Abstract
Purpose
Patients with gestational diabetes mellitus (GDM) have been reported to exhibit the same genetic susceptibility as that observed in those with type 2 diabetes mellitus (T2DM). Recent polymorphism studies have shown that several genes are related to T2DM and GDM. The aim of this study was to examine whether certain candidate genes, previously shown to be associated with T2DM, also offer a specific genetic predisposition to GDM.
Materials and Methods
The current study was conducted in 136 Korean pregnant women, who gave birth at Gil Hospital, from October 2008 to May 2011. These study subjects included 95 subjects with GDM and 41 non-diabetic controls. We selected the specific genes of PPARγ2, IGF2BP2, and KCNQ1 for study and amplified them using the polymerase chain reaction. This was followed by genotyping for single nucleotide polymorphisms. We then compared the genotype frequencies between patients with GDM and non-diabetic controls using the χ2 test. We obtained and analyzed clinical information using Student's t-test, and statistical analyses were conducted using logistic regression with SPSS Statistics software, version 19.0.
Results
Significant differences were observed in maternal age, body mass index, weight gain and weight at time of delivery between the groups compared. Among pregnant women, polymorphisms in PPARγ2 and IGF2BP2 were shown to be highly correlated with GDM occurrence, whereas no correlation was found for KCNQ1 polymorphisms.
Conclusion
Our results indicated that genetic polymorphisms could also be of value in predicting the occurrence and diagnosis of GDM.
Keywords: Gestational diabetes mellitus, type 2 diabetes mellitus, gene, single nucleotide polymorphism
INTRODUCTION
Gestational diabetes mellitus (GDM) is defined as glucose intolerance of varying degrees of severity, and is first detected during pregnancy.1 When pregnant women present with clinical characteristics that are consistent with a high risk of GDM (marked obesity, a personal history of GDM, glycosuria or a significant family history of diabetes), they should immediately undergo a 50-g oral glucose challenge test. In addition, at risk pregnant women should also receive attentive care after giving birth, as they have increasingly been shown to involve a high risk of type 2 diabetes mellitus (T2DM) following pregnancy, in addition to a number of pregnancy complications.2
The genetic background of T2DM may also be applied in GDM because significant evidence has demonstrated the presence of T2DM in women with GDM.3 Moreover, the prevalence of T2DM has been shown to be relatively higher in mothers with GDM after pregnancy.4 Genome-wide association studies have recently identified new genetic variants which have reproducible associations with a susceptibility to T2DM.5-9 Of these, PPARγ2 has been proposed to play an essential role in glucose homeostasis, and IGF2BP2 in insulin function. Additionally, KCNQ1, present in adipose tissue, has been shown to be associated with susceptibility to T2DM.10-12 If it is true that GDM and T2DM share a common genetic background, then genetic variants used to determine the risk of developing T2DM may also be associated with the prevalence of GDM.13
In the present study, we compared the frequency of single nucleotide polymorphisms (SNPs) between GDM patients with diabetogenic genes and non-diabetic controls. The aim of this study was to examine whether certain candidate genes previously shown to be associated with T2DM also offer a specific genetic predisposition to GDM.
MATERIALS AND METHODS
Study subjects
This study included 136 pregnant, ethnic Korean women, 95 of whom were diagnosed with GDM (n=95) and 41 of whom were healthy, non-diabetic controls (n=41). All subjects gave birth at our institution between September 2008 and May 2011. They were all screened for GDM between 24 to 28 weeks of pregnancy using a 50-g, 1-h glucose challenge test (a plasma glucose concentration of ≥140 mg/dL was considered to be positive for diagnosis of GDM) as recommended by the Third International Workshop-Conference on GDM. This was followed by a 100-g oral glucose tolerance test (OGTT).
Threshold glucose values were rated as follows: fasting ≥95 mg/dL, 1 h ≥180 mg/dL, 2 h ≥155 mg/dL and 3 h ≥140 mg/dL. We also obtained demographic and clinical data including age, height, body mass index (BMI), weight gain throughout pregnancy, weight at delivery, and gestational age (GA) at delivery. In the present study, the exclusion criteria were twin pregnancy and other complicated pregnancies, such as pregnancy-induced hypertension. The study protocol was approved by the Institutional Review Board of Gil Hospital, and written informed consent was obtained from all subjects.
Genetic analysis
We selected PPARγ2 rs1801282, IGF2BP2 rs4402960, and KCNQ1 rs151290 as polymorphisms for this study. Total DNA was prepared from peripheral blood leukocytes by the proteinase K-phenol-chloroform extraction method. Blood samples were sequentially treated two times each with buffer (10 mM Tris-HCl, 0.1 M EDTA, 1% SDS, proteinase K), phenol (Bioneer, Daejeon, Korea), PCI solution (Phenol, Chloroform, Isoamyl alcohol), chloroform (Junsei Chem. Co., Tokyo, Japan), isopropanol (Junsei Chem.Co., Tokyo, Japan) and ethanol (Dae-Jung chemicals and metals co. Ltd., Siheung, Korea). Then, isolated DNA was dissolved in distilled water, and the resulting genomic DNA samples were stored at -80℃ until ready for analysis.
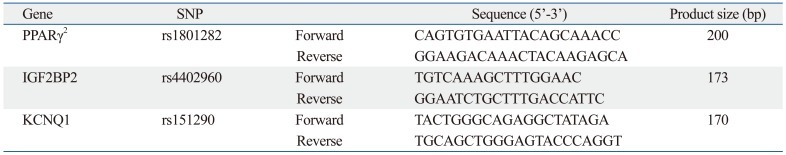
Genes for the assay were amplified with polymerase chain reaction (PCR) using forward and reverse primers (Table 1). PCR was performed in a 20 µL volume containing 100 ng of genomic DNA with 10 pmol of each primer, 10× EX Taq buffer (100 mM Tris-HCl, 500 mM KCl and 15 mM MgCl2), 2.5 mM dNTP mixture and 1U EX Taq polymerase (TaKaRa, Otsu, Shiga, Japan), for which a PCR thermal cycler (TaKaRa, Otsu, Shiga, Japan) was used. The PCR conditions included initial denaturation at 94℃ for 30 sec, annealing at 56℃ for 30 sec, and final extension at 72℃ for 5 min. DNA amplified with PCR was purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. PCR products were separated on a 2% agarose gel electrophoresis (SeaKem, Rockland, ME, USA), stained with ethidium bromide and visualized using a UV transilluminator (Vilber Lourmat, Marne-la-vallée Ceedex, France).
Table 1.
Sequencing of Each PCR Product, Genotyped for Single Nucleotide Polymorphisms (SNPs)
PCR, polymerase chain reaction.
All PCR products were genotyped for SNPs. Sequencing reactions were performed in sequencing kits using AmpliTaq DNA polymerase according to the manufacturer's protocol. Single-pass sequencing was performed on each template using the primer. The fluorescent-labeled fragments were purified from unincorporated terminators using an ethanol precipitation protocol. The samples were resuspended in distilled water and then subjected to electrophoresis in an ABI 3730xl sequencer (Applied Biosystems Inc., Foster City, CA, USA). The graphed genotype sequences were compared (Fig. 1) and the frequency of SNPs found in patients with GDM was compared with those in the non-diabetic controls. PPARγ2 and IGF2BP2 were interpreted in a forward manner, whereas KCNQ1 was interpreted in a reverse one.
Fig. 1.
Analysis of each gene sequence (A) PPARγ2, (B) IGF2BP2, (C) KCNQ1. PPARγ2 and IGF2BP2 were interpreted in a forward manner whereas KCNQ1 was interpreted in a reverse manner.
Statistical analyses
Statistical analysis was conducted utilizing SPSS software (SPSS, Inc., Chicago, IL, USA) for Windows (Microsoft, Redmond, WA, USA), version 19.0. All data were expressed as mean±standard deviation or as the number and proportion (%) of subjects. Continuous data such as maternal age, height, BMI, weight gain throughout pregnancy, weight at delivery and GA at delivery were analyzed using Student's t-test. A χ2 test was used to compare differences in genotype frequencies between patients with GDM (n=95) and non-diabetic controls (n=41). In addition, logistic regression analysis was used to calculate the odds ratios (ORs), 95% confidence intervals (CI), and corresponding p-values, with regard to the number of risk alleles, using an additive model. p-values <0.05 were considered statistically significant.
RESULTS
Clinical parameters
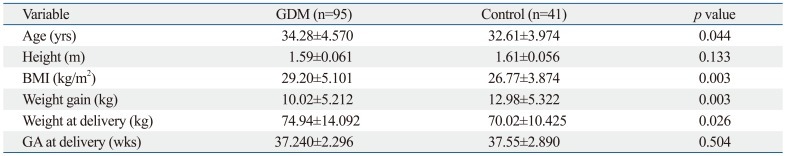
Phenotypic characteristics of the study subjects were collected and analyzed. As shown in Table 2, mean age, mean BMI, and mean weight at the time of delivery were slightly higher in patients with GDM, compared to non-diabetic controls, and found to be statistically significant [(34.28±4.57 vs. 32.61±3.97 years, p=0.044), (29.20±5.10 vs. 26.77±3.87, p=0.003), (74.94±14.09 vs. 70.02±10.42 kg, p=0.026)]. However, mean weight gain throughout pregnancy was significantly smaller in patients with GDM than in non-diabetic controls (10.02±5.21 vs. 12.98±5.32 kg, p=0.003) (Table 2).
Table 2.
Clinical Characteristics of the Study Subjects
GDM, gestational diabetes mellitus; BMI, body mass index; GA, gestational age.
Of the patients with GDM (n=95), 51.5% (49/95) controlled their plasma glucose levels only with diet and exercise, 45.3% (43/95) received an insulin injection and 1.1% (1/95) took oral anti-hyperglycemic agents. In addition, 2.1% (2/95) of the patients with GDM neglected to manage their blood glucose level.
The frequency of cesarean section was significantly higher than normal spontaneous vaginal delivery in patients with GDM than in non-diabetic controls (OR 2.401, 95% CI 1.137-5.070, p=0.022). In addition, there was also a significant difference in the frequency of cesarean section between patients with GDM and non-diabetic controls (p=0.020).
Genetic analysis
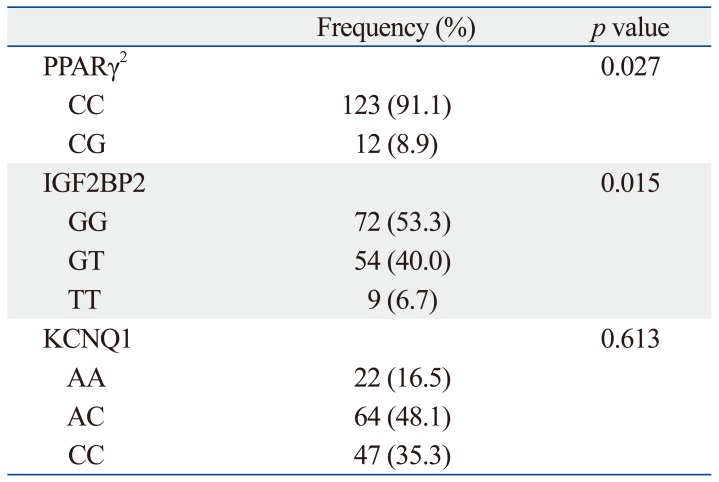
We compared genotypes between patients with GDM (n=95) with non-diabetic controls (n=41). In doing so, we found that CC in PPARγ2, GG in IGF2BP2 and AC in KCNQ1 were the most frequent genotypes in patients with GDM, similar to the control group. There were, however, significant differences in genotypes between patients with GDM and non-diabetic controls in PPARγ2 (CC, CG; p=0.027) and IGF2BP2 (GG, GT, TT; p=0.015). Despite the fact that AC was the most frequently observed genotype for KCNQ1 in both GDM and control groups, there was no significant difference in each genotype (AA, AC, CC) between patients with GDM and non-diabetic controls (p=0.613) (Table 3).
Table 3.
Frequencies of Specific Genotypes among the Study Subjects
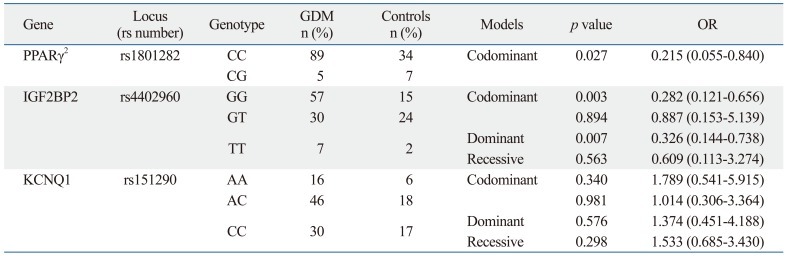
Based on these findings, we performed a logistic regression analysis for each gene to examine whether a certain genotype contributes to the manifestation of GDM and, if so, to what extent. In regards to PPARγ2, study subjects with the CC genotype exhibited a 78% higher risk of developing GDM than those with CG, a finding which was statistically significant (OR 0.215, 95% CI 0.055-0.840; p=0.027). For IGF2BP2, subjects with the GG genotype were found to involve a 72% higher risk of developing GDM than those with GT, which was statistically significant (OR 0.282, 95% CI 0.121-0.656, p=0.003). Moreover, upon further analysis of the IGF2BP2 data, as shown in a dominant model, study subjects with the GG genotype demonstrated a 67% higher risk of developing GDM than those with both GT or TT together (OR 0.326, 95% CI 0.144-0.738, p=0.007). However, there was no significant difference in the risk of developing GDM between the genotypes of KCNQ1 (Table 4).
Table 4.
Genotype and Allele Distributions and Corresponding ORs for Developing GDM
GDM, gestational diabetes mellitus; OR, odds ratio.
DISCUSSION
Although the impact of gestational diabetes on maternal and fetal health has garnered increasing recognition, there remains a lack of universal consensus on the appropriate diagnostic methods and thresholds for diagnosing gestational diabetes. We therefore set out to examine the association of variations in certain genes with a susceptibility to GDM as a diagnostic clue applicable in Korean populations. Another recently published study, the Hyperglycemia and Adverse Pregnancy Outcomes study, evaluated the treatment of maternal hyperglycemia, and confirmed the findings of smaller, nonrandomized studies, solidifying the link between maternal hyperglycemia and adverse perinatal outcomes. In response to these studies, the International Association of Diabetes and Pregnancy Study Groups formulated new guidelines for the screening and diagnosis of diabetes in pregnancy, and recommended that high-risk women be screened universally at 24-28 weeks of gestation using a 75-g OGTT, at 1-hour and 2-hours.14 Although the diagnostic threshold value of fasting plasma glucose was set at 75 mg/dL, the category of "impaired fasting glucose" was included in the diagnosis of diabetes, making the diagnostic thresholds arbitrary.
Several studies have uncovered a significant correlation between maternal hyperglycemia and perinatal morbidity. Moreover, increases in adverse maternal-fetal outcomes have been documented across the spectrum of carbohydrate intolerance, even for glucose values below the currently recommended cut-offs for the diagnosis of gestational diabetes. Previous studies have documented that an abnormal glucose screening test is solely an independent predictor of macrosomia,15 and advocate the usefulness of follow-up diagnostic testing or close surveillance for fetal overgrowth during the third trimester in women with abnormal screening tests between 24 and 28 weeks of gestation.16
Many studies have revealed that genetic variants are associated with a susceptibility to GDM and T2DM. These two disease entities share common pathophysiological background, including β-cell dysfunction and insulin resistance.17 Based on this background, PPARγ2, IGF2BP2 and KCNQ1 were selected in this study because they have been reported to exhibit SNPs in patients with T2DM. Impaired β-cell function (IGF2BP2), insulin resistance (PPARγ2) and obesity are major pathophysiological features known to be associated with loci variants and susceptibility to T2DM. PPARγ2 is a nuclear receptor and is an important regulator of adipocyte differentiation. Specific mutations in the ligand binding domain of PPARγ2 have been proposed to play an essential role in glucose, lipid and blood pressure homeostasis.10 Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) belongs to a family of IGF2 mRNA-binding proteins that play an important role in embryogenesis and pancreatic development.18 In addition, IGF2BP2 is known to regulate the transcription of IGF2, which in turn is involved in insulin development and function.11 KCNQ1 encodes the pore-forming subunit of a voltage-gated K+ channel (KvLQT1) which is essential for the repolarization phase of the action potential in cardiac muscle.19 K+ channel is also expressed in other tissues including brain, adipose and pancreas, and many variants of the KCNQ1 gene have been shown to have an association with susceptibility to T2DM.12,20,21
To date, no published methods have provided the key to diagnosing GDM. Other aspects including the human gene have therefore been considered. Previous published genotyping studies have shown that there is a strong correlation between GDM and T2DM. We therefore evaluated CC in PPARγ2 and GG in IGF2BP2, and thereby, observed higher correlations (78% and 72%, respectively) therein with a higher prevalence of GDM than other genotypes. Accordingly, it might be necessary to develop a kit for identifying polymorphisms of a certain gene in a simpler, more generalized manner.
The limitations of the current study include a relatively smaller number of enrolled subjects than previous studies. We are therefore in the process of collecting more data from a larger sample size, and hope to obtain more exact results therefrom. Another limitation is that we were only able to select a specific SNP from one gene each. This is because no previous studies have attempted to evaluate and analyze other SNPs in the same gene. Moreover, we conducted the current study with a single gene and did not consider interactions between the gene and its proteins, which would require further studies.
To summarize, our results showed that not only clinical factors, but also polymorphisms in PPARγ2 and IGF2BP2 were associated with a higher prevalence of GDM. In addition, our results also showed that genetic variants associated with T2DM, as shown in recent published studies, were also associated with GDM in a Korean population.
In conclusion, we propose that genetic polymorphisms could also be of value in predicting the occurrence and diagnosis of GDM.
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Buchanan TA, Xiang AH. Gestational diabetes mellitus. J Clin Invest. 2005;115:485–491. doi: 10.1172/JCI24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feig DS, Palda VA. Type 2 diabetes in pregnancy: a growing concern. Lancet. 2002;359:1690–1692. doi: 10.1016/S0140-6736(02)08599-9. [DOI] [PubMed] [Google Scholar]
- 3.McLellan JA, Barrow BA, Levy JC, Hammersley MS, Hattersley AT, Gillmer MD, et al. Prevalence of diabetes mellitus and impaired glucose tolerance in parents of women with gestational diabetes. Diabetologia. 1995;38:693–698. doi: 10.1007/BF00401841. [DOI] [PubMed] [Google Scholar]
- 4.Martin AO, Simpson JL, Ober C, Freinkel N. Frequency of diabetes mellitus in mothers of probands with gestational diabetes: possible maternal influence on the predisposition to gestational diabetes. Am J Obstet Gynecol. 1985;151:471–475. doi: 10.1016/0002-9378(85)90272-8. [DOI] [PubMed] [Google Scholar]
- 5.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 7.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 9.Zeggini E, Wäedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumvoll M, Häring H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes. 2002;51:2341–2347. doi: 10.2337/diabetes.51.8.2341. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 13.Cho YM, Kim TH, Lim S, Choi SH, Shin HD, Lee HK, et al. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia. 2009;52:253–261. doi: 10.1007/s00125-008-1196-4. [DOI] [PubMed] [Google Scholar]
- 14.Leary J, Pettitt DJ, Jovanovic L. Gestational diabetes guidelines in a HAPO world. Best Pract Res Clin Endocrinol Metab. 2010;24:673–685. doi: 10.1016/j.beem.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Landon MB, Mele L, Spong CY, Carpenter MW, Ramin SM, Casey B, et al. The relationship between maternal glycemia and perinatal outcome. Obstet Gynecol. 2011;117(2 Pt 1):218–224. doi: 10.1097/aog.0b013e318203ebe0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 17.Shaat N, Ekelund M, Lernmark A, Ivarsson S, Almgren P, Berntorp K, et al. Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia. 2005;48:2544–2551. doi: 10.1007/s00125-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 18.Christiansen J, Kolte AM, Hansen TO, Nielsen FC. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol. 2009;43:187–195. doi: 10.1677/JME-09-0016. [DOI] [PubMed] [Google Scholar]
- 19.Hu C, Wang C, Zhang R, Ma X, Wang J, Lu J, et al. Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia. 2009;52:1322–1325. doi: 10.1007/s00125-009-1335-6. [DOI] [PubMed] [Google Scholar]
- 20.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 21.Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, et al. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]