Abstract
Sortase enzymes are vitally important for the virulence of gram-positive bacteria as they play a key role in the attachment of surface proteins to the cell wall. These enzymes recognize a specific sorting sequence in proteins destined to be displayed on the surface of the bacteria and catalyze the transpeptidation reaction that links it to a cell wall precursor molecule. Because of their role in establishing pathogenicity, and in light of the recent rise of antibiotic-resistant bacterial strains, sortase enzymes are novel drug targets. Here, we present a study of the prototypical sortase protein Staphylococcus aureus Sortase A (SrtA). Both conventional and accelerated molecular dynamics simulations of S. aureus SrtA in its apo state and when bound to an LPATG sorting signal (SS) were performed. Results support a binding mechanism that may be characterized as conformational selection followed by induced fit. Additionally, the SS was found to adopt multiple metastable states, thus resolving discrepancies between binding conformations in previously reported experimental structures. Finally, correlation analysis reveals that the SS actively affects allosteric pathways throughout the protein that connect the first and the second substrate binding sites, which are proposed to be located on opposing faces of the protein. Overall, these calculations shed new light on the role of dynamics in the binding mechanism and function of sortase enzymes.
Keywords: sortase enzymes, Staphylococcus aureus, molecular dynamics, induced-fit binding, allostery
Introduction
Attached to the cell walls of Gram-positive bacteria, surface proteins play key roles in multiple pathogenic mechanisms such as invasion of host cells, cellular adhesion, and evasion of the immune response, in addition to more benign pathways that include cell division, cell wall maintenance, and acquisition of nutrients from the environment.1–3 Many of these proteins are covalently linked to the peptidoglycan molecules that compose the cell wall by sortase enzymes. These transpeptidases recognize a conserved pentapeptide motif in the target protein and form an amide bond between it and a nucleophile in the cell wall.4–7 Class A sortase enzymes (SrtA), which are commonly referred to as “housekeeping” sortases due to the large number of proteins they anchor to the cell wall, target an LPXTG (where X is any amino acid) sorting signal (SS) substrate motif, cleave it between the threonine and the glycine residues, and catalyze its amide-linkage to a lipid II molecule.8–11 The most widely studied SrtA enzyme is from Staphylococcus aureus, a pathogen responsible for a multitude of ailments such as meningitis, septicaemia, and toxic shock syndrome.4, 12 Although it is not essential for survival, the direct link of S. aureus SrtA to bacterial virulence makes it a novel and interesting target for the development of antibiotic drugs.13–15
S. aureus SrtA is composed of 206 residues that are divided into two domains: a membrane-spanning N-terminal domain (residues 1–59) and an autonomously folding catalytic domain (residues 60–206).9, 16–18 NMR and crystallography experiments have shown that the catalytic domain adopts a unique eight-stranded β-barrel fold with individual strands that are connected by two short helices and several loop regions (see Fig. 1).19, 20 Residues within the loop connecting the β6 and β7 strands exhibited resonance line broadening in the NMR experiments and were poorly resolved with high B-factors in the crystallography experiments, both of which indicate that this loop exhibits increased dynamics relative to the remainder of the protein. Motions of the β6/β7 loop are particularly interesting, given that many of the residues that compose it are positioned adjacent to the SS active site, notably residues 164–169. Although the binding site for the second substrate of catalysis, lipid II, has not been conclusively determined, chemical shift perturbation experiments indicate that it is located on the protein face opposite to the SS active site in the region around the β4/H2 loop, between H1 and the β7/β8 loop.21 However, crystallographic experiments on a related protein have suggested an alternative lipid II binding location.20
Figure 1.
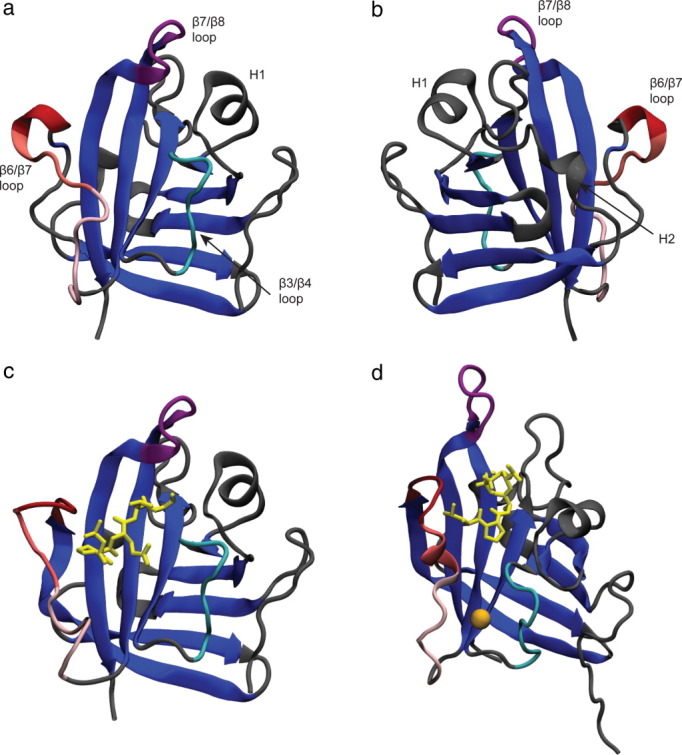
SrtA apo crystal structure (a)20; apo crystal structure rotated 180° about the y-axis (b); crystal structure of C184A SrtA mutant bound to LPETG peptide sequence (yellow; c)20; NMR SrtA structure covalently bound to a modified sorting signal (d).21 The unique central β-barrel structure is colored blue. Large differences exist in the positioning of the β6/β7 and β7/β8 loops, with the β6/β7 loop (red) more open in both the crystal structures and the β7/β8 loop (purple) more open in the NMR structure. The catalytically important calcium ion (orange) interacts with residues in the β3/β4 loop (cyan) and lower β6/β7 loop. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The active sites of all sortases contain a conserved catalytic triad that consists of residues H120, C184, and R197, mutations to each of which have been shown to severely reduce SrtA's catalytic activity.9, 22–25 C184 has been established as the active-site nucleophile that covalently attaches to the carbonyl carbon of the SS threonine residue through a ping-pong mechanism.17, 18 R197 appears to stabilize the binding of an SS or an oxyanion intermediate to SrtA,20, 21, 24, 26 although it has also been suggested to play a role in the deprotonation of C184 or lipid II.20, 24 The function of H120 is less well understood, but it has been proposed that the rare, charged form protonates the substrate leaving group during catalysis.18
To further elucidate the mechanism of SS binding and hydrolysis, a pair of experiments was previously performed to resolve the binding mode of a pentapeptide to SrtA. In the first, Zong et al.20 diffused LPETG peptides into crystals of C184A mutant SrtA molecules. In this holo crystal structure, the SS adopted an elongated form while the β6/β7 loop remained in an “open” conformation. R197 was observed to make contact with the SS threonine residue; however, the side chain of the catalytic H120 was located more than 10 Å away from the peptide, the β6/β7 loop was in an “open” state, and the side chain of the leucine residue in the SS protruded into solution and did not interact with the protein, factors that are all incompatible with biochemical studies. These factors suggest that this may be a nonspecific binding pose and not the conformation that promotes catalysis. In the second study, Suree et al.21 synthesized an LPAT analog that formed a disulfide linkage between the threonine derivative and C184, resulting in a structure similar to the catalytic thioacyl intermediate. In this structure, the SS adopted an “L-shape” configuration in which there was a bend of ∼90° between the alanine and proline residues, and the β6/β7 loop not only contained a short 310 helix spanning residues V166–L169, but was also far less mobile and in a “closed” configuration. To accommodate the covalently bound peptide in the active site, the β7/β8 loop transitioned to a more open conformation, suggesting an “induced-fit” mechanism for SS binding. In addition, each of the catalytic triad members was observed in close proximity to residues within the SS.
Results from simulations have begun to clarify the role of dynamics on the mechanism of SrtA. In a series of quantum mechanical/molecular mechanical simulations, Tian and Eriksson27, 28 addressed the multiple roles of R197 by showing that it stabilizes both SS binding and the catalytically active charged states of H120 and C184. Recently, Moritsugu et al.29 used a series of Hamiltonian replica exchange simulations to probe the free energy landscape of the β6/β7 and β7/β8 loops upon binding of a SS and a catalytically important calcium ion. These results demonstrated the essential interplay between the binding of these two ligands to sortase and highlighted the importance of the bound calcium ion to the structural stability of the SS active site.
Despite these efforts, several questions remain concerning the dynamics and mechanism of SS binding by SrtA: How does an unmodified SS noncovalently associate with native SrtA? Are there multiple binding poses for the SS in the active site? How does SS binding affect the presumed lipid II binding site? To address these questions, we report on a series of all-atom molecular dynamics (MD) simulations of SrtA bound to, and free from, an LPATG SS motif. By using a combination of conventional MD (cMD) and accelerated MD (aMD) simulations, it is observed that the SS likely possesses multiple metastable states, binds through a mechanism that may be described as conformational selection followed by induced fit, and has a dramatic influence on the allosteric networks that link the SS and lipid II binding sites. Our results are consistent with the previous experimental and computational findings and shed new light on the mechanism of SrtA.
Results
cMD simulations of apo state
The overall conformation of SrtA was stable in the three 100-ns cMD simulations initiated from the crystal structure that was resolved without a bound SS. In each simulation, the heavy-atom root mean square deviations (RMSDs) relative to the initial structure stabilized at ∼2.5 Å in the first 10 ns of simulation (Supporting Information Fig. S1, top row). Root mean square fluctuations (RMSFs) of individual Cα atoms were low (below 1 Å) for residues within the β-sheets and elevated in many of the loop regions, particularly the β5/β6, β6/β7, and β7/β8 loops (Fig. 2).
Figure 2.
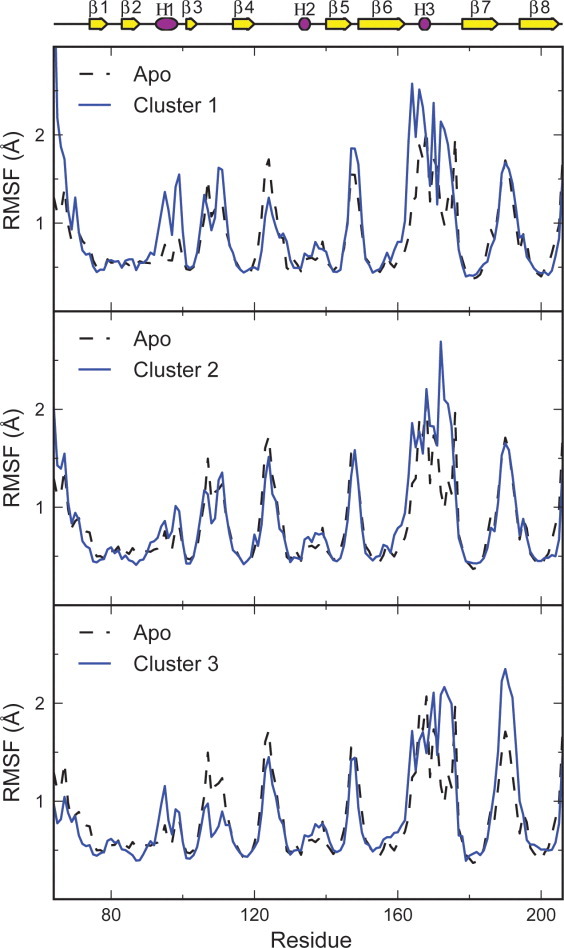
Root mean square fluctuations over the final 80 ns of each of the apo cMD simulations and for structures in clusters 1–3, representing the different sorting signal binding locations. Trends in flexibility are similar for all simulations, with residues in the loops exhibiting higher flexibility than residues in the β-strands. Notably, the dynamics of the β6/β7 loop appear to be affected by the sorting signal binding location. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary. com.]
The dominant, low-frequency motions that contribute to biologically relevant conformational changes in SrtA were determined by a full-correlation analysis (FCA), which improves upon the more widely used principal component analysis by taking into account not only linear, but also nonlinear and higher order correlations (details in Methods). The mode corresponding to the largest structural change, FCA mode 1, corresponds to a combination of the closing motion of the β6/β7 loop and an opening of the β7/β8, whereas FCA mode 2 primarily describes motions of the β4/H2 loop [see Fig. 3(a)]. The projections of the apo simulations along the first two FCA modes sampled only the region of the mode 1/2 landscape in which the simulations were initiated, indicating that there was little change along these dimensions on the 100-ns timescale [Fig. 3(b)].
Figure 3.
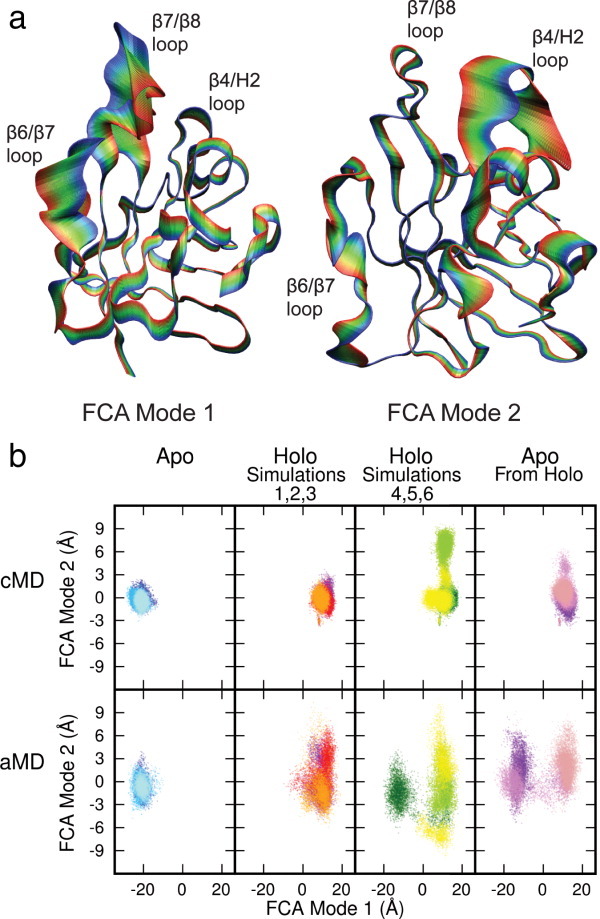
Superposed projections of the Cα atoms onto the first and second FCA modes (a) and projections of simulation trajectories onto FCA mode 1/2 landscape (b). FCA mode one predominantly describes the opening and closing of the β6/β7 and β7/β8 loops, while FCA mode two largely describes motions of the β4/H2 loop. The holo simulations were divided into two groups, simulations 1–3 in which the sorting signal remained in the active site and 4–6 in which it did not. Projections of the cMD trajectories indicate that the overall SrtA structures were stable and the apo and holo states remained conformationally distinct on the 100-ns timescale. Projections of the aMD trajectories show no significant increase in sampling, with the exception of holo aMD simulation 4 (colored dark green), in which a transition from the holo to a more apo-like structure occurred. The apo from holo structure was stable on the 100-ns timescale, but transitioned to a structure closer to that of the apo state in two of the three aMD simulations. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The motions of the β6/β7 loop are of particular interest due to its proximity to the active site. At the top of the loop in the apo enzyme, residues 163–165 intermittently formed a short 310 helix for a total of 23.3% of the simulations, and residues 163–166 adopted an alpha helix for 6.3% of the simulations. At the beginning of each of the apo simulations, the lower part of this loop (residues 166–173) transitioned to a more open state. In simulations 1 and 3, the β6/β7 loop returned to its original semi-closed state as residue D170 made contact with the calcium ion. However, in simulation 2, D170 did not make contact with the calcium ion, the loop did not close, and it remained more mobile throughout the simulation (Supporting Information Fig. S2). Residue E171, in addition to D170, in the β6/β7 loop intermittently interacted with the calcium ion, stabilizing the semi-closed conformation of the loop. Throughout the simulations, the calcium ion remained stably bound in a pocket formed by residues E105, E108, D112, and N114, with a RMSF value of 1.3 Å.
cMD simulations of holo state
Based upon the covalently bound holo NMR structure, a model for the noncovalently bound SrtA/SS complex was generated by replacing the covalently attached SS analog with a noncovalently bound LPATG sequence (see Methods for details). This structure represents the enzyme-substrate michaelis complex that forms before catalysis. It was chosen over the substrate bound crystal structure as, in the latter, the polypeptide did not interact with two of the three catalytic triad residues, suggesting that it was bound in a nonspecific binding location and was only resolved in one of the three SrtA proteins in the asymmetric unit. The overall conformation of these holo SrtA structures remained stable and distinct from that of the apo state throughout each of the six 100-ns cMD simulations. RMSDs stabilized around 3 Å within the first 20 ns of each simulation (relative to its initial conformation; Supporting Information Fig. S1, bottom 2 rows). Projections of the trajectories onto the FCA mode 1/2 subspace show that the protein remained close to its initial conformation throughout the simulations (Fig. 3); however, the distinct location of the holo simulations relative to the apo simulations along the first mode demonstrates that FCA mode 1 describes the structural transition that takes places upon substrate binding, with positive projection values indicating a “holo-like” structure and negative values indicating an “apo-like” structure. Throughout the simulations, residues exhibited the same trends in flexibility as observed in the apo simulations. RMSFs for residues within the loops were higher than for those in the beta strands. However, the location of the SS did appear to impact the dynamics of SrtA (see below).
As in the apo simulations, the calcium ion remained stable, with an RMSF value of 1.2 Å. Again, the binding pocket was formed by residues E105, E108, D112, and N114. However, interaction of the β6/β7 loop with the calcium ion was limited to a contact with residue E171 that was more stable than in the apo simulations and did not include interaction with residue D170, in agreement with the previous site-directed mutagenesis experiments.30
Distinct SS binding conformations
Although the protein remained close to its initial conformation throughout the simulations, the sorting signal (SS) adopted multiple distinct bound states. Clustering analysis revealed six main conformations, altogether encompassing 81.6% of all the cMD simulations (Fig. 4 and Table I). In the first cluster, the SS adopted a “U-shaped” conformation in the active site, much like that observed in the holo NMR structure. This conformation was stable, with an MM/GBSA binding free energy estimate of −21.9 ± 3.7 kcal/mol (Table II, see Methods for calculation details).31, 32 Specifically, the SS was stabilized by interaction with residues near the top of the β6/β7 loop and with residues of the catalytic triad (H120, C184, and R197), particularly with R197 (Table III). Additionally, hydrogen bonds formed between R197 and the Pro and Gly residues of the SS (Fig. 5). The MM/GBSA method oftentimes results in binding free energy estimates with a dynamical range much larger than observed in experiments33, 34; therefore, we emphasize that these binding free energies should be taken as relative binding values (to the energies reported below) and not the absolute binding free energies.
Figure 4.
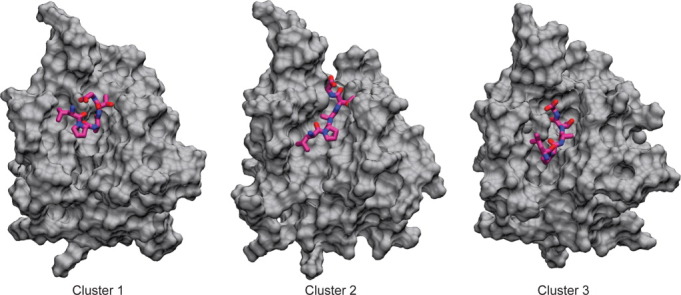
Distinct binding conformations of the sorting signal identified through clustering analysis. The sorting signal position in cluster 1 resembles the NMR structure, while the binding pose in cluster 3 resembles the crystal structure. Cluster 2 represents a more elongated conformation of the sorting signal in the active site. For populations and binding energies of these clusters, see Tables I and II. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table I.
Percentage of Each Simulation Contained in Clusters 1–6
| MD 1 | MD 2 | MD 3 | MD 4 | MD 5 | MD 6 | |
|---|---|---|---|---|---|---|
| Cluster 1 (21.7%) | 1.9% | 33.6% | 94.5% | — | — | — |
| Cluster 2 (20.9%) | 95.5% | 29.9% | — | — | — | — |
| Cluster 3 (14.8%) | — | — | — | — | — | 88.6% |
| Cluster 4 (9.0%) | — | — | — | 54.2% | — | — |
| Cluster 5 (8.1%) | — | — | — | — | 48.8% | — |
| Cluster 6 (7.1%) | — | — | — | 42.5% | — | — |
Clustering analysis of sorting signal position revealed the main binding modes. Percentages in parentheses indicate the total percentage of holo cMD simulation frames contained in the cluster. Clusters are numbered in order of decreasing population.
Table II.
MM/GBSA Binding Free Energies (kcal/mol)
| ΔEElectrostatics | ΔEvdw | ΔGpolar | ΔGnonpolar | −TΔSvib | ΔGbinding | |
|---|---|---|---|---|---|---|
| Cluster 1 | −55.9 ± 7.3 | −35.5 ± 1.7 | 68.7 ± 6.0 | −4.8 ± 0.2 | 5.7 ± 2.9 | −21.9 ± 3.7 |
| Cluster 2 | −65.7 ± 9.1 | −41.5 ± 2.1 | 88.7 ± 8.0 | −5.9 ± 0.2 | 4.0 ± 3.2 | −20.5 ± 3.9 |
| Cluster 3 | −116.2 ± 12.9 | −41.6 ± 2.2 | 132.6 ± 12.0 | −5.9 ± 0.2 | 4.9 ± 3.8 | −26.3 ± 4.5 |
| Cluster 4 | −117.4 ± 15.2 | −33.6 ± 4.3 | 139.0 ± 15.1 | −4.9 ± 0.5 | 6.7 ± 4.9 | −10.2 ± 6.7 |
| Cluster 5 | −80.6 ± 10.4 | −19.2 ± 2.3 | 81.5 ± 9.8 | −3.4 ± 0.2 | 8.4 ± 4.6 | −13.3 ± 5.3 |
| Cluster 6 | −100.0 ± 18.3 | −24.1 ± 2.4 | 111.4 ± 16.4 | −4.0 ± 0.3 | 7.1 ± 5.3 | −9.5 ± 5.9 |
MM/GBSA binding free energies indicate relative stability. Energies are decomposed into electrostatic, van der Waals, polar, nonpolar, and entropic contributions. All binding poses were stable, although the sorting signal was most stable in clusters 1–3 in which it was in the active site.
Table III.
Interaction Energies Between Sorting Signal and Select Residues (kcal/mol)
| H120 | C184 | R197 | Top of β6/β7 loopa | β6/β7 helixb | Bottom of β6/β7 loopc | β3/β4 loopd | |
|---|---|---|---|---|---|---|---|
| Cluster 1 | −2.1 ± 0.3 | −1.8 ± 0.2 | −8.6 ± 0.6 | −1.5 ± 0.3 | −3.1 ± 0.4 | −0.28 ± 0.03 | −1.0 ± 0.1 |
| Cluster 2 | −4.2 ± 0.4 | −2.7 ± 0.2 | −5.8 ± 0.5 | −2.2 ± 0.3 | −2.9 ± 0.3 | −0.7 ± 0.1 | −2.6 ± 0.2 |
| Cluster 3 | −1.1 ± 0.2 | −2.5 ± 0.2 | −8.1 ± 0.7 | −1.3 ± 0.4 | −3.7 ± 0.5 | −1.7 ± 0.4 | −7.3 ± 0.6 |
| Cluster 4 | −0.06 ± 0.02 | −0.04 ± 0.01 | −2.4 ± 0.4 | −0.3 ± 0.2 | −7.4 ± 1.8 | −4.3 ± 0.5 | −12.8 ± 0.9 |
| Cluster 5 | −0.01 ± 0.04 | −0.04 ± 0.04 | −2.5 ± 0.8 | −0.3 ± 0.4 | −0.5 ± 0.1 | 0.44 ± 0.05 | 0.3 ± 0.1 |
| Cluster 6 | −0.03 ± 0.02 | −0.04 ± 0.01 | −3.0 ± 0.7 | 0.3 ± 0.1 | −2.6 ± 0.6 | −0.28 ± 0.05 | −8.0 ± 0.8 |
Interaction with R197 was favorable in all binding poses. Residues in the β6/β7 loop interacted favorably with the sorting signal in clusters 1–4, with the strongest interaction with residues nearest the respective sorting signal binding site. In clusters 3 and 4, interaction with the β3/β4 loop was particularly favorable.
Residues 164–165.
Residues 166–169.
Residues 170–175.
Residues 105–111.
Figure 5.
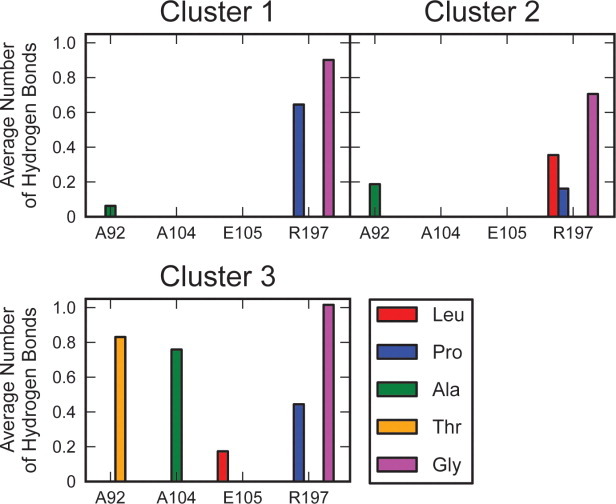
Average number of hydrogen bonds formed between residues of the sorting signal and SrtA for each bound conformation of the sorting signal. In each of the spatially distinct binding conformations, the sorting signal formed hydrogen bonds with R197 in the active site. In cluster 3, the sorting signal also formed hydrogen bonds to residues in the β3/β4 loop.
The SS was also observed to adopt a second, more outstretched configuration in the active site (cluster 2). This conformation was also stable, with an MM/GBSA binding free energy estimate of −20.5 ± 3.9 kcal/mol. The specific interactions of the SS with SrtA were similar to those in cluster 1; interaction energies were favorable between the SS and H120, C184, R197, and residues near the top of the β6/β7 loop. SS interaction with H120 was particularly favorable in this cluster, with an average interaction energy of −4.2 ± 0.4 kcal/mol. Hydrogen bonds between R197 and Leu, Pro, and Gly also stabilized the SS in this conformation.
When the SS was bound in either conformation in the active site, two distinct states of the β6/β7 loop were observed. In the first, the loop was shortened by the interaction of residues 167–169 closer to the top of the loop with residues 170–172 near the middle of the loop. The absence of these interactions characterized the second state of the loop, which was consequently slightly longer and more outstretched. Between the two states, Q172 was the maximally displaced residue, differing in position by ∼4 Å. RMSFs computed from structures in cluster 2 reflect this movement, as residue Q172 had a value of 2.7 Å, which is remarkably higher than in the apo simulations and higher than the surrounding residues. In both clusters, the shorter conformation of the β6/β7 loop correlated with the formation of a short 310 helix composed of residues 166–169.
In cluster 3, the SS remained outstretched, but moved down the groove adjacent to the β6/β7 loop, leading into the active site. The location of the peptide relative to SrtA is reminiscent of the observed binding mode in the crystal structure; however, the conformation of the β6/β7 loop remained closed and the β7/β8 loop remained open, resulting in positional differences of the SS in the simulations relative to the crystal structure. This conformation appeared stable, given the MM/GBSA binding free energy of −26.3 ± 4.5 kcal/mol. Hydrogen bonds between the SS and R197 in the active site and A104 and E105 at the top of the β3/β4 loop contributed to this stability. SS interactions with H120, C184, R197, residues 164–171 in the β6/β7 loop, and residues 105–108 in the β3/β4 loop were also favorable.
When the SS was bound in this third conformation, the β6/β7 loop was observed to exist in the outstretched and shortened conformations observed in clusters 1 and 2. However, in addition to the formation of a 310 helix spanning residues 166–169, these residues also adopted an alpha helix. Furthermore, the correlation between helix formation and the shorter conformation of the β6/β7 loop was lost.
In the fourth through sixth main binding conformations, which constitute 24.2% of all the holo simulations and were only present during holo cMD simulations 4 and 5, the SS was located away from the active site. Discussion of these modes is presented in SI text for the interested reader.
Allosteric networks in SrtA
The correlation between residues was assessed through a MutInf analysis for all the apo simulations and the holo simulations in which the SS remained in the active site (holo simulations 1–3, Fig. 6, see Methods section). In the apo state, the lower half of the β6/β7 loop (residues 168–178) was correlated with residues in the β3/β4 loop around the calcium binding site and with residues in H1 and part of the β4/H2 loop (residues 120–126), which is near the active site. Additionally, the residues within the β4/H2 loop were highly correlated with each other and with Q96, located in H1. In the holo simulations, these correlations were largely replaced with ones that were generally different from those observed in the apo simulations. Notably, the β3/β4 loop (residues 103–113) was correlated with the entire β6/β7 loop (residues 159–173), and several residues throughout the protein showed high correlation with D170. Residues with mutual information scores above 1.5 kT to D170 appeared to form a network that went across the protein, from the SS binding site, through the SS peptide, and to the presumed lipid II binding site (Fig. 7).
Figure 6.
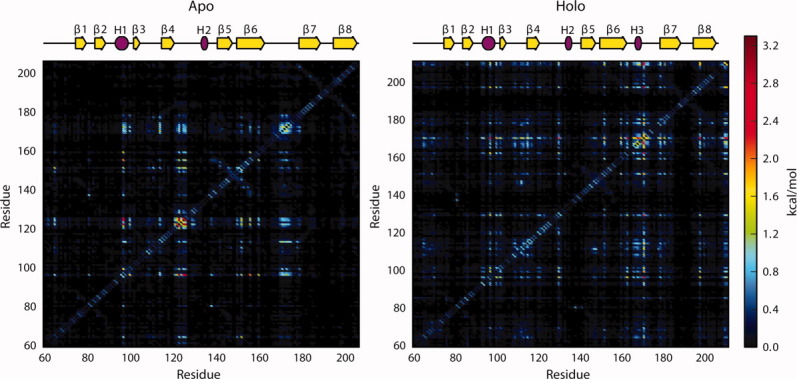
Mutual information scores indicating correlation between residues in the apo simulations and holo simulations in which the sorting signal remained in the active site. In the apo simulations, the lower β6/β7 loop was correlated with H1 and the β4/H2 loop. Additionally, residues in the β4/H2 loop were correlated with Q96 in H1. There was a general increase in correlation between residues in the holo simulations. Notably, the β3/β4 loop was correlated with the entire β6/β7 loop, and many residues throughout the protein were highly correlated with D170 in the β6/β7 loop.
Figure 7.
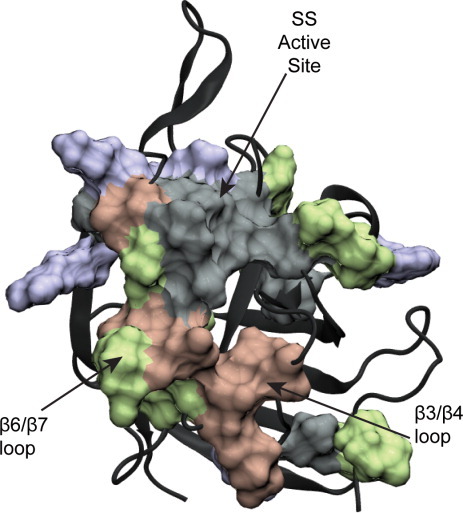
Residues with MutInf scores above 1.5 kT to residue D170 in the holo simulations in which the sorting signal remained bound in the active site, shown as a surface on SrtA. These residues form a network that goes through the sorting signal connecting the sorting signal binding site to the lipid II binding site.
Using the mutual information scores, community structure graphs were created, in which highly correlated residues were grouped into communities that were subsequently connected by edges of widths proportional to the strength of correlation between them, thus providing a coarse-grain representation of the two-dimensional correlation plots.35–38 This approach effectively creates a graph in which the protein/SS complex is divided into regions that move as a single unit, with the widths of the lines connecting them indicating the degree to which motions in one group of residues directly influence the motions in another group of residues. Fortuitously, communities were defined similarly in both the apo and holo states, allowing for direct comparison of correlations between regions (color-coded in Fig. 8). In the holo state, the β6/β7 loop region (pink) was correlated with the β3/β4 loop region (maroon); however, this correlation was lost in the apo state. Additionally, the community colored dark purple encompassed much more of the active site region in the holo state than it did in the apo state, indicating that the active site itself was more correlated when the SS was bound to SrtA. This community was correlated with the β3/β4 loop (maroon) in the holo state, but not in the apo state. In the holo state, this purple community encompassed the leucine and proline residues of the SS, while the alanine, threonine, and glycine residues were part of the green community, which encompassed the β7/β8 loop. These two communities were, however, highly correlated. In both the apo and the holo states, the β7/β8 loop (green) was correlated with the β4/H2 loop (tan) and the community that encompassed H1 and the surrounding residues (orange).
Figure 8.
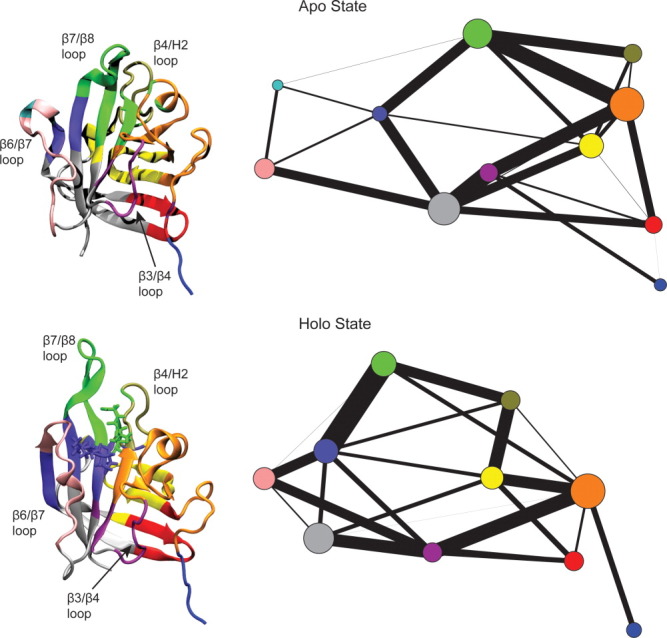
Community structure graphs (right) and images of SrtA with residues colored according to community (left) for the apo and holo states (top and bottom, respectively). The thickest lines, indicating greatest correlation, going around the holo community structure graph suggest a network that extends from the β3/β4 and β6/β7 loops, to the β7/β8 loop, through the sorting signal and the active site, and to the lipid II binding site.
Overall, the apo community structure graph depicts a network that primarily connects regions of the central β-barrel. On the other hand, following the thickest edges around the holo community structure graph suggests an allosteric network much like the one identified in the holo MutInf analysis. This network goes from the β3/β4 loop (maroon) to the β6/β7 loop (pink), through the active site and the L and P residues of the SS (purple), to the β7/β8 loop and the A, T, and G residues of the SS (green), to the residues around the lipid II binding site, and back to the β3/β4 loop.
aMD simulations
Accelerated molecular dynamics (aMD) simulations introduce an additional potential energy term to the simulation that lowers the energy barriers between local minima, thus allowing the sampling of long-timescale events that are beyond the current scope of conventional simulations.39 Throughout the three 40-ns apo aMD simulations, the overall conformation of SrtA was stable, with RMSDs around 3 Å and FCA projections sampling little phase space. In five of the six 40-ns holo aMD simulations, SrtA appeared only marginally more dynamic than in the cMD simulations, as shown by the slightly increased fluctuation of the RMSD values. However, in simulation 4, with the SS bound away from the active site, the holo structure underwent a large-scale conformational transition to more closely resemble SrtA in its apo state, as is reflected in the FCA projections [Fig. 3(b), colored dark green]. The SS was observed to move away from the active site in two of the three aMD simulations in which it started in the active site, in one at 27 ns and in the other at 28 ns.
To further investigate the propensity of the holo state to transition to the apo conformation when the SS was not bound in the active site, a set of three aMD simulations of the initial holo structure with the SS removed were run. Projections of the trajectories onto the FCA space indicate that the initial structure did transition to a conformation more similar to that of the apo state in two of the three simulations. cMD simulations of these same constructs showed that this transition did not occur on the 100-ns timescale (Fig. 3).
Because motions of the β6/β7 and β7/β8 loops, which are responsible for the most significant differences between the apo and holo states, are encompassed in FCA mode one, normalized RMSD differences for these loops were considered (Supporting Information Fig. S3). Of particular interest were the simulations with FCA projections indicative of a conformational transition to a structure closer to that of the apo state, namely holo aMD simulation 4, and apo from holo aMD simulations 1 and 2. In each of these three simulations, the β7/β8 loop moved to become more apo-like near the beginning of the simulation and remained that way until the end of the simulation. In contrast, the β6/β7 loop transitioned to a more apo-like state in apo from holo aMD simulation 2 only to return to a more holo-like state. The β6/β7 loop failed to become more apo-like in the other two simulations. Thus, the significant motion along FCA mode one in all three of these simulations was largely due to the conformational transition of the β7/β8 loop.
Discussion
Results presented here provide several key insights into the mechanism of SrtA binding to a SS motif. In the absence of a bound peptide, cMD and aMD simulations indicate that the most significant motions occur in the β6/β7 loop, as it samples configurations ranging from an open state that creates a large cleft between it and the central β-barrel, and a semi-closed configuration in which two residues (D170 and E171) may each individually interact with the stably bound calcium ion. Upon binding to an LPXTG sequence, the β6/β7 loop further closes and samples multiple states in which the section closest to the active site transiently samples alpha and 310 helices, along with disordered states (see Fig. 9). The β7/β8 loop also opens to accommodate the SS in the active site. The fact that, even in the longer timescales accessible to the aMD simulations, transitions between the holo and apo states only occurred when the protein began in the holo state and the SS dissociated or was removed from the active site suggests that the free energy minima of the holo conformation is partially stabilized by SS binding. This is consistent with an induced-fit mechanism. However, the observed fluctuations of the β6/β7 loop between open and semi-closed states, and the previously reported NMR data on micro to millisecond timescale dynamics of these same residues,30 point toward a conformational selection mechanism. Taken together, experiments and simulations therefore suggest a recognition model of conformational selection followed by induced fit, consistent with the emerging paradigm for a diverse range of recognition processes.40, 41
Figure 9.
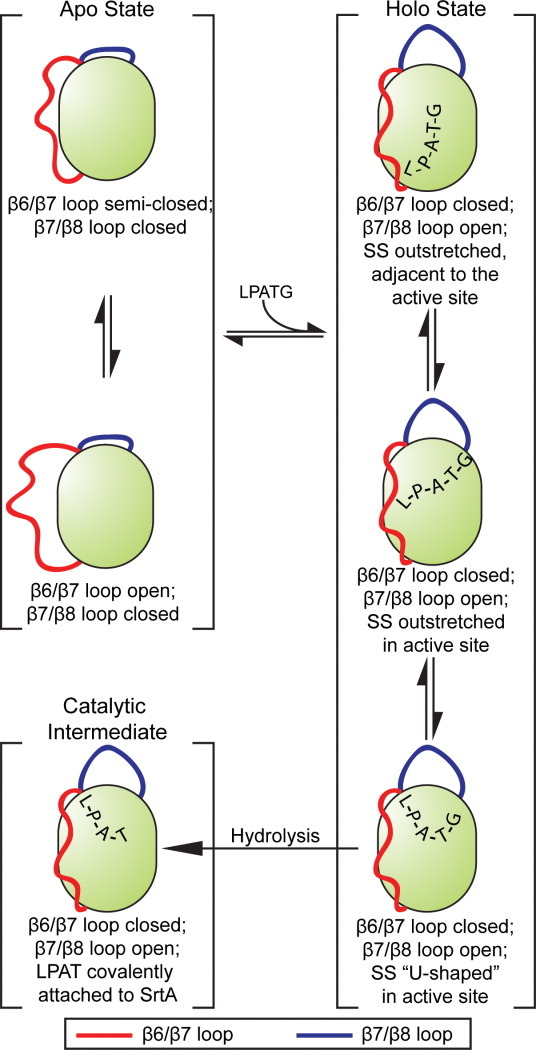
The mechanism of SrtA. In the apo state, the β6/β7 loop fluctuates between open and semi-closed states, while the β7/β8 loop remains closed. When the sorting signal binds, the β7/β8 loop stays open, and the β6/β7 loop remains closed. In this holo state, the sorting signal binds in three distinct configurations: in the first, which is likely the precursor to the catalytic intermediate, the sorting signal adopts a “U-shaped” conformation in the active site; in the second, the sorting signal is more outstretched in the active site; and in the third, the sorting signal remains outstretched, but is located in the groove leading into the active site adjacent to the β6/β7 loop. These structures are represented by clusters 1–3 (see Fig. 4). The catalytic intermediate in this figure is based on the 2KID NMR structure.21 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Simulation results also indicate that the pentapeptide likely exists in an equilibrium of bound conformations. One conformation (cluster 1) resembles the previously reported NMR structure, with the exception that in our simulations, the LPATG sequence is noncovalently bound to the active site cysteine. The location of the SS in the cluster 3 conformation resembles that of the previously reported crystal structure; however, it differs for multiple reasons, including the fact that the β6/β7 loop is in a closed state. The third conformation (cluster 2) represents a previously unobserved elongated form in which the SS is centrally located in the active site. These structures may represent metastable states along the binding and hydrolysis pathway, where cluster 3 is a location of nonspecific SS binding, cluster 2 is a more specific active-site location, and cluster 1 is a prehydrolysis configuration (Fig. 9). Each of these states appears to be stabilized by a distinct series of protein interactions: cluster 1 makes strong contacts with R197, cluster 2 with H120, C184, and R197, and cluster 3 with R197 and residues 164–171 in the β6/β7 loop and 105–108 in the β3/β4 loop. In cluster 3, this movement of the SS away from the center of the active site allows for increased flexibility in the β7/β8 loop and slightly lowers fluctuations in the β6/β7 loop relative to clusters 1 and 2. It is interesting to note that although all these binding poses are spatially separated from one another, R197 makes at least one strong interaction to the SS backbone in each of them. In addition, H120, the role of which is still unclear, makes stabilizing contacts to the SS in cluster 2, suggesting that it may have a role in the binding process that has yet to be identified.
The process of binding also appears to have a profound effect on the presence of allosteric networks within SrtA. In the apo form, we identified correlations primarily between regions of the central β-barrel, but we were unable to detect long-range networks that strongly link the SS and lipid II binding regions. However, when bound to the peptide, two analysis methods (which were both based upon a mutual information analysis of side-chain motions) indicate a pair of possible allosteric pathways that link the SS-binding site to residues in H1 and the β4/H2 loop, in the presumed lipid II binding region. Remarkably, both these pathways include residues within the SS, suggesting that if SrtA binds to the SS before its association with lipid II, as experimental evidence suggests, the enzyme uses its first substrate to transmit information to the region in which it binds its second substrate. In vivo, this may be a mechanism that prohibits premature binding of SrtA to a lipid II molecule until it has bound to a SS. Should the in vivo catalytic cycle of SrtA involve the binding of lipid II molecules before SS association, the allosteric network observed in these holo simulations may play a role in the SS/lipid II linkage mechanism.
Simulation results presented herein complement and extend previous computational studies of SrtA. By performing QM/MM calculations, Tian and Eriksson27 demonstrated that R197 plays multiple roles in stabilization of the SS, which is compatible with the observed hydrogen bonding and interaction energies we report. In their study, Moritsugu et al.29 observed that association of the sorting sequence restricts the movement of the β6/β7 loop to the closed state, in agreement with our cMD and aMD results. However, in neither of these studies was it observed that the SS adopted distinct, stable conformational states differing from the NMR structure, which we attribute to the shorter sampling time in the QM/MM studies, and the simulation of a four-peptide sorting sequence that lacked the terminal glycine residue of the LPATG motif in the latter. Furthermore, the analyses of allosteric interactions throughout the protein resulting from this work are novel findings that enhance our understanding of SrtA function.
Although simulations were performed on a short sorting sequence, their results have interesting implications for the more physiologically relevant case of SrtA binding to a much longer peptide. In our simulations, the SS adopted multiple metastable states that were stabilized by distinct contacts with the protein. However, given the strong similarities between the SrtA conformations for these peptide locations, it is likely that a longer SS would bind to multiple of these sites simultaneously, for example, by positioning the LPXTG motif in the active site (as in clusters 1 and 2) and residues upstream of it in the cleft between the β6/β7 and β3/β4 loops (as in clusters 3 and 4). By satisfying multiple of these sets of contacts simultaneously, the probability of binding a target protein to SrtA, positioning the LPXTG motif in the active site and biasing SrtA into the holo “closed” form, may all be increased. Therefore, when considering the mechanism of SrtA binding to a target, it may be important to take into account not only residues within the active site, but also those within the cleft formed between the β6/β7 and β3/β4 loops.
Methods
The cMD simulations of apo SrtA were started from the 1T2P crystal structure, which was solved in the absence of a calcium ion.20 Because it has been shown to be important for the catalytic activity of the enzyme, a calcium ion was added to the structure, positioned in its known binding site.20, 30 Initial coordinates for the holo SrtA simulations were generated from the 2KID NMR structure in which an LPAT analog is covalently attached to the catalytic C184 residue in the enzyme's active site.21 For the holo structure, an unmodified LPATG sequence was subsequently docked into this site using Glide with the XP scoring function,42 and the structure in which the positions of the L, P, and A residues most closely matched those in the NMR structure was chosen for simulation. Simulations were not performed from the peptide-bound crystal structure as the discrepancies between it and the experimental data (particularly the orientation of protein side chains relative to the SS and the conformation of the β6/β7 loop) suggest that crystal artifacts may have influenced the structure. The “apo from holo” structure was created from the identical NMR coordinates; however, the SS was removed in this case. Each structure was solvated in an orthorhombic box containing an ∼150 mM NaCl solution, with at least 10 Å of solvent between the surface of the protein and the edges of the box. All simulations were performed with NAMD,43 using the AMBER99SB-ILDN force field.44 To allow the use of a 2-fs timestep, the SHAKE algorithm was used to constrain all hydrogen-containing bonds.45 Long-range electrostatics were treated using the particle-mesh Ewald summation method with 1-Å grid spacing and cubic B-spline,46 while short-range nonbonded interactions were truncated at 10 Å, with the introduction of a smoothing function at 9 Å. The NPT ensemble was implemented using Langevin dynamics with a damping coefficient of 2 ps−1 to maintain the system at 300 K and the Nosé-Hoover constant pressure method with a target pressure of 1 atm, a piston period of 100 fs, and a damping time scale of 50 fs.47, 48 Following 5000 steps of minimization, heavy atom restraints were incrementally reduced over the timespan of 500 ps. A total of twelve 100-ns cMD simulations were performed: three of the apo state, six of the holo state, and three of the apo from holo state. The final snapshots of each of these simulations were used to initialize a 40-ns dual-boost aMD simulation.49–51 The aMD parameters were chosen based upon the system size and average dihedral and potential energies (for a full discussion, see Ref.52): Edih = 2260 kcal mol−1, αdih = 120 kcal mol−1, Etot = −71,630 kcal mol−1, and αtot = 4727 kcal mol−1.
Frames from the first 20 ns of each of the cMD simulations were excluded from analysis to allow for equilibration. RMSDs and RMSFs were computed using the positions of the Cα atoms, excluding those of the first six residues for the RMSDs. In addition, to specifically consider the conformational similarity of the β6/β7 and β7/β8 loops at each frame with respect to the apo and holo states, normalized RMSD differences (Supporting Information Fig. S3) were computed as follows:
| (1) |
where RMSDholo and RMSDapo are the RMSDs with respect to the average holo and apo structures, respectively. RMSDapo-to-holo is the RMSD of the average apo structure with reference to the average holo structure (5.17 Å for the β6/β7 loop and 13.35 Å for the β7/β8 loop). Average structures were determined from the cMD simulations, and all calculations were performed using the Gromacs analysis tools.53
Following the alignment of the stable Cα atoms in and around the active site, clustering analysis was performed on the Cα atoms in the SS using the gromos clustering algorithm in Gromacs with a 2.2 Å cutoff.54 Because of computational limitations, clustering was performed on structures chosen every 15 ps from each of the holo cMD simulations. Clusters are numbered according to their populations; cluster 1 contains the greatest number of frames.
Binding free energies of the SS in each of the clusters were estimated using the MM/GBSA approach on every fifth cluster member using the “mm_pbsa.pl” script available in Amber 11.31, 32 For generalized Born calculations, an effective salt concentration of 0.15M, a surface tension of 0.0072 kcal/(mol·Å2), and a surface offset term of 0 kcal/mol were used, while vibrational entropy differences were computed by a normal-mode analysis.55, 56 Hydrogen bonds were computed with the HBonds plugin in VMD57 and defined to exist when the donor and acceptor atoms were within 3.5 Å of each other, and the donor-hydrogen-acceptor angle was below 30°. Other interactions in these clusters were considered by calculating the interaction energies between the SS and residues shown to be important to the activity of the enzyme using NAMD with electrostatics scaled by a dielectric constant of 20 (Table III). Standard errors reported in Tables II and III were computed by dividing the standard deviations of the reported quantities by the number of independent data points sampled. The statistical inefficiency of the data set was estimated by computing the mean statistical inefficiencies of the MM/GBSA data for each of the holo simulations, which resulted in a value of 8300 ps.
Dominant, low-frequency protein motions were determined by performing FCA in Gromacs on combined trajectories from the apo and holo cMD simulations (a total of nine simulations). This method focuses on minimizing the system's mutual information and offers measurable improvements over the more commonly used principal component analysis by taking into account nonlinear and higher order correlations to identify biologically relevant motions.58 All cMD and aMD trajectories were projected onto the first and second FCA modes (Fig. 3). By performing FCA analysis on the combined apo and holo cMD simulations, the first FCA mode corresponds to the largest variance in the data as a whole and thus describes (to a large degree) the apo to holo transition. In these simulations, this is desirable, as it quantifies not only the phase space corresponding to the end states, but also that pertaining to the transition state regions.
Correlations between individual residues were investigated through the application of the MutInf method to the apo cMD simulations and the holo cMD simulations 1–3 (in which the SS remained in the active site).59 MutInf is an entropy-based approach that provides improvements over other methods by capturing anharmonic correlations, correcting for undersampling, and testing the statistical significance of the correlations. Each simulation was divided into two simulation blocks, for a total of six holo and six apo simulation blocks of 40 ns each. MutInf scores were then used for community analysis,35–38 in which highly correlated residues are grouped into communities, which are connected by edges of widths proportional to the correlation between them. For the Girvan-Newman algorithm that created the community-analysis graphs, initial edges in the graphs were created for residues that were within 7.5 Å of each other for a minimum of 75% of the simulations.35–38
Acknowledgments
We extend our thanks to A. Chan, P. Gasper, S. Nichols, and members of the McCammon lab for valuable discussion concerning the work presented here.
Glossary
Abbreviations
- aMD
accelerated molecular dynamics
- cMD
conventional molecular dynamics
- MM/GBSA
molecular mechanics/generalized born surface area
- SrtA
Sortase A
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Paterson GK, Mitchell TJ. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004;12:89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Scott JR, Barnett TC. Surface proteins of gram-positive bacteria and how they get there. Ann Rev Microbiol. 2006;60:397–423. doi: 10.1146/annurev.micro.60.080805.142256. [DOI] [PubMed] [Google Scholar]
- 3.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clancy KW, Melvin JA, McCafferty DG. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers. 2010;94:385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickx APA, Budzik JM, Oh S-Y, Schneewind O. Architects at the bacterial surface—sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 6.Ton-That H, Marraffini A, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 8.Mazmanian SK, Liu G, Hung TT, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 9.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ton-That H, Schneewind O. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J Biol Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- 12.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 13.Walsh C. Where will new antibiotics come from? Nat Rev Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 14.Cossart P, Jonquieres R. Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc Natl Acad Sci USA. 2000;97:5013–5015. doi: 10.1073/pnas.97.10.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharm Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 16.Ton-That H. Anchoring of surface proteins to the cell wall of Staphylococcus aureus—sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 17.Huang XY, Aulabaugh A, Ding WD, Kapoor B, Alksne L, Tabei K, Ellestad G. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry. 2003;42:11307–11315. doi: 10.1021/bi034391g. [DOI] [PubMed] [Google Scholar]
- 18.Frankel BA, Kruger RG, Robinson DE, Kelleher NL, McCafferty DG. Staphylococcus aureus sortase transpeptidase SrtA: insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry. 2005;44:11188–11200. doi: 10.1021/bi050141j. [DOI] [PubMed] [Google Scholar]
- 19.Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci USA. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong YN, Bice TW, Ton-That H, Schneewind O, Narayana SVL. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 21.Suree N, Liew CK, Villareal VA, Thieu W, Fadeev EA, Clemens JJ, Jung ME, Clubb RT. The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J Biol Chem. 2009;284:24465–24477. doi: 10.1074/jbc.M109.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankel BA, Tong Y, Bentley ML, Fitzgerald MC, McCafferty DG. Mutational analysis of active site residues in the Staphylococcus aureus transpeptidase SrtA. Biochemistry. 2007;46:7269–7278. doi: 10.1021/bi700448e. [DOI] [PubMed] [Google Scholar]
- 23.Liew CK, Smith BT, Pilpa R, Suree N, Ilangovan U, Connolly KM, Jung ME, Clubb RT. Localization and mutagenesis of the sorting signal binding site on sortase A from Staphylococcus aureus. FEBS Lett. 2004;571:221–226. doi: 10.1016/j.febslet.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 24.Marraffini LA, Ton-That H, Zong YN, Narayana SVL, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus—a conserved arginine residue is required for efficient catalysis of sortase A. J Biol Chem. 2004;279:37763–37770. doi: 10.1074/jbc.M405282200. [DOI] [PubMed] [Google Scholar]
- 25.Ton-That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus—cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem. 2002;277:7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- 26.Bentley ML, Lamb EC, McCafferty DG. Mutagenesis studies of substrate recognition and catalysis in the sortase A transpeptidase from Staphylococcus aureus. J Biol Chem. 2008;283:14762–14771. doi: 10.1074/jbc.M800974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian BX, Eriksson LA. Catalytic mechanism and roles of Arg197 and Thr183 in the Staphylococcus aureus sortase A enzyme. J Phys Chem B. 2011;115:13003–13011. doi: 10.1021/jp2058113. [DOI] [PubMed] [Google Scholar]
- 28.Tian B, Eriksson SA. Structural changes of Listeria monocytogenes Sortase A: a key to understanding the catalytic mechanism. Proteins. 2011;79:1564–1572. doi: 10.1002/prot.22983. [DOI] [PubMed] [Google Scholar]
- 29.Moritsugu K, Terada T, Kidera A. Disorder-to-order transition of aniIntrinsically disordered region of sortase revealed by multiscale enhanced sampling. J Am Chem Soc. 2012;134:7094–7101. doi: 10.1021/ja3008402. [DOI] [PubMed] [Google Scholar]
- 30.Naik MT, Suree N, Ilangovan U, Liew CK, Thieu W, Campbell DO, Clemens JJ, Jung ME, Clubb RT. Staphylococcus aureus sortase A transpeptidase. J Biol Chem. 2006;281:1817–1826. doi: 10.1074/jbc.M506123200. [DOI] [PubMed] [Google Scholar]
- 31.Kollman PA, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 32.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 33.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. I. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guimaraes CR, Mathiowetz AM. Addressing limitations with the MM-GB/SA scoring procedure using the WaterMap method and free energy perturbation calculations. J Chem Inf Model. 2010;50:547–559. doi: 10.1021/ci900497d. [DOI] [PubMed] [Google Scholar]
- 35.Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys Rev E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 37.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce LCT, Salomon-Ferrer R, Augusto F deOliveiraC, McCammon JA, Walker RC. Routine access to millisecond time scale events with accelerated molecular dynamics. J Chem Theory Comp. 2012;8:2997–3002. doi: 10.1021/ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 43.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kräutler V, van Gunsteren WF, Hunenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comp Chem. 2001;22:501–508. [Google Scholar]
- 46.Darden T, York D, Pedersen L. Particle mesh Ewald: an N · log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 47.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 48.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 49.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Harrison CB, Schulten K, McCammon JA. Implementation of accelerated molecular dynamics in NAMD. Comput Sci Discov. 2011;4(pii):015002. doi: 10.1088/1749-4699/4/1/015002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamelberg D, de Oliveira CA, McCammon JA. Sampling of slow diffusive conformational transitions with accelerated molecular dynamics. J Chem Phys. 2007;127:155102. doi: 10.1063/1.2789432. [DOI] [PubMed] [Google Scholar]
- 52.Wereszczynski J, McCammon JA. Accelerated molecular dynamics in computational drug design. Methods Mol Biol. 2012;819:515–524. doi: 10.1007/978-1-61779-465-0_30. [DOI] [PubMed] [Google Scholar]
- 53.Lindahl E, Hess B, Van Der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 54.Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide folding: when simulation meets experiment. Angew Chem Intl Ed. 1999;38:236–240. [Google Scholar]
- 55.Brooks BR, Janezic D, Karplus M. Harmonic-analysis of large systems. I. Methodology. J Comp Chem. 1995;16:1522–1542. [Google Scholar]
- 56.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman P. Amber, a package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular-dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comp Phys Commun. 1995;91:1–41. [Google Scholar]
- 57.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 58.Lange OF, Grubmuller H. Full correlation analysis of conformational protein dynamics. Proteins. 2008;70:1294–1312. doi: 10.1002/prot.21618. [DOI] [PubMed] [Google Scholar]
- 59.McClendon CL, Friedland G, Mobley DL, Amirkhani H, Jacobson MP. Quantifying correlations between allosteric sites in thermodynamic ensembles. J Chem Theory Comp. 2009;5:2486–2502. doi: 10.1021/ct9001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


