Abstract
Amyloid fibrils are the pathological hallmark of a large variety of neurodegenerative disorders. The structural characterization of amyloid fibrils, however, is challenging due to their non-crystalline, heterogeneous, and often dynamic nature. Thus, the structure of amyloid fibrils of many proteins is still unknown. We here show that the structure calculation program CS-Rosetta can be used to obtain insight into the core structure of amyloid fibrils. Driven by experimental solid-state NMR chemical shifts and taking into account the polymeric nature of fibrils CS-Rosetta allows modeling of the core of amyloid fibrils. Application to the Y145X stop mutant of the human prion protein reveals a left-handed β-helix
Keywords: fibril, NMR, structure, prion
Introduction
Protein aggregates are the pathological hallmark of a large variety of neurodegenerative disorders.1 Increasing evidence suggests that a variety of aggregation intermediates are the primary toxic species, while amyloid fibrils might have protective functions.2 At the same time, different fibril morphologies can result in different degrees of toxicity3 and fibril fragmentation can enhance amyloid cytotoxicity.4 Thus, detailed knowledge about the structure of amyloid fibrils is required to obtain insight into the toxicity of protein aggregates.
X-ray crystallography has provided unique insights into the structural properties of amyloid fibrils formed by short peptides.5 The structural characterization of protein aggregates, however, is challenging due to their non-crystalline, heterogeneous, and often dynamic nature. This made NMR spectroscopy a key method for the study of protein aggregation.6–8 H/D exchange coupled to solution-state NMR spectroscopy has provided residue-specific insight into the backbone hydrogen-bonding of a variety of protein aggregates (e.g. see Refs.9–13). Solid-state NMR spectroscopy resolved the sequence specific assignment of backbone resonances of several proteins in their amyloid state (for a review see Refs.6–8). In addition, for some amyloid fibrils intra- and intermolecular contacts were detected by solid-state NMR. So far the structurally best characterized system is the prion protein (PrP) HET-s(218–289), for which a well-defined structure of amyloid fibrils was determined on the basis of a large number of intra- and intermolecular distance restraints.14 However, in many cases the detection and interpretation of intra- and intermolecular contacts is complicated by internal dynamics and the heterogeneous nature of amyloid fibrils. In addition, the polymeric nature of amyloid fibrils complicates the interpretation of intermolecular and interfilament contacts.
Because of the difficulty to obtain a sufficiently large number of intra- and intermolecular contacts by solid-state NMR spectroscopy, the structure of amyloid fibrils of many proteins is still not known despite the availability of backbone chemical shifts (e.g. see Refs.15 and16). Here, we demonstrate that the CS-Rosetta methodology17–20 can be used to obtain insight into the core structure of amyloid fibrils. Our study shows that CS-Rosetta—driven by experimental solid-state NMR chemical shifts and taking into account the polymeric nature of fibrils—allows modeling of the core of amyloid fibrils.
Results and Discussion
Calculations were performed using the CS-Rosetta chemical shift-based structure calculation program.17 To reflect the polymeric nature of amyloid fibrils, the CS-Rosetta protocol was modified in the following way: (i) Calculations are performed on multiple (five in the calculations reported below) identical segments that are connected by glycine-serine linkers. (ii) Distance restraints enforce interstrand distances between different layers orthogonal to the fibril axis to 4.8 Å consistent with X-ray diffraction patterns of amyloid fibrils. (iii) Hydrogen-bond distance restraints for parallel in-register alignment are used. (iv) The location of β-strands as identified by solid-state NMR chemical shifts is enforced using Hα(i)–HN(i+1) distance restraints.
The amyloid-specific CS-Rosetta protocol was tested on the PrP HET-s(218–289). Using a large number of experimental intra- and intermolecular distance restraints, the 3D structure of HET-s(218–289) amyloid fibrils was determined.14 The 3D structure revealed a left-handed β-solenoid with each molecule forming two helical windings (Fig. 1). The two helical windings are formed by residues G225-L250 and its pseudo-repeat T260-F286, respectively. Due to the polymeric nature of amyloid fibrils the core structure of the backbone of the two layers is highly similar.
Figure 1.
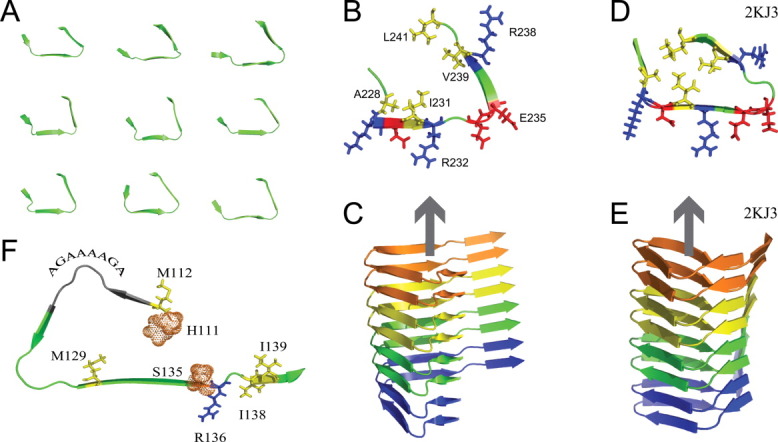
Determination of amyloid core structure using chemical shifts. A: Backbone conformation of 9 of the 19 lowest energy CS-Rosetta structures of the fibrillar core of HET-s(218–289). The view is along the fibril axis. B: Lowest energy conformer of the fibrillar core of HET-s(218–289) obtained by CS-Rosetta. Side chains of selected residues are shown (hydrophobic: yellow; positive: blue; negative: red). C: Side view of an 8-mer based on the β-helix motif shown in (B). D, E: Fibril core and side view of the solid-state NMR structure of amyloid fibrils of the PrP HET-s(218–289) (PDB code: 2KJ3). F: Lowest energy CS-Rosetta conformer of residues 111–141 of the Y145X stop mutant of the human PrP. Selected residues are highlighted. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
CS-Rosetta calculations were performed for a molecule containing three segments of G225-L250 interleaved by two segments of T260-F286. G225-L250 and T260-F286 are connected by a linker that remains—according to solid-state NMR and hydrogen/deuterium-exchange coupled to solution NMR—disordered and flexible in the amyloid structure. This HET-s specific linker was replaced by a 12-residue glycine-serine linker. The connection of T260-F286 to G225-L250 of the next molecule was also made by a 12-residue glycine-serine linker. As solid-state NMR and hydrogen/deuterium-exchange data demonstrated that residues 247AATL250 and 283GKGF286 are also unstructured,12, 14 the total length of the linkers between different layers along the fibril axis was 16 residues. CS-Rosetta fragment selection was performed using the previously reported experimental solid-state NMR chemical shifts.14, 21 To minimize the contribution of the flexible linkers to structure calculation, the conformation of the glycine and serine residues in the linker was driven toward the disordered state by providing random coil chemical shifts for these residues during CS-Rosetta fragment selection.
Fold assembly was driven by a total of 733 distance restraints enforcing the cross-β structure of HET-s(218–289) fibrils and the location of β-strands indicated by chemical shifts (Table I). Hydrogen bond restraints enforced intermolecular, parallel, in-register β-sheets (Table I).14 Distance restraints were supplied eight times to CS-Rosetta. Supplying the distance restraints only six times, that is, increasing the weight effectively sixfold, resulted in similar folds for HET-s(218–289) fibrils. Supplying the distance restraints only once, that is, not increasing their weight, did not result in a polymeric arrangement. Changing the distance restraints for interstrand H-bonds to O–HN 2.1 Å/O–N 3.5 Å and for interstrand polymer to 5.0 Å/10.0 Å/20.0 Å resulted in similar lowest energy CS-Rosetta structures for HET-s(218–289).
Table I.
Restraints Used in CS-Rosetta Calculations of HET-s(218–289) Fibrils
| Restraints | Type | Upper distance limits | No. of restraints |
|---|---|---|---|
| Chemical shifts | Experimentala | 508 | |
| GS-linkerb | 168 | ||
| Total | 676 | ||
| Distance restraints | Interstrand H-bonds | O–HN 1.8 Å/O–N 2.7 Å | 160 |
| Intra β-strand Hα(i)-HN(i+1) | 2.5 Å | 80 | |
| Interstrand polymer | 4.8 Å/9.6 Å/19.2 Åc | 493 | |
| Total | 733 |
Taken from Ref.21.
Random coil values for N, CA, C of Gly and N, CA, C, CB for Ser.
Upper distance limits for strands i+1, i+2, and i+3 along the fibril axis enforcing the polymeric nature of the fibrils and in agreement with their cross β-structure.
Out of the 100 lowest energy CS-Rosetta structures only those structural models were retained for which the root-mean-square difference in the ϕ/Ψ backbone torsion angles between the angles observed in the model and those predicted by the program TALOS+22 on the basis of the experimental NMR chemical shifts was less than 80°. For the TALOS+ selection only those residues were considered for which the TALOS+ prediction were labeled as good. This final step ensures that the selected models are in agreement with dihedral angles predicted on the basis of the experimental chemical shifts.
Inspection of the 19 lowest energy Rosetta structures of the HET-s amyloid core revealed three β-strands with β-strands 2 and 3 assuming two sides of a triangle. β-strand 1 (residues 226NSA228) has a more variable position including conformers in which the triangle is closed as well as ones with more open conformations. In all the 19 lowest energy Rosetta structures, the core is stabilized by hydrophobic interactions between I231, V239, and L241 [Fig. 1(B)]. The CS-Rosetta model with the lowest chemical shift score is shown in more detail in Figure 1(B). The triangular core [Fig. 1(B,C)] is similar to the conformation seen in the solid-state NMR structure (PDB code: 2KJ3) [Fig. 1(D,E)]. In both structures the hydrophobic side chains of residues I231, V239, and L241 point to the interior while polar side chains are on the surface of the fibrillar core. Importantly, no distance restraints except for those reflecting the polymeric nature of amyloid fibrils were used in the CS-Rosetta calculations, whereas the solid-state NMR structure was calculated with a large number of experimental intra- and intermolecular distance restraints.
The CS-Rosetta structure shown in Figure 1(B) only represents the fold and not a high-resolution structure. In applications to globular proteins, CS-Rosetta was demonstrated to result in backbone structures that differ by 1–4 Å from a high-resolution structure depending on the complexity and the size of the protein. In the case of HET-s(218–289) amyloid fibrils, the hydrophobic core of the CS-Rosetta model is less compact when compared to the solid-state NMR structure. In addition, the compactness of this core varies in the 19 lowest energy CS-Rosetta conformers [Fig. 1(A)]. The different degree of compactness is associated with different twists of the three strands and the β-helix [Fig. 1(A,C)]. In line with variable twists in the hydrophobic core, the turn connecting the hydrophobic core to the β-strand formed by residues N243-T246 is less bent [Fig. 1(C,E)]. Keeping the twist of the β-helix small—as suggested by electron microscopy—might further improve modeling of the amyloid core.
Next we applied the CS-Rosetta protocol to amyloid fibrils of the Y145X stop mutant of the human PrP, for which the structure is not known. Misfolding of the natively α-helical PrP into a β-sheet rich isoform is related to various human diseases such as Creutzfeldt–Jakob disease.23 Currently, little is known about the structure of PrPSc.24, 25 Instead, a variety of studies have investigated the structure of amyloid fibrils produced in vitro from recombinant PrP.26–35 Hereditary prion diseases include C-terminally truncated variants of the PrP, Y145X, Q160X, Y226X, and Q227X. Previously, we showed that the β-sheet content is highly similar in amyloid fibrils of the Y145X and Q160X stop mutants of human PrP.36 In addition, solid-state NMR spectroscopy demonstrated that residues 112–140 assume extended β-sheet conformation in a parallel, in register alignment in amyloid fibrils of the Y145X stop mutant.29–31
We subjected the solid-state NMR chemical shifts reported by Helmus et al.29 for amyloid fibrils of the Y145X prion stop mutant to the adapted CS-Rosetta protocol.17 Calculations were performed on five identical segments that were connected by 16-residue glycine-serine linkers. Distance restraints (Table II) enforced interstrand distances between different layers orthogonal to the fibril axis to 4.8 Å, in agreement with X-ray diffraction patterns of mammalian prions37 and amyloid fibrils of PrP(82–146).38 Hydrogen-bond distance restraints for parallel, in-register alignment—as shown by solid-state NMR spectroscopy of humPrP(23–144)31—were used. In addition, the location of β-strands as identified by solid-state NMR chemical shifts (Supporting Information Fig. S1) was enforced using Hα(i)–HN(i+1) distance restraints.
Table II.
Restraints Used in CS-Rosetta Calculations for Residues 111–141 of Amyloid Fibrils of the Y145X Stop Mutant of the Human Prion Protein
| Restraints | Type | Upper distance limits | No. of restraints |
|---|---|---|---|
| Chemical shifts | Experimentala | 510 | |
| GS-linkerb | 224 | ||
| Total | 734 | ||
| Distance restraints | Interstrand H-bondsc | O–HN 1.8 Å/O–N 2.7 Å | 176 |
| Intra β-strand Hα(i)–HN(i+1)d | 2.5 Å | 85 | |
| Interstrand polymer | 4.8 Å/9.6 Å/19.2 Åe | 714 | |
| Total | 975 |
Taken from Ref.29.
Random coil values for N, CA, C of Gly and N, CA, C, CB for Ser.
H-bond restraints enforce a parallel, in register alignment in agreement with solid-state NMR data for fibrils of the Y145X stop mutant of the human prion protein.31
Residues 112MAGA115, 120AVVG123, 128YMLGSAMSR136, 138IIHF141 are in a β-strand conformation according to fragments selected by CS-Rosetta (Supporting Information Fig. S1).
Upper distance limits for strands i+1, i+2, and i+3 along the fibril axis enforcing the cross-β structure of the fibrils.
The lowest energy CS-Rosetta amyloid structure of the amyloid core of the Y145X stop mutant is shown in Figure 1(F). It revealed a left-handed β-helix comprising three β-strands that are formed by residues 111HMAGA115, 120AVVG123, and 128YMLGSAMSR136. At P137 the third β-strand twists such that residues 138IIHF141 extend the β-helix. The palindromic sequence 113AGAAAAGA120 encompasses a loop that connects two β-strands. The fibrillar core is formed by the most hydrophobic part of the human prion sequence from H111 to S135. The β-strand conformation of residues 128YMLGSAMSRPIIHF141 shown in Figure 1(F) is highly similar to the structure of amyloid fibrils of humPrP(127–147) as determined by solid-state NMR spectroscopy.39 In addition, electron microscopy of 2D crystals of the 27–30 kDa infectious fragment, PrP27–30, suggested that PrP amyloid fibrils might consist of left-handed β-helices with the core formed by residues 89–140.24, 40
In summary, we show that the CS-Rosetta methodology allows modeling of the core of amyloid fibrils. In line with the application of CS-Rosetta to globular proteins, the calculated conformations are not high-resolution structures. At the same time, the backbone folds can be useful for designing additional experiments and to obtain insights into the mechanism of protein aggregation. Additional experimental information such as spatial proximity between side chains based on mutations and experimental distance restraints obtained from solid-state NMR spectroscopy can be readily included into the calculations. The combination of CS-Rosetta modeling with intra- and intermolecular medium- and long-range distance restraints is expected to improve the accuracy of the structure of amyloid fibrils.
Materials and Methods
CS-Rosetta calculations were performed according to the authors' manual.17 Note, that chemical shifts and distance restraints (hydrogen bonds, distances) are used in distinct stages of the CS-Rosetta protocol. Chemical shifts are only used during fragment selection and rescoring of CS-Rosetta models, while distance restraints (hydrogen bonds, distances) are only used during the fragment assembly stage of CS-Rosetta (see http://spin.niddk.nih.gov/bax/software/CSROSETTA/). Hydrogen bonds are enforced as distance restraints (Tables I and II) and have the same weight as all other distance restraints during CS-Rosetta fragment assembly. To ensure that the distance restraints are the major driving force during CS-Rosetta fold assembly, they were supplied eight times to CS-Rosetta (see Supporting Information). Thus, the weight of the distance restraints was effectively increased eightfold compared to the intrinsic CS-Rosetta energy terms. Alternatively, the CS-Rosetta distance restraint force constant might be increased by a factor of eight. About 2500 structures were calculated. For further details please see the main text.
Acknowledgments
Authors thank Dr. Min-Kyu Cho, Dr. Yang Shen, and Dr. Ad Bax for assistance with the CS-Rosetta package, and Dr. Stefan Becker and Luis Fonseca for useful discussions.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 2.Benilova I, Karran E, De Strooper B. The toxic Abeta oligomer and Alzheimer's disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 3.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 4.Xue WF, Hellewell AL, Gosal WS, Homans SW, Hewitt EW, Radford SE. Fibril fragmentation enhances amyloid cytotoxicity. J Biol Chem. 2009;284:34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldus M. Solid-state NMR spectroscopy: molecular structure and organization at the atomic level. Angew Chem Int Ed Engl. 2006;45:1186–1188. doi: 10.1002/anie.200503223. [DOI] [PubMed] [Google Scholar]
- 7.Heise H. Solid-state NMR spectroscopy of amyloid proteins. Chembiochem. 2008;9:179–189. doi: 10.1002/cbic.200700630. [DOI] [PubMed] [Google Scholar]
- 8.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino M, Katou H, Hagihara Y, Hasegawa K, Naiki H, Goto Y. Mapping the core of the beta(2)-microglobulin amyloid fibril by H/D exchange. Nat Struct Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 10.Cho MK, Kim HY, Fernandez CO, Becker S, Zweckstetter M. Conserved core of amyloid fibrils of wild type and A30P mutant alpha-synuclein. Protein Sci. 2011;20:387–395. doi: 10.1002/pro.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olofsson A, Ippel JH, Wijmenga SS, Lundgren E, Ohman A. Probing solvent accessibility of transthyretin amyloid by solution NMR spectroscopy. J Biol Chem. 2004;279:5699–5707. doi: 10.1074/jbc.M310605200. [DOI] [PubMed] [Google Scholar]
- 12.Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen-deuterium (H/D) exchange mapping of Abeta 1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 14.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 15.Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M. Characterization of Alzheimer's-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. J Am Chem Soc. 2008;130:5922–5928. doi: 10.1021/ja7100517. [DOI] [PubMed] [Google Scholar]
- 16.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowers PM, Strauss CE, Baker D. De novo protein structure determination using sparse NMR data. J Biomol NMR. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- 19.Rohl CA, Baker D. De novo determination of protein backbone structure from residual dipolar couplings using Rosetta. J Am Chem Soc. 2002;124:2723–2729. doi: 10.1021/ja016880e. [DOI] [PubMed] [Google Scholar]
- 20.Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins. 1999;37(Suppl 3):171–176. doi: 10.1002/(sici)1097-0134(1999)37:3+<171::aid-prot21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- 21.Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Bockmann A, Meier BH. Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguzzi A, Sigurdson C, Heikenwaelder M. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- 24.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz-Espinoza R, Soto C. High-resolution structure of infectious prion protein: the final frontier. Nat Struct Mol Biol. 2012;19:370–377. doi: 10.1038/nsmb.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X, Wintrode PL, Surewicz WK. Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci USA. 2007;104:1510–1515. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nazabal A, Hornemann S, Aguzzi A, Zenobi R. Hydrogen/deuterium exchange mass spectrometry identifies two highly protected regions in recombinant full-length prion protein amyloid fibrils. J Mass Spectrom. 2009;44:965–977. doi: 10.1002/jms.1572. [DOI] [PubMed] [Google Scholar]
- 28.Damo SM, Phillips AH, Young AL, Li S, Woods VL, Jr, Wemmer DE. Probing the conformation of a prion protein fibril with hydrogen exchange. J Biol Chem. 2011;285:32303–32311. doi: 10.1074/jbc.M110.114504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmus JJ, Surewicz K, Apostol MI, Surewicz WK, Jaroniec CP. Intermolecular alignment in Y145Stop human prion protein amyloid fibrils probed by solid-state NMR spectroscopy. J Am Chem Soc. 2011;133:13934–13937. doi: 10.1021/ja206469q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerum C, Silvers R, Wirmer-Bartoschek J, Schwalbe H. Unfolded-state structure and dynamics influence the fibril formation of human prion protein. Angew Chem Int Ed Engl. 2009;48:9452–9456. doi: 10.1002/anie.200903771. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler J, Viehrig C, Geimer S, Rosch P, Schwarzinger S. Putative aggregation initiation sites in prion protein. FEBS Lett. 2006;580:2033–2040. doi: 10.1016/j.febslet.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Tycko R, Savtchenko R, Ostapchenko VG, Makarava N, Baskakov IV. The alpha-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel beta-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry. 2010;49:9488–9497. doi: 10.1021/bi1013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar J, Sreeramulu S, Schmidt TL, Richter C, Vonck J, Heckel A, Glaubitz C, Schwalbe H. Prion protein amyloid formation involves structural rearrangements in the C-terminal domain. Chembiochem. 2010;11:1208–1213. doi: 10.1002/cbic.201000076. [DOI] [PubMed] [Google Scholar]
- 36.Watzlawik J, Skora L, Frense D, Griesinger C, Zweckstetter M, Schulz-Schaeffer WJ, Kramer ML. Prion protein helix1 promotes aggregation but is not converted into beta-sheet. J Biol Chem. 2006;281:30242–30250. doi: 10.1074/jbc.m605141200. [DOI] [PubMed] [Google Scholar]
- 37.Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, Stubbs G. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci USA. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmona M, Morbin M, Massignan T, Colombo L, Mazzoleni G, Capobianco R, Diomede L, Thaler F, Mollica L, Musco G, Kourie JJ, Bugiani O, Sharma D, Inouye H, Kirschner DA, Forloni G, Tagliavini F. Structural properties of Gerstmann-Straussler-Scheinker disease amyloid protein. J Biol Chem. 2003;278:48146–48153. doi: 10.1074/jbc.M307295200. [DOI] [PubMed] [Google Scholar]
- 39.Lin NS, Chao JC, Cheng HM, Chou FC, Chang CF, Chen YR, Chang YJ, Huang SJ, Chan JC. Molecular structure of amyloid fibrils formed by residues 127 to 147 of the human prion protein. Chemistry. 2010;16:5492–5499. doi: 10.1002/chem.200903290. [DOI] [PubMed] [Google Scholar]
- 40.Wille H, Govaerts C, Borovinskiy A, Latawiec D, Downing KH, Cohen FE, Prusiner SB. Electron crystallography of the scrapie prion protein complexed with heavy metals. Arch Biochem Biophys. 2007;467:239–248. doi: 10.1016/j.abb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


