Abstract
Significant health benefits have been demonstrated for certain probiotic strains through intervention studies; however, there is a shortage of experimental evidence relative to the mechanisms of action. Here, noninvasive experimental procedure based on a colon organ culture system has been used that, in contrast to most experimental in vitro models reported, can preserve natural immunohistochemical features of the human mucosa. This system has been used to test whether commensal lactobacilli (Lactobacillus paracasei BL23, Lactobacillus plantarum 299v and L. plantarum 299v (A−)) were able to hinder inflammation-like signals induced by phorbol 12-myristate 13-acetate (PMA)/ionomycin (IO). Whole genome microarrays have been applied to analyze expression differences, from which mRNA markers could be inferred to monitor the effect of putative probiotic strains under such conditions. Regarding the gene expression, PMA/IO treatment induced not only interleukin (IL)-2 and interferon gamma (IFN-γ), as expected, but also other relevant genes related to immune response and inflammation, such as IL-17A, chemokine (C-X-C motif) ligand (CXCL) 9 and CXCL11. The ex vivo culturing did not modify the pattern of expression of those genes or others related to inflammation. Interestingly, this study demonstrated that lactobacilli downregulated those genes and triggered a global change of the transcriptional profile that indicated a clear homeostasis restoring effect and a decrease in signals produced by activated T cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-012-0301-y) contains supplementary material, which is available to authorized users.
Keywords: Colonic explants, PMA/ionomycin, Lactobacillus paracasei, Lactobacillus plantarum, Human microarrays, Inflammation, Homeostasis, Functionality markers
Introduction
Some commensal bacteria isolated from body fluids or feces with beneficial effects for the host are used as probiotics in food or nutritional supplements preparation. Their effect on human health has been frequently demonstrated through properly designed intervention studies that made them clinically reliable for the prevention or symptom attenuation of specific diseases (Gill and Guarner 2004). However, their mechanisms of action are largely unknown and a great amount of experimental work is currently devoted to this aim through the application of powerful molecular techniques in different experimental models. Human intervention trials and studies using animal models could show that certain probiotic strains exert protective effects in chronic intestinal inflammation (reviewed in Hormannsperger and Haller 2010; Mileti et al. 2009). Different strains may have different forms of interaction with immune and nonimmune cells (Latvala et al. 2008; Mileti et al. 2009), and therefore, their appropriateness has to be specifically determined. Some probiotic strains maintain or restore the epithelial barrier function after pathogen challenge (Otte and Podolsky 2004) or proinflammatory cytokine stimulus (Resta-Lenert and Barrett 2006), and others alter the capacity of intestinal epithelial cells (IEC) to release proinflammatory mediators or directly modify immune cell activity. Studies using IEC cultures have shown the downregulation of the secretion of the proinflammatory chemokine IL-8 (Frick et al. 2007; Kamada et al. 2008; Ma et al. 2004; O’Hara et al. 2006) and other NF-κB regulated chemokines, such as MCP-1 (Petrof et al. 2009) or CCL-20 (Sibartie et al. 2009), as well as a mechanism for the inhibition of IP-10 secretion. Also, a number of works demonstrated that probiotic bacteria directly interacting with immune cells elicit a differential cytokine production, thereby driving the polarization of T cells toward Th1, Th2, Th17 or regulatory T cells (Foligne et al. 2007a, b; Kekkonen et al. 2008; Latvala et al. 2008).
With the aim of disclosing underlying in vivo mechanisms, transcriptomic approaches have been recently applied to study signaling pathways stimulated in the gut mucosa by probiotics in animal models (reviewed in Hormannsperger and Haller 2010). Important information has also been obtained in studies that used human biopsies after probiotic administration for transcriptomic analyses. The administration of Lactobacillus rhamnosus GG affected mainly the expression of genes involved in the immune and inflammatory response, as well as genes related to apoptosis, cell growth and differentiation, signaling and adhesion (di Caro et al. 2005). In a different study, volunteers received Lactobacillus plantarum WCFS1 through an intraduodenal cannula from where biopsies were taken after 6 h. Authors found differences in the modulation of NF-κB-related pathways (Troost et al. 2008; van Baarlen et al. 2009). In a similar experiment performed with other probiotic species (Lactobacillus casei, Lactobacillus acidophilus and L. rhamnosus), the same authors suggested that mucosal responses related to immune response, homeostasis, cell proliferation, Th1–Th2 balance, wound healing and TNF response are differentially induced by the different probiotic strains (van Baarlen et al. 2011). Doubtlessly, experiments carried out in humans provided a lot of information about the mechanisms of action of intestinal probiotics; however, the difficulties in the experimental setups and the need to obtain biopsies from volunteers are major drawbacks. This prompts to the implementation of new experimental models. The implementation of a human colonic organ culture system allows the use of human material obtained from diagnostic biopsies or after surgery. This system has favorable features for laboratory assays: the tissue preserves histological and immunohistochemical features for up to 2 days, and histiotypic relationships and interactions between different cell types remain intact (Dame et al. 2010; Resau et al. 1991). In fact, previous works demonstrated the responsiveness of cultured ileal fragments in coculture with probiotic and commensal bacteria (Borruel et al. 2002; Llopis et al. 2009). Here, in order to get a controlled stimulus that would reproduce proinflammatory responses, human colon explants were incubated with phorbol 12-myristate 13-acetate (PMA)/ionomycin (IO). PMA/IO stimulation has been for long time a standard protocol for general activation of T cells through protein kinase C (PKC) (Kemp and Bruunsgaard 2001), as it has shown to be a potent stimulator of interleukin-2 (IL-2) and interferon gamma (IFN-γ), hence inducing proinflammatory Th1 cytokine profiles (Baran et al. 2001; Heckford et al. 1986; Rostaing et al. 1999; Takahama and Nakauchi 1996).
In this study, colon fragments were incubated with Lactobacillus strains, L. paracasei BL23 (BL23) and L. plantarum 299v (LP) after PMA/IO stimulation. These strains were selected because they displayed in vivo immunomodulatory activity and showed high adhesion ability to extracellular matrix proteins and intestinal tissue (Mangell et al. 2006; Muñoz-Provencio et al. 2009; Rochat et al. 2007; Schultz et al. 2002). Also a nonadherent spontaneous mutant of L. plantarum 299v (LP(A−)) has been included in this study, which had lost the ability to agglutinate Saccharomyces cerevisiae cells in a mannose-sensitive manner and showed reduced adherence to intestinal epithelial cells. In a rat model, LP strain prevented bacterial translocation through the intestinal gut barrier, whereas the mutant strain LP(A−) could not do it (Mangell et al. 2006). Results reported here will show that these probiotic strains restore homeostasis after PMA/IO stimulation and decrease signaling events occurring during T cell activation.
Materials and methods
Bacterial strains and culture conditions
Lactobacillus paracasei BL23, Lactobacillus plantarum 299v and Lactobacillus plantarum 299v (A−) were cultured O/N at 37 °C in de Man-Rogosa-Sharpe liquid medium (Oxoid) under static conditions. Bacteria were harvested after 15-h incubation at early stationary phase (OD600 = 1.0–1.2), and bacterial cell counts were estimated by linear regression to a standard curve correlating colony forming unit (cfu) counts to optical density (600 nm) for each strain. Then, bacteria were added to the tissue culture wells at the appropriate dilution to reach a final concentration of 106 cfu/ml of incubation medium.
Organ culture of human colonic mucosa
Macroscopically normal colonic tissue from a distal region of resected intestine was obtained after adenocarcinoma surgery from 6 patients. Full-thickness colonic wall specimens distant from the tumor and without macroscopic or microscopic lesions were collected. Preparation for surgery was similar for all patients and included gut washes with electrolyte–polyethylene glycol solution and broad-spectrum antibiotic therapy. Specimens were rinsed under a jet of saline and gently washed twice in sterile saline. Multiple mucosal samples, weighing 25–35 mg each, were separated from the underlying tissue. Some of the samples were directly frozen at −80 °C (DF group) and the rest were placed on culture filter plates (15-mm-diameter wells with 500-μm bottom-mesh, Netwell culture systems, Costar, Cambridge, MA) with the epithelial surface uppermost. Filters were placed into wells containing 1,500 μl medium, consisting of RPMI 1640 (CanSera, Rexdale, Ontario, Canada) supplemented with 10 % fetal calf serum (Gibco BRL, Eggenstein, Germany), 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μg/ml gentamycin. Samples were preincubated with antibiotics for 1 h at 37 °C in humidified 5 % CO2 atmosphere to eradicate indigenous flora—total endogenous bacterial counts are always below 102—and equilibrate the tissue to the culture conditions.
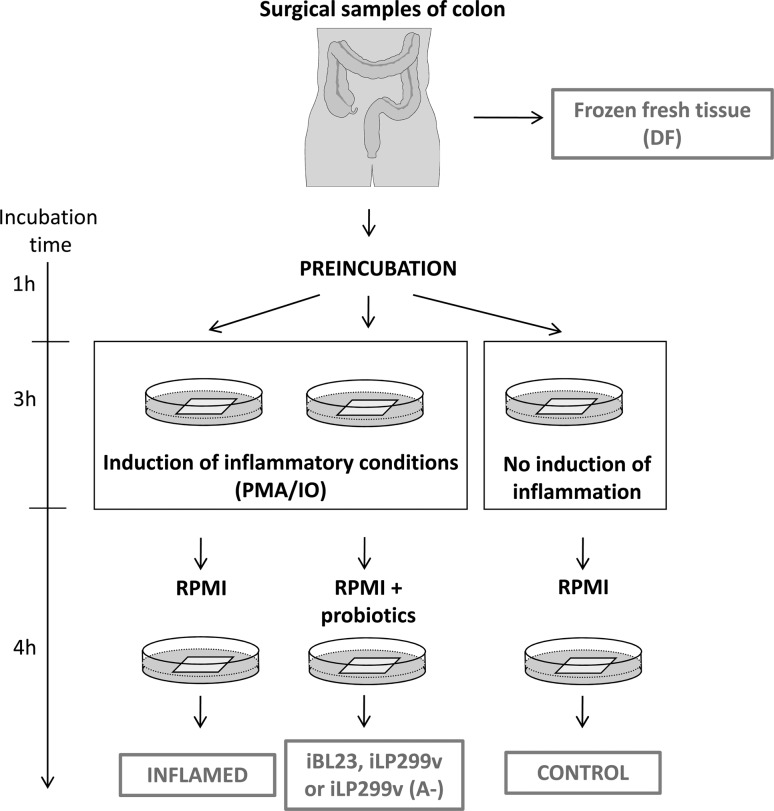
Thereafter, tissues were stimulated for 3 h by the addition of phorbol 12-myristate 13-acetate (PMA) (10 ng/mL, Sigma, Saint Louis, MO) and ionomycin (100 ng/mL, Sigma, Saint Louis, MO). Then, the medium was replaced with fresh medium (Inflamed group) or fresh medium containing L. paracasei BL23 (iBL23 group), L. plantarum 299v (iLP group) or L. plantarum 299v (A−) (iLP(A−) group) (106 cfu/ml), and incubation was continued for 4 h. In addition to these samples, tissue fragments were incubated for the same periods without stimulation by PMA/IO or probiotic bacteria (Control group). Three technical replicates per patient were used for each experimental condition. The supernatant pH was monitored showing small changes during the process (7.4 ± 0.1 pH units). A scheme of the experimental procedure is reproduced in Fig. 1.
Fig. 1.
Experimental design. Macroscopically normal colonic tissue was obtained at surgery from 6 patients with neoplasm. Some tissue fragments were directly frozen (DF) at −80 °C to monitor gene expression changes due to the culture conditions. Colonic tissue fragments were preincubated for 1 h with antibiotics to eradicate indigenous microbiota. Subsequently, the culture medium was replaced with fresh medium containing phorbol 12-myristate 13-acetate (PMA)/ionomycin (IO) to induce inflammation or without PMA/IO (Control sample). Then, the samples were incubated with plain culture medium (Inflamed sample) or culture medium containing 1 × 106 cfu/ml of L. paracasei BL23 (iBL23), L. plantarum 299v (iLP) or the mutant of L. plantarum 299v (iLP(A−)), respectively. During the procedure, bacterial counts (106 cfu/ml) of all three strains had no significant variation and only a small proportion of cell lysis occurred in the tissue fragments, as determined by LDH released to the supernatant in each sample (RSL-1, 30 %; RSL-2, 10.2 %; RSL-6, 9.9 %; RSL-7, 10.0 %; RSL-10, 24.4 %; RSL-21, 10.4 %). Tissues were harvested and stored at −80 °C until total RNA was isolated
LDH activity assessment
Tissue viability was assessed by measuring the release of lactate dehydrogenase into the supernatant according to Llopis et al. (2009). Briefly, tissue samples were homogenized in Tris/HCl (100 mmol/L, pH 7.4) and LDH activity was analyzed by the spectrophotometric method (Pyruvate–Lactate) recommended by the Scandinavian Society of Enzymes. The ratio of LDH activity in the culture supernatant over total LDH activity in tissue homogenates was used to estimate the percentage of viable tissue.
RNA isolation
After incubation, tissues of six different volunteers were harvested in RNAlater (Ambion) and stored at −80 °C for later RNA isolation. Total RNA was isolated using RNeasy mini kit (Qiagen GmbH, Hilden) following the manufacturer’s instructions. Total RNA quantity and integrity were evaluated using RNA 6000 Nano Chip in a Bioanalyzer 2100 (Agilent Technologies). Total RNA was judged to be acceptable if the ribosomal fragments were present in a 28S/18S ratio over 1.5 and RNA integrity number (RIN) to be over 6 (Fleige et al. 2006). RNA samples were stored at −80 °C until use for microarray hybridizations and qPCR analysis.
Microarray analysis
A search for potential candidate genes involved in the modulation of an inflammatory response by probiotic bacteria was performed using Affymetrix microarrays (GeneChip Human Genome U133A Plus 2.0). From three individuals chosen at random (RSL-6, RSL-10 and RSL-21), total RNA of the three replicated samples of each experimental treatment was pooled, and 100 ng was amplified and labeled according to the manufacturer’s two-cycle protocol. The concentration of biotinylated and fragmented cRNA was measured using the 2100 Bioanalyzer (Agilent Technologies), and twenty micrograms were used in the hybridization cocktail. cRNA was hybridized for 16 h to GeneChip arrays. Then, chips were washed, stained with streptavidin/phycoerythrin and scanned with a GeneChip scanner 3,000 (Affymetrix). Affymetrix’s GeneChip Operating Software (GCOS, Affymetrix) was used to obtain and analyze images. After scanning, the obtained files were used to analyze significant changes in gene expression profiles using the dCHIP analysis software (Li and Hung Wong 2001; Li and Wong 2001). Data were normalized using the invariant set method and modeled using the PM/MM model. Global differences between samples were determined by principal component analysis (PCA). To identify significant changes between directly frozen tissue (DF group) and the Control group, dChip software was used to compute 90 % confidence intervals around estimates of the fold change. Genes were selected as differentially expressed if the lower bound of fold change was greater than 1.5. Next, two-sample t test statistics (p < 0.01) was applied to filter significant differences between DF and Control group. All data discussed in this publication was MIAME compliant and the raw data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE23630.
Analysis of variance (ANOVA) (p < 0.01) was applied to find significant changes among the experimental groups: Control, PMA/IO treated (Inflamed) and the three groups incubated with probiotics after PMA/IO treatment (BL23, LP and LP(A−)). Genes exceeding fold changes of ±1.5 were considered for hierarchical cluster analysis. Given the small changes in gene expression expected from probiotic action and the variability of gene expression between individuals (van Baarlen et al. 2009), restrictive statistical criteria, such as very low p values or FDR filtering, were omitted to avoid the loss of important biological information.
Functional annotation analysis
Gene lists corresponding to the different clusters or to significantly up- and downregulated genes (p < 0.01 and fold change exceeding ± 1.5) in each experimental condition were analyzed using D.A.V.I.D. (Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/)). D.A.V.I.D. is based on the established biomedical literature and uses specific approaches to estimate significance (p values) (Huang et al. 2009). We focused our analysis on annotations and pathways derived from the Gene Ontology (GO) database and KEGG, respectively. Gene Ontology data are represented according to the categories of “biological process,” “cell component” and “molecular function.” The p values for a given category and term provide an estimate of the likelihood that a given annotation is enriched in a given gene list by chance alone when compared to the human genome. All of the p values presented in the D.A.V.I.D.-based analyses have been corrected for multiple comparisons using false discovery rate (FDR) value ≤5 %.
qRT-PCR
Total RNA (1 μg) used for reverse transcription was obtained from the six individuals: three samples used in microarray hybridizations (RSL-6, RSL-10 and RSL-21), plus three additional ones (RSL-1, RSL-2 and RSL-7) which were used for additional qPCR validation of selected genes. The reactions were performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the instructions of the manufacturer. Then, quantitative real-time PCR was performed using 96-well plates with the LightCycler 480 SYBR Green I Master Mix (Roche) and a LightCycler 480 apparatus (Roche). PCR cycling conditions comprised an initial polymerase activation step at 95 °C for 10 min followed by 40 cycles of 10 s at 95 °C, 10 s at 59 °C and 12 s at 72 °C with a single fluorescence measurement. For the commercial primer pair for granzyme B (GZMB) (SA Biosciences), an annealing temperature of 60 °C was used according to the manufacturer’s instructions. After each run, a dissociation curve was performed to further confirm specificity of the product and the absence of primer-dimers. Agarose gel electrophoresis was run to demonstrate that qPCR yielded a unique band. The primer sequences were as follows (sense and antisense, respectively): interleukin 2 (IL2): 5-TCCCAAACTCACCAGGATGCTCACA-3, 5-GCACTTCCTCCAGAGGTTTGAGTTCT-3; interleukin 17A (IL17A): 5-TTCCCCCGGACTGTGATGGT-3, 5-CGGCACTTTGCCTCCCAGAT-3; guanylate-binding protein 1, interferon-inducible (GBP1): 5-TCCCAGCCCTACAACTTCGG-3, 5-TGCCATGTCCAGGCTGTTCC-3. The primer sequences for interleukin 2 receptor α (IL2RA), chemokine (C-X-C motif) ligand (CXCL) 11, ubiquitin D (UBD), tumor necrosis factor receptor superfamily member 9 (TNFRSF9), interferon γ (IFN-γ) and CXCL9 have been previously described (Nishioka et al. 2007; Pattyn et al. 2003; Spandidos et al. 2008; Wang and Seed 2003).
After validation of seven housekeeping genes (β-actin (ACTB), β-2 microglobulin (B2M), glyceraldehyde-3P dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L13a (RPL13A), succinate dehydrogenase complex, subunit A (SDHA) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ)) using the GeNorm application (Vandesompele et al. 2002), B2M was chosen due its high expression stability and the absence of pseudogenes in the human genome. Reactions were performed in triplicate, and relative quantification of gene expression was achieved using the efficiency-corrected method according to Pfaffl (2001). Primer sequences of the housekeeping genes ACTB, GAPDH, HPRT1, RPL13A, SDHA, B2M and YWHAZ were used as reported elsewhere (Chiu et al. 2005; Pattyn et al. 2003).
Derivation of gene networks
Gene lists of differentially expressed genes (p < 0.01 and fold change exceeding ±1.5) were also analyzed for the presence of gene networks using the Ingenuity Pathways Analysis (IPA) application (http://www.ingenuity.com) (Ingenuity Systems, Redwood City, CA). The IPA application provides a tool for the discovery of gene networks within the uploaded gene lists. It is a database integrating this information with other relevant databases such as EntrezGene and Gene Ontology. IPA computes a score for each network according to the fit of the set of uploaded focus genes. The score is used to rank networks according to their relevance to the genes in the input dataset. These scores, derived from p values, indicate the likelihood of focus genes to belong to a network versus those obtained by chance. A high number of focus genes within a dataset leads to a higher network score. Mathematically, the score is simply the negative exponent of the p-value calculation. For example, a score of three indicates that there is a 1/1,000 chance that the focus genes are in a network due to random chance. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge. In our gene network analysis, we focused on direct relationships between proteins. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation.
Ethical considerations
The project was approved by the local ethics committee (Comité Ético de Investigación Clínica, Hospital Vall d′Hebron), and a written informed consent approved by the ethics committee was obtained from all participants involved in the study.
Results
Principal component analysis (PCA)
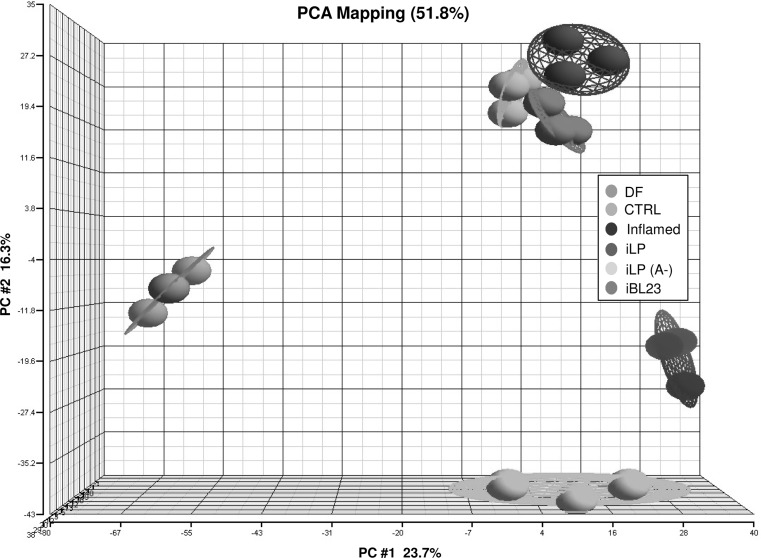
The suitability of a model system to study a given biological process relies on its reproducible responses to stimuli, but most important, it must be as close as possible to the in vivo conditions. In this work, a colon organ culture system has been used as model to study gene expression changes in response to probiotic bacteria after an inflammation-like stimulus. An amount of fresh tissue was preserved frozen (DF) before starting all the incubations to monitor alterations caused by the experimental protocol. Then, colon fragments treated with PMA/IO were incubated with plain culture medium (Inflamed sample), or with lactobacilli, L. paracasei BL23 (sample iBL23) and L. plantarum 299v (sample iLP) and with a nonadherent mutant of L. plantarum 299v (sample iLP(A−)). To study the global differences and correlations between the samples, PCA was performed using all probe sets on the microarray. Figure 2 shows a PCA with the five experimental groups of samples (Control, Inflamed, iLP, iLP(A−) and iBL23), as well as the fresh directly frozen tissue (DF), each one consisting of three biological replicates. Samples treated with probiotics grouped together and were separated from both Control and PMA/IO-treated tissue. PCA demonstrates that the set of directly frozen explants is different to the rest of the samples. This difference is observed in the main principal component analysis (explaining 23.7 % of variability) and it is due to the set of genes differentially expressed (see below) as consequence of the experimental setup.
Fig. 2.
Supervised principal component analysis (PCA). The gene expression profiles of DF, Control, stimulated with PMA/IO (Inflamed), iBL23, iLP and iLP(A−) samples from 3 different patients (RSL-6, RSL-10 and RSL-21) were compared using PCA. The three-dimensional (3D) plot view of gene expression data (including all probe sets on U133 Plus 2.0 GeneChip) is shown with respect to their correlation to the first three principal components
Gene expression changes due to the organ culture system
In order to certify the usefulness of the model, it was first necessary to evaluate whether the complete experimental procedure severely affected gene expression. Therefore, differences in gene expression were determined between fresh human mucosa directly frozen after extirpation (DF) and the Control sample in the organ culture model system without any treatment. Using the filtering criteria described in the Materials and Methods section, we identified 283 genes differentially expressed between the two groups; 229 genes were upregulated and only 54 genes were downregulated due to the experimental setup. In order to infer the biological meaning of up- and downregulated genes, gene ontology and pathway analysis (KEGG) were performed (Table 1). Incubation of intestinal colon fragments induced not only genes that predominantly corresponded to apoptosis and cell death, but also response to stress, wound healing and blood coagulation. A significant portion of downregulated genes were located to the mitochondrion or mitochondrion membrane and contained genes like cytochrome c oxidase subunit Va (COX5A) and cytochrome c oxidase assembly protein (yeast) COX15 homolog, both involved as terminal components of the mitochondrial respiratory chain. In summary, these gene expression patterns indicate that the incubation of tissue fragments have triggered wound response mechanisms. Only a few genes related to inflammation and immune response were found among upregulated genes, such as IL-8 and tumor necrosis factor alpha-induced protein 6 (TNFAIP6) (complete information Online Resource 1). They are two relevant mediators of the inflammatory response (Milner et al. 2006), which together with the aforementioned genes related to apoptosis and cell death situate the cultured tissue fragments in a surgical stress frame (Chachkhiani et al. 2005).
Table 1.
Functional annotation analysis (DAVID) for up- and downregulated genes in the comparison of Control sample to directly frozen human mucosa (DF)
| Category | Annotation term | Significance (FDR) | Molecules |
|---|---|---|---|
| Upregulated genes | |||
| Biological processes (top ten regulated terms) | Apoptosis | 1.53E−07 | 30 |
| Death | 1.54E−07 | 31 | |
| Cell death | 1.54E−07 | 31 | |
| Programmed cell death | 1.86E−07 | 30 | |
| Biological regulation | 3.21E−07 | 94 | |
| Cell development | 3.71E−06 | 36 | |
| Developmental process | 4.20E−06 | 66 | |
| Cell differentiation | 6.70E−06 | 45 | |
| Cellular developmental process | 6.75E−06 | 45 | |
| Negative regulation of biological process | 1.02E−05 | 34 | |
| Cellular components | Intracellular part | 3.29E−02 | 130 |
| Molecular functions | Protein binding | 1.27E−07 | 113 |
| Binding | 1.18E−02 | 153 | |
| KEGG pathway | Complement and coagulation cascades | 4.83E−03 | 7 |
| Downregulated genes | |||
| Cellular components | Organelle membrane | 2.36E−04 | 14 |
| Mitochondrial inner membrane | 9.46E−03 | 6 | |
| Organelle inner membrane | 1.28E−02 | 6 | |
| Cytoplasmic part | 2.34E−02 | 19 | |
| Mitochondrial part | 2.48E−02 | 7 | |
| Mitochondrial membrane | 2.84E−02 | 6 | |
| Mitochondrion | 3.13E−02 | 9 | |
| Organelle envelope | 3.45E−02 | 7 | |
| Mitochondrial envelope | 3.50E−02 | 6 | |
| Envelope | 3.52E−02 | 7 | |
| Integral to organelle membrane | 3.97E−02 | 4 | |
| Membrane | 4.01E−02 | 28 | |
Since genes related to inflammation were not significantly enriched by the experimental processing of the explants, differential gene expression will be investigated in the following experiments, to determine the effect of different commensal probiotic strains on colonic explants subjected to a PMA/IO treatment that mimics some inflammatory conditions which involves T cell activation.
Identification of gene expression changes in PMA/IO-treated explants and PMA/IO-treated explants after incubation with Lactobacillus strains
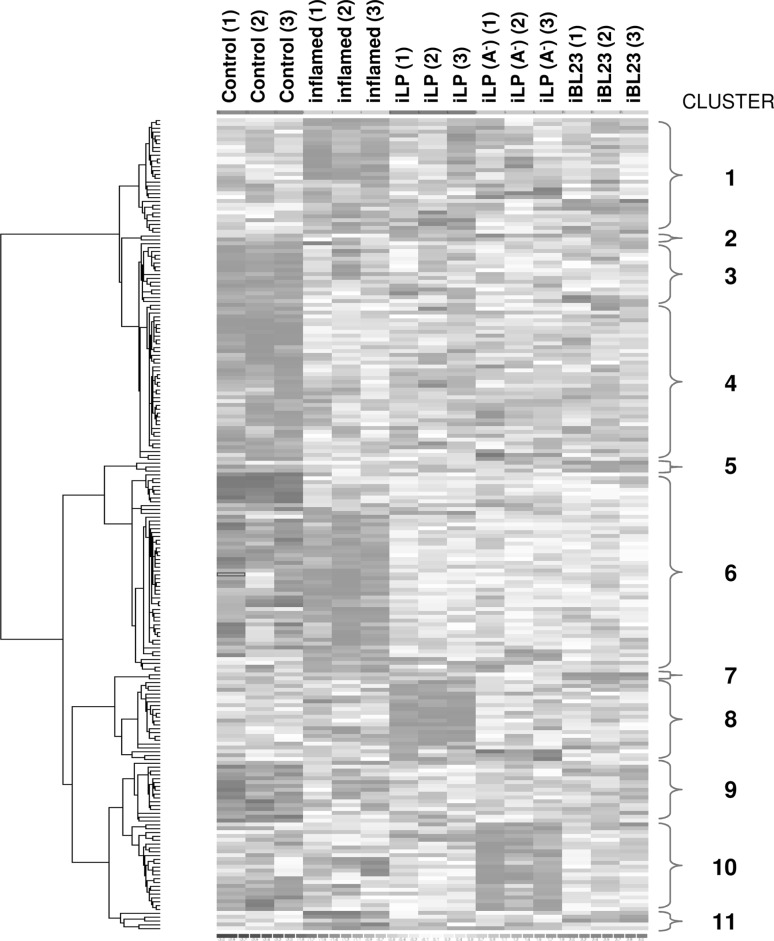
After ANOVA analysis (p ≤ 0.01 and fold change ≥ ±1.5), 199 differentially expressed genes were identified among the five experimental groups. Then, cluster analysis was performed to classify those genes according to their expression profiles, giving rise to 11 subclusters that differed in their size ranging from 2 to 52 genes (Fig. 3). Mean expression levels were mostly between ±1.5-fold and ±3-fold, although some genes exhibited inductions of 40-fold and downregulations of almost 10-fold. Genes related to inflammation and other relevant processes were found in clusters 1, 3, 4 and 6. Analysis of clusters containing less than 5 genes will not be described. A list of all genes belonging to the eleven clusters is provided in Online Resource 2.
Fig. 3.
Supervised hierarchical cluster analysis. Significantly regulated genes (p < 0.01) were used for 2D hierarchical clustering of Control, stimulated with PMA/IO (Inflamed), iLP, iLP(A−) and iBL23 samples from 3 different patients ((1)RSL-6, (2) RSL-21 and (3) RSL-10)). Each row displays expression of an individual probe set and each column of a biological replicate. The red color represents an expression level above the mean expression of a gene across all samples, and the blue color represents an expression level lower than the mean. The brighter the color, the stronger the factor of gene regulation. White indicates no changes
Cluster 1 includes genes slightly upregulated (average 1.28-fold change) in response to PMA/IO (Inflamed group) and repressed after treatment with the three probiotic strains (iLP, iLP(A−) and iBL23 groups). This cluster contains genes important in inflammatory processes such as signal transducer and activator of transcription 4 (STAT4), interleukin 7 receptor (IL7R), interferon-stimulated transcription factor 3 γ (ISGF3G) or genes that are implicated in nuclear factor κ-light-chain-gene enhancer of activated B cells (NF-κB) signaling, like receptor-interacting serine/threonine-protein kinase 1 (RIPK1). Cluster 3 includes genes that were downregulated after PMA/IO treatment when compared to Control tissue, but on which probiotics did not show any influence. Members of this cluster are genes relevant to signal transduction, such as mitogen-activated protein kinase kinase kinase 12 (MAP3K12) and phosphoinositide-3-kinase, catalytic, β polypeptide (PIK3CB), or which are related to major histocompatibility complexes (MHC) I and II, like MHC-related protein 1 (MR1) and class II MHC transactivator (CIITA). Cluster 4 contains genes that were slightly downregulated (average 1.56-fold change) after PMA/IO treatment and on which subsequent probiotic incubations provoked a more pronounced downregulation. Some genes of this cluster are relevant due to their importance in inflammatory processes, such as serpin peptidase inhibitor 2, clade B (SERPINB2), interleukin 13 receptor alpha (IL13RA), chemokine (C-X-C motif) ligand (CXCL)5, CXCL6 and protein inhibitor of activated STAT2 (PIAS2), which is a transcriptional regulator of STAT4, and other transcription factors involved in MAP kinase signaling.
Cluster 6 encompassed genes known to be specifically induced by PMA/IO (IFN-γ and IL-2), as well as other proinflammatory cytokines and chemokines, such as IL-17A, CXCL9, CXCL11 and chemokine (C motif) ligand 1 (XCL1). These genes were highly upregulated (fold change from 7.7 to 14.3) after PMA/IO treatment (Inflamed group) (Table 2). Most notably, their expression after treatment with the three probiotic strains (iLP, iLP(A−) and iBL23 groups) was significantly downregulated with respect to PMA/IO-stimulated samples. Besides, 2 genes of the tumor necrosis factor receptor superfamily (TNFRSF), TNFRSF9 and TNFRSF4, as well as members of the family of guanylate-binding proteins (GBP) also belong to this cluster. Due to their biological interest and to the relevance for the objectives of this work, differential expression of some of these genes (IL-2, IL2RA, IFN-γ, IL-17A, GBP1, CXCL11, CXCL9, GZMB, UBD and TNFRSF9) was used for the validation of the microarray data (see below).
Table 2.
Differentially expressed genes in Cluster 6
| Symbol | Gene | Fold changea | |||
|---|---|---|---|---|---|
| Inflamed | iLP | iLP(A−) | iBL23 | ||
| IL17A | Interleukin 17A | 14.3 | 6.9 | 5.6 | 5.9 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 10.9 | 4.2 | 6.6 | 3.1 |
| IFNG | Interferon, gamma | 10.5 | 5.9 | 4.8 | 4.4 |
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 9.9 | 3.0 | 3.5 | 2.4 |
| IL2 | Interleukin 2 | 9.5 | 2.2 | 2.2 | 2.8 |
| XCL1 | Chemokine (C motif) ligand 1 | 7.7 | 4.3 | 3.3 | 4.1 |
| GZMB | Granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | 6.5 | 2.3 | 3.2 | 2.6 |
| TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 | 4.5 | 1.9 | 2.3 | 1.6 |
| INDO | Indoleamine-pyrrole 2,3 dioxygenase | 4.5 | 1.8 | 1.3 | 1.6 |
| XIRP1 | Xin actin-binding repeat containing 1 | 4.4 | 2.8 | 2.7 | 2.2 |
| RNF19B | Ring finger protein 19B | 4.2 | 1.3 | 2.2 | 1.6 |
| ZBED2 | Zinc finger, BED-type containing 2 | 3.7 | 2.4 | 1.7 | 1.9 |
| GBP1 | Guanylate-binding protein 1, interferon-inducible, 67 kDa | 3.5 | 2.0 | 1.6 | 2.0 |
| IRF1 | Interferon regulatory factor 1 | 3.2 | 2.5 | 2.9 | 2.4 |
| CRTAM | Cytotoxic and regulatory T cell molecule | 3.2 | 2.3 | 1.7 | 2.1 |
| CD226 | CD226 molecule | 3.0 | 1.4 | 1.6 | 1.5 |
| ENPP3 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | 3.0 | 1.4 | 1.9 | 1.6 |
| GBP4 | Guanylate-binding protein 4 | 2.9 | 1.6 | 1.6 | 1.3 |
| UBD | Ubiquitin D | 2.9 | 2.2 | 2.4 | 1.9 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 2.9 | 1.8 | 1.3 | 2.2 |
| IL2RA | Interleukin 2 receptor, alpha | 2.7 | 2.1 | 1.7 | 2.0 |
| TNFRSF4 | Tumor necrosis factor receptor superfamily, member 4 | 2.7 | 1.8 | 1.5 | 1.6 |
| ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 2.7 | 1.6 | −1.1 | 1.6 |
| EGR2 | Early growth response 2 | 2.6 | 2.1 | 2.3 | 2.0 |
| GBP2 | Guanylate-binding protein 2, interferon-inducible | 2.1 | 1.9 | 2.0 | 1.8 |
| IRF1 | Interferon regulatory factor 1 | 2.4 | 2.3 | 2.6 | 2.3 |
| GJA4 | Gap junction protein, alpha 4, 37 kDa | 2.3 | 2.0 | 2.7 | 2.1 |
| DUOX1 | Dual oxidase 1 | 2.1 | 1.3 | 1.4 | 1.8 |
| STK19 | Serine/threonine kinase 19 | 2.1 | 1.3 | 1.2 | 1.4 |
| CPN1 | Carboxypeptidase N, polypeptide 1 | 2 | 1.5 | 1.3 | 1.7 |
| CTSS | Cathepsin S | 2.0 | 1.3 | 1.3 | 1.1 |
| CYB561 | Cytochrome b-561 | 1.9 | 1.3 | 1.9 | 1.4 |
| PPP1R11 | Protein phosphatase 1, regulatory (inhibitor) subunit 11 | 1.8 | 1.0 | 1.2 | 1.1 |
aFold changes are expressed relative to Control
Gene expression of the genes in Clusters 8, 9 and 10 remained unchanged after PMA/IO treatment, but signal intensities were higher in the probiotic groups: Cluster 8 represents genes upregulated in iLP explants, Cluster 10 in the iLP(A−) samples and Cluster 9 encompasses genes which, compared to Control samples, were upregulated in iLP, iLP(A−) and iBL23 samples. Cluster 8 contains genes that were highly upregulated in the iLP group: thyroid adenoma associated (THADA) (fold change (FC) +41.7), beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) (B3GAT1) (FC +23.2) and PHD finger protein 8 (PHF8) (FC +20.8). B3GAT1 codes for a glucuronosyltransferase, a key enzyme in the biosynthesis of the carbohydrate epitope CD57, present in natural killer (NK) cells and subsets of B cells, T cells and monocytes. Another gene related to carbohydrate interaction is sialic acid-binding Ig-like lectin 8 (SIGLEC8), also upregulated in the iLP samples. Interestingly, in the iLP group two enzymes known to be involved in epigenetic modifications were upregulated: PHF8 (FC +20.8) and euchromatic histone–lysine N-methyltransferase 2 (EHMT2) (FC +1.9). EHMT2 was downregulated by PMA/IO (FC-1.9), and expression levels in iBL23 (FC +1.1) and iLP(A−) (FC +1.1) groups were similar to Control tissue, indicating that all 3 strains were able to reverse the effect of PMA/IO treatment on that gene to a different extent. Cluster 10 contains genes specifically upregulated in the iLP(A−) group, such as low-density lipoprotein receptor-related protein 6 (LRP6), a co-receptor involved in Wnt/beta catenin signaling; B cell CLL/lymphoma 10 (BCL10), which is known to activate NF-κB and induce apoptosis, and trinucleotide repeat containing 6C protein (TNRC6C), implicated in the modulation of gene expression by repression of miRNAs.
Microarray validation and characterization of likely markers
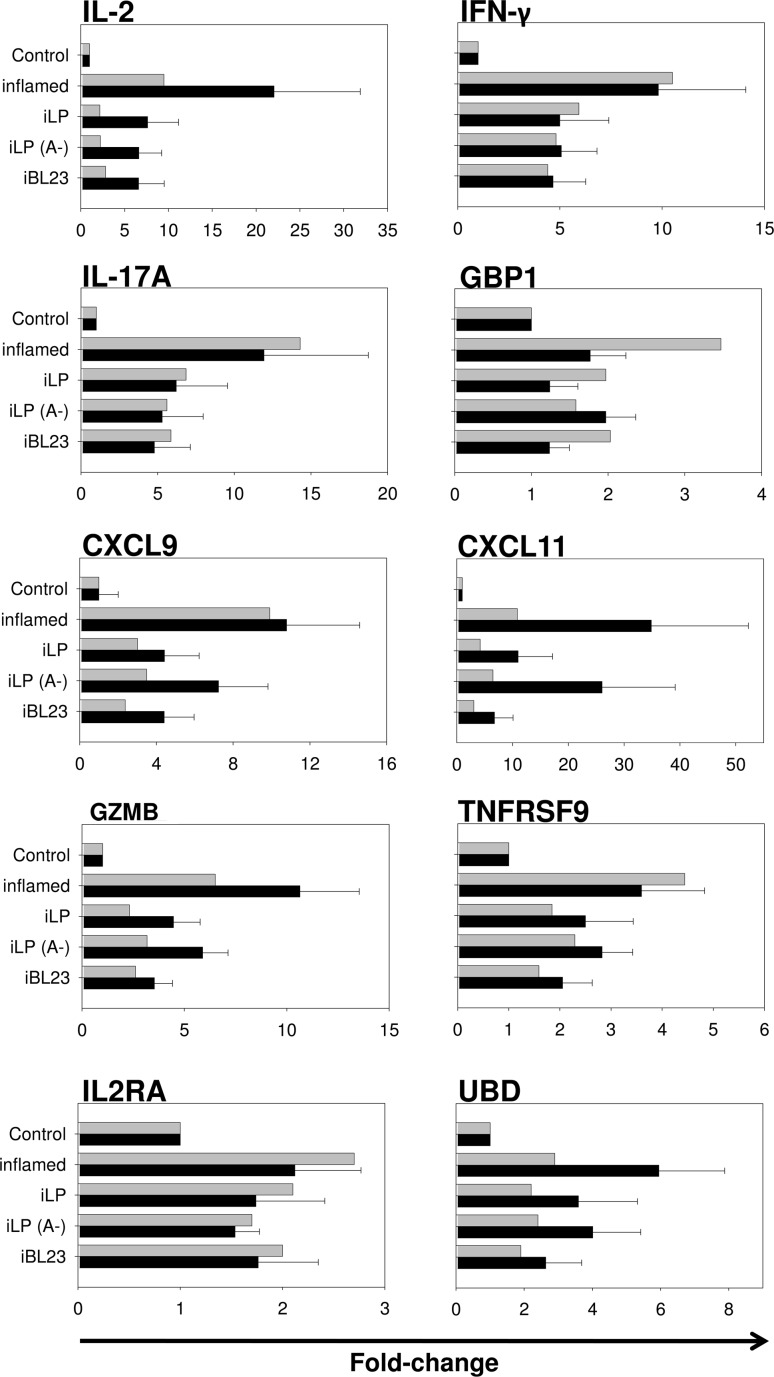
In order to test the results obtained from the microarrays, differential expression of 10 relevant genes was checked by real-time qPCR from the mRNA samples used for the microarray assays. Random distribution of the probe sets on the Affymetrix GeneChips ® allowed to assay genes of particular relevance for the objectives of this work (Fig. 4, Online Resource 3). The trend perfectly reproduced fold change differences found in microarrays. The group of selected genes (IL-2, IL2RA, IFN-γ, IL-17A, GBP1, CXCL11, CXCL9, GZMB, UBD and TNFRSF9) are directly related to inflammatory response pathways, they were induced by the PMA/IO treatment and all of them were downregulated to different degrees after treatment with lactobacilli. Therefore, they could constitute appropriate indicators of probiotic effect in this model system if data were confirmed in additional biological samples. Hence, expression of those 10 genes was tested on samples from three additional volunteers subject to PMA/IO induction and followed by the incubation with the same Lactobacillus strains (Fig. 5, Online Resource 4). The pattern of expression was remarkably reproduced, showing that all the genes tested were downregulated by lactobacilli after PMA/IO treatment. As in previous data (Fig. 4, Online Resource 3), differences in relative expression was more pronounced in IL-2, IFN-γ, CXCL9, CXCL11 and IL17A, which suggested that expression of those genes could be used to monitor the probiotic modulation of inflammatory pathways after the PMA/IO stimulus in cultured mucosal fragments.
Fig. 4.
Validation of microarray data. qPCR results (black bars) of selected genes using total RNA from colonic explants (pooled data of samples from 3 patients used in microarray hybridizations (RSL-6, RSL-10 and RSL-21)) and comparison with microarray data (gray bars). Fold changes are calculated relative to Control tissue and normalized to B2M expression. Expression values of each individual patient obtained by qPCR and microarray hybridization are shown in Online Resource 3
Fig. 5.
Expression ratios of validated genes in additional samples. Data obtained by real-time qPCR from RNA of colonic explants of 3 additional individuals (RSL-1, RSL-2 and RSL-7). Expression ratios of ten relevant marker genes in the samples treated with PMA/IO, and LP (black bars), LP(A−) (gray bars) and BL23 (hatched bars) are represented relative to PMA/IO-stimulated explants (expression ratio 1.0) and normalized to B2M expression. Expression values of each individual patient are shown in Online Resource 4
Identification of significant pathways and gene ontology annotations
Gene lists of each of the eleven clusters were uploaded to the D.A.V.I.D. database and analyzed for the presence of significant signaling pathways and gene ontology annotation terms grouped into the categories signaling pathway, biological process, cellular component and molecular function. For genes in clusters 1, 4 and 6, at least one significant annotation term was identified (Online Resource 5). Cluster 1 was only enriched in genes belonging to the annotation term regulation of primary metabolic process. Interestingly, cluster 4, representing genes particularly downregulated in iLP, iLP(A−) and iBL23 explants, is enriched in genes belonging to the annotation terms heparin, glycosaminoglycan, polysaccharide, pattern and carbohydrate binding (CXCL6, FGF2, FGFR1, SERPINE2, PRG2). Cluster 6 shows significant enrichment in genes related to immune response, regulation of the immune system, regulation of different subtypes of immune cells and was significantly overrepresented in genes involved in cytokine–cytokine receptor interaction. Cluster 6 is of particular interest for our study, since it includes proinflammatory key genes that were upregulated in the Inflamed explants and whose expression was downregulated after incubation with probiotics.
In addition to the functional annotation analysis of the eleven subclusters, the lists of up- and downregulated genes, in Inflamed versus Control samples and probiotic (iLP, iLP(A−) and iBL23) versus Inflamed samples, were uploaded to D.A.V.I.D. database. This analysis showed that genes upregulated in Inflamed samples were significantly related to immune response, immune system process and cytokine–cytokine receptor interaction (Table 3, complete information Online Resource 6). The same annotation terms could be retrieved in the lists of downregulated genes in each probiotic group when compared to the Inflamed samples, thereby confirming the results of the D.A.V.I.D. analysis for cluster 6. This indicates again that the three tested strains selectively modulate the expression of genes related to the immune system in the intestinal mucosa. Although probiotic treatment of the Inflamed explants provoked mainly a downregulation of genes, upregulated genes were found in the three strains tested that could not be significantly related to any annotation term (iLP samples: 30 genes up-/86 downregulated; iLP(A−) samples: 32 genes up-/78 downregulated; and iBL23 samples: 23 genes up-/79 downregulated).
Table 3.
Functional annotations analysis corresponding to up- and downregulated genes in Inflamed compared to Control and to downregulated genes in iBL23, iLP and iLP(A−) compared to Inflamed sample (The analysis is based on the default parameters in D.A.V.I.D. and uses FDR correction for multiple testing)
| Inflamed versus CTRL | iLP versus Inflamed | iLP(A−) versus Inflamed | iBL23 versus Inflamed | ||
|---|---|---|---|---|---|
| Category | ↑ Regulated genes | ↓ Regulated genes | ↓ Regulated genes | ↓ Regulated genes | ↓ Regulated genes |
| Biological Processes | Immune response | Response to wounding | Immune response | Immune response | Immune response |
| Immune system process | Wound healing | Taxis | Immune system process | Immune system process | |
| Positive regulation of immune system process | Regulation of localization | Chemotaxis | Response to external stimulus | Regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | |
| Regulation of lymphocyte proliferation | Regulation of transport | Response to external stimulus | Response to wounding | Regulation of adaptive immune response | |
| Regulation of leukocyte proliferation | Response to external stimulus | Immune system process | Response to stimulus | Response to external stimulus | |
| Cellular components | ns | ns | Extracellular space | Extracellular space | Extracellular space |
| Extracellular region part | Extracellular region part | Extracellular region part | |||
| Extracellular region | Extracellular region | ||||
| Molecular functions | Cytokine activity | Heparin binding | Cytokine activity | Cytokine activity | Cytokine activity |
| Glycosaminoglycan binding | Chemokine activity | Receptor binding | Receptor binding | ||
| Pattern binding | Chemokine receptor binding | Chemokine activity | Chemokine activity | ||
| Polysaccharide binding | G-protein-coupled receptor binding | Chemokine receptor binding | G-protein-coupled receptor binding | ||
| Receptor binding | G-protein-coupled receptor binding | Chemokine receptor binding | |||
| KEGG pathways | Cytokine–cytokine receptor interaction | ns | Cytokine–cytokine receptor interaction | Cytokine–cytokine receptor interaction | Cytokine–cytokine receptor interaction |
| Jak-STAT signaling pathway | Jak-STAT signaling pathway | ||||
KEGG Kyoto encyclopedia of genes and genomes; ns none significant
Comparison of gene networks of PMA/IO-treated explants and PMA/IO-treated explants after incubation with Lactobacillus strains
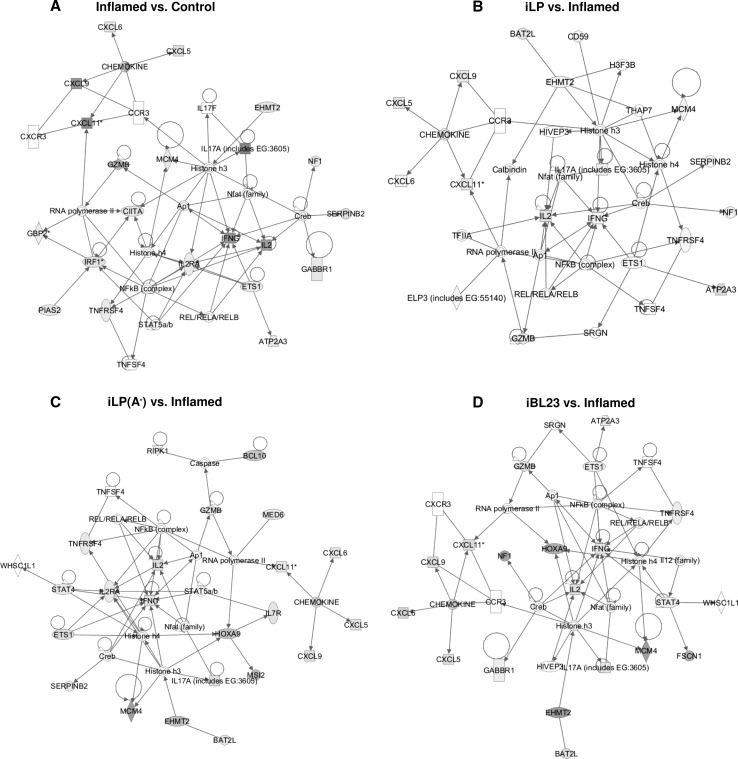
Gene networks were derived by introducing gene lists of Inflamed compared to Control samples and iLP, iLP(A−), iBL23 samples compared to Inflamed explants into the IPA database as focus molecules. Several high-scoring networks were identified (Online Resource 7). The networks with the highest scores are represented in Fig. 6. The top functions of all networks include either cell-to-cell signaling and interaction, inflammatory response or both. Consequently, in all of the four networks cytokines IL-2 and IFN-γ are target of several transcription factors (activator protein 1 (AP1), cAMP response element-binding (Creb), nuclear factor of activated T cells (NFAT), NF-κB, STAT4) and are highly upregulated in Inflamed tissue and downregulated when Inflamed tissue is incubated with the three Lactobacillus strains, evidencing the downregulation of proinflammatory gene expression by probiotic bacteria.
Fig. 6.
Ingenuity protein–protein interaction networks. The top-scoring network of genes induced in Inflamed compared to Control includes 21 focus molecules and has an assigned score of 39. This network correlates with the following functions: cell-to-cell signaling and interaction, inflammatory response, hematological system development and function (a). The top-scoring network of genes induced in iLP compared to Inflamed includes 18 focus molecules and has an assigned score of 31. This network correlates with the following functions: cell-to-cell interaction and signaling, hematological system development and function, immune cell trafficking (b). The top-scoring network of genes induced in iLP(A−) compared to Inflamed includes 22 focus molecules and has an assigned score of 41. This network correlates with the following functions: cellular development, cellular growth and proliferation, and inflammatory response (c). The top-scoring network of genes induced in iBL23 compared to Inflamed includes 18 focus molecules and has an assigned score of 32. This network correlates with the following functions: cell-to-cell signaling and interaction, inflammatory response, hematological system development and function (d). The intensity of the node color indicates the degree of up-(red) or down-(green) regulation. See Online Resource 8 for network legend
Discussion
Several in vitro or ex vivo model systems have been used to study the interactions between commensal bacteria and the host. They are particularly relevant to the study of likely probiotic effects, which, as other healthy foods and ingredients, have been commercially successful during the last years (Perez Martinez et al. 2012). In general, some evidence obtained by intervention or observational studies needs to be supported by the description of their mechanism of action and the discovery of biomarkers, in order to comply with the regulations of governmental and supranational organizations, as occurs in Japan (Yamada et al. 2008), U.S. FDA (Office of Nutrition 2009), EU (Regulation 1924/2006) and FAO report on Functional Foods (Food Quality and Standards Service 2007). During the past twenty years, cultured intestinal fragments from patients suffering inflammatory bowel disease (IBD) were used to determine their cytokine profile or the effects of several drugs, through the detection of inflammatory mediators (Borruel et al. 2002; Mahida et al. 1991), yet transcriptomic studies on cultured intestinal explants are scarce. Here, our main aim was to determine whether strains of the genus Lactobacillus could attenuate inflammatory processes in a colon organ culture model. The comparison of gene expression of colonic mucosa subjected to the experimental process (untreated Control) with directly frozen fresh mucosa suggested a certain degree of tissue decay. Nevertheless, the experimental procedure did not enhance the expression of proinflammatory or immune system–related genes, which let us consider the colon organ culture model suitable for the purpose of this study. In this approximation, we tested a combination of phorbol 12-myristate 13-acetate and ionomycin (PMA/IO). PMA/IO exposure is a commonly used procedure for the activation of T cells and for the simulation of inflammatory conditions in human colonic explants, since PMA/IO act as potent inducers of Th1-type cytokines IL-2 and IFN-γ, which are found upregulated in inflammatory bowel diseases (Niedel et al. 1983; Silva et al. 2005).
The analysis of the data in our experiments confirmed that PMA/IO provoked an inflammation-like pattern. A set of important genes related to inflammation (mostly grouped in cluster 6, Fig. 3) were upregulated after PMA/IO treatment and, interestingly, all the Lactobacillus strains used reduced their expression level, supporting the concept that probiotics can influence immune homeostasis (Hill and Artis 2010). Cluster 6 grouped a significant number of genes coding for proinflammatory cytokines, chemokines and other inflammatory mediators, including IL-2, IFN-γ, IRF-1, IL-17A, CXCL9, CXCL11, TNFRSF4 or GZMB, that were upregulated by PMA/IO, but markedly reduced by all the lactobacilli, LP, LP(A−) and BL23. Among them, a number of cytokine genes were upregulated by PMA/IO, which suggested T cell proliferation (IL-2, IFN-γ and IL-17A) (Murphy et al. 2007) and T cell activation (IL2RA) (Gao et al. 2010; Ledger et al. 2004). The reduction in the expression of those genes after treatment with the lactobacilli suggests that all three strains probably counteract the molecular events leading to T cell activation, as shown before for the probiotic strain L. casei DN 114-001 (Carol et al. 2006). Reduction of IFN-γ could be a very interesting probiotic feature since, besides its crucial role for innate and adaptive immunity against intracellular pathogens, prolonged high levels of IFN-γ lead to chronic autoimmune and inflammatory diseases, such as multiple sclerosis and IBD (Bouma and Strober 2003; Rovedatti et al. 2009; Schoenborn and Wilson 2007). High levels of IL-17A are found in IBD; it is secreted by Th17 cells, CD8+ T cells, natural killer cells and neutrophils and acts on both immune and nonimmune cells inducing a number of cytokines, chemokines, inflammatory effector molecules and antimicrobial proteins (Onishi and Gaffen 2010). Other genes of this cluster known to be overexpressed in IBD were the chemokines CXCL9 and CXCL11 (Egesten et al. 2007; Singh et al. 2007). Both are chemotactic for leukocytes as well as activated T cells that are expressing chemokine (C-X-C motif) receptor 3 (CXCR3), hence playing an essential role in the recruitment of immune cells to sites of inflammation. Furthermore, a strong downregulation by probiotic strains of Th1 cytokines, such as IL-2, IFN-γ and the proinflammatory IL-17, has been reported in other studies using different model systems (Lee et al. 2009; Llopis et al. 2009; Reiff et al. 2009; Schultz et al. 2002). Two other genes relevant to IBD that followed this pattern were TNFRSF9 (also referred to as 4-1BB) and TNFRSF4 (Ox-40) (Lee et al. 2005; Stuber et al. 2000; Totsuka et al. 2003). They are members of the tumor necrosis factor receptor (TNFR) family that sustain T cell numbers and responses after initial CD28-dependent T cell activation (Croft 2009). Differences in expression of the genes studied in detail here reached fold changes greater than 2, and even higher than or close to 10 (PMA/IO-stimulated versus control sample), and repression imposed by lactobacilli was two to fourfold. Ten genes were selected for qPCR validation of the microarray results and the reproducibility of the model system. Further testing of additional biological replicates rendered very reproducible results, and lactobacilli showed a remarkable downregulation, particularly of IL-2, IFNG, CXCL9, CXCL11 and IL-17A. Those were also the five genes most highly induced by PMA/IO with respect to the control, for which they are suitable to be used as markers for the antiinflammatory properties of lactobacilli in such organ culture system. Other members of cluster 6 that follow the same expression pattern have also relevance. Lymphotactin, encoded by XCL1/XCL2, is a chemotactic molecule related to the attraction of CD8+ T and CD4+ T cells (Hedrick and Zlotnik 1998). Ubiquitin D (UBD), also referred to as FAT10, is a cytokine-inducible ubiquitin-like protein that modifies proteins for proteosomal degradation and it is essential for TNF-α-induced NF-κB activation (Gong et al. 2010; Kalveram et al. 2008). Granzyme B (GZMB) is one of five granzymes expressed in humans; it is a serine protease expressed by CD8+ T cells that plays an important role in T cell mediated cytotoxicity. Granzymes, once delivered into the cytosol of an infected cell, rapidly induce apoptosis (Bots and Medema 2006).
Of note, some of the genes described above can be induced by IFN-γ, for example, CXCL9, CXCL11, IRF1 and each of the GBPs. It is thus possible that downregulation of these important inflammatory mediators was due to a primary reduction of IFN-γ. Moreover, functional annotation analysis revealed the implication of the Janus kinase (JAK)/STAT signaling pathway in downregulated genes in Lactobacillus-treated samples. Some regulated genes related to JAK/STAT signaling, for example IFN-γ, IL-13RA, IL-2, IL-11 and STAT4, showed a clear downregulation when compared to PMA/IO-treated samples. Downregulation of STAT4 is of particular interest, since it mediates most biological actions of IL-12 and is therefore required for the normal differentiation in Th1 cells and expression of Th1-specific genes such as IFN-γ (Kaplan 2005). Therefore, STAT4 represents a candidate gene possibly responsible for the inhibition of IFN-γ and IFN-γ inducible genes and should be further investigated.
Cluster 8 and cluster 10 illustrate genes that remained unchanged after PMA/IO treatment, but were upregulated after LP and LP(A−) treatment, respectively, evidencing that the putatively different surface characteristics of both strains had a differential impact on tissue gene expression. However, genes of both clusters were not significantly enriched in any signaling pathway or GO annotation term. Interestingly, LP strongly upregulated the gene expression of two chromatin modifying enzymes: EHMT2, a histone–lysine N-methyltransferase, and PHF8, a histone demethylase, which were shown to act on histone 3 (Fortschegger et al. 2010; Fritsch et al. 2010). Upregulation of EHMT2 could be observed after treatment with all three strains compared to Inflamed tissue. This upregulation was most pronounced in LP, whereas the induction of PHF8 was LP specific. In iLP(A−)-treated mucosa, the gene TNRC6C was upregulated, while it was downregulated in iLP. The protein encoded by TNRC6C regulates gene expression post-transcriptionally by repression of microRNAs (Zipprich et al. 2009). Although there was an important number of genes in clusters 8 and 10, no great differences could be found in the gene expression profile of key inflammatory effector molecules nor clear pathways indicating likely receptors involved that could be related to the different adhesion properties of LP and LP(A−). Also, as mentioned above, all three strains were able to downregulate proinflammatory cytokines and chemokines to a similar extent in our model system. Therefore, we could not evidence putative strain-specific immunomodulatory mechanisms.
In conclusion, the analysis of gene expression changes that occurred during the culture of intestinal mucosal fragments treated with PMA/IO showed activation of T cells and the upregulation of numerous proinflammatory effector molecules, such as IL-17A, IFN-γ, CXCL9 and CXCL11, GZMB, as well as TNRFSF4 and TNRFSF9, partially resembling gene expression patterns found in IBD patients. A general downregulation of genes encoding those factors could be observed after probiotic treatment and likely markers could be identified to monitor it. Some of the changes in gene expression could be explained by primary downregulation of IFN-γ and the JAK/STAT signaling pathway by the probiotic lactobacilli. Reduction in gene expression in IL-2 and IL-2RA suggests that these lactobacilli are also counteracting the molecular events leading to T cell activation and proliferation.
Electronic supplementary material
Acknowledgments
This work was supported by the grant AGL2010-22211 and by the Fun-C-Food CSD2007-00063 project from the Consolider-Ingenio programme, both from the Spanish Ministry of Science and Innovation.
References
- Baran J, Kowalczyk D, Ozog M, Zembala M. Three-color flow cytometry detection of intracellular cytokines in peripheral blood mononuclear cells: comparative analysis of phorbol myristate acetate-ionomycin and phytohemagglutinin stimulation. Clin Vaccine Immunol. 2001;8:303–313. doi: 10.1128/CDLI.8.2.303-313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borruel N, Carol M, Casellas F, Antolin M, de Lara F, Espin E, Naval J, Guarner F, Malagelada JR. Increased mucosal tumour necrosis factor alpha production in Crohn’s disease can be downregulated ex vivo by probiotic bacteria. Gut. 2002;51:659–664. doi: 10.1136/gut.51.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bots M, Medema JP. Granzymes at a glance. J Cell Sci. 2006;119:5011–5014. doi: 10.1242/jcs.03239. [DOI] [PubMed] [Google Scholar]
- Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- Carol M, Borruel N, Antolin M, Llopis M, Casellas F, Guarner F, Malagelada JR. Modulation of apoptosis in intestinal lymphocytes by a probiotic bacteria in Crohn’s disease. J Leukoc Biol. 2006;79:917–922. doi: 10.1189/jlb.0405188. [DOI] [PubMed] [Google Scholar]
- Chachkhiani I, Gurlich R, Maruna P, Frasko R, Lindner J. The postoperative stress response and its reflection in cytokine network and leptin plasma levels. Physiol Res. 2005;54:279–285. [PubMed] [Google Scholar]
- Chiu ST, Hsieh FJ, Chen SW, Chen CL, Shu HF, Li H. Clinicopathologic correlation of up-regulated genes identified using cDNA microarray and real-time reverse transcription-PCR in human colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:437–443. doi: 10.1158/1055-9965.EPI-04-0396. [DOI] [PubMed] [Google Scholar]
- Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol. 2009;9:271–285. doi: 10.1038/nri2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame MK, Bhagavathula N, Mankey C, Dasilva M, Paruchuri T, Aslam MN, Varani J. Human colon tissue in organ culture: preservation of normal and neoplastic characteristics. In Vitro Cell Dev Biol Anim. 2010;46:114–122. doi: 10.1007/s11626-009-9247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Caro S, Tao H, Grillo A, Elia C, Gasbarrini G, Sepulveda AR, Gasbarrini A. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig Liver Dis. 2005;37:320–329. doi: 10.1016/j.dld.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Egesten A, Eliasson M, Olin AI, Erjefalt JS, Bjartell A, Sangfelt P, Carlson M. The proinflammatory CXC-chemokines GRO-alpha/CXCL1 and MIG/CXCL9 are concomitantly expressed in ulcerative colitis and decrease during treatment with topical corticosteroids. Int J Colorectal Dis. 2007;22:1421–1427. doi: 10.1007/s00384-007-0370-3. [DOI] [PubMed] [Google Scholar]
- Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, Dewulf J, Brassart D, Mercenier A, Pot B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol. 2007;13:236–243. doi: 10.3748/wjg.v13.i2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foligne B, Zoumpopoulou G, Dewulf J, Ben Younes A, Chareyre F, Sirard JC, Pot B, Grangette C. A key role of dendritic cells in probiotic functionality. PLoS ONE. 2007;2:e313. doi: 10.1371/journal.pone.0000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FMA, Timmers HT, Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010;30:3286–3298. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick JS, Schenk K, Quitadamo M, Kahl F, Koberle M, Bohn E, Aepfelbacher M, Autenrieth IB. Lactobacillus fermentum attenuates the proinflammatory effect of Yersinia enterocolitica on human epithelial cells. Inflamm Bowel Dis. 2007;13:83–90. doi: 10.1002/ibd.20009. [DOI] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Meyar-Zazoua M, It-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Gao Y, Flori L, Lecardonnel J, Esquerre D, Hu ZL, Teillaud A, Lemonnier G, Lefevre F, Oswald IP, Rogel-Gaillard C. Transcriptome analysis of porcine PBMCs after in vitro stimulation by LPS or PMA/ionomycin using an expression array targeting the pig immune response. BMC Genomics. 2010;11:292. doi: 10.1186/1471-2164-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill HS, Guarner F. Probiotics and human health: a clinical perspective. Postgrad Med J. 2004;80:516–526. doi: 10.1136/pgmj.2003.008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P, Canaan A, Wang B, Leventhal J, Snyder A, Nair V, Cohen CD, Kretzler M, D’Agati V, Weissman S, Ross MJ. The ubiquitin-like protein FAT10 mediates NF-{kappa}B activation. J Am Soc Nephrol. 2010;21:316–326. doi: 10.1681/ASN.2009050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckford SE, Gelmann EP, Agnor CL, Jacobson S, Zinn S, Matis LA. Distinct signals are required for proliferation and lymphokine gene expression in murine T cell clones. J Immunol. 1986;137:3652–3663. [PubMed] [Google Scholar]
- Hedrick JA, Zlotnik A. Lymphotactin. Clin Immunol Immunopathol. 1998;87:218–222. doi: 10.1006/clin.1998.4546. [DOI] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormannsperger G, Haller D. Molecular crosstalk of probiotic bacteria with the intestinal immune system: clinical relevance in the context of inflammatory bowel disease. Int J Med Microbiol. 2010;300:63–73. doi: 10.1016/j.ijmm.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kalveram B, Schmidtke G, Groettrup M. The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition. J Cell Sci. 2008;121:4079–4088. doi: 10.1242/jcs.035006. [DOI] [PubMed] [Google Scholar]
- Kamada N, Maeda K, Inoue N, Hisamatsu T, Okamoto S, Hong KS, Yamada T, Watanabe N, Tsuchimoto K, Ogata H, Hibi T. Nonpathogenic Escherichia coli strain Nissle 1917 inhibits signal transduction in intestinal epithelial cells. Infect Immunol. 2008;76:214–220. doi: 10.1128/IAI.01193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH. STAT4: a critical regulator of inflammation in vivo. Immunol Res. 2005;31:231–242. doi: 10.1385/IR:31:3:231. [DOI] [PubMed] [Google Scholar]
- Kekkonen RA, Kajasto E, Miettinen M, Veckman V, Korpela R, Julkunen I. Probiotic leuconostoc mesenteroides ssp. Cremoris and streptococcus thermophilus induce IL-12 and IFN-gamma production. World J Gastroenterol. 2008;14:1192–1203. doi: 10.3748/wjg.14.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp K, Bruunsgaard H. Identification of IFN- + -producing CD4 + T cells following PMA stimulation. J Interferon Cytokine Res. 2001;21:503–506. doi: 10.1089/10799900152434376. [DOI] [PubMed] [Google Scholar]
- Latvala S, Pietila TE, Veckman V, Kekkonen RA, Tynkkynen S, Korpela R, Julkunen I. Potentially probiotic bacteria induce efficient maturation but differential cytokine production in human monocyte-derived dendritic cells. World J Gastroenterol. 2008;14:5570–5583. doi: 10.3748/wjg.14.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger TN, Pinton P, Bourges D, Roumi P, Salmon H, Oswald IP. Development of a macroarray to specifically analyze immunological gene expression in Swine. Clin Diagn Lab Immunol. 2004;11:691–698. doi: 10.1128/CDLI.11.4.691-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee EN, Kim EY, Park HJ, Chang CY, Jung DY, Choi SY, Lee SK, Lee KW, Kwon GY, Joh JW, Kim SJ. Administration of agonistic anti-4-1BB monoclonal antibody leads to the amelioration of inflammatory bowel disease. Immunol Lett. 2005;101:210–216. doi: 10.1016/j.imlet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Lee H, Ahn YT, Lee JH, Huh CS, Kim DH. Evaluation of anti-colitic effect of lactic acid bacteria in mice by cDNA microarray analysis. Inflammation. 2009;32:379–386. doi: 10.1007/s10753-009-9146-y. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2:research0032.1-0032.11 [DOI] [PMC free article] [PubMed]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. PNAS. 2001;98:31–36. doi: 10.1073/pnas.98.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopis M, Antolin M, Carol M, Borruel N, Casellas F, Martinez C, Espin-Basany E, Guarner F, Malagelada JR. Lactobacillus casei downregulates commensals’ inflammatory signals in Crohn’s disease mucosa. Inflamm Bowel Dis. 2009;15:275–283. doi: 10.1002/ibd.20736. [DOI] [PubMed] [Google Scholar]
- Ma DL, Forsythe P, Bienenstock J. Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor alpha-induced interleukin-8 expression. Infect Immun. 2004;72:5308–5314. doi: 10.1128/IAI.72.9.5308-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahida YR, Lamming CE, Gallagher A, Hawthorne AB, Hawkey CJ. 5-Aminosalicylic acid is a potent inhibitor of interleukin 1 beta production in organ culture of colonic biopsy specimens from patients with inflammatory bowel disease. Gut. 1991;32:50–54. doi: 10.1136/gut.32.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangell P, Lennernas P, Wang M, Olsson C, Ahrne S, Molin G, Thorlacius H, Jeppsson B. Adhesive capability of Lactobacillus plantarum 299v is important for preventing bacterial translocation in endotoxemic rats. Apmis. 2006;114:611–618. doi: 10.1111/j.1600-0463.2006.apm_369.x. [DOI] [PubMed] [Google Scholar]
- Mileti E, Matteoli G, Iliev ID, Rescigno M. Comparison of the immunomodulatory properties of three probiotic strains of Lactobacilli using complex culture systems: prediction for in vivo efficacy. PLoS ONE. 2009;4:e7056. doi: 10.1371/journal.pone.0007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner CM, Higman VA, Day AJ. TSG-6: a pluripotent inflammatory mediator? Biochem Soc Trans. 2006;34:446–450. doi: 10.1042/BST0341261. [DOI] [PubMed] [Google Scholar]
- Muñoz-Provencio D, Llopis M, Antolin M, de Torres I, Guarner F, Perez-Martinez G, Monedero V. Adhesion properties of Lactobacillus casei strains to resected intestinal fragments and components of the extracellular matrix. Arch Microbiol. 2009;191:153–161. doi: 10.1007/s00203-008-0436-9. [DOI] [PubMed] [Google Scholar]
- Murphy K, Travers P, Walport M (2007) Janeway’s immunobiology. Garland Science Seventh Edition
- Niedel JE, Kuhn LJ, Vandenbark GR. Phorbol diester receptor copurifies with protein kinase C. PNAS. 1983;80:36–40. doi: 10.1073/pnas.80.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y, Manabe K, Kishi J, Wang W, Inayama M, Azuma M, Sone S. CXCL9 and 11 in patients with pulmonary sarcoidosis: a role of alveolar macrophages. Clin Exp Immunol. 2007;149:317–326. doi: 10.1111/j.1365-2249.2007.03423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara AM, O’Regan P, Fanning A, O’Mahony C, Macsharry J, Lyons A, Bienenstock J, O’Mahony L, Shanahan F. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology. 2006;118:202–215. doi: 10.1111/j.1365-2567.2006.02358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology. 2010;129:311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613–G626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- Pattyn F, Speleman F, De Paepe A, Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Martinez G, Bäuerl C, Collado MC (2012) Selection and evaluation of probiotics. In: Nollet LML, Toldra F (eds) Handbook of analysis of active compounds in functional foods. Taylor & Francis Group, CRC Press, Boca Raton, pp 607–637
- Petrof EO, Claud EC, Sun J, Abramova T, Guo Y, Waypa TS, He SM, Nakagawa Y, Chang EB. Bacteria-free solution derived from Lactobacillus plantarum inhibits multiple NF-kappaB pathways and inhibits proteasome function. Inflamm Bowel Dis. 2009;15:1537–1547. doi: 10.1002/ibd.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff C, Delday M, Rucklidge G, Reid M, Duncan G, Wohlgemuth S, Hormannsperger G, Loh G, Blaut M, Collie-Duguid E, Haller D, Kelly D. Balancing inflammatory, lipid, and xenobiotic signaling pathways by VSL#3, a biotherapeutic agent, in the treatment of inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1721–1736. doi: 10.1002/ibd.20999. [DOI] [PubMed] [Google Scholar]
- Resau JH, Sakamoto K, Cottrell JR, Hudson EA, Meltzer SJ. Explant organ culture: a review. Cytotechnology. 1991;7:137–149. doi: 10.1007/BF00365924. [DOI] [PubMed] [Google Scholar]
- Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130:731–746. doi: 10.1053/j.gastro.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Rochat T, Bermudez-Humaran L, Gratadoux JJ, Fourage C, Hoebler C, Corthier G, Langella P. Anti-inflammatory effects of Lactobacillus casei BL23 producing or not a manganese-dependant catalase on DSS-induced colitis in mice. Microb Cell Fact. 2007;6:22. doi: 10.1186/1475-2859-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostaing L, Tkaczuk J, Durand M, Peres C, Durand D, de Préval C, Ohayon E, Abbal M. Kinetics of intracytoplasmic Th1 and Th2 cytokine production assessed by flow cytometry following in vitro activation of peripheral blood mononuclear cells. Cytometry. 1999;35:318–328. doi: 10.1002/(SICI)1097-0320(19990401)35:4<318::AID-CYTO4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Rovedatti L, Kudo T, Biancheri P, Sarra M, Knowles CH, Rampton DS, Corazza GR, Monteleone G, Di Sabatino A, MacDonald TT. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut. 2009;58:1629–1636. doi: 10.1136/gut.2009.182170. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. Lactobacillus plantarum 299 V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71–80. doi: 10.1097/00054725-200203000-00001. [DOI] [PubMed] [Google Scholar]
- Sibartie S, O’Hara AM, Ryan J, Fanning A, O’Mahony J, O’Neill S, Sheil B, O’Mahony L, Shanahan F. Modulation of pathogen-induced CCL20 secretion from HT-29 human intestinal epithelial cells by commensal bacteria. BMC Immunol. 2009;10:54. doi: 10.1186/1471-2172-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MA, Menezes J, Deslandres C, Seidman EG. Anti-inflammatory role of interleukin-15 in Crohn’s disease. Inflamm Bowel Dis. 2005;11:219–230. doi: 10.1097/01.MIB.0000160804.52072.6a. [DOI] [PubMed] [Google Scholar]
- Singh UP, Venkataraman C, Singh R, Lillard JW. CXCR3 axis: role in inflammatory bowel disease and its therapeutic implication. Endocr Metab Immune Disord Drug Targets. 2007;7:111–123. doi: 10.2174/187153007780832109. [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber E, Buschenfeld A, Luttges J, Von Freier A, Arendt T, Folsch UR. The expression of OX40 in immunologically mediated diseases of the gastrointestinal tract (celiac disease, Crohn’s disease, ulcerative colitis) Eur J Clin Invest. 2000;30:594–599. doi: 10.1046/j.1365-2362.2000.00658.x. [DOI] [PubMed] [Google Scholar]
- Takahama Y, Nakauchi H. Phorbol ester and calcium ionophore can replace TCR signals that induce positive selection of CD4 T cells. J Immunol. 1996;157:1508–1513. [PubMed] [Google Scholar]
- Totsuka T, Kanai T, Uraushihara K, Iiyama R, Yamazaki M, Akiba H, Yagita H, Okumura K, Watanabe M. Therapeutic effect of anti-OX40L and anti-TNF-alpha MAbs in a murine model of chronic colitis. Gastrointest Liver Physiol. 2003;284:G595–G603. doi: 10.1152/ajpgi.00450.2002. [DOI] [PubMed] [Google Scholar]
- Troost FJ, van Baarlen P, Lindsey P, Kodde A, de Vos WM, Kleerebezem M, Brummer RJ. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics. 2008;9:374. doi: 10.1186/1471-2164-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarlen P, Troost FJ, van Hemert S, van der Me-er C, de Vos WM, de Groot PJ, Hooiveld GJEJ, Brummer RJ, Kleerebezem M. Differential NF-{kappa}B pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. PNAS. 2009;106:2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJM, Kleerebezem M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. PNAS. 2011;108:4562–4569. doi: 10.1073/pnas.1000079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1-0034.11 [DOI] [PMC free article] [PubMed]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Sato-Mito N, Nagata J, Umegaki K. Health claim evidence requirements in Japan. J Nutrit. 2008;138:1192S–1198S. doi: 10.1093/jn/138.6.1192S. [DOI] [PubMed] [Google Scholar]
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.