged at 4°C for at least 10 min at 13.2 krpm to pellet genomic DNA and other insoluble materials. The supernatant was transferred to a new chilled tube, adjusted to a final glycerol concentration of 20% v/v, aliquoted and stored at −80°C. The protein concentration was determined by Bradford assay (BioRad cat # 500-0006) and ranged from 2 to 6 µg/µl depending on the amount of lysis buffer used.
In vitro transcription
In vitro transcription (IVT) using whole cell extract (WCE), uniform labeling with [α-32P]-UTP and the linear or circular (coligo) templates was performed as follows. A typical 20 µl reaction mixture contained 25 µg total WCE protein, 20 units RNase inhibitor (Promega), 1.25 mM each adenosine triphosphate (ATP), cytidine triphosphate (CTP), guanosine triphosphate (GTP), 0.2 mM uridine triphosphate (UTP), [except Figure 1B, which contained 1.25 mM each nucleotide triphosphate (NTP)], ∼2 μCi [α-32P]-UTP, 40 mM Tris–HCl pH 7.9, 6 mM MgCl2, 10 mM dithiothreitol (DTT), 2 mM spermidine, 100 µM NaCl, 100 nM coligo template unless otherwise indicated. IVT using E. coli RNAP (USB) was carried out in 40 mM Tris–HCl pH 8.0, 10 mM MgCl2, 5 mM DTT, 50 mM KCl, 50 μg/ml bovine serum albumin, 1 µM coligo template, nucleotide triphosphate, 1.25 mM NTP, ∼2 μCi [α-32P]-UTP, 1 U/μl RNAse inhibitor. IVT with yeast RNAP II was performed with 2.5 µg purified enzyme in 40 mM Tris–HCl pH 7.9, 6 mM MgCl2, 10 mM DTT, 2 mM spermidine, 1.25 mM NTP with 1 µM coligo template. Transcription reaction mixtures were incubated for 90 min at 37°C, after which time the RNA was extracted with 150 μl TriReagent (Invitrogen) per 20 µl reaction volume, according to the manufacturer’s instructions, with 10 µg glycogen added. After ethanol precipitation, radiolabeled transcripts were separated over 9% DPAGE, dried on Whatman paper and exposed to a Molecular Dynamics Phosphorimager Screen. Images and quantitation were performed using MD ImageQuant software. In Figure 1B, the optimal coligo concentration for each RNAP was used to show each enzyme at its most processive and productive.
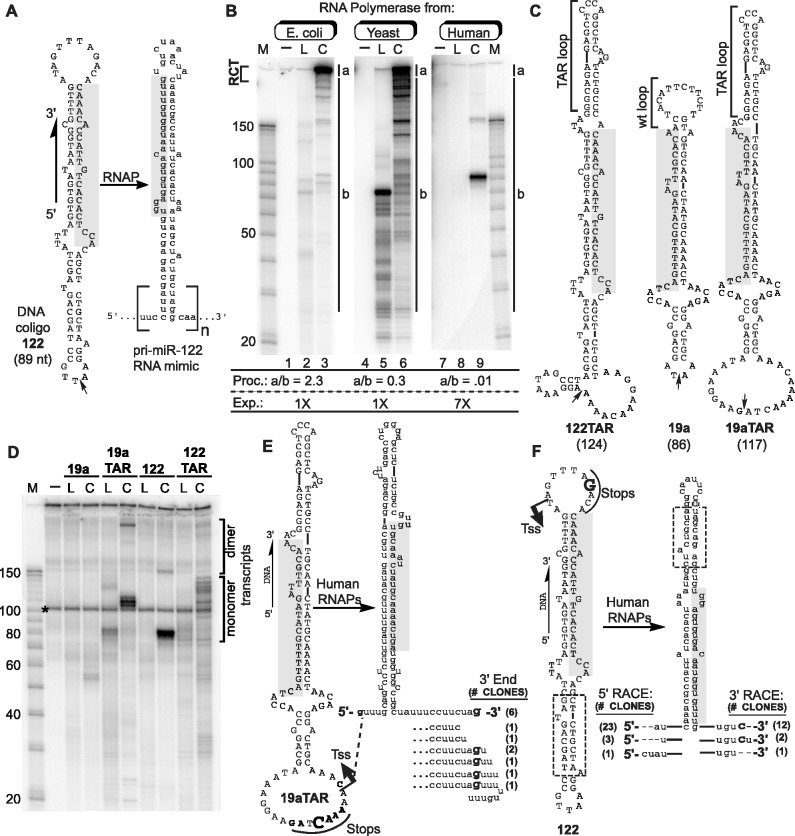
Figure 1.
Transcription of coligos by RNAPs. (A) Schematic illustration showing the sequence and predicted secondary structure of a coligo and its RNA. Coligo 122 was taken from the template strand of the human gene encoding miR-122. The subscript n denotes number of tandem repeats. RCT produces n » 1; n ∼ 1 indicates single round transcription leading to monomer transcripts. Small arrow: circularization site, linear forms are discontinuous here with 5′ phosphate. Shaded region, mature miR-122 or its cDNA. nt, size in nucleotides. (B) IVT of coligo 122 by RNAPs of varying evolutionary age and complexity. E. coli RNAP, yeast RNAP II and human RNAPs from HEK293T WCE were used for IVT. RNA was visualized on a denaturing polyacrylamide gel (DPAGE) by uniform [α-32P]-UTP incorporation. L, linear; C, coligo. Lanes 1, 4, 7: no template. M, RNA marker. Relative processivity (Proc.) defined by the a/b ratio, and relative exposure (Exp., i.e. Phosphorimager grayscale setting difference) are indicated. (C) Sequences and predicted secondary structures of coligos 122TAR, 19a and 19aTAR. (D) IVT using HEK293T WCE followed by DPAGE analysis of uniformly labeled transcripts. Dimer transcripts read through the termination site one time to produce tandem dimer transcripts. (E) Sequence analysis of transcripts made by human WCE from coligo template 19aTAR. The 19aTAR IVT products were isolated and their cDNA sequenced. Tss, transcription start site. (F) Sequence analysis of transcripts made by human WCE from coligo template 122. The 122 IVT product was isolated and sequenced using 5′ and 3′ RLM-RACE protocols.
19aTAR RNA whole transcript cDNA sequencing
WCE IVT was scaled up 10-fold, and coligo-dependent transcripts were separated from cellular sRNA using a biotinylated probe chosen based on a preliminary 5′ Rapid Amplification of cDNA Ends (RACE) experiment. Linkers were added to the 3′ and 5′ ends, followed by reverse transcription and nested polymerase chain reaction (PCR). The PCR products were cloned into pBluescriptSK. Miniprep DNA was isolated and sequenced by Macrogen. See the Supplementary Data for the detailed step-by-step procedure and selector, linker and primer sequences.
122 RLM-RACE 5′ end sequencing
WCE IVT was scaled up 10-fold and the single resolvable 83 nt transcript was located using film exposure, excised and treated with 5′ polyphosphatase. A 5′ RACE adaptor was added using Ambion’s RLM-RACE kit according to the manufacturer’s instructions. Reverse transcription was primed using random hexamers, followed by PCR using 5′ linker-specific forward primer and coligo-specific reverse primers. The PCR product was cloned into pBluescript II SK, and miniprep DNA was sequenced by Macrogen. See the Supplementary Data for the detailed step-by-step procedure.
122 RLM-RACE 3′ end sequencing
WCE IVT was scaled up 10-fold and the isolated transcript was 3′ extended using 5 units E. coli poly(A) polymerase, followed by reverse transcription using Ambion’s 3′ RACE kit adaptor. The resulting cDNA was PCR amplified using the Ambion 3′ RACE inner primer and a coligo-specific forward primer. The PCR product was cloned into pBluescriptSK, and miniprep DNA was sequenced by Macrogen. See the Supplementary Data for the detailed step-by-step procedure.
Quantitative northern blotting of in vitro transcription products
To make RNA quantitation standards and northern probes, the linear precursors of coligos 122 and 19aTAR were PCR amplified with primers containing BamHI and EcoRI restriction sites, then directionally cloned into pBluescript II SK, forming plasmids p122 and p19aTAR, both of which have the ds form of the coligo sequence under the control of the T3 and T7 RNAP promoters in opposite directions. Uniformly labeled probes complementary to the coligo transcripts were made from linearized plasmids using T7 RNAP. Unlabeled RNA standards complementary to the probe were made from linearized plasmids using T3 RNAP in a 3-h 100-µl IVT. Further details are provided in the Supplementary Data.
Template recovery from IVT reactions
In Figure 4A, each reaction was scaled up 4-fold to ensure visualization of the templates by Stains-All dye. Here, 1 µl of a one-tenth dilution of an RNase A/T1 mix (RNase cocktail, Ambion) was added to each reaction and incubated for 30 additional min at 37°C. Nucleic acids were isolated by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. After DPAGE, the gel was soaked for 5 min in ddH2O and stained with Stains-All plus 1.25 ml of formamide (diluted to 20 ml with ddH2O) for 30 min, destained in ddH2O under incandescent light, dried on Whatman paper and scanned.
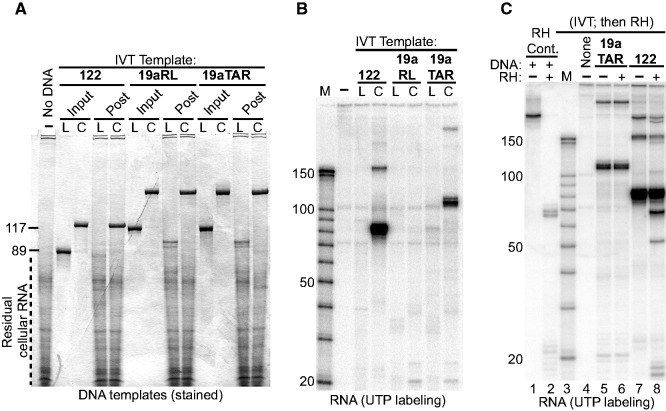
Figure 4.
Coligo topology is necessary but not sufficient to template the synthesis of stable released sRNA transcripts in human WCE. (A) Circularization stabilizes oligonucleotides in human WCE. Circular (C) or linear (L) templates (Input) were recovered (Post) from HEK293T WCE IVT, digested with RNase cocktail to reduce cellular RNA, and stained after DPAGE. Linear forms were degraded during IVT; coligos were stable. Coligo 19aRL sequence is shown in Supplementary Figure S2. (B) DPAGE separation of HEK293T WCE IVT of the three coligos and linear precursors from the reactions shown in panel A. (C) Transcripts are released from the coligo template during IVT. RNase H (RH) was added to (+) or withheld from (−) the indicated coligo IVT reactions at the end of a typical 90-min incubation period. Following additional incubation, the RNA products were separated by DPAGE. Lanes 1 and 2, validation of exhaustive RNase H activity on a 32P-RNA:DNA hybrid. Reaction in lane 2 was supplemented with total HEK293T cellular RNA to normalize non-specific competing RNAs among all RNase H reactions. The result shows that the coligo 19aTAR’s transcripts do not remain hybridized to the coligo template, while ∼20% of coligo 122’s transcripts do remain bound to the coligo template.
RNase H probing
At the end of a standard 90-min IVT, 1 unit of RNase H (Promega) was added and the incubation was continued for an additional hour. For the RNase H positive control lane, the linear form of coligo 19aTAR was mixed with the p19aTAR plasmid T3 RNAP run-off transcript, annealed by heating 4 min in 93°C followed by slow-cooling. Isolated total RNA from the same amount of WCE used in the IVT reaction was added to normalize non-specific competition, and RNAse H was added as above. RNA was isolated by extraction with TriReagent, resolved by DPAGE and exposed to a Phosphorimager screen.
Transfection of HEK293T cells and northern blotting
Twelve hours before transfection, HEK293T cells were seeded in 12-well plates [1 ml 1× DMEM with 10% fetal bovine serum (FBS)], and then grown to 90% confluency. Here, 46 µl of 1× DMEM was mixed with 4 µl of 10 µM template stock in 10 mM Tris-HCl/1 mM EDTA (TE), and 10 µl of PolyFect (Qiagen) was added and complexed for 8 min at room temperature. The non-transfected control contained 46 µl 1× DMEM, 4 µl TE and 10 µl PolyFect. From each well, 700 µl of the media was removed, and the DNA complexes were added with 640 µl of fresh 1× DMEM supplemented with 10% FBS (without Penn./Strep). After 24 h, or as indicated, the RNA was harvested by lysing the cells of each well with 800 µl TriReagent and isolating the RNA according to the manufacturer’s (Invitrogen) instructions. The pellet was resuspended in 50 µl 1× DNase I buffer, 2 µl were removed for OD260 measurement and the RNA was digested with 4 U of DNase I for 1 h at 37°C. A volume containing ∼12 µg of RNA was ethanol precipitated and separated over 10% DPAGE (0.75 or 0.4 mm gel thickness), blotted onto BrightStar®-Plus Positively Charged Nylon Membrane (Ambion AM10102), baked for 20 min at 55°C, UV-crosslinked, pre-hybridized in Church hybridization buffer (0.25 M Na2HPO4, 7% sodium dodecyl sulphate, 1 mM Ethylenediaminetetraacetic acid) and hybridized in the same buffer at 55°C overnight to a uniformly labeled RNA probe transcribed from the appropriate linearized plasmid (p19aTAR or p122). For the LNA northern blots, 20 pmol of the miR-122 or -19a LNA (Exiqon), complementary to the mature 122 and 19a human miRNAs, were 5′ end-labeled with [γ-32P]-ATP/T4 Polynucleotide kinase, and used in place of the uniformly labeled probe described above. Locked Nucleic Acid (LNA) hybridization and washing were done also as described above, except at 42°C. Final northern blots were washed twice for 30 min each with Church wash buffer (1% sodium dodecyl sulphate, 20 mM Na2HPO4) and exposed to a PhosphorImager screen (Molecular Dynamics).
RNAP inhibitor use in vitro and in tissue culture
ML-60218 (Maybridge) was dissolved in dimethylsulfoxide (DMSO) at a final concentration of 135 mM and stored at −20°C. Just before use, the stock was diluted to 9 mM and 67% DMSO with ddH2O. Only at this dilution could ML-60218 be added to an IVT reaction or transfection without precipitation of the inhibitor.
α-Amanitin (Sigma-Aldrich) was dissolved in water at 1 mg/ml and stored at −20°C. For transfection experiments, the inhibitor was added to the cells together with the PolyFect-DNA complex and incubated for 9 h to minimize cell death by prolonged exposure. Final concentrations were as follows: α-amanitin: low = 0.12 µg/ml, medium = 1.2 µg/ml, high = 40 µg/ml; ML60218: 67.5 µM (∼2× IC50 RNAP III).
RESULTS
Coligo transcription: RNA polymerase processivity and evolutionary age
Before the discovery of miRNA and siRNA, coligos were serendipitously found to undergo rolling circle transcription (RCT; e.g. Figure 1A, n ≅ 12–260) (20) by isolated RNAPs from bacteriophage and E. coli (20,21). However, it has not been reported whether eukaryotic RNAPs also transcribe coligos. The therapeutic potential of sRNA makes this a timely and important question. To investigate whether human RNAPs also carry out RCT, we designed a coligo to code for a minimized primary (pri)-miR-122 stem loop RNA [Figure 1A, and (19)]. In the form of rolling circle transcripts (Figure 1A, n = large number), coligo transcripts should fold into tandemly arrayed multimers resembling naturally occurring pri-miRNA from clustered miRNA genes. Indeed, we previously found that human Drosha could process RCT transcripts made by purified E. coli RNAP (19).
Coligo 122 (Figure 1A) was made by circularizing a synthetic 89 nt genomic sequence encompassing human miR-122’s non-coding strand (19). Figure 1B shows an IVT comparison of 122 with RNAPs from bacteria, yeast (RNAP II) and human (HEK293T WCE). Transcripts were visualized by [α-32P]-UTP incorporation. RCT is recognized by the characteristic pattern of large transcripts that enter the denaturing polyacrylamide gel without migrating further (20) (Figure 1B, gel region ‘a’). As a relative measure of transcriptional processivity, we calculated the ratio of the radioactivity incorporated in the rolling circle transcripts (Figure 1B, gel region ‘a’ in lanes 3, 6, 9) to the sum of all shorter transcripts (gel region ‘b’). Lower a/b ratios indicate lower transcriptional processivity. A comparison of lanes 3, 6 and 9 reveals a clear trend in which a/b declined from E. coli to human RNAPs. Thus, the ability to initiate promoterless transcription on coligo templates appears to have been retained during RNAP evolution, whereas the processivity needed for RCT appears to have been lost.
Precise coligo transcription by a human RNA Polymerase
Although no RCT was observed, transcription of coligo 122 by the human WCE was remarkable for having produced a single, well-defined and relatively abundant in vitro transcript, possibly indicating transcription initiation and termination at specific sites. The size similarity between coligo 122 (89 nt) and its transcript (∼83 nt) indicated that the putative initiation and termination sites were close. About 4% of transcription events read through one time and terminated on the second pass, producing a tandem dimer transcript at ∼160 nt. Interestingly, although no RCT treadmill action on the coligo was needed to produce the single round (monomer) transcript, the circular topology of the coligo sequence was required (Figure 1B, lane 8 versus 9). Thus, the circular topology of coligo 122 appeared to promote transcription by ≥1 human RNAPs, and termination, or termination followed by rapid processing occurred non-randomly just upstream of the site of initiation.
In an initial test for generality, we made three additional coligos (Figure 1C) similar in size and design to 122, including one based on another human pre-miRNA, miR-19a (19). Figure 1D shows the RNA products formed during IVT in HEK293T WCE using these templates in both linear (L) and circular (C) form. Coligo 19a was a poor substrate, but 19aTAR, which contains the same 19a miRNA stem but different terminal loops, templated the synthesis of three resolvable and relatively abundant transcripts. Coligo 122TAR, which contains the 122 stem but different loops, produced six resolvable transcripts. As observed for 122, the transcripts from 19aTAR and 122TAR were slightly shorter than their template, low levels of dimer transcripts resulted from read through and the linear forms did not template productive transcription. Taken together, these results showed that single round transcription was not peculiar to coligo 122, and that sequence variations outside of the miRNA-encoding stem at least in part determine transcription efficiency and the sites of initiation and termination.
We probed the 5′ end of the 19aTAR transcripts using 5′ end modifying enzymes (Supplementary Figure S1). Neither a 5′ phosphate-dependent 5′→3′ exonuclease (‘terminator 5′-phosphate-dependent exonuclease’, Epicentre), nor sequential treatment with tobacco acid pyrophosphatase, which reduces 5′ cap structures to a 5′ phosphate, followed by the terminator exonuclease, altered the pattern of coligo-dependent transcripts. This result indicated that the transcripts are not 5′ capped or mono-phosphorylated. Two different enzymatic treatment sequences provided evidence that the transcripts are, as expected, 5′ triphosphorylated, but also have terminator exonuclease-impeding secondary structure. Polyphosphatase, which reduces triphosphate tails to single phosphates, followed by the terminator 5′ exonuclease, shortened but did not fully degrade the transcripts. The same result was obtained when complete dephosphorylation was followed by 5′ mono-phosphorylation, and then terminator exonuclease treatment. While the incomplete digestion that was observed could signal the presence of an unexpected 5′ modification in some transcripts, or no modification at all, the simplest explanation is that the 19aTAR transcripts have, as expected, a 5′ triphosphate and secondary structure near the 5′ end that stops the 5′→3′ terminator exonuclease from fully degrading the transcripts.
Adding linkers to, and sequencing the cDNA of, the ∼110 nt 19aTAR transcripts revealed a single transcription start site (13/13 clones) in the larger ss loop (Tss, Figure 1E). About half (6/13) of the cloned transcripts appeared to terminate precisely at a dC just upstream of Tss. This putative termination site immediately preceded an A5 run, a sequence that in a ds DNA context is a strong RNAP III termination signal (25). All other transcripts appeared to terminate either just before or just after this position. The 3′ ends may result from termination with or without subsequent 3′→5′ processing. Abundant cellular RNA in the same size range made cDNA sequencing difficult. We therefore used 5′ and 3′ RACE procedures to obtain a composite sequence for the ∼83 nt 122 transcript (Figure 1F). This transcript also began exclusively in the larger of the two terminal loops of the coligo and, similar to the 19aTAR case, mainly at one pyrimidine nt near the helix stem (Figure 1F, Tss, 23/27 clones). Transcription termination (or 3′ end formation) of coligo 122 occurred just after a known RNAP III termination signal (AAACA) (26). Thus, although ss and promoterless, coligos 19aTAR and 122 both underwent precise transcription initiation to produce stable hairpin RNAs with low 3′ end heterogeneity.
Transcription of hairpin-encoding coligos is a general property of human cellular extract
We made additional coligos to probe the structural determinants of monomer circumtranscription. (See Supplementary Figure S2 for these coligo sequences and predicted secondary structures.) First, in an effort to produce a transcript more closely resembling a typical pre-miRNA, we removed an 18 nt section of the coligo 122 stem, shown boxed in Figure 1F. Removal of this section did not appear to change the sites of initiation and termination, as the resulting coligo, 122 s, was transcribed into the 65 nt transcript expected if the 122 start and stop sites were used, although the amount of the monomer transcript was significantly lower after taking into consideration the labeling difference (Figure 2A, Gel 1). This result showed the stem could be altered while maintaining loop-directed start and stop sites, but also that the stem length and/or sequence somehow contributes to transcription efficiency. Coligo 19aTAR contains a terminal stem–loop encoding the HIV trans-activation response element TAR (Figure 1C). The TAR loop increases transcriptional processivity in HIV RNAP II transcripts (27), and was added to 19a in an effort to stimulate transcriptional elongation before we learned that RNAP III was responsible for coligo transcription (see below). To gauge the effect of the TAR sequence on coligo transcription efficiency, we changed its sequence entirely, while keeping a similar secondary structure. The resulting coligo, 19am3 (Supplementary Figure S2), was transcribed similarly to 19aTAR, showing that the TAR loop encoding sequence was not important for transcription, but that its secondary structure, including its small terminal loop, might contribute to transcription (Figure 2A, Gel 2). This conclusion was further supported by coligo 19aRL (Supplementary Figure S2), where the TAR loop was changed to a large unstructured sequence. This coligo, which has two large ss terminal loops capping the miRNA-encoding stem, produced no observable transcripts (see below). These results suggest that, for transcription to occur in human cell extract, a coligo should have an imperfect ds stem and one large and one small terminal ss loop, the larger of which, where transcription is likely to initiate, should not be pyrimidine-rich (e.g. coligo 19a).
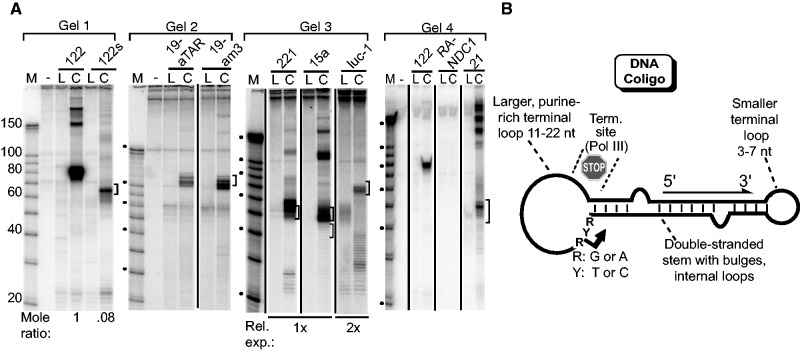
Figure 2.
Coligo secondary structure contributes to transcription by human WCE. (A) IVT of indicated coligos, as in Figure 1. Coligo sequences and predicted secondary structures are shown in Supplementary Figure S2. Brackets indicate predicted monomer transcript size (10 nt window) based on sequencing data shown in Figure 1E and F. Gel 1: The boxed sequence shown in Figure 1F was removed from coligo 122 to produce 122s, whose transcript size changed accordingly. Ratio is for monomer transcripts at 83 and 65 nt, adjusted for label content. Gel 2: The TAR loop encoding sequence in coligo 19aTAR was changed while keeping a similar secondary structure to produce coligo 19am3, which was similarly transcribed. Gel 3: Coligos 221 and 15a are based on the human miRNAs of the same number. Luc-1 encodes a pre-miRNA complementary to a luciferase 3′ UTR sequence. Each coligo was transcribed by WCE to predicted monomer-size transcripts (brackets). Gel 4: RANDC1 encodes a largely unstructured transcript. Coligo 21 is based on human miR-21 and contains no RNAP III termination sites. It produces less monomer transcript and more tandem multimer transcripts. (B) Schematic summary of sequence and secondary coligo features that appear to promote discrete transcription events in WCE.
We tested four additional coligos, three based on randomly selected human miRNAs (hsa miR-221, -15 and -21) and one on an siRNA complementary to the luciferase 3′ UTR, luc-1 (28). In a manner similar to coligos 122, 122s, 19am3 and 19aTAR, we designed each to have a larger purine-rich terminal loop at one end with a pyrimidine near the stem–loop junction, and a smaller terminal loop at the other end. The genomic template strand sequences used in the design of coligos 221 and 15a, like those of 122 and 19aTAR, contained RNAP III termination sequences near the stem–larger loop junction (bold in Supplementary Figure S2). In coligo luc-1, two artificial A5 sequences were added to the larger loop. No RNAP III termination sequences were found or put in coligo 21. As shown in Figure 2A (Gels 3 and 4), all four coligos were transcribed in HEK293T WCE to monomer transcripts having a size approximately that predicted by initiation and termination similar to those found in 19aTAR and 122 (see Supplementary Figure S2 for expected start and stop sites). However, there were some differences. Coligo 21 gave rise to significant amounts of larger tandem transcripts, possibly explained by its complete lack of any sequence resembling a RNAP III termination sequence. Coligo luc-1’s monomer transcript was significantly less abundant than the others, which may be the result of the A5 sequence present between the putative starting pyrimidine and the stem (Supplementary Figure S2). Overall, the transcriptional behavior of these additional coligos was similar to that of 19aTAR and 122, indicating that coligo circumtranscription is a general activity of HEK293T WCE. Although there are differences in efficiency, precision and read through, and more examples will need to be studied to attain consistent control, our most precise example, coligo 122, shows that the transcriptional precision from these chemically synthesized templates can approach that of canonical ds RNAP promoters.
The results from the tested coligos allowed us to formulate a preliminary description of the features that lead to successful circumtranscription resulting in an RNA hairpin (Figure 2B). They include one large relatively purine-rich loop and one smaller loop flanking an imperfect ds stem; a pyrimidine close to the 3′ end of the stem/larger loop junction where in the two sequenced cases transcription initiated; an RNAP III termination sequence either in the larger loop or near the 5′ end of the stem–large loop junction. On this last point, we speculate that, based on coligo 21, which has no such sequence, the junction structure itself may contribute to termination even when no RNAP III termination signal is present. Indeed, the RNAP III termination sequences were merely associated with nearby transcription termination, and we cannot be certain they function as they do in a ds DNA context. (Exonuclease trimming in the 3′→5′ direction may also contribute to 3′ end formation.) With these features in mind, we made and tested a coligo that had none of these characteristics, coligo RANDC1 (Supplementary Figure S2). This coligo has the same G+C and A+T content as 19aTAR. It was produced by a random sequence generator and chosen because it had as little predicted structure as could be generated. This coligo produced no detectable transcripts (Figure 2A), supporting the hypothesis that there are sequence and secondary structure rules that must be followed for successful coligo transcription to occur.
Nanomolar coligo levels lead to biologically relevant transcript levels
We used quantitative northern blotting to estimate the amount of single round transcripts formed as a function of coligo concentration (Figure 3A and B). Single round transcripts from 19aTAR were detectable using as little as 10 nM coligo, and increased to an RNA concentration of ∼0.6 nM transcribed from 100 nM coligo. Raising the coligo concentration eventually reduced the amount of transcript. In the case of coligo 122, single round transcripts were detectable at 100 pM coligo, and a plateau of ∼6 nM transcript was reached at 50 nM coligo. A time course showed that RNA levels continued to increase up to 2.5 h, the longest time point monitored (Figure 3C). No trace of longer transcripts were seen at early time points, indicating either that 3′ exonuclease trimming does not contribute to transcript 122’s 3′ end, or that such trimming is too fast to observe by this method.
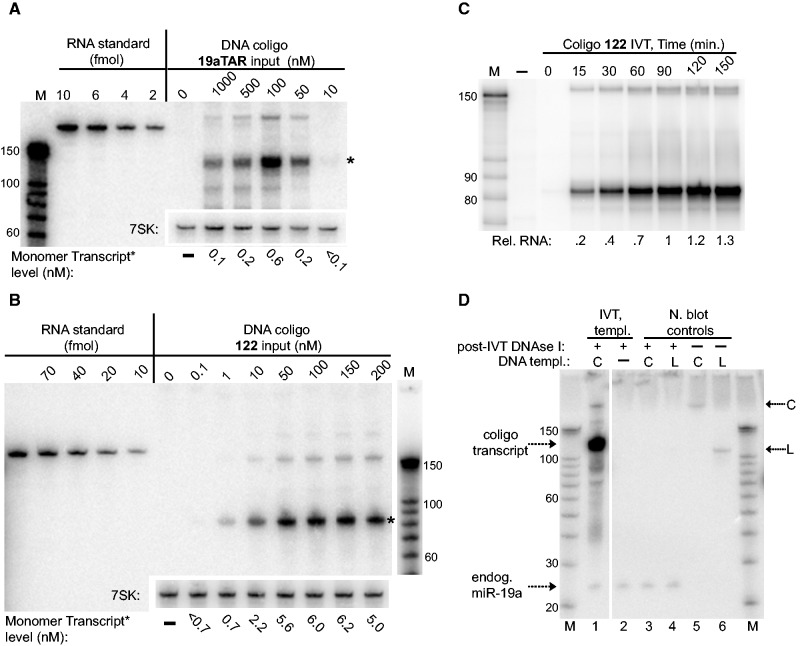
Figure 3.
Coligo concentration dependence and RNA transcript quantitation for HEK293T WCE IVT. (A) Quantitative northern blotting of coligo 19aTAR IVT transcripts. The indicated range of coligo concentrations was used in unlabeled HEK293T WCE IVT reactions and the transcript levels were compared with known amounts of RNA containing the same northern probe recognition sequence. Numbers below blot indicate transcript molarity estimated after 90 min IVT. Endogenous 7SK RNA was probed separately as a loading control. Asterisk (*) indicates the single round transcript whose concentration estimates are listed below gels. (B) Quantitative northern blotting of coligo 122 IVT transcripts, as in panel A. (C) Time course for 100 nM coligo 122 IVT, visualized by [α-32P]-UTP incorporation. Rel. RNA: relative amount of RNA. (D) Quantitative LNA northern blot comparison of endogenous HEK293T miR-19a with the 90 min IVT transcripts made from coligo 19aTAR (100 nM), in the same extract. The ratio of the 19aTAR in vitro single-round transcript (∼110 nt) to endogenous miR-19a (23 nt) is ∼65. Full LNA northern signal cross-reacting with the DNA input templates, in absence of DNase treatment, can be seen in lanes 5 and 6. In lanes 3 and 4, the DNA templates were added to HEK293T extract and immediately processed without IVT incubation period to show that DNase I treatment of templates ensures they have no detectable signal in the northern blots shown in panels (A–C).
To put coligo transcript levels into perspective, we compared the single round transcript level from 19aTAR with the mature miR-19a in HEK293T extract. Based on RNA-Seq experiments in the closely related HEK293 cell line, miR-19a is moderately to highly expressed in HEK cells (29). Endogenous miR-19a in the HEK293T WCE (Figure 3D) was visualized using a standard LNA northern probe visualizing both transcripts with equal sensitivity. The coligo 19aTAR transcript at 90 min was ∼65-fold higher than the endogenous miR-19a in WCE. Thus, a coligo template can quickly be transcribed by HEK293T WCE to a level beyond that of a moderately expressed endogenous miRNA.
Coligo and transcript are stable to cellular nucleases, but do not remain hybridized
Figures 1 and 2 showed that the coligo’s circular topology is required for productive transcription, although—and in contrast to RCT—no treadmill action should be required to produce single round transcripts. To test whether transcribability correlated with stability against cellular nucleases, we attempted to recover linear and coligo templates at the end of a WCE IVT reaction. Figure 4A shows that while coligos were stable in HEK293T WCE, their linear forms were degraded. Coligo 19aRL (Supplementary Figure S2), where the structured TAR loop of 19aTAR was replaced by an unstructured loop, was stable but not transcribed (Figure 4B). Thus, a circular topology appears necessary but not sufficient for an oligonucleotide to undergo specific transcription and, as stated above, certain sequence and secondary structure rules must be met for transcription to occur.
Our data show that both coligo and transcript are stable in WCE. Promoterless transcription from 3′ tailed templates can lead to transcription events ending in a stable RNA:DNA hybrid (30). While the fact that stable coligo transcripts are observed argues against the presence of extensive RNA:DNA hybrids, whose RNA would be subject to cellular RNase H degradation (31), we further tested for RNA:DNA hybrid formation by adding supplemental RNase H to an IVT reaction (Figure 4C). Exhaustive RNase H activity (lanes 1 and 2) had no effect on the 19aTAR transcripts (having 38 predicted bp, Figure 1E), while ∼20% of the 122 transcripts (having 32 predicted bp, Figure 1F) were susceptible to added RNase H. We conclude that the strong intramolecular secondary structure of coligo and transcript favor release of the transcript, although a fraction can remain hybridized when there are relatively few bp in the stem. This result also rules out the possibility that the RNAP terminates after single round transcription because it encounters—and cannot unwind—a persisting RNA:DNA hybrid at the transcription start site. Instead, termination must be caused by other factors.
Coligo transcription in human cells
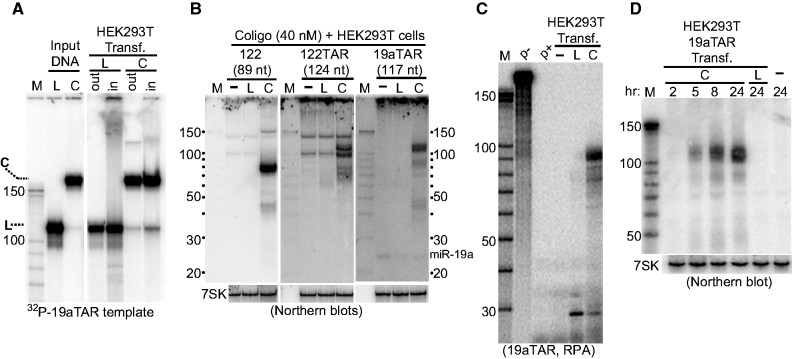
We next asked whether coligos can enter and undergo transcription in human cells. Coligo 19aTAR and its linear counterpart were transfected into HEK293T cells at 40 nM. Including 32P-labeled tracers of the same DNA revealed that ∼50% of the linear and coligo templates entered the cells and remained stable during a 24-h experiment (Figure 5A). Three coligos that were transcribed in vitro were transfected into HEK293T cells and assayed by northern blotting of total cellular RNA after 24 h. In each case, transcripts the sizes of those seen in vitro were detected (Figure 5B). After 24 h transfection, the 19aTAR transcript was found at 26-fold greater abundance than the mature miR-19a (Figure 5B, right panel). Although apparently stabilized by the transfection reagent (Figure 5A), the linear forms produced no detectable transcripts, underscoring the strict requirement of the circular topology for productive transcription in cells, apart from the stability conferred by circularization. The importance of the circular topology in vivo was further supported by an RNase protection assay (RPA) performed on total RNA isolated from transfected cells (Figure 5C). Here, the temperature of the assay was lower than the northern hybridization temperature, enabling the detection of aborted transcripts. Both the linear and coligo forms of 19aTAR templated a small fragment at ∼30 nt, but only the coligo underwent circumtranscription. A time course furthermore showed that the 19aTAR transcript continued to increase over a 24-h transfection period (Figure 5D). Overall, Figure 5 shows that transcription in cells parallels transcription in vitro: a circular template is required, single round transcripts slightly smaller than the coligo are made and the coligos template the accumulation of stable transcripts over time.
Figure 5.
Coligo transcription in human cells. (A) Fate of transfected coligo and linear templates. 32P-labeled DNA tracers were spiked into unlabeled templates and transfected at 40 nM into HEK293T cells using PolyFect transfection reagent. Templates recovered from harvested cells (‘in’) and media supernatant (‘out’) were adjusted to equal percentages of total fractions, and separated by DPAGE. C, coligo; L, linear; M, RNA marker. About half of the DNA entered cells during transfection. (B) Coligos are transcribed in human cells. Templates (C or L) were transfected into HEK293T cells for 24 h after which time total RNA was assayed by northern blotting using a 5′ labeled template-specific LNA. The ratio of the 19aTAR single round transcript (∼110 nt) to endogenous miR-19a (23 nt) is ∼26. Endogenous 7SK RNA was probed separately as a loading control. (C) RPA on total RNA isolated from cells transfected with coligo or linear 19aTAR. p−, probe input; p+, probe alone digested with RNase cocktail. (D) Coligo transcripts accumulate during transfection period. Total RNA of cells transfected with coligo 19aTAR harvested after the indicated transfection times and probed by northern blotting using a uniformly labeled in vitro RNA transcript complementary to one complete coligo transcript sequence. 7SK, loading control.
Coligos are transcribed in vitro and intracellularly by RNAP III
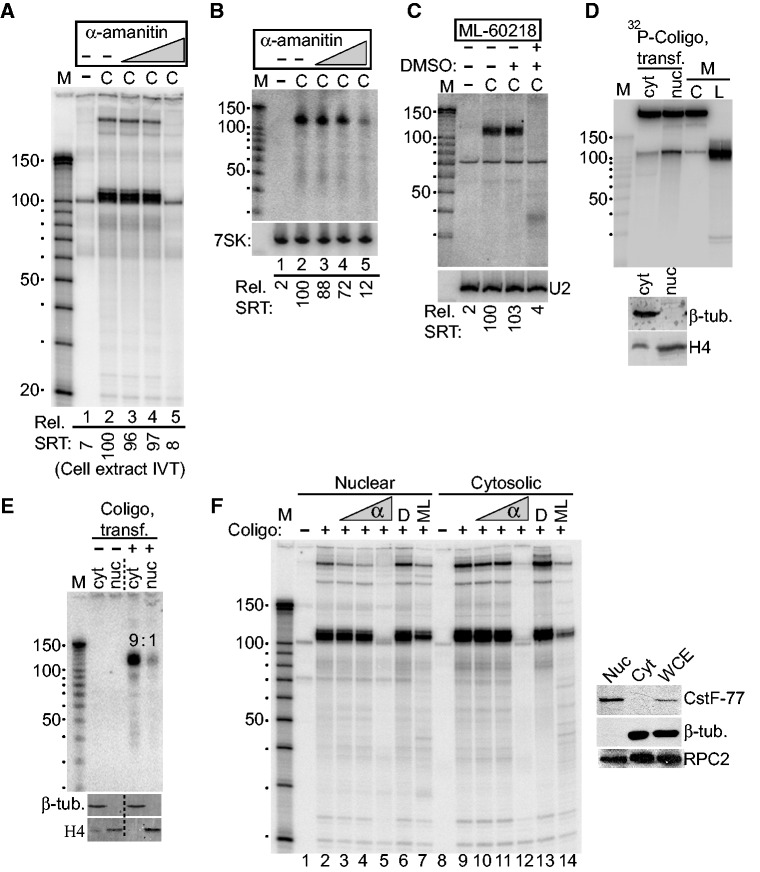
To learn which of the human RNAPs is responsible for coligo transcription, we took advantage of their different susceptibilities to the transcription inhibitor α-amanitin (32). IVT of coligo 19aTAR was carried out using HEK293T WCE in the presence of increasing concentrations of α-amanitin. At 0.12 µg/ml, a concentration that only inhibits RNAP II (32), coligo transcription was unaffected, ruling out RNAP II (Figure 6A). At 120 µg/ml, which completely inhibits RNAP III but not RNAP I, no coligo transcripts were produced, implicating RNAP III. Similar results were obtained when coligo 19aTAR was transfected into HEK293T cells in the presence of increasing amounts of α-amanitin, followed by northern blotting of total cellular RNA with a 19aTAR-specific probe (Figure 6B). To further test the identity of the RNAP, we treated HEK293T cells with ML-60218, an RNAP III-specific inhibitor (33). Northern blotting showed that no single round transcripts were produced in ML-60218-treated cells transfected with coligo 19aTAR (Figure 6C). These results demonstrated that RNAP III is responsible for producing all transcripts from coligo templates in vitro and in human cells.
Figure 6.
RNAP III is responsible for coligo transcription, which appears to take place mainly in the cytosol of transfected cells. (A) α-Amanitin inhibits coligo transcription at concentrations consistent with RNAP III transcription. HEK293T WCE IVT was carried out with increasing concentrations of α-amanitin. C, coligo 19aTAR, 100 nM. Lanes 3–5: 0.12, 1.2, 120 µg/ml α-amanitin. Rel. SRT: relative amount of single round transcript (aka monomer transcript). (B) Northern blot of total RNA from HEK293T cells transfected with 40 nM coligo 19aTAR with concurrent α-amanitin treatment (Lanes 3–5: 0.12, 1.2, 40 µg/ml). 7SK RNA was probed as a loading control. (C) Northern blot of total RNA from HEK293T cells transfected with 40 nM coligo 19aTAR with concurrent RNAP III-specific inhibitor ML-60218 treatment at 68 µM. DMSO, inhibitor solvent. U2 snRNA was probed as loading control. (D) DPAGE of 32P tracer-labeled coligo 19aTAR recovered from HEK293T transfection after separation of nuclear and cytosolic fractions. Equal percentages of the nuclear and cytosolic fractions were loaded. Inset: western blot assessment of fractionation, β-tub., β-tubulin (cytosolic protein). H4, histone H4 (nuclear protein). M, markers and input templates. (E) Northern blot of RNA isolated from HEK293T nuclear and cytosolic fractions 24 h after transfection with 40 nM coligo 19aTAR. Equal percentages of the nuclear and cytosolic fractions were loaded. Inset: western blot assessment of fractionation, as in panel D. (F) HEK293T nuclear and cytosolic extract IVT in the presence of increasing α-amanitin (α, lanes 3–5 and 10–12: 0.12, 1.2 and 120 µg/ml), DMSO (D) or ML-60218/DMSO solution (ML). Inset: western blot assessment of fractionation, β-tub., β-tubulin (cytosolic protein). CstF-77, (nuclear protein); RPC2, RNAP III subunit.
Subcellular fractionation of transfected coligos and coligo-derived transcripts
Promoter-independent RNAP III activity was recently found in the cytosol of HEK293 cells, where its transcription of poly(dA-dT) revealed an unexpected role for the polymerase in the innate immune system (34,35). RNAP III was found to transcribe poly(dA-dT), a viral DNA surrogate, into ds RNA, which in turn activated an interferon response through the RIG-I helicase. Because coligos also undergo promoterless transcription, and because A/T-rich DNA is prone to forming transient loops that might resemble A-rich coligo loops, we wondered if coligos might be transcribed in the cytosol by engaging this natural process. We therefore tracked the sub-cellular location of the coligo and its transcripts during subcellular fractionation. A radiolabeled coligo was transfected into HEK293T cells and found to be evenly distributed between nuclear and cytosolic fractions (Figure 6D). In contrast, northern blotting of the fractionated extracts from coligo 19aTAR-transfected cells showed that ∼90% of the coligo transcripts were in the cytosolic fraction (Figure 6E). To verify that the cytosol fraction contained RNAP III capable of coligo transcription, we fractionated untransfected HEK293T WCE, verified that a significant amount of RNAP III was present in both nuclear and cytosolic fractions (RPC2 western blotting, Figure 6F) and carried out IVT with and without RNAP III inhibitors. The results showed clearly that RNAP III in both cytosolic and nuclear fractions carried out strong coligo transcription similar to the pattern generated by WCE (Figure 6F). While we cannot rule out the possibility that the cytosolic transcripts in Figure 6E were made in the nucleus and exported to the cytosol, or that half of the nuclear RNAP III leaked into the cytosol during the fractionation procedure, until a promoter-independent role for RNAP III in the nucleus is found, the simplest explanation is that the coligo was transcribed mainly by RNAP III in the cytosol. These data support the hypothesis that coligos are primarily transcribed in the cytosol through fortuitous engagement of RNAP III’s pathogen-recognition role in innate immunity.
DISCUSSION
Here we report the unexpected finding that circularizing synthetic oligodeoxynucleotides can result in their use as transcription templates by RNAP III in vitro and in cultured human cells. Promoter-independent transcription by mammalian RNAPs is known to initiate at internal ss regions such as nicks and gaps in otherwise ds DNA (18), and site-specific promoterless transcription has been regularly used on 3′ ss tailed ds DNA (30,36). Our templates, however, are ss and circular, and thus without free 3′ ends, nicks or gaps, and reveal a previously unrecognized means to initiate site-specific de novo promoterless transcription by a mammalian RNAP in vitro and in human cells. Because biologically active sRNA falls squarely within the size range of currently accessible synthetic DNA, our findings have the potential to form the basis of a new class of sRNA delivery vectors made by the chemical synthesis of promoterless DNA templates able to be read by human RNAP III. In addition, we find that the circular topology required for single round transcription of the coligos protects them from nuclease degradation in WCE. Circularization of oligonucleotides has previously been shown to stabilize ss DNA against degradation in human serum (37). Thus, one of the defining structural determinants promoting coligo transcription stabilizes the coligo in a biological context.
Although we found that circularization conferred stability against intracellular nucleases, it was not sufficient to trigger productive transcription; a hairpin-encoding secondary structure in the coligo was also required, possibly to mimic a natural transcription intermediate (see below), and possibly to favor intramolecular base-pairing over persistent RNA:DNA hybrid formation, which would lead to RNase H susceptibility. Hairpins constitute the basic unit of sRNA secondary structure. As such, coligos may prove generally suitable for the transient ectopic expression of a wide variety of functional sRNA in human cells. Random sequences with insignificant stem lengths, like RANDC1, or coligos with a highly pyrimidine-rich larger loop, like 19a, or two large terminal loops, like 19aRL, appear not to fulfill the requirements for transcription or the transcription of stable transcripts.
Our choice of pre-miRNA–like stem loop RNA as proof of principle transcription targets called for DNA coligos consisting of an imperfect stem capped by terminal loops, including one small loop to encode the pre-miRNA terminal loop. Productive synthesis of single round transcripts resulted when the loop at the other end of the imperfect stem, which in most cases was derived from pri-miRNA flanking sequence, was larger (11–22 nt) and adenine rich (33–68%), but not larger and pyrimidine rich (e.g. 19a) or if there were two large loops (e.g. 19aRL). Transcript sequencing of 122 and 19aTAR showed nearly homogeneous transcription initiation at a specific pyrimidine, flanked by purines, in the larger loop 2–4 nt from the 3′ end of the stem, a structural feature that may prove to be a general coligo initiation motif. A loop directly adjacent to a stem may recruit RNAP III by locally mimicking an intermediate structure on the path to promoter-dependent transcription. The greater degree of conformational freedom in the linear templates may fail to stabilize such a structure long enough for transcription to begin, at least in the time before the linear templates are degraded by cellular exonucleases. Furthermore, in a cellular context and in cellular extracts, other ss DNA-binding proteins may compete with RNAP III for the coligo loops, reducing RNAP III initiations—a potential limitation that might be overcome by loop sequence optimization. Such ss DNA-binding proteins might also contribute to transcriptional pausing and termination in the absence of a RNAP III termination signal (e.g. 21) by obstructing processive transcription. Given an imperfect ds stem capped by one large and one small loop, the larger loop’s size, sequence and secondary structure appear to control initiation and termination (or 3′ end formation), and efficient larger loops, like natural promoters, may be transferable to other hairpin stems.
Our identification of RNAP III as the enzyme responsible for coligo transcription has the potential to lead to general applicability because termination appears to be controlled, in an as yet undefined manner, by this enzyme’s simple termination signals (possibly followed by exonuclease processing). The need for these sequences might be considered a limitation because they should not occur in the middle of a desired sRNA sequence. However, we note that the relatively strong RNAP III termination sequence A4 occurs in the stem of 19aTAR, yet no termination was found there, whereas the weaker -AAACA- signal (26) at the stem–loop junction appeared to direct strong termination in the larger loop of coligo 122. It is possible that termination signal strength can be modulated by the coligo’s secondary structure context. We also acknowledge that RNAP III termination signals as they occur in ss coligos may not function the same way they do in natural ds templates, as the coding strand is not present in the coligo. Nevertheless, the modest 3′ end heterogeneity that resulted from termination on the 122 and 19aTAR templates in vitro is not different from that seen in the deep sequencing of naturally occurring miRNA (38) and pre-miRNA (39,40), though the 3′ ends of endogenous pre-miRNA result from Dicer processing. Because some, if not most, coligo transcription appears to occur in the cytosol, where Dicer functions, it remains possible that the 5′ and 3′ ends of coligo transcripts may be suitably prepared for Dicer by initiation and termination on a coligo template, as opposed to the natural case of pri-miRNA Drosha processing in the nucleus.
Promoterless RNAP III activity was previously found in cytosolic extracts (18,32). The recent discovery that RNAP III functions in the innate immune system as a sensor of pathogenic ds DNA invading the cytosol may explain its location there (34,35). Transfected poly(dA-dT) was found during the innate immune system studies to be transcribed in the cytosol by RNAP III (35). Though intracellular leakage is always a possibility in fractionation-based experiments such as ours, coligos appear to be transcribed by cytosolic and nuclear RNAP III. Nevertheless, the transcripts from transfected coligos localized mainly to the cytosol, an asymmetry that cannot be explained by leakage. Taken together, our data support a model in which transfected coligos distribute throughout cells but are transcribed mainly in the cytosol, by RNAP III presumably diverted from its innate immunity role. Promoterless nuclear transcription followed by export is less likely, given that pre-miRNAs with ss extensions suffer from poor export (41,42). Transcription of poly(dA-dT) makes self-complementary poly(rU-rA), which in turn activates interferon production via RIG-I, an RNA-binding protein preferring fully base-paired, blunt-ended ds RNA with a 5′ triphosphate (43). We found evidence that coligo transcripts are not capped or 5′ mono-phosphorylated, and that at least some bear a 5′ triphosphate. Learning the rules governing the precise coligo transcription initiation and termination observed here may allow RNAP III’s innate immunity activity to be engaged without triggering an immune response, as two of the three RIG-I recognition features (blunt ends, and fully base-paired stems) can be avoided when designing coligo templates.
More work will be needed to predictably control RNAP III initiation and termination on coligo templates encoding any desired RNA hairpin sequence, but coligos 122 and 19aTAR demonstrate that precise control is attainable. Our findings therefore have the potential to recast the sRNA delivery problem into a more tractable one in which the synthetic informational molecule that requires intracellular delivery is more stable and simpler to synthesize than synthetic RNA, and likely free of the dangers posed by viral vectors. Although lacking canonical transcriptional promoters, coligos having certain features can promote oligonucleotide transcription by RNAP III, and provide an alternative approach to synthetic RNA and viral vectors as a means to generate biologically active sRNA in human cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2 and Supplementary Materials and Methods.
FUNDING
National Institutes of Health (NIH) [R21GM073944 and SC1GM083754 to K.R.]; NIH National Center for Research Resources [2G12RR03060-26A1]; National Institute on Minority Health and Health Disparities [8G12MD007603-27]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank D. Bushnell and R. Kornberg for yeast RNAP II. The authors also thank G. H. Williams for making possible the completion of this work.
REFERENCES
- 1.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr. Opin. Chem. Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA versus shRNA: similarities and differences. Adv. Drug Deliv. Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug. Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 8.Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr. Opin. Chem. Biol. 2006;10:607–614. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J. Intern. Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couto LB, High KA. Viral vector-mediated RNA interference. Curr. Opin. Pharmacol. 2010;10:534–542. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Manjunath N, Wu H, Subramanya S, Shankar P. Lentiviral delivery of short hairpin RNAs. Adv. Drug Deliv. Rev. 2009;61:732–745. doi: 10.1016/j.addr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev. Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 14.Wilson JM. Lessons learned from the gene therapy trial for ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2009;96:151–157. doi: 10.1016/j.ymgme.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 16.Somoza A. Protecting groups for RNA synthesis: an increasing need for selective preparative methods. Chem. Soc. Rev. 2008;37:2668–2675. doi: 10.1039/b809851d. [DOI] [PubMed] [Google Scholar]
- 17.Sabel J. Ultramer Oligonucleotides Mutagenesis Application Guide. An Integrated DNA Technologies User Guide. 2012 http://www.idtdna.com/pages/support/technical-vault/reading-room/user-guides-protocols (8 December 2012, date last accessed) [Google Scholar]
- 18.Roeder RG. Eukaryotic Nuclear RNA Polymerases. In: Losick R, Chamberlin M, editors. RNA Polymerase. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1976. p. 285ff. [Google Scholar]
- 19.Seidl CI, Ryan K. Circular single-stranded synthetic DNA delivery vectors for microRNA. PLoS One. 2011;6:e16925. doi: 10.1371/journal.pone.0016925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daubendiek SL, Ryan K, Kool ET. Rolling-circle RNA synthesis: circular oligonucleotides as efficient Substrates for T7 RNA Polymerase. J. Am. Chem. Soc. 1995;117:7818–7819. doi: 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daubendiek SL, Kool ET. Generation of catalytic RNAs by rolling transcription of synthetic DNA nanocircles. Nat. Biotechnol. 1997;15:273–277. doi: 10.1038/nbt0397-273. [DOI] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2001. [Google Scholar]
- 25.Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 26.Orioli A, Pascali C, Quartararo J, Diebel KW, Praz V, Romascano D, Percudani R, van Dyk LF, Hernandez N, Teichmann M, et al. Widespread occurrence of non-canonical transcription termination by human RNA polymerase III. Nucleic Acids Res. 2011;39:5499–5512. doi: 10.1093/nar/gkr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karn J. Tackling tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 28.Chang K, Elledge SJ, Hannon GJ. Lessons from nature: microRNA-based shRNA libraries. Nat. Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 29.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. Table S1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dedrick RL, Chamberlin MJ. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry. 1985;24:2245–2253. doi: 10.1021/bi00330a019. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Lima WF, Crooke ST. Molecular cloning and expression of cDNA for human RNase H. Antisense Nucleic Acid Drug Dev. 1998;8:53–61. doi: 10.1089/oli.1.1998.8.53. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz LB, Sklar VE, Jaehning JA, Weinmann R, Roeder RG. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J. Biol. Chem. 1974;249:5889–5897. [PubMed] [Google Scholar]
- 33.Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, Bulawa CE, Gould AE, Ocain TD, Dick LR, Errada P, et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot. Cell. 2003;2:256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bardeleben C, Kassavetis GA, Geiduschek EP. Encounters of Saccharomyces cerevisiae RNA polymerase III with its transcription factors during RNA chain elongation. J. Mol. Biol. 1994;235:1193–1205. doi: 10.1006/jmbi.1994.1073. [DOI] [PubMed] [Google Scholar]
- 37.Rumney S, Kool ET. DNA recognition by hybrid oligoether-oligodeoxynucleotide macrocycles. Angew. Chem. Int. Ed Engl. 1992;31:1617–1619. doi: 10.1002/anie.199216171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burroughs AM, Kawano M, Ando Y, Daub CO, Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic Acids Res. 2012;40:1424–1437. doi: 10.1093/nar/gkr903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 43.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.