Abstract
In this study we provide a molecular and biochemical identification of two arylalkylamine N-acetyltransferases (aaNAT) from Aedes aegypti mosquitoes. N-acetyldopamine, the enzyme product of aaNAT, was detected in Ae. aegypti, indicating the presence of an aaNAT in this mosquito. A BLAST search of the Ae. aegypti genome, using sequence information from an activity-verified Drosophila aaNAT, identified thirteen putative aaNAT sequences sharing 13-48% sequence identity with the Drosophila enzyme. Eight of the thirteen putative aaNAT proteins were expressed using a bacterial expression system. Screening of purified recombinant proteins against 5-hydroxytryptamine, dopamine, methoxytryptamine, norepinephrine, octopamine, tryptamine, and tyramine substrates, established that two of the putative aaNATs are active to the tested arylalkylamines. We therefore named them aaNAT1 and 2, respectively. Analysis of the transcriptional profiles of the two aaNAT genes from Ae. aegypti revealed that aaNAT1 is more abundant in the whole body of larvae and pupae, and aaNAT2 is more abundant in the head of adult mosquitoes. Based on their substrate and transcriptional profiles, together with previous reports from other insects, we suggest that the two aaNATs play diverse roles in Ae. aegypti, with aaNAT1 primarily involved in sclerotization and aaNAT2 mainly in neurotransmitter inactivation. Our data provide a beginning to a more comprehensive understanding of the biochemistry and physiology of aaNATs from the Ae. aegypti and serve as a reference for studying the aaNAT family of proteins from other insect species.
Keywords: N-acetyltransferase, arylalkylamine, Dopamine, serotonin, tryptamine, Aedes, aaNAT
Introduction
Proteins that catalyze the transacetylation of acetyl coenzyme A (Acetyl-CoA) to arylamines and arylalkylamines are commonly referred to as arylamine N-acetyltransferases and arylalkylamine N-acetyltransferases (aaNAT), respectively (Evans, 1989). In mammals, aaNAT is primarily involved in the synthesis of N-acetylserotonin. The N-acetylation of serotonin (5-hydroxytryptamine) is a rate-limiting step for the synthesis of melatonin in the pineal gland. Additionally, in the pineal gland aaNAT is transcriptionally regulated in a manner that reflects daily physiological requirements for the release of melatonin (Klein, 2006, 2007). Because the concentrations of melatonin coincide with internal biological clocks, mammalian aaNAT ultimately regulates circadian rhythm and is considered one of the circadian proteins (Klein, 2006, 2007).
It has been proposed that invertebrate aaNATs play a role in the inactivation of arylalkylamines (Amherd et al., 2000; Brodbeck et al., 1998). Arylalkylamines, such as octopamine, dopamine, and serotonin function as key neurotransmitters, and the levels of these compounds need to be regulated in order to prevent neurotoxicity and prolonged signaling. In mammals, excessive dopamine or other aromatic amines are inactivated by monoamine oxidases (MAO) (Bortolato et al., 2008). This may also be true in other vertebrate species. However, a database search of currently available invertebrate genome sequences indicates that MAO homologs are absent from invertebrates (Tsugehara et al., 2007). This provides a basis for the argument that invertebrate aaNATs may play a role in preventing the over-accumulation of certain neurotransmitters.
In insects, aaNAT enzymes are considered to be involved in cuticle sclerotization, aromatic neurotransmitter inactivation, and melatonin synthesis (Smith, 1990). Based on the physiological requirements of insects, all of these proposed physiological functions regarding insect aaNATs are reasonable predictions. aaNAT proteins have been studied in Drosophila melanogaster (Amherd et al., 2000; Hintermann et al., 1996) and Periplaneta americana (Ichihara et al., 1997; Ichihara et al., 2001). And, recent studies dealing with an aaNAT from Bombyx mori clearly established its function in adult cuticle sclerotization through the production of N-acetyldopamine, a key cuticle protein crosslinking precursor (Dai et al., 2010; Zhan et al., 2010).
The available genome sequences for a number of insect species make it possible to predict aaNAT sequences via a bioinformatic approach. A BLAST search using the activity verified D. melanogaster aaNAT sequence against three currently available mosquito species (Aedes aegypti, Anopheles gambiae and Culex quinquefasciatus) identified eight sequences from An. gambiae, thirteen from Ae. aegypti and fifteen from C. quinquefasciatus showing recognizable similarity (Fig. S1 and S2). This suggests the presence of multiple aaNATs in mosquitoes and the importance of arylalkylamine acetylation in these species. However, no single aaNAT sequence from mosquitoes has ever been identified at biochemical level. The ambiguity in substrate specificity and overall biochemical properties of these putative aaNAT enzymes has become a major barrier to understanding the physiology and biochemistry of these proteins. The potential biological significance of these mosquito aaNATs warrants an extensive effort to unambiguously establish their biochemical identity.
In this study, we used Ae. aegypti to begin a biochemical characterization of aaNAT enzymes from mosquitoes. We successfully expressed eight recombinant proteins out of the thirteen putative aaNATs in a bacterial protein expression system and screened the substrate specificity for each of the expressed recombinant proteins. From these studies we identified two aaNATs. Transcriptional studies also showed the two aaNATs have different stage and tissue patterns in transcription, suggesting the diverse roles they might play. These data provide a basis to suggest the possible functions for aaNAT1 and aaNAT2 proteins.
2. Materials and Methods
2.1. Rearing of mosquitoes
Ae. aegypti mosquitoes were reared at 27 °C with 75% relative humidity and maintained in the insectary of the Department of Biochemistry at Virginia Tech. Generation time was about 10 days under these conditions.
2.2. Identification of N-acetyldopamine in the brain and body extracts using positive-ion electrospray tandem mass spectrometry
Adult, female mosquitoes were fed on dopamine at 5 mM in the sucrose solution and the female heads were separated from the bodies and each was placed in 0.4 M formic acid and 0.05 M Na2-EDTA. Tissues were sonicated and centrifuged at 14,000 g for 20 min at 4 °C, and then the supernatant was collected and stored at -20 °C prior to analysis. Agilent LC pump with a reverse phase column (C18, 2.1x50 mm) was used to separate the N-acetyldopamine and dopamine. LC separation was done using organic solvent comprised of 0.1% formic acid, 60% methanol, and 40% water as a mobile phase. The system was equilibrated for 2 min. 100 μL of prepared samples were injected for LC-MS analysis. N-acetyldopamine and dopamine were introduced into the 3200 Q-TRAP (Applied Biosystems/MDS SCIEX). Tandem mass spectrometry was used in the positive-ion mode interfaced with a TurboIonSpray™. An enhanced product ion (EPI) scan was used to trace N-acetyldopamine. The collision gas was nitrogen with a cell pressure of 1.1 Pa. The mass transition-dependent collision energy was 20 V for transitions m/z 154-137, 196-137 for the semiquantitative analysis of N-acetylated compounds in the head and bodies of Ae. aegypti. Data were processed using Analyst 1.4.2.
2.3. Identification of putative aaNAT sequences in Ae. aegypti genome
A protein-specific BLAST search combined with a Position-Specific Iterated BLAST search (Altschul et al., 1997) using a Drosophila aaNAT (GeneBank accession no, Y07964) revealed thirteen sequences from Ae. aegypti with reasonable similarities (Table 1). For convenience, they were sequentially referred to as putative aaNAT1 to aaNAT12 based on their level of sequence identity with the D. melanogaster aaNAT. Table 1 lists essential information for the these putative aaNATs (PaaNAT), including GenBank accession numbers, chromosomal location, start and end in the genome, presence of exons, amino acid length, and protein sequence identity with the Drosophila aaNAT.
Table 1.
Putative aaNAT proteins found in the Ae. aegypti genome as a result of a BLAST search using the D. melanogaster aaNAT1 (GeneBank accession no, Y07964)
| Protein named in this report | Gene ID (GeneBank accession no) | Location | No of Exons | Length (number of aa) | Identity with Dm-aaNAT (%) | ||
|---|---|---|---|---|---|---|---|
| Supercontig location | Start | End | |||||
| PaaNAT1 | AaeL_AAEL011088/(XP_001661400) | Supercontl.540 Contig_20021-3 | 459,031 | 509,488 | 3 | 288 | 48 |
| PaaNAT2 | AaeL_AAEL012952/(XP_001663122) | Supercontl.766 Contig_24253 | 154,320 | 167,075 | 4 | 222 | 30 |
| PaaNAT3 | AaeL_AAEL004847/(XP_001649915) | Supercontl.132 Contig_7561 | 1,935,510 | 1,936,184 | 1 | 224 | 26 |
| PaaNAT4 | AaeL_AAEL002255/(XP_001661173) | Supercontl.52 Contig_3460 | 1,754,718 | 1,755,380 | 1 | 220 | 24 |
| PaaNAT5a | AaeL_AAEL014713 (XP_001649422) | Supercontl.1233 Contig_30084 | 148,688 | 149,574 | 1 | 217 | 23 |
| PaaNAT5b | AaeL_AAEL004827/(XP_001649916) | Supercontl.132 Contig_7562 | 1,997,239 | 1,998,124 | 1 | 217 | 22 |
| PaaNAT6 | AaeL_AAEL012866/(XP_001663012) | Supercontl.752 Contig_24066 | 209,570 | 210,139 | 2 | 168 | 20 |
| PaaNAT7 | AaeL_AAEL012870/(XP_001663019) | Supercontl.752 Contig_24070 | 362,533 | 363,312 | 2 | 238 | 17 |
| PaaNAT8 | AaeL_AAEL004659 (XP_001649572.1) | Supercontl. 126 Contig_7273-4 | 1,515,283 | 1,516,986 | 5 | 270 | 17 |
| PaaNAT9 | AaeL_AAEL012860/(XP_001663014) | Supercontl.752 Contig_24066 | 227,851 | 247,030 | 3 | 238 | 16 |
| PaaNAT10 | AaeL_AAEL012859 (XP_001663018) | Supercontl.752 Contig_24070 | 361,424 | 362,375 | 3 | 240 | 16 |
| PaaNAT11 | AaeL_AAEL012864 (XP_001663020) | Supercontl.752 Contig_24072 | 384,934 | 386,013 | 3 | 240 | 13 |
| PaaNAT12 | AaeL_AAEL012863 (XP_001663015) | Supercontl.752 Contig_24066 | 273,694 | 274,762 | 2 | 237 | 13 |
aa: amino acid; PaaNAT: putative aaNAT from Ae. aegypti; Dm-aaNAT: D. melanogaster aaNAT; PaaNATs whose recombinant proteins were expressed in this study ware written in bold font.
2.4. Sequence comparison of putative aaNATs from Ae. aegypti
A multiple sequence alignment of the thirteen protein sequences was constructed using the MEGA4-ClustalW alignment program. A maximum likelihood phylogenetic tree was constructed using the neighbor joining method and bootstrap analysis (Tamura et al., 2008).
2.5. Expression and purification of eight putative aaNATs from Ae. aegypti
Amplification of cDNA for PaaNATs was achieved using the forward and reverse primers corresponding to 5’- and 3’- regions of the coding sequences of PaaNATs (Table 2). Their amplified cDNA sequences were cloned into an Impact™-CN plasmid (New England Biolabs) for expression of their recombinant proteins. Escherichia coli cells transformed with individual PaaNAT recombinant plasmids were cultured at 37 °C. After induction with 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG), the cells were cultured at 15 °C for 24 hours. Four to 20 liters (depending upon the level of the expressed recombinant protein) of cell cultures were prepared and the harvested cells were lysed by sonication on ice in a lysis buffer containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 1 mM EDTA. Supernatants were obtained by centrifugation at 37,500 × g (4 °C for 30 minutes) and applied to a chitin bead column that had been equilibrated using the lysis buffer. The column was washed extensively with the lysis buffer. The column with the associated recombinant protein was equilibrated with the hydrolysis buffer containing 50 mM Tris (pH 8.0) and 50 mM β-mercaptoethanol and incubated for 24 hours at 4 °C. The protein was eluted using 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonate (HEPES) buffer (pH 7.5) and concentrated using a membrane concentrator with a molecular weight cutoff at 10,000 kDa (Millipore Corp). The concentrated protein samples were further purified by ion exchange (Mono-Q column, GE Health) and gel-filtration (Superose 12 column, GE Health) chromatographies. The purity of the PaaNATs was assessed by the presence of a single band between 25 kDa and 30 kDa on an SDS-PAGE gel. Protein concentrations were determined by a Bio-Rad protein assay kit using bovine serum albumin as a standard.
Table 2.
Primers used for recombinant protein expression. Restriction enzyme sites used for the cloning are underlined. PaaNAT: putative aaNAT; F: forward; R: reverse.
| Genes | Cloning primers |
|---|---|
| Paanat1 | F-5’-CATATGGCTTCGAAGATGTCGACCGTT -3’ R-5’-GAATTCAGGCCAGTCTTTTTGTTAGAATAC 3’ |
| Paanat2 | F-5’ CATATGTTGGACAGCAAGCTCAACAAC 3’ R-5’ GAATTCAGTTGATCACTTTGCACATAATTTT-3’ |
| Paanat3 | F-5’-CATATGGAAGAGTCAACCTCTCGTAA-3’ R-5’-GAATTCATACAATCAATAGTTTAACAA-3’ |
| Paanat4 | F-5’-CATATGGGAGCCACGCCGTTGA-3’ R-5’-GAATTCATTGCAATATCTTAACCATCC-3’ |
| Paanat5b | F-5’-GAATTCTCAGCAGTTTAACACATGT-3’ R-5’-CATATGGTCGCCCCCGAAAGCAT-3’ |
| Paanat7 | F-5’-CATATGAAATGGACAAGGTCGGTG-3’ R-5’-GAATTCAATCAACTCTCAAGCTCATA-3’ |
| Paanat9 | F-5’-CATATGGTTTGGACGCGACC-3’ R-5’-CTCGAGTTATTCGACTTTCAAGCTCAT-3’ |
| Paanat11 | F-5’-CATATGGGTTGGCAAAGACTTTC-3’ R-5’-CATATGGGTTGGCAAAGACCTTC-3’ |
2.6. aaNAT activity assays
Two different methods were used to assay the activity of the eight putative aaNATs. Method 1 was based on the hydrolysis of Acetyl-CoA by aaNATs. 5′-dithio-bis (2-nitrobenzoic acid) (DTNB) was used as a colorimetric developing agent for detection of CoA (Kawamura et al., 2005). The chemicals tested as a potential substrate in the assay include dopamine, octopamine, tyramine, tryptamine, nor-epinephrine, methoxytryptamine, and 5-hydroxytryptamine. The substrate (2 mM) and 0.4 μg purified recombinant PaaNAT were mixed and pre-incubated at room temperature for 5 min in a 250 μL 96-well plate. Acetyl-CoA (0.4 mM) was added to start the reaction in a final volume of 100 μL. The final mixture contained 50 mM Tris-HCl, and 1.0 mg mL-1 bovine serum albumin (for stabilizing the enzymes). The mixture was continually incubated at room temperature for 5 min. The reaction was stopped with 25 μL guanidine hydrochloride solution (6.4 M guanidine-HCl, 0.1 M Tris-HCl, pH 7.3) containing 5 mM DTNB and the absorbance at 405 nm was measured on a plate-reader (SpectraMax M5e Multi-Mode Microplate Reader) within 5 min. Reaction mixtures wherein substrate, Acetyl-CoA or PaaNAT were omitted respectively were used as controls. The amount of CoA produced was determined from a standard curve generated using CoA standards. Method 2 was used to detect the acetylated arylalkylamines to confirm the acetyl product formation. Each of the purified recombinant proteins (1 μg) was mixed with Acetyl-CoA at a final concentration of 0.4 mM and dopamine or tryptamine at a final concentration of 5 mM in a total volume of 100 μL prepared in 50 mM Tris-HCl buffer (pH 7.3). All substrates were freshly prepared and used immediately (to avoid oxidation of the monoamines). The reaction mixtures were incubated for 10 minutes at 37°C and the reaction was stopped by addition of an equal volume of 800 mM formic acid. The acidified reaction mixtures were centrifuged for 10 minutes at 15,000g at 4 °C. Supernatants were chromatographed by HPLC with a reverse-phase column (C18 of 5 μm particles, 4.6 × 100 mm) and resolved substrate and product by reverse-phase HPLC were detected by electrochemical detection. Because some of the acetylation products of arylalkylamines are not available as standards for use in quantifying enzyme activity, we only used method 1 to quantify the enzyme activity and to conduct kinetic studies.
2.7. Kinetic analysis
PaaNAT1 and PaaNAT2 were active to the screened arylalkylamines and their kinetic properties to the eight substrates listed above were further assessed by measuring their specific activity in the presence of varying concentrations (0.016 to 2 mM) of each arylalkylamine and a fixed concentration of acetyl-CoA (0.4 mM). The data were fitted to the Michaelis–Menten equation by the non-linear regression method. Estimation of apparent Km values was obtained by SigmaPlot Enzyme Kinetics Module (SPSS, San Jose, CA). Data are the means ± SEM of three experiments.
2.8. Expression profiling of aaNAT1 & 2 based on real time quantitative reverse transcriptase PCR (qRT-PCR)
2.8.1. RNA extraction and cDNA synthesis
The third day larvae and pupae were collected for qRT-PCR analysis. Adult females used were 5-6 days old. The heads were dissected on ice from live adult mosquitoes after being kept at 4 °C for 10 min. The head-less mosquitoes were collectively termed body. Both head and body of adults were collected for analysis. Total RNA extraction was carried out following the manufacturer's protocol using the mirVana kit (Ambion). Tissue samples in microcentrifuge tubes were disrupted in a lysis/binding buffer at 10 to 1 ratio between lysis buffer and tissue mass and were placed into a homogenization vessel on ice. Homogenization of tissues was conducted using digital benchtop homogenizers (PRO Scientific Inc). One-tenth volume of miRNA homogenate additive (Ambion. Inc) was added to the lysate, and the mixture was incubated on ice for 10-30 min. The samples were extracted with acidified-phenol:chloroform equal to the initial lysate volume. Samples were then centrifuged for 10 minutes at 14,000 g at room temperature to separate the aqueous and organic phase. After centrifugation, the aqueous phase was removed without disturbing the lower organic phase and applied to normal phase columns (Ambion. Inc). The columns were washed twice with approximately 70% ethanol and 30% gaunidinium thiocyanate. Further, RNA was eluted with nuclease free water. The concentration and purity of the RNA samples in water was evaluated using a NanoDrop Spectrophotometer (Nanodrop Technology).
The cDNA synthesis was done following the manufacturer's protocol (SuperScript® III First-Strand Synthesis System, Invitrogen) for all the samples. The synthesized cDNA was diluted (1: 5) before the qRT-PCR test and stored at -20 °C until used.
2.8.2. Transcription profiles and temporal expression analysis of aaNAT1 and aaNAT2 using qRT-PCR
qRT-PCR was performed in a qRT-PCR detection system (Applied biosystem 7300, Foster city, CA, USA) using SYBR Green, qRT-PCR master mix (Platinum SYBR Green qRT-PCR master mix, Invitrogen) in a 96 well format. Each pair of PCR primers for aaNAT1 and aaNAT2 (Table 3) was designed to span a cDNA exon-exon or intron-exon gene wherever possible and amplify 202 bp individual aaNAT fragments from cDNA. A primer pair that amplifies a 202 bp fragment of Ae. aegypti ribosomal binding protein (Ae-rpS7) gene, a relatively abundant and constitutively expressed gene, was used to normalize the results of variable target genes and to correct for sample-to-sample variations. SYBR green assays were carried out in parallel for the control Ae-rpS7 gene. All test samples and the controls were performed in triplicate. For every sample, an amplification plot was generated showing the reporter dye fluorescence (ΔRn) at each PCR cycle. A threshold cycle (CT) was determined for each amplification cycle, representing the cycle number at which the fluorescence passes the threshold. This was the method by which the CT values of aaNAT1 and 2, as well as for the control gene, Ae-rpS7, were determined. The CT value of the control gene was then subtracted from the CT value of the aaNAT genes. Finally, 2-ΔΔCT values were calculated to estimate the fold changes in the mRNA levels of aaNATs in different samples. The controls, without reverse transcriptase, showed CT values of 40, indicating no amplification of product. The interpretation of the results for transcription patterns is based on two trials.
Table 3.
Primers for the gene expression analysis.
| Genes | Forward Primer | Reverse Primer | Product size of cDNA (bp) |
|---|---|---|---|
| aaNAT1 | 5' AGGATGTCCTGAAATTGCTGA 3' | 5' TCAGTCTCTTTCGGCCTGTT 3' | 202 |
| aaNAT2 | 5’ CAACATCCGTTTCGAGACAA 3’ | 5’ GTCGCCATCGTTGGAAATAG 3’ | 202 |
| Ae-rpS7 | 5' ATCTGTACATCACCCGCGCT 3' | 5' GATCGTGGACGCTTCTGCTT 3' | 202 |
qRT-PCR was performed to compare the transcriptional levels of aaNAT1 and aaNAT2 from adult female heads and body, and from whole larva and pupa. The cDNA for all samples were synthesized from 5 μg of mRNA. The comparative 2-ΔΔct method was used to quantify aaNAT1 and 2 transcripts. All the samples were normalized with respect to Ae-rpS7.
3. Results
3.1. Identifcation of N-acetyldopamine in the brain and body extracts
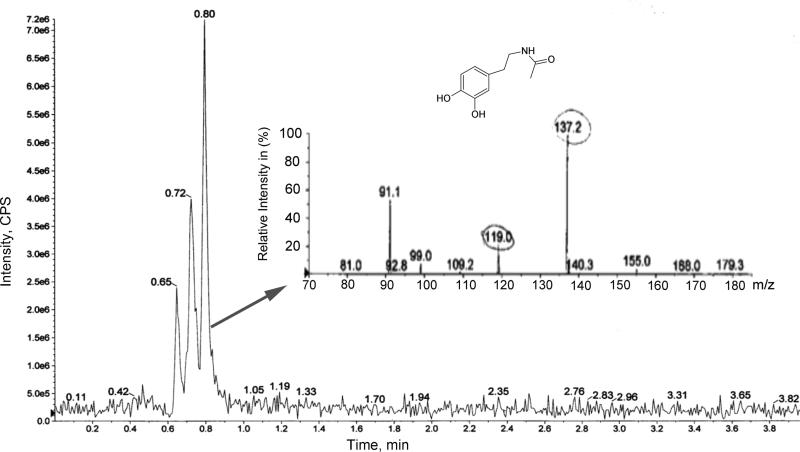
N-acetyldopamine is an aaNAT-catalyzed enzymatic product from dopamine. Detection of this compound in the mosquito provides evidence of the presence of aaNAT. N-acetyldopamine was detected in the brain and body extracts of mosquitoes fed with dopamine using LC-MS/MS in positive mode (Fig. 1), indicating the operation of the N-acetylation pathway of dopamine in Ae. aegypti mosquitoes.
Figure 1.
Analysis of N-acetyldopamine from Aedes aegypti by LC/MS. LC-MS-TIC chromatogram of N-acetyldopamine and LC/MS ESI mass spectrum of N-acetyldopamine fragments from head samples of mosquitoes fed with dopamine. CPS: Counts per seconds, ESI, electrospray ionization, TIC, total ion current.
3.2. Putative aaNAT proteins from Ae. aegypti
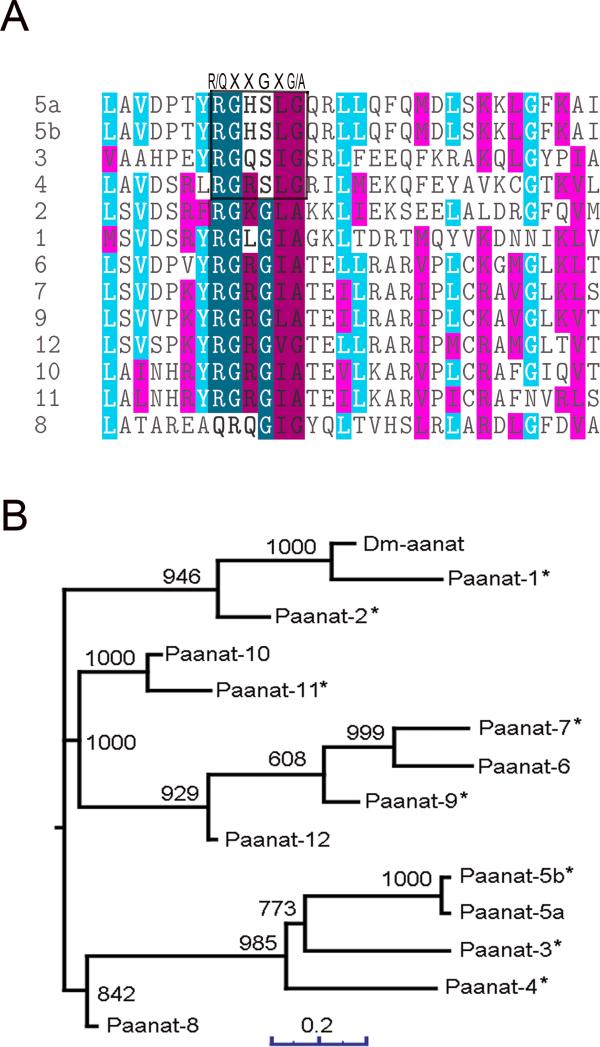
D. melanogaster aaNAT (gene name: Dat), which is capable of catalyzing the acetylation of tyramine, octopamine, dopamine, and serotonin (Hintermann et al., 1996; Hintermann et al., 1995), was used as a query to search the Ae. aegypti proteins in the National Center of Biotechnology and Information (NCBI) database using the BLAST and PSI BLAST search programs (Altschul et al., 1997). As described in the Method section, thirteen individual sequences were obtained (Table 1). Analysis of these selected Ae. aegypti proteins determined that they contain two motifs (commonly named Motif A and Motif B), which are characteristic of the N-acyltransferase superfamily (Bembenek et al., 2005). All the putative aaNATs except 3, 4, 5a and 5b have the R/QXXGXG/A fragment, where X is any amino acid residue. This fragment is considered to be an Acetyl-CoA binding site (Cort, 2008 #11854} (Fig. 2A). To understand the phylogenetic relationship among the 13 putative aaNATs, a phylogenetic tree was constructed (Fig. 2B). Phyletic distribution analysis confirmed three major clusters.
Figure 2.
Multiple sequence alignment and phylogenetic analyses of putative aaNATs. Sequences were aligned using ClustalW and the dendrogram was generated using the neighbor joining method. A) Part of the alignment is shown. The Acetyl-CoA binding site, R/QXXGXG/A is highlighted. B) The phylogenetic tree of 13 putative aaNATs. The number on the internal branches shows the bootstrap values based on 1000 replicates. The aaNATs, of which the recombinant proteins were expressed in later studies, are labeled with a star.
3.3. Protein expression and purification
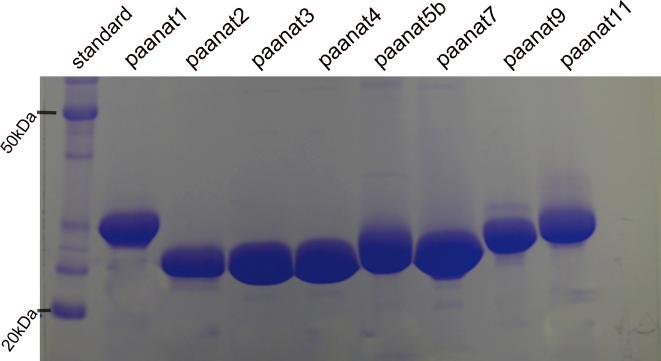
The putative aaNATs were selected for recombinant protein expression based on the level of identity with the Drosophila aaNAT and the position in the phylogenetic tree. Eight selected putative aaNATs are shown in table 1 and Fig. 2B. To determine whether the selected Ae. aegypti sequences are true aaNATs, their coding sequences were amplified from Ae. aegypti cDNA preparations and recombinant proteins were expressed using the bacterial protein expression system. Soluble recombinant proteins for eight individual putative aaNATs were successfully expressed and purified sequentially by chitin affinity, ion exchange, and gel filtration chromatography. The final preparation contained the major single band for all the putative aaNATs between 20 - 35 kDa (Fig. 3).
Figure 3.
Purified recombinant proteins on SDS-polyacrylamide gel. The purified recombinant putative aaNATs and protein standards were run in 10% SDS-polyacrylamide gel. After electrophoresis, the gel was stained with Coomassie blue. kDa: kilodalton, standard, protein standard, paanat: putative aaNAT.
3.4 Substrate specificity and kinetic analysis of aaNAT1 and aaNAT2
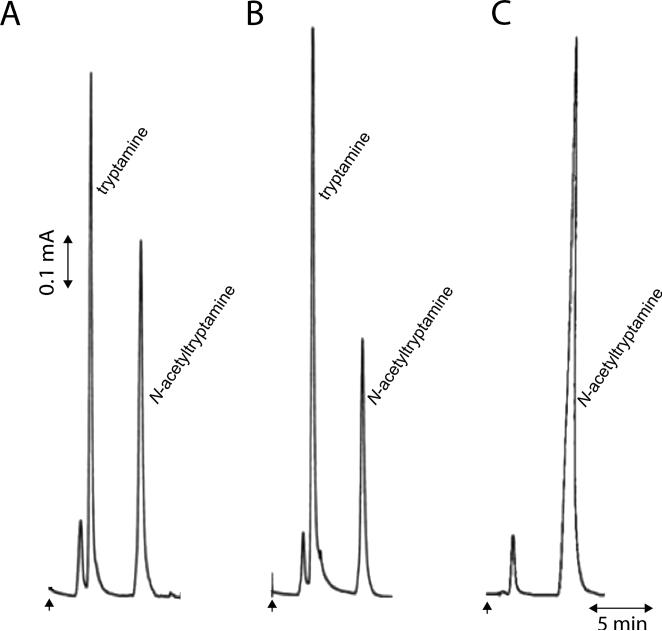
Each recombinant protein was screened for aaNAT activity using 5-hydroxytryptamine, dopamine, methoxytryptamine, norepinephrine, octopamine, tryptamine, and tyramine as the acetyl group acceptor and Acetyl-CoA as the acetyl group donor using method 1 described in the Method section. Among them, aaNAT1 and aaNAT2 recombinant proteins displayed aaNAT activity to all tested substrates. Others did not show detectable aaNAT activity to the tested arylalkylamines. The colorimetric assay detects the amount of CoA released into the reaction mixture. To further confirm the proportionality of CoA release to the formation of the acetylated arylalkylamine product, aaNAT1 and aaNAT2 were further tested using method 2 described in the Method section. Figure 4 illustrates the production of acetyltryptamine by aaNAT1 and aaNAT2 in their corresponding reaction mixtures by HPLC-ED analysis. The acetyl product was eluted after arylalkylamine (its precursor) during HPLC separation because the acetylated arylalkylamine is more hydrophobic.
Figure 4.
Activity assay using tryptamine as a substrate. aaNAT1 and aaNAT2 were tested using tryptamine as the acetyl group acceptor and Acetyl-CoA as acetyl group donor using method 2 described in the Method section. N-acetyltryptamine was detected in the reaction mixture. X-axis represents elution time (min) and Y-axis indicates electric current generated during oxidation of tryptamine and N-acetyltryptamine at the working electrode. In chromatograms, arrow indicates time of sample application. Chromatograms illustrate the production of N-acetyltryptamine in a tryptamine and Acetyl-CoA mixture in the presence of aaNAT1 (A), the production of N-acetyltryptamine in a tryptamine and Acetyl-CoA mixture in the presence of aaNAT2 (B), and the retention time of tryptophan and N-acetyltryptamine standards in the absence of aaNAT1 or aaNAT2 under identical analysis conditions by HPLC with electrochemical detection (C).
Ae. aegypti aaNAT1 and aaNAT2 showed a broad substrate specificity and their affinity and catalytic efficiency to each of the seven arylalkylamines were tested. The kinetic parameters are given in table 4. The affinity between aaNAT1 and aaNAT2 to most of the arylalkylamines is similar except that aaNAT2 has less affinity than aaNAT1 to norepinephrine. Also aaNAT1 is more efficient in catalyzing all the tested substrates than that of aaNAT2.
Table 4.
Kinetic analysis of aaNAT1 and aaNAT2. The activities were measured as described in the Materials and Methods section. The parameters were calculated by fitting the Michaelis–Menten equation to the experimental data using the enzyme kinetics module. Results are means ± SE.
| Km mM | kcat min-1 | kcat/Km min-1mM- | |
|---|---|---|---|
|
aaNAT1
|
|||
| octopamine | 0.14±0.08 | 435.5±67.0 | 3111 |
| norepinephrine | 0.16±0.04 | 465.5±34.6 | 2909 |
| tryptamine | 0.16±0.04 | 440.7±31.7 | 2754 |
| methoxy-tryptamine | 0.18±0.06 | 461.6±44.8 | 2564 |
| tyramine | 0.17±0.04 | 406.5±24.8 | 2391 |
| dopamine | 0.19±0.07 | 438.0±44.5 | 2305 |
| serotonin | 0.23±0.06 | 452.7±35.7 | 1968 |
|
aaNAT2
|
|||
| dopamine | 0.13±0.05 | 153.5±15 | 1181 |
| tyramine | 0.17±0.03 | 190.1±11 | 1118 |
| tryptamine | 0.17±0.04 | 183±11.8 | 1076 |
| methoxy-tryptamine | 0.23±0.04 | 224.4±11 | 976 |
| serotonin | 0.23±0.05 | 189.7±11.9 | 825 |
| octopamine | 0.27±0.04 | 218.8±10.5 | 810 |
| norepinephrine | 0.42±0.06 | 208.2±11.5 | 496 |
3.5. Gene expression analysis of aaNAT1 and 2
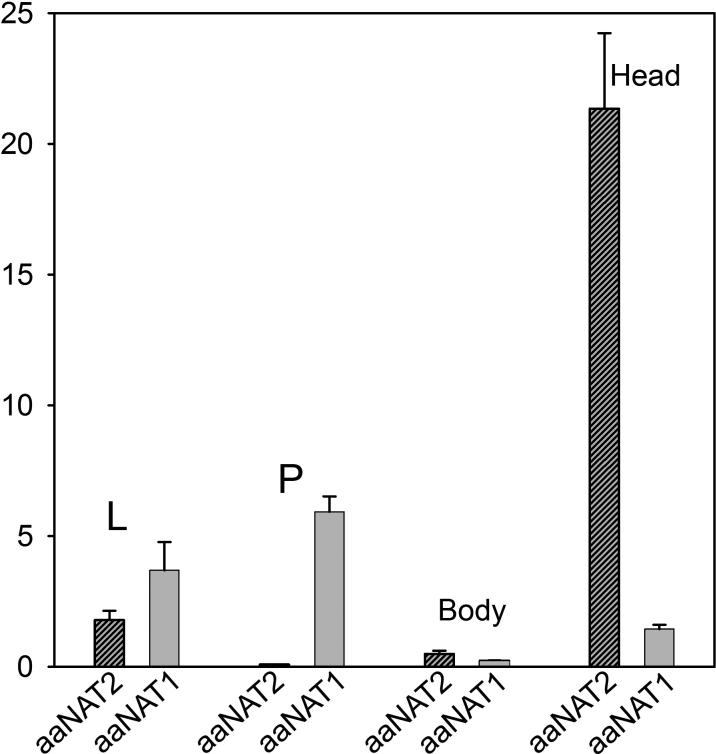
Both aaNAT1 and aaNAT2 transcripts were detected throughout the lifecycle based on the qRT-PCR results in larvae, pupae, and female adults. However, aaNAT1 transcripts were more abundant in larvae and pupae and aaNAT2 transcripts were more abundant in the adult female head (Fig. 5).
Figure 5.
Relative quantification of transcription of aaNAT1 and 2 by qRT-PCR. X-axis shows developmental stages and tissues examined, while Y-axis shows the relative transcript quantity compared with the Ae-rpS7 gene. Panels A and B represent transcription of aaNAT1 and 2, respectively. L: Three day old larvae, P: Pupae, Body: Female adult body, Head: Female adult head.
4. Discussion
The available genome sequence for Ae. aegypti mosquito (Nene et al., 2007) allowed us to select thirteen putative aaNATs for further bioinformatic and biochemical analysis. Although all putative aaNAT sequences have motifs A and B, which are characteristics of N-acyltransferase superfamily proteins (Bembenek et al., 2005) and most of them have an Acetyl-CoA binding sequence, only two of them are true aaNATs. We did not detect aaNAT activity for the other six recombinant putative aaNATs. Based on our biochemical analysis and sequence comparison, we predict that putative aaNATs, 5a, 6, 8, 10 and 12 likely are not aaNAT and no effort was made to produce their recombinant proteins.
Similar to Ae. aegypti, analysis of other insects, such as Cx. quinquefasciatus, An. gambiae, D. malenogaster, P. americana and B. mori indicates they also have multiple putative aaNAT sequences. Ae. aegypti aaNAT1 shares the highest sequence identity (48%) with the D. melanogaster aaNAT1 (Hintermann et al., 1996), and Ae. aegypti aaNAT2 shares the highest sequence identity with an un-identified D. melanogaster putative aaNAT (CG13759) (43%), but only shares 23% sequence identity with D. melanogaster aaNAT2 (CG9486) (Amherd et al., 2000). The information here together with the information from other studies of insect aaNATs (Hintermann et al., 1996; Tsugehara et al., 2007) could be used to predict true aaNATs in other insect species.
Broad substrate specificity of aaNAT1 and aaNAT2 to different arylalkylamines suggests that they have multiple functions depending on the availability of the substrate and on their localization. Other insect aaNATs, such as D. melanogaster aaNAT1, P. americana aaNAT, and B.mori aaNAT have shown broad substrate specificity similar to Ae. aegypti aaNAT1 and aaNAT2 (Brodbeck et al., 1998; Hintermann et al., 1996; Ichihara et al., 2001; Tsugehara et al., 2007). However, Ae. aegypti aaNAT1 and aaNAT2 are different in their turnover numbers and catalytic efficiencies for all substrates (aaNAT1 showed much higher turn-over numbers than aaNAT2). The biological significance and mechanism of this difference needs to be further investigated.
Km values of Ae. aegypti aaNAT1 and aaNAT2 for most tested arylalkylamines were similars. However, the kinetic parameters are quite variable in earlier studies of aaNATs from other insects. For example, D. melanogaster aaNAT1 has Km values of 1.15 mM for dopamine, 1.62 mM for serotonin, and 0.19 for tryptamine (Hintermann et al., 1996) and 0.89 to 0.97 mM for tryptamine, dopamine and serotonin (Hintermann et al., 1995); D. melanogaster aaNAT2 has two Km values of 0.0072 and 0.6 mM for tryptamine (Amherd et al., 2000); P. americana aaNAT has Km values of 0.02 - 0.05 mM for tryptamine, serotonin, dopamine, octopamine, norepinephrine, tyramine and methoxytryptamine (Ichihara et al., 1997); B. mori aaNAT has a Km value of 0.0017 mM for tryptamine (Tsugehara et al., 2007). The very low Km values might be caused by the low Acetyl-CoA concentrations used in the assays in some of these studies.
Our data suggest there are several possible functions of aaNAT1 and aaNAT2 in Ae. aegypti. In terms of their biochemical activity, one possible role for these two enzymes may be connected to neurotransmitter inactivation. This possibility is supported by the observations that (1) they are able to acetylate most tested arylalkylamines/neurotransmitters, (2) N-acetyldopamine was detected in the head of mosquitoes fed with dopamine, and (3) these genes are expressed in the head. Neurotransmitters, such as octopamine, dopamine, and serotonin are thought to play a role in neurotransmission in mosquitoes (Lee and Pietrantonio, 2003; Pratt and Pryor, 1986). An accumulation of neurotransmitters such as dopamine can cause severe damage to an insect because MAOA and MAOB (major neurotransmitter inactivators) are absent in insects, including Ae. aegypti (Meyer et al., 2006), and constant firing of the neurotransmitters may be fatal. From our transcriptional study, aaNAT2 has high mRNA levels in the head, indicating it may play a major role in the deactivation of the neurotransmitters in the mosquito brain.
Another possible function of aaNAT1 and aaNAT2 may be associated with the process of sclerotization. Sclerotization is an important event that occurs when new cuticle forms in connection with each molt, and N-acetyldopamine is an important component in the sclerotization process (Andersen, 2008; Brodbeck et al., 1998). Drosophila aaNAT was considered to have a major role in sclerotization (Wittkopp et al., 2003), and the enzyme product, N-acetyldopamine is involved in forming a clear sclerotin (True et al., 2005). N-acetyldopamine metabolism is originated from tyrosine. First, tyrosine is hydroxylated to DOPA by tyrosine hydroxylase (Neckameyer et al., 2005), DOPA can then be transformed into dopamine by the action of DOPA decarboxylase (Han et al., 2010; Hirsh and Davidson, 1981) or alternatively to 3,4-dihydroxylphenylacetaldehyde by 3,4-dihydroxylphenylacetaldehyde synthase (Vavricka et al., 2011), and finally dopamine is converted to N-acetyldopamine by aaNAT (Hintermann et al., 1996). Two recent studies demonstrated that a loss of function for an aaNAT resulted in the formation of cuticle with strong black coloration in B. mori (Dai et al., 2010; Zhan et al., 2010), indicating its involvement in the formation of clear sclerotin. We predict that both aaNAT1 and aaNAT2 play a role in sclerotization, but based on the high catalytic efficiency of Ae. aegypti aaNAT1 to dopamine and its high expression level in larval and pupal stages, aaNAT1 may play a major role in sclerotization than that of aaNAT2 in Ae. aegypti.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant AI 19769. The authors are grateful to Haizhen Ding for helping with protein expression, and to Kim Harich for helping with LC-MS/MS analysis of the aaNAT product.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amherd R, Hintermann E, Walz D, Affolter M, Meyer UA. Purification, cloning, and characterization of a second arylalkylamine N-acetyltransferase from Drosophila melanogaster. DNA Cell Biol. 2000;19:697–705. doi: 10.1089/10445490050199081. [DOI] [PubMed] [Google Scholar]
- Andersen SO. Quantitative determination of catecholic degradation products from insect sclerotized cuticles. Insect Biochem Mol Biol. 2008;38:877–882. doi: 10.1016/j.ibmb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Bembenek J, Sakamoto K, Takeda M. Molecular cloning of a cDNA encoding arylalkylamine N-acetyltransferase from the testicular system of Periplaneta americana: primary protein structure and expression analysis. Arch Insect Biochem Physiol. 2005;59:219–229. doi: 10.1002/arch.20070. [DOI] [PubMed] [Google Scholar]
- Bortolato M, Chen K, Shih JC. Monoamine oxidase inactivation: from pathophysiology to therapeutics. Adv Drug Deliv Rev. 2008;60:1527–1533. doi: 10.1016/j.addr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck D, Amherd R, Callaerts P, Hintermann E, Meyer UA, Affolter M. Molecular and biochemical characterization of the aaNAT1 (Dat) locus in Drosophila melanogaster: differential expression of two gene products. DNA Cell Biol. 1998;17:621–633. doi: 10.1089/dna.1998.17.621. [DOI] [PubMed] [Google Scholar]
- Dai FY, Qiao L, Tong XL, Cao C, Chen P, Chen J, Lu C, Xiang ZH. Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J Biol Chem. 2010;285:19553–19560. doi: 10.1074/jbc.M109.096743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DA. N-acetyltransferase. Pharmacol Ther. 1989;42:157–234. doi: 10.1016/0163-7258(89)90036-3. [DOI] [PubMed] [Google Scholar]
- Han Q, Ding H, Robinson H, Christensen BM, Li J. Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase. PLoS One. 2010;5:e8826. doi: 10.1371/journal.pone.0008826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermann E, Grieder NC, Amherd R, Brodbeck D, Meyer UA. Cloning of an arylalkylamine N-acetyltransferase (aaNAT1) from Drosophila melanogaster expressed in the nervous system and the gut. Proc Natl Acad Sci U S A. 1996;93:12315–12320. doi: 10.1073/pnas.93.22.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermann E, Jeno P, Meyer UA. Isolation and characterization of an arylalkylamine N-acetyltransferase from Drosophila melanogaster. FEBS Lett. 1995;375:148–150. doi: 10.1016/0014-5793(95)01198-n. [DOI] [PubMed] [Google Scholar]
- Hirsh J, Davidson N. Isolation and characterization of the dopa decarboxylase gene of Drosophila melanogaster. Mol Cell Biol. 1981;1:475–485. doi: 10.1128/mcb.1.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara N, Okada M, Nakagawa H, Takeda M. Purification and characterization of arylalkylamine N-acetyltransferase from cockroach testicular organs. Insect Biochem Mol Biol. 1997;27:241–246. doi: 10.1016/s0965-1748(96)00091-4. [DOI] [PubMed] [Google Scholar]
- Ichihara N, Okada M, Takeda M. Characterization and purification of polymorphic arylalkylamine N-acetyltransferase from the American cockroach, Periplaneta americana. Insect Biochem Mol Biol. 2001;32:15–22. doi: 10.1016/s0965-1748(01)00075-3. [DOI] [PubMed] [Google Scholar]
- Kawamura A, Graham J, Mushtaq A, Tsiftsoglou SA, Vath GM, Hanna PE, Wagner CR, Sim E. Eukaryotic arylamine N-acetyltransferase. Investigation of substrate specificity by high-throughput screening. Biochem Pharmacol. 2005;69:347–359. doi: 10.1016/j.bcp.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Klein DC. Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int. 2006;23:5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- Klein DC. Arylalkylamine N-acetyltransferase: “the Timezyme”. J Biol Chem. 2007;282:4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- Lee DW, Pietrantonio PV. In vitro expression and pharmacology of the 5-HT7-like receptor present in the mosquito Aedes aegypti tracheolar cells and hindgut-associated nerves. Insect Mol Biol. 2003;12:561–569. doi: 10.1046/j.1365-2583.2003.00441.x. [DOI] [PubMed] [Google Scholar]
- Meyer JH, Ginovart N, Boovariwala A, Sagrati S, Hussey D, Garcia A, Young T, Praschak-Rieder N, Wilson AA, Houle S. Elevated monoamine oxidase a levels in the brain: an explanation for the monoamine imbalance of major depression. Arch Gen Psychiatry. 2006;63:1209–1216. doi: 10.1001/archpsyc.63.11.1209. [DOI] [PubMed] [Google Scholar]
- Neckameyer WS, Holt B, Paradowski TJ. Biochemical conservation of recombinant Drosophila tyrosine hydroxylase with its mammalian cognates. Biochem Genet. 2005;43:425–443. doi: 10.1007/s10528-005-6781-3. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt S, Pryor SC. Dopamine- and octopamine-sensitive adenylate cyclase in the brain of adult Culex pipiens mosquitoes. Cell Mol Neurobiol. 1986;6:325–329. doi: 10.1007/BF00711117. [DOI] [PubMed] [Google Scholar]
- Smith TJ. Phylogenetic distribution and function of arylalkylamine N-acetyltransferase. Bioessays. 1990;12:30–33. doi: 10.1002/bies.950120107. [DOI] [PubMed] [Google Scholar]
- Tamura H, Takayama H, Nakamura Y, Reiter RJ, Sugino N. Fetal/placental regulation of maternal melatonin in rats. J Pineal Res. 2008;44:335–340. doi: 10.1111/j.1600-079X.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou SR, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugehara T, Iwai S, Fujiwara Y, Mita K, Takeda M. Cloning and characterization of insect arylalkylamine N-acetyltransferase from Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol. 2007;147:358–366. doi: 10.1016/j.cbpb.2006.10.112. [DOI] [PubMed] [Google Scholar]
- Vavricka C, Han Q, Huang Y, Erickson SM, Harich K, Christensen BM, Li J. From L-dopa to dihydroxyphenylacetaldehyde: a toxic biochemical pathway plays a vital physiological function in insects. PLoS One. 2011;6:e16124. doi: 10.1371/journal.pone.0016124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- Zhan S, Guo Q, Li M, Li J, Miao X, Huang Y. Disruption of an N-acetyltransferase gene in the silkworm reveals a novel role in pigmentation. Development. 2010;137:4083–4090. doi: 10.1242/dev.053678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.