Abstract
Neuronal activity induces the post-translational modification of synaptic molecules, promotes localized protein synthesis within dendrites and activates gene transcription, thereby regulating synaptic function and allowing neuronal circuits to respond dynamically to experience. Evidence indicates that many of the genes that are mutated in autism spectrum disorders are crucial components of the activity-dependent signalling networks that regulate synapse development and plasticity. Dysregulation of activity-dependent signalling pathways in neurons may therefore,have a key role in the aetiology of autism spectrum disorders.
About 1 in 100 children display signs and symptoms that lead to a diagnosis of autism spectrum disorder (ASD)1. This debilitating developmental disorder is characterized by impairments in social interaction and communication, and by restricted, repetitive and stereotyped behaviour and interests. In addition, individuals with ASD often have a seizure disorder and intellectual disability2. Most of these features of ASD manifest in the first few years of life, at the time of brain development when sensory experience is modifying excitatory synapse maturation and elimination, and promoting the development of inhibitory synapses3. This has led to the hypothesis that ASD may be due to a disruption of the normal process of experience-dependent synaptic development, resulting in an imbalance between excitation and inhibition in the developing brain.
Research has shown unequivocally that ASD is largely a genetic disorder4. Advances in human genetics and next-generation sequencing have identified a stunning number of newly arising mutations that are linked to ASD. In some cases, the mutations are limited to a specific gene and result in syndromic neurodevelopmental disorders with highly penetrant features of autism. These include Fragile X syndrome, tuberous sclerosis complex, Angelman syndrome, Timothy syndrome and Rett syndrome2. In addition, an impressive array of human genetic studies, including studies of families with several children with ASD and more recent exome sequencing of trios of individuals (mother, father and child with ASD), has surprisingly shown that ASD is often due to newly arising gene copy number variants (CNVs) — such as a deletion or duplication of a region of a chromosome — or a rare mutation that arises in the germ cell, particularly in sperm of older fathers5–10. In cases with CNVs, ASD is hypothesized to be a result of the increased or decreased expression of one, or several, genes that lie within the region of the genome in which the CNV mutation resides.
A future challenge is to make biological sense of the large number, and diversity, of genes that are associated with ASD. Identifying convergent molecular pathways in which multiple candidate genes are involved may be an effective way to advance our understanding of the underlying molecular basis for ASD, as well as a way to develop treatments for a broad range of forms of this disorder. Recent studies suggest that a convergent molecular pathway dysregulated in ASD is the signalling network that controls synapse development and function. An interesting feature of this signalling network is that it is composed of many proteins for which expression or function is regulated by neuronal activity, suggesting that one cause of ASD may be the dysregulation of neuronal activity-dependent synapse development and function.
A cardinal feature of human brain development is that sensory, cognitive and emotional experiences shape synapse and neural-circuit development. Neuronal activity triggers local changes at the synapse, altering the composition, shape and strength of the synapse. Synaptic stimulation both induces specific changes in messenger RNA translation near synapses and sends signals to the nucleus to induce gene transcription programs that control synaptic maturation and function. These neuronal activity-dependent pathways are crucial to learning and memory and for adaptive behavioural responses11–13.
In this Review, we explore the hypothesis that the dysregulation of activity-dependent signalling networks that control synapse development and function may be an important component of the molecular basis of ASD. For instance, ASD may arise from dysregulation of activity-dependent signalling pathways locally at a synapse, including changes in the post-translational modifications of synaptic proteins and the local translation of mRNAs at synapses, as well as from the dysregulation of activity-dependent gene transcription. Alternative explanations for the molecular and cellular basis of ASD must also be considered, including the possibility that it is a result of dysregulation of synaptic function in general, impairment of neurotransmission (which secondarily may alter activity-dependent signalling) and defects in earlier steps in nervous-system development. To begin to evaluate these hypotheses, we review studies of the molecular function of the most studied genes that are associated with ASD, including those for syndromic disorders with penetrant autism spectrum features and rare mutations associated with ASD. The current weight of evidence does not conclusively demonstrate that dysregulation of activity-dependent neuronal signalling is the singular cause of ASD. Indeed, impairment of multiple, different molecular pathways that are not mutually exclusive, probably contributes to the development of various subsets, or aspects, of ASD. However, as we review here, there is emerging evidence that genes and proteins associated with ASD are both regulated by and control activity-dependent pathways that modulate synaptic function. Many of the mutations associated with ASD lead to alterations in excitatory or inhibitory neurotransmission that disrupt activity-dependent signalling and disrupt activity-dependent synapse development, maturation and refinement. In addition, neuronal activity clearly regulates the function, localization and expression of many of the proteins that are associated with ASD. Taken together, these findings are beginning to provide compelling evidence that disruption of activity-dependent molecular programs that control synaptic function significantly contributes to the molecular basis of ASD.
Neuronal activity regulates synapses and circuits
Experience drives the neuronal activity that regulates synaptic development and plasticity, thereby altering the structure and function of neural circuits (Fig. 1). In a classic set of experiments done nearly 50 years ago14, David Hubel and Thorsten Weisel discovered that occluding one eye during a crucial period of development disrupts the formation of ocular dominance columns in the visual cortex, demonstrating a central role for experience in the development of neural circuits. We now know from studies in hundreds of laboratories that synapses are stabilized, matured and eliminated in response to neuronal activity during post-natal development, and the molecular underpinnings of these processes have been significantly detailed15.
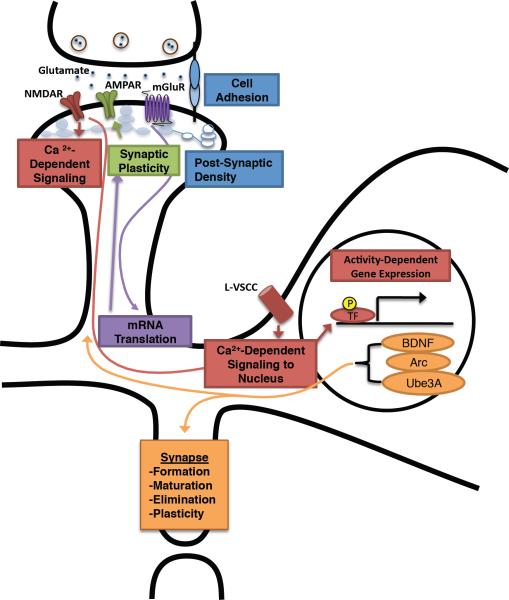
Figure 1. Regulation of synaptic development and function by neuronal activity.
Cell adhesion molecules and components of the postsynaptic density organize and regulate the formation of excitatory synapses on dendritic spines and can stimulate activity-dependent signalling networks within the postsynaptic neuron. At the synapse, the excitatory neurotransmitter glutamate can bind several glutamate receptors — NMDA receptor (NMDAR), AMPA receptor (AMPAR) and mGlu receptor (mGluR). Signalling from stimulated mGluR regulates mRNA translation, which is required for long-lasting forms of synaptic plasticity. Modification and cell-surface expression of AMPA receptors underlies many aspects of synaptic plasticity. Stimulated NMDA receptors flux calcium, inducing calcium-dependent signalling networks at the synapse that regulate AMPA receptor function and actin reorganization. In addition, calcium influx through NMDA receptor and L-type voltage-sensitive calcium channels (L-VSCC) triggers calcium-dependent signalling to the nucleus, leading to the modification of transcriptional regulators and resulting in the induction of activity-dependent gene expression. Genes induced by neuronal activity — including Bdnf, Arc, and Ube3A — function to regulate synapse formation, maturation, elimination and plasticity. P, phosphorylation.
The structure and protein composition of excitatory synapses are crucial for synaptic function and the ability of the organism to respond to its environment. Synapses are highly specialized compartments within the neuron comprised of pre- and post-synaptic regions and a synaptic cleft for which function is regulated both spatially and temporally. Synaptic cell-adhesion molecules modulate specific steps in synapse formation and function, including the specialization of pre- and post-synaptic compartments that occurs during synapse development, the recruitment of molecules and vesicles into synapses, the modulation of trans-synaptic signalling, and the synaptic plasticity that is induced in response to sensory stimuli16. Excitatory synapses form mostly on dendritic spines — protrusions from dendrites that function as a finely tuned morphological and functional compartment. The postsynaptic density (PSD) forms at the centre of the dendritic spine head. The PSD contains structural proteins and signalling molecules as well as receptors that respond to the excitatory neurotransmitter glutamate. PSD signalling proteins include the calcium-dependent kinases CaMK2α and CaMK2β, and the scaffold protein PSD-95 that helps to organize the synapse, connect glutamate receptors to downstream signalling complexes, and modulate synaptic plasticity. Other PSD scaffolding proteins include GKAP (alternatively named SAPAP), SHANK and HOMER proteins that promote dendritic spine growth, regulate synaptic activity and anchor signalling complexes that mediate aspects of the synaptic response to sensory stimuli (Fig. 2)17–19.
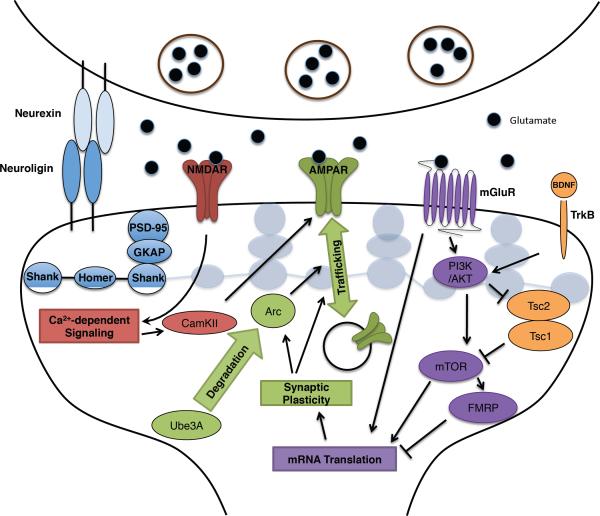
Figure 2. Neuronal activity regulates mRNA translation and synaptic plasticity.
At the excitatory synapse, the cell adhesion molecules neurexin and neuroligin and structural proteins in the postsynaptic density, including Shank proteins, regulate, and are regulated by, neuronal activity-dependent signalling networks. The excitatory neurotransmitter glutamate binds NMDA receptors (NMDAR), AMPA receptors (AMPAR) and mGlu receptors (mGluRs). AMPA receptors mediate the fast excitatory neurotransmission. UBE3A degrades Arc, which regulates trafficking of AMPA receptors. During plasticity, activation of NMDA receptors triggers calcium-dependent signalling at the synapse, stimulating CaMKII and modifying AMPA receptor function and actin reorganization. Activation of mGluR triggers several signalling cascades, including the PI(3)K–AKT–mTOR pathway, the Ras–MAPK pathway, and PLC to regulate mRNA translation. FMRP inhibits mRNA translation, and mGluR signalling can regulate FMRP activity. Growth factors, including BDNF, whose expression is induced by neuronal activity, bind receptor tyrosine kinases (including TrkB) that activate multiple signalling pathways, including the PI(3)K–AKT pathway. The PI(3)K–AKT pathway when activated leads to phosphorylation of the TSC1–TSC2 complex to control mTOR activity. Mutations in neurexins, neuroligins, SHANK, GKAP, UBE3A, FMRP and TSC1–TSC2 are associated with ASD.
Experience leads to the release of neurotransmitters at specific synapses. Glutamate — the most common excitatory neurotransmitter — binds to the receptors AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) and NMDA (N-methyl-d-aspartate). These ionotropic receptors then flux Na+ and K+, leading to membrane depolarization. AMPA receptors mediate fast excitatory neurotransmission and can be dynamically inserted and removed from the synaptic membrane, thus affecting the strength of the postsynaptic response to glutamate released at the synapse. NMDA receptors open up and conduct current only when both the postsynaptic membrane is depolarized and when glutamate binds the receptor, providing coincidence detection and a signal that induces forms of synaptic plasticity20. Activated NMDA receptors flux calcium, thereby inducing multiple calcium-dependent signalling pathways within dendrites, including calcium/calmodulin-dependent kinases (CaMKs) and the Ras– mitogen-activated protein kinase (MAPK) pathway21, which alter the synapse locally and stimulate gene transcription in the nucleus. For instance, activated CaMK2 phosphorylates AMPA receptors, altering their conductivity, and triggering reorganization of actin cytoskeleton to regulate dendritic growth15. Neuronal activity also induces long-lasting changes in the efficacy of synaptic transmission, including long-term potentiation (LTP) and long-term depression (LTD), that involve the strengthening or weakening of synaptic transmission, respectively, and are thought to be molecular correlates of learning and memory11,20,22. Depending on the nature of the stimulus, different forms of synaptic plasticity are induced at synapses. LTP or LTD are mediated, at least in part, via the insertion or removal, respectively, of AMPA receptors from the synaptic membrane, leading to alterations in synaptic strength (Fig. 2).
Glutamate release at synapses can also induce longer lasting forms of synaptic plasticity that are mediated by NMDA-receptor-dependent calcium influx and typically require activity-dependent protein synthesis that is a consequence of the local mRNA translation within the dendrite or gene transcription within the nucleus. In a similar manner, glutamate binding to metabotropic glutamate receptors (mGluR) induces protein-synthesis-dependent forms of synaptic plasticity by activating the mechanistic target of rapamycin (mTOR) and extracellular signal-regulated kinase (ERK) pathways11,20,22,23.
Neuronal activity also regulates programs of gene transcription that affect synapses, including key steps in synaptic development and forms of synaptic plasticity12,15. Typically, activity-dependent gene transcription requires calcium-dependent signaling, and, depending on the route and dynamics of calcium entry (either through NMDA receptors or through voltage-gated L-type calcium channels), can trigger distinct gene expression responses24,25. Neuronal activity-induced elevations in intracellular calcium lead to the activation of multiple signalling molecules, including CaMKII, CaMKIV, protein kinase A (PKA), MAPK pathways, ribosomal s6 kinase (RSK), mitogen- and stress-activated kinase (MSK), and the calcium-dependent phosphatase calcineurin, each of which phosphorylate or dephosphorylate transcriptional regulators within the nucleus. For instance, multiple stimuli induce the phosphorylation of the cyclic AMP-responsive element-binding protein (CREB) at Ser-133, which recruits the CREB-binding protein (CBP) to gene promoters. CBP catalyses the acetylation of histones, leading to changes in chromatin structure that facilitate the recruitment of RNA polymerase II and the activation of gene-transcription programs that promote synapse development12,15.
In addition to stimulating CREB-dependent gene transcription, neuronal activity-dependent calcium influx into neurons triggers the dephosphorylation of the transcription factor myocyte enhancer factor 2 (MEF2) by calcineurin, disrupting the association of MEF2 with histone deacetylases and leading to the recruitment of CBP and the stimulation of gene transcription26. Neuronal activity that functions through CREB, MEF2 and multiple other activity-regulated transcription factors — including SRF, Fos27 and NPAS428 — induces the transcription of the genes encoding proteins that function directly at synapses, including Bdnf, Arc and Ube3A12,15. For example, activity-regulated MEF2 by activating Arc suppresses the number of excitatory synapses26,29, whereas CREB and NPAS4, by activating Bdnf transcription, control the number of inhibitory synapses that form on excitatory neurons28,30.
Many of the proteins that form the activity-dependent signalling network that controls gene transcription are implicated in disorders of cognition and behaviour, including ASD (Fig. 3). For example, mutations in the voltage-gated L-type calcium channels that flux calcium to initiate activity-dependent gene transcription are associated with Timothy syndrome, which is characterized by impairments in cardiac and immune function and has a significant association with ASD phenotypes31. Likewise, mutations in RSK2, CBP, the methyl-binding protein MECP2, and the ubiquitin ligase UBE3A are causes of Coffin–Lowry syndrome, Rubinstein–Taybi syndrome, Rett syndrome and Angelman syndrome, respectively3. The finding that many of the genes that are involved in activity-dependent gene transcription are associated with human disorders of cognitive function, including ASD, provides support for the hypothesis that dysregulation of activity-dependent signalling networks in general may contribute significantly to the synaptic dysfunction that occurs in ASD.
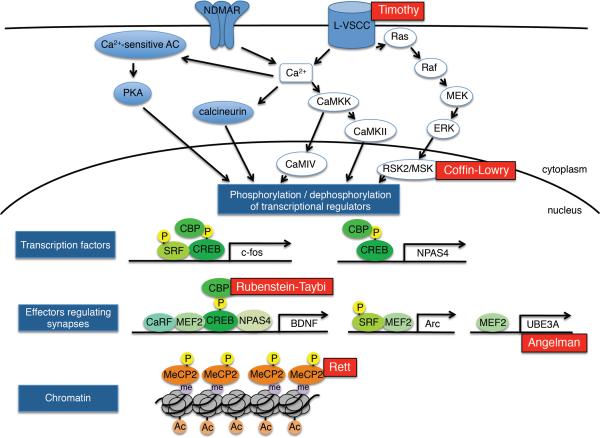
Figure 3. Activity-dependent gene expression and ASD.
Activation of NMDA receptors and L-type voltage-sensitive calcium channels (L-VSCC) trigger calcium influx and induction of calcium-dependent signalling molecules, including CaMKK that activates the kinases CaMKII and CaMKIV, calcium-sensitive adenylyl cyclases (AC) that activate PKA, and the calcium dependent phosphatase calcineurin. In addition, the RAS–ERK–RSK/MSK pathway is activated upon stimulation of L-VSCC. These signalling cascades phosphorylate or dephosphorylate transcriptional regulators in the nucleus, leading to the expression of transcription factors (shown as c-Fos and NPAS4). These transcription factors contribute to the regulation of activity-dependent gene transcription and to the expression of effectors that function to modify synapses (shown as BDNF, Arc and Ube3A). In addition, changes in chromatin modifications (including histone acetylation), and the phosphorylation of chromatin-associated proteins (for example, MECP2) may modulate activity-dependent gene transcription. Mutations of proteins involved in activity-dependent gene expression are associated with some forms of ASD including L-VSCC in Timothy syndrome, RSK2 in Coffin-Lowry, CBP in Rubenstein-Taybi, Ube3A in Angelman syndrome and MeCP2 in Rett syndrome. Ac, acetylation; CaRF, calcium response factor; Me, methylation; P, phosphorylation
Genetic architecture of ASD
ASD is caused predominantly by genetic mutations4. Recent evidence suggests that de novo mutations that arise in the germline are a significant cause of ASD, although it is important to keep in mind that the genetic aetiology of about 70% of the forms of ASD is unknown and it remains perplexing why ASD is at least four times more prevalent in males than in females. Higher age of the father is positively correlated with the higher rate of de novo genetic mutations and an increased risk of ASD9,10. ASD is polygenic in aetiology; recent studies of de novo exonic mutations suggest the involvement of as many as 400 to 1,000 genetic loci in ASD9. The complexity of the genetic aetiology of ASD may complicate understanding its molecular mechanisms. However, as already suggested, many of the genes implicated in ASD could function in convergent molecular pathways, allowing for the formulation of specific mechanistic hypotheses that could explain significant portions of ASD.
Rare genetic variations play a significant part in the aetiology of ASD. Several de novo CNVs that are associated with ASD have been detected multiple times, including CNVs at 16p11.2, 15q11–13, 22q11.2, deletions at NRXN1, and duplications at 7q11.235,32. The fact that these CNVs have been detected in multiple individuals with ASD strongly suggests that mutations in these regions of the genome are contributing to ASD. More subtle, rare mutations in NLGN4, NLGN3, NRXN1, SHANK2, SHANK3, CNTNAP2 (ref. 5) have also been identified in multiple individuals with ASD, suggesting that these mutations also contribute these disorders. In addition, very recent exome sequencing of parent-offspring trios has identified de novo exonic mutations in SCN2A, KATNAL2, CHD8, FOXP1, NTNG1, GRIN2B, LAMC3(refs 7–9), extending further the number of genes for which mutation may contribute to ASD. As de novo mutations within gene-coding regions occur with reasonable frequency and do not necessarily result in a deleterious phenotype, additional evidence is required before each of these mutations can unequivocally be said to contribute to ASD. In the future, the keys to understanding how mutations in these proteins lead to ASD will be to identify additional individuals with ASD that carry mutations in these genes, and to obtain a more thorough understanding of the molecular functions of each of these proteins in neurons and how their mutation leads to defects in synaptic function.
In addition to these rare mutations, several widely studied syndromic disorders, such as Fragile X, with penetrant autism spectrum features have provided useful insight into the pathophysiology of ASD. These syndromic disorders are caused by single gene mutations and have phenotypes that have allowed the disorder to be readily diagnosed; thus, the mechanistic understanding of these syndromes is more advanced than for most ASD. For example, Fragile X syndrome — characterized by macroorchidism, connective-tissue disease, motor abnormalities, language deficits and impaired social interactions — is the leading cause of inherited intellectual disability and ASD23. Fragile X syndrome results from a trinucleotide expansion (CGG) in FMR1, which encodes fragile X mental retardation protein (FMRP). The CGG expansion occurs within the 5′ untranslated region of the FMRP mRNA, resulting in decreased translation of the FMRP mRNA and a decrease in the amount of FMRP expressed in neurons. FMRP has recently been reported to modulate the rate of translational elongation of mRNAs that encode synaptic proteins33, suggesting that the decrease in FMRP expression that occurs in Fragile X syndrome may result in the dysregulation of synaptic protein expression owing to an impairment in translational control23. Thus, as described later, studying the genes that are mutated in these widely studied syndromes has the potential to provide insight into the molecular and cellular basis of ASD.
Several genetic loci (UBE3B, CLTCL1, NCKAP5L and ZNF18) that are associated with ASD include genes for which expression is induced by neuronal activity34,35. In addition, knockdown of several transcription factors that mediate activity-regulated gene transcription, including MEF2 and NPAS4, led to decreased expression of multiple genes that are linked to ASD (c3orf58, SLC9A9, and PCDH10)34. Only about 3% of the neuronal transcriptome is altered when the membrane is depolarized, but a higher percentage of the genes that are implicated in ASD have been shown to be regulated by neuronal activity, it has been proposed, therefore, that ASD may result, at least in part, from the disruption of the activity-dependent signalling networks that control gene transcription34,35. Thus, it will be important to determine if the expression of RNAs that are encoded by other genetic loci associated with ASD are regulated by neuronal activity. A crucial consideration is that genetic loci associated with ASD may not only be found to disrupt gene function through mutations within gene-coding sequences, but may also be found to disrupt the function of the regulatory regions that control these genes. Of note, therefore, is a recent study that identified thousands of neuronal activity-regulated enhancers of transcription that could potentially be sites of human variation that lead to ASD36.
Synaptic proteins implicated in ASD
Mutations in multiple synaptic cell adhesion molecules and components of the PSD are associated with ASD. The fact that these proteins have key roles in regulating synaptic responses to experience, and are themselves modulated by neuronal activity supports the hypothesis that ASD may result from the disruption of the normal process of activity-dependent synapse development (Fig. 2).
Neuroligins and neurexins
Rare mutations in multiple members of the neuroligin and neurexin families of synaptic adhesion molecules have repeatedly been found to be associated with ASD5. Neurexins are presynaptic membrane proteins that are encoded by mRNAs, which have undergone extensive altnerative splicing. Neuroligins are postsynaptic membrane proteins that function as neurexin receptors and, working together with neurexins, modulate the formation and function of synapses16,37. Particular neuroligin family members are localized specifically to inhibitory or excitatory synapses, suggesting that individual neuroligins may play a part in specifying the formation or function of excitatory and inhibitory synapses. In addition to binding neuroligins, neurexins bind other postsynaptic transmembrane proteins, including leucine-rich repeat (LRR) transmembrane proteins, adding to the number of different potential trans-synaptic connections16. There is increasing evidence for the involvement of neurexins, neuroligins and LRR transmembrane proteins in activity-dependent programs that modulate synapse function.
Several of the neuroligin mutations found in individuals with ASD have been modelled in mice, and these mice have been shown to have alterations in excitatory or inhibitory neurotransmission. For example, knock-in mice that harbour the neuroligin-3 ASD missense mutation Nlgn3(R451C) display impaired social interactions, recapitulating a key feature of ASD38. These knock-in mice also have enhanced inhibitory neurotransmission with no alterations in excitatory neurotransmission, resulting in a defect in excitatory–inhibitory balance in the brain38. Knock-in mice with another ASD-associated missense mutation, Nlgn3(R704C) displayed a decrease in AMPA receptor-mediated synaptic transmission in the hippocampus, but no alteration in NMDA or GABA (γ-aminobutyric acid)-receptor mediated neurotransmission39. In addition to the neuroligins, mutations in NRXN1 are also associated with ASD in humans. Nrxn1 knock-out mice have defects in miniature excitatory postsynaptic current (EPSC) frequency and evoked postsynaptic potentials, but show no change in inhibitory neurotransmission in the hippocampus40. These studies indicate that particular ASD mutations in neurexins or neuroligins disrupt excitatory or inhibitory neurotransmission in the brain in specific ways and suggest that modelling ASD-associated mutations in mice will be important for elucidating how each mutation affects synaptic function and give rise to ASD.
Notably, neuroligins have been found to regulate the activity-dependent maturation and modulation of synapses. In cultured neurons of rodents, overexpression of Nlgn1 increases the number of functional excitatory synapses, an effect that is dependent on NMDA receptors, as well as activity-regulated CaMKII that functions downstream of NMDA receptor activation to regulate of synaptic plasticity41. In contrast with overexpression of wild-type Nlgn1, overexpression of Nlgn1 containing an ASD-associated missense mutation leads to a decrease in the number and strength of excitatory synapses41. This suggests that activity-dependent functions of Nlgn-1 may be disrupted in ASD. In another study42, knockdown of Nlgn1, delivered by a viral vector, in the amygdala of adult rats led to the selective reduction of NMDA-receptor-mediated currents, and impairments in LTP and in the storage of associative fear memory. This suggests a role for neuroligins in experience-regulated learning and memory which may be altered by ASD-associated mutations in NLGN1.
Similarly, using sensitization of the gill-withdrawal reflex in the sea hare Aplysia californica as a simple model of learned fear and long-term memory showed that depletion of the Aplysia homologue of neuroligin in the postsynaptic motor neuron, or neurexin in the presynaptic sensory neuron, impaired initiation, stabilization and persistence of long-term facilitation of the sensory-to-motor neuron synaptic connection. Relative to wild-type neuroligin, overexpression of neuroligin with an ASD-associated mutation blocked intermediate and long-term facilitation at the sensory-to-motor neuron synapse43. Taken together, these findings suggest that neuroligins and neurexins modulate activity-dependent synaptic remodelling and long-term memory, and that ASD-associated mutations in neuroligins or neurexins may disrupt these processes.
The interaction between neurexins and neuroligins is regulated by neuronal activity, providing further evidence that mutations that cause ASD might be disrupting activity-dependent processes that control synaptic function. The dynamic nature of the neuroligin–neurexin interaction was studied using the bacterium Escherichia coli enzyme biotin ligase (BirA), which catalyzes the biotinylation of an acceptor peptide only when BirA and the acceptor peptide are in close proximity44. When neurexin-1 fused to BirA, and neuroligin fused to an acceptor peptide are expressed in neurons, biotinylation of neuroligin–acceptor protein occurs when neurexin-1–BirA and neuroligin–acceptor protein interact across the synapse. Using this approach, neuronal activity was found to acutely promote an increase in the interaction of neurexins and neuroligins across the synapse in an NMDA-receptor-dependent manner44. Neuronal activity induced both the insertion of neurexin and neuroligin into the synaptic membrane and suppressed internalization of neurexin and neuroligin from the synaptic membrane, leading to an overall increase in the trans-synaptic neurexin–neuroligin interaction. Disruption of this activity-regulated enhancement impaired the recruitment of AMPA receptors to the synaptic membrane, thereby altering synaptic function44. It will be important to determine if ASD-associated mutations in neurexins and neuroligins affect these activity-dependent changes in their trans-synaptic interaction.
Neuronal activity also regulates alternative splicing of neurexin mRNA and, thus, alters trans-synaptic signalling and presynaptic differentiation45. It will be interesting to determine if ASD-associated mutations disrupt activity-dependent alternative splicing of neurexins and if disruption of this process contributes to the development of ASD.
Shanks
Shank proteins function as scaffolds in the PSD of excitatory synapses that regulate the organization of postsynaptic signalling complexes, as well as the morphology and function of synapses17–19. Rare mutations associated with ASD have been found in SHANK2 and SHANK3 (ref. 5), and recently in SHANK1 (ref. 46). In particular, Phelan-McDermid syndrome is an ASD that results from a microdeletion that includes heterozygous loss of SHANK3. Knockout mice with deletions in members of the Shank family of genes exhibit behaviours similar to those observed in ASD. For example, Shank3 knockout mice have deficits in social interactions and engage in repetitive behaviours, such as excessive grooming, that lead to self injury47. In addition, mice lacking Shank2 have abnormalities in motor behaviour, vocalization and socialization — phenotypes typically observed in ASD48,49.
These Shank2 and Shank3 knockout mice display alterations in excitatory neurotransmission that correlate with ASD behavioural phenotypes. The Shank3 knockout mice have deficits in striatal glutamatergic synapse structure and function, including reduced cortico-striatal synaptic transmission47, whereas Shank2 knockout mice models show defects in NMDA-receptor-dependent excitatory neurotransmission and synaptic plasticity in the hippocampus48,49. In one study of Shank2 knockout mice, by using either a partial agonist of NMDA-receptor function (D-cycloserine) or a positive allosteric modulator of mGluR5, it was possible to normalize NMDA receptor function and decrease autistic behaviours, suggesting that impairment of NMDA receptor functioning may be a key mechanism through which mutations in genes that encode the Shank family of proteins lead to ASD49. Taken together, these studies have revealed how the Shank family of proteins modulate excitatory synapse function and begin to explain how mutations in the genes that encode them might lead to ASD.
One possibility is that Shank mutations that cause ASD disrupt activity-dependent regulation of Shank function. Neuronal activity has been shown to affect the molecular composition of the post-synaptic compartment by inducing the ubiquitination and proteasome-mediated degradation of a subset of PSD proteins, including those of the Shank family, thus leading to changes in synaptic signalling50,51. The TRIM3 ubiquitin ligase mediates the activity-dependent degradation of Shank1 and another PSD-scaffolding protein GKAP, by a ubiquitin–proteasome dependent mechanism52. In mice, the retrieval of fear memory was found to lead to the polyubiquitination and degradation of Shank proteins and to correlate with a requirement for protein degradation in memory extinction53. These findings suggest that the activity-dependent degradation of synaptic proteins, including those of the Shank family, regulates the reorganization of synapses and memories. Taken together, these studies begin to explain how Shank proteins control synaptic function and plasticity, and how their mutation might contribute to autism spectrum phenotypes.
Activity-dependent regulation of mRNA translation and ASD
Long-lasting forms of synaptic plasticity that are elicited by glutamate binding to NMDA receptors or group 1 mGluRs, typically require protein synthesis that is mediated by mRNAs and ribosomes that are localized near synapses. Long-lasting NMDA receptor-dependent LTP and LTD have a delayed requirement for protein synthesis, whereas mGluR-dependent LTD requires protein synthesis that occurs within minutes. Several genes that are mutated in ASD — FMR1, TSC1, TSC2 and PTEN — have key roles in protein synthesis-dependent plasticity of synapses (Fig. 2)13,23.
Fragile X syndrome
Fragile X syndrome is caused by mutations in FMR1, resulting in decreased expression of the RNA-binding protein FMRP. This protein negatively regulates the translation of specific mRNAs at the synapse23,54. FMRP has been suggested to function by blocking the initiation of mRNA translation, or stalling movement of the ribosome along the mRNA, leading to decreased protein synthesis33; however, the precise molecular mechanisms by which FMRP regulates translation remain unclear23. Fmr1 knockout mice exhibit abnormally high levels of protein synthesis and enhanced LTD in response to mGluR stimulation, suggesting that under normal conditions FMRP suppresses mGluR-dependent protein synthesis and LTD55. Remarkably, the excessive protein synthesis observed in Fmr1 knockout mice is reversed by antagonists of mGluR5. In addition, both genetic and pharmacological inhibition of mGluR5 reverses synaptic and behavioural impairments in these mice. These findings are the basis of human clinical trials that aim to test the efficacy of mGluR5 antagonists for the treating Fragile X syndrome23,56–58.
Arc is one of the best-studied mRNA targets of FMRP. The protein encoded by this activity-regulated gene promotes the internalization of AMPA receptors at excitatory synapses. Translation of Arc mRNA is enhanced during, and crucial for, mGluR-dependent LTD59,60. In Fmr1 knockout mice, the dynamic translation of Arc mRNA in response to stimulation is impaired59,61, and the basal level of the Arc protein in dendrites is elevated61. This abnormally high level of Arc expression in Fmr1 knockout mice may be responsible for the enhanced mGluR–LTD observed in these animals and might explain, in part, why their LTD is independent of new mRNA translation62.
During early post-natal brain development, neuronal activity promotes changes in glutamate receptor subtype that are required for proper maturation of excitatory thalamocortical synapses in the somatosensory cortex. In Fmr1 knockout mice, this activity-dependent maturation of the excitatory thalamocortical synapses is dysregulated, resulting in a persistence of silent NMDA-receptor-only synapses at times during brain development when these synapses would normally express both NMDA and AMPA receptors63. The absence of AMPA receptors at these synapses may be due, in part, to the dysregulation of Arc mRNA translation that occurs in Fmr1 knockout mice.
FMRP has also been suggested to be crucial for activity-dependent synapse elimination, a key process during post-natal brain development64 that may be defective in Fragile X syndrome. The transcription factor MEF2 controls a program of gene expression that has a role in developmental synapse elimination26,29,65,66. Notably, activation of MEF2 leads to a decrease in the number of excitatory synapses in wild-type neurons, but not in Fmr1 knockout neurons, suggesting that the effects of MEF2 on synapse number may be FMRP dependent64. Arc may be the link between MEF2 and FMRP. MEF2 seems to restrict excitatory synapse number, at least in part, by activating Arc transcription. Once Arc transcription is induced, translation of the Arc mRNA is kept in check by FMRP. However, in the absence of FMRP, the level of Arc protein is abnormally high, so that further activation of Arc transcription by MEF2 may not have much of an effect on synapse number.
However, Arc is almost certainly not the only target of FMRP that mediates the effects of FMRP on LTD, excitatory synapse number and ASD. Many other FMRP targets have been shown to be involved in synaptic plasticity, and there is significant overlap between FMRP targets and ASD candidate genes, including NLGN3, NRXN1, SHANK3, PTEN, TSC1 and NF123,33. This suggests that these ASD candidate genes and FMRP may function in overlapping pathways. Taken together, these studies implicate FMRP as a key regulator of activity-dependent mRNA translation and synaptic plasticity and suggest that dysregulation of this pathway may contribute significantly to ASD phenotypes.
Tuberous sclerosis complex
Tuberous sclerosis complex is the other main syndromic ASD that seems to be a consequence of a dysregulation of activity-modulated mRNA translation. TSC is characterized by benign tumours in multiple organs as well as a high penetrance of ASD. This disorder is caused by mutations in TSC1 or TSC2 that encode tumour suppressor proteins which form the complex TSC1–TSC2 that regulate protein synthesis in a wide range of mammalian cell types, including neurons67. The binding of growth factors to receptor tyrosine kinases leads to phosphorylation of the TSC complex and subsequent activation of the mTOR, a positive regulator of protein synthesis67.
One major activator of TSC1–TSC2 signalling in neurons is brain-derived neurotrophic factor (BDNF), a secreted protein that binds to the receptor tyrosine kinase TrkB on the surface of neurons and thereby mediates many aspects of brain development and function. Neuronal activity potently induces Bdnf transcription in excitatory neurons. Newly synthesized BDNF is then secreted from excitatory neurons and functions near the site of release to regulate synaptic development and plasticity, including the development of inhibitory synapses30. Several studies suggest that the effect of BDNF on these processes is mediated by the local control of protein translation68. Specifically, BDNF binding to TrkB activates the phosphatidylinositol-3-OH kinase (PI(3)K), which phosphorylates phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) at the 3′ position to generate phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3). This then binds and activates phosphoinositide-dependent protein kinase (PDK1), which in turn contributes to activation of the serine/threonine protein kinase Akt proteins. Activated Akt phosphorylates TSC2, reducing its GTPase activating protein (GAP) activity for the Ras family GTPase, Rheb. Thus, phosphorylation of TSC2 leads to Rheb activation, which in turn triggers activation of the protein kinase mTOR, contributing to the activation of protein synthesis (Fig. 4)67. Loss of function mutations in TSC1 or TSC2 that occur in tuberous sclerosis complex, result in deregulated mTOR signalling and abnormal neuronal protein synthesis, suggesting that tuberous sclerosis complex may be a result of the dysregulation of these processes13,69. Although it remains to be demonstrated that the disruption of BDNF-mediated protein synthesis has a key role in TSC1-TSC2-dependent ASD, the observations of BDNF's widespread role in nervous system development, that Bdnf transcription is robustly induced by neuronal activity and that BDNF regulates protein translation through inhibition of TSC1–TSC2, suggest a role for BDNF in tuberous sclerosis complex. Signalling by TSC1–TSC2 downstream of other neurotrophic factors is also likely to contribute to tuberous sclerosis complex.
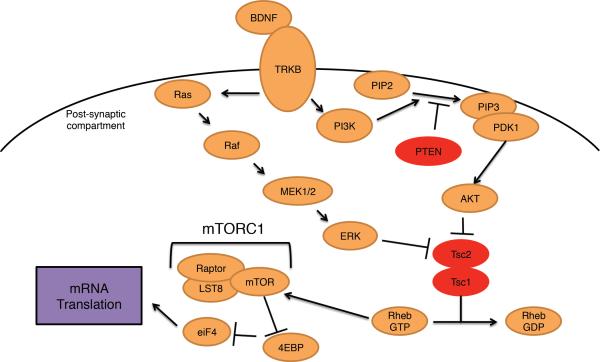
Figure 4. BDNF regulates mRNA translation.
Neuronal activity potently induces expression of the neurotrophic factor BDNF. BDNF binds the receptor TrkB, a receptor tyrosine kinase, triggering several signalling cascades, including the PI(3)K–AKT and Ras–MAPK pathways. PI(3)K is a lipid phosphatase, converting PtdIns(4,5)P2 to PtdIns(3,4,5)P3, which can be reversed with the phosphatase PTEN. PDK1 together with PtdIns(3,4,5)P3 activates AKT, which inhibits TSC1–TSC2. TSC1–TSC2 serves as a GAP for the GTPase Rheb, promoting the conversion of active GTP bound Rheb to the inactive GDP bound form. Rheb-GTP activates mTOR to induce mRNA translation. PTEN and TSC1–TSC2 are associated with ASD, as are mutations in Ras, Raf, MEK1–MEK2.
Further evidence that a defect in neurotrophic-factor-regulated protein synthesis contributes to ASD is provided by the finding that mutations in PTEN, a negative regulator of PI(3)K–AKT signalling upstream of TSC1–TSC2, also result in syndromic ASD. The lipid phosphatase PTEN catalyses the dephosphorylation of PtdIns(3,4,5)P3 at the 3′ position to generate PtdIns(4,5)P2 thus antagonizing the activity of PI(3)K70. Mutations that inactivate PTEN lead to excessive PI(3)K–AKT and mTOR signalling and in this way may lead to abnormal protein synthesis, and ultimately synaptic dysfunction and ASD. Thus, characterization of the role of PTEN in neurotrophic factor regulated protein synthesis may help to uncover the mechanisms that contribute to ASD.
The study of mice that carry Pten, Tsc1, or Tsc2 mutations has the potential to provide insight into how defects in the PTEN–PI(3)K–AKT–TSC signalling lead to ASD. Loss of function of TSC1 and TSC2 leads to deregulated activation of mTOR13,69. As TSC1–TSC2 are negative regulators of protein synthesis, it was reasonable to hypothesize that — similar to loss of FMRP — loss of TSC1 or TSC2 would lead to increased protein synthesis and enhanced mGluR–LTD. Interestingly, the opposite has been found in recent studies71,72. Mice with a heterozygous loss of function mutation in Tsc2 (Tsc2+/−) have deficient mGluR–LTD in the hippocampus, but no defect in basal synaptic transmission. Compared with wild-type mice, Tsc2+/− mice have decreased translation of Arc mRNA71. Similarly, in another study, post-natal deletion of Tsc1 in hippocampal CA1 mouse neurons by viral delivery of Cre recombinase led to loss of mGluR–LTD72. These findings of deficient mGluR–LTD in mice with deletions in Tsc1 or Tsc2 are in contrast with Fmr1 knockout mice, which have increased mGluR–LTD and excessive protein synthesis57.
Inhibition of mTOR in Tsc2+/− mice restored mGluR–LTD and normalized protein synthesis rates to that of wild-type mice rates, suggesting that excessive mTOR activity leads to decreased protein synthesis and LTD deficiency in Tsc2+/− mice71. Whereas inhibition of mGluR5 in Fmr1 knockout mice corrects excessive protein synthesis and mGluR5-dependent LTD, augmentation of mGluR5 signalling by a positive allosteric modulator restored protein synthesis and mGluR-dependent LTD in Tsc2+/− mice71. In a fear conditioning behavioural model, Tsc2+/− mice have a defect in distinguishing familiar and new contexts, a defect that can also be corrected by augmentation of mGluR5 signalling71. These findings suggest that, for proper synaptic function in response to neuronal activity, protein synthesis at synapses is tightly regulated, and TSC1 and TSC2 as well as Fmr1 mutations disrupt this balance in opposite directions, leading to defects in synaptic plasticity that may contribute to ASD phenotypes.
Despite recent progress, major gaps in knowledge remain about how neuronal activity dynamically regulates protein synthesis to control synaptic function and how disruption of TSC1–TSC2 or FMRP alters these processes to lead to ASD. Indeed, how loss of function of FMRP or TSC1–TSC2 in the two disorders leads to differential dysregulation of protein synthesis and synaptic plasticity is not yet understood. One possibility is that enhanced mTOR activity in tuberous sclerosis complex results in increased FMRP phosphorylation, which in turn functions to suppress the translation of specific mRNAs. Indeed, stimulus-regulated phosphorylation or dephosphorylation of FMRP alters its ability to suppress mRNA translation61,73–75. Alternatively, TSC1–TSC2 and FMRP may regulate the translation of different pools of mRNAs, so that abnormal translation of the two pools of mRNAs has different outcomes. Improved understanding of the activity-dependent programs that control protein synthesis and synaptic plasticity will probably help to clarify the molecular mechanisms that underlie these two forms of ASD.
Activity-dependent gene transcription and ASD
In addition to its affects on protein synthesis at synapses, neuronal activity regulates programs of gene expression in the nucleus that modulate synaptic development and plasticity. Mutations associated with ASD disrupt multiple features and components of the signalling network that controls activity-dependent gene transcription. These include effects on the route and dynamics of calcium entry into neurons, effects on transcriptional regulators that control gene expression in response to neuronal activity, and an impact on effector genes that encode proteins that regulate synaptic development and function (Fig. 3).
Timothy syndrome
The route and dynamics of calcium entry into the neuron determines the program of activity-dependent gene expression that is activated24,25. In excitatory neurons, activity-dependent gene transcription is mediated, at least in part, by calcium entry through L-type calcium channels25. The L-type calcium channels are voltage-gated and thus open upon membrane depolarization. Mutations in CACNA1C, which encodes the α1 pore-forming subunit of the L-type calcium channel Cav1.2, has been associated with multiple neuropsychiatric disorders, including ASD76. In addition, Timothy syndrome, a disorder characterized by cardiac arrhythmia, congenital heart disease, immune deficiency, intermittent hypoglycaemia, global development delay and ASD, is caused by a missense mutation in CACNA1C31. Studies in mice indicate that this missense mutation leads to increased Ca2+ currents and loss of voltage-dependent channel inactivation31,77. To determine the effect of the Timothy syndrome mutation on human neurons, induced pluripotent stem cells were derived from fibroblasts of individuals with Timothy syndrome and differentiated into neurons78. Compared with wild-type neurons, the Timothy syndrome neurons had impaired Ca2+ channel inactivation and a significant increase in sustained intracellular Ca2+ elevation after membrane depolarization. Activity-dependent gene expression was also altered in the Timothy syndrome neurons, compared with control neurons, with more than two hundred genes displaying altered expression following membrane depolarization78. These findings raise the possibility that the ASD phenotypes associated with Timothy syndrome may be the result of the deregulation of activity-dependent gene transcription.
Rett syndrome
Mutations in MECP2 lead to Rett syndrome, one form of ASD characterized by impaired language development, loss of social engagement, stereotyped hand movements, seizures and motor-system disabilities. MECP2 binds methylated cytosines within DNA and seems to function mainly as a transcriptional repressor. The level of MECP2 expression in post-mitotic neurons is about tenfold higher than in other cell types, suggesting that MECP2 may have a unique function in neurons. Notably, the level of MECP2 expression in mature neurons is similar to that of core histones, and MECP2 is bound broadly across the genome. These findings suggest that MECP2 may have a histone-like role in regulating chromatin structure and gene expression79,80. Neuronal activity induces the phosphorylation of MECP2 at Ser421 (refs 81–83) broadly across the genome, raising the possibility that activity-dependent phosphorylation of MECP2 mediates a genome-wide chromatin response to neuronal activity80. The fact that Rett syndrome manifests itself in the first year of life — a time when experience is shaping synapse development — together with the observation that MECP2 is a chromatin regulator that is modified in response to neuronal activity, suggests that Rett syndrome may be due to a defect in the process of activity-dependent chromatin regulation or gene transcription that controls synaptic development, plasticity and behaviour.
To test the role of activity-dependent phosphorylation of MECP2 in synapse development and behaviour, knock-in mice were generated in which Ser421 was converted to an alanine (Ser421Ala), preventing this residue from being phosphorylated80,84. The Ser421Ala knock-in mice and wild-type mice express equivalent levels of MECP2, but the Ser421Ala knock-in mice display increased dendritic complexity and increased inhibitory synaptic strength in the cortex80. Behaviourally, the Ser421Ala knock-in mice had deficits in their response to new objects or mice80. These findings indicate that activity-dependent phosphorylation of MECP2 regulates synapse development and function and behavioural responses to environmental stimuli. However, extensive analysis of activity-dependent gene transcription in wild-type and MECP2 Ser421Ala knock-in neurons failed to detect any effect of the Ser421Ala mutation. Thus, it remains to be determined if activity-dependent phosphorylation of MECP2 affects gene transcription and chromatin structure and, if so, how the disruption of these processes contributes to Rett syndrome.
MECP2 has recently been shown to be required for experience-dependent synaptic remodelling of the retinogeniculate synapse85. In Mecp2 knockout mice, the initial development and refinement of the retinogeniculate circuit proceeds without impairment, during a phase of development that does not depend on visual experience. However, during a late stage of development when visual experience is required for modulation and maintenance of the retinogeniculate circuit, the synapses that form on the neurons within this circuit are abnormal in Mecp2 knockout mice, resulting in impaired synaptic plasticity on visual deprivation85. This raises the possibility that during the late phase of development of the retinogeniculate circuit, MECP2 may regulate an experience-dependent gene expression program that controls synaptic remodelling, a program that could potentially be controlled by activity-dependent phosphorylation of MECP2.
There is also evidence that loss of MECP2 changes the balance between inhibition and excitation in the brain, altering the neuronal response to sensory experience. In Mecp2 knockout mice, whole-cell patch clamp recordings in the somatosensory cortex revealed decreased excitatory synaptic drive and increased inhibitory drive onto L5 pyramidal cells86. Hippocampal neurons from Mecp2 knockout mice, both grown in culture and in vivo, have reduced glutamatergic synapse number and density with reduced excitatory synapse strength87. In addition, mice expressing a truncated Mecp2 variant, lacking the C-terminal region, have multiple defects in learning and memory, as well as synaptic plasticity88. Together with the molecular studies of MECP2 function, these electrophysiological experiments suggest that MECP2 is a key regulator of chromatin architecture and gene expression that modulates synaptic development, function and plasticity. It is likely that the dysregulation of at least some of MECP2 functions lead to Rett syndrome and possibly other ASD.
Angelman syndrome
Angelman syndrome is characterized by motor dysfunction, speech impairment, seizures and high prevalence of autism, and is caused by deletions of the maternally inherited allele of chromosomal region 15q11–q13. The loss of UBE3A function in this chromosomal region is by itself sufficient to cause Angelman syndrome. Notably, CNVs that lead to duplications and triplications of the same chromosomal region are associated with ASD. Ube3A expression is up-regulated by neuronal activity, it has therefore been suggested that a loss of experience-dependent induction of the UBE3A mRNA during post-natal development may contribute to Angelman syndrome, and increased activity-dependent UBE3A expression may underlie some forms of ASD89. Support for the idea that fine-tuning the level of UBE3A expression may be important for normal cognitive function comes from the observation that the paternally inherited allele of UBE3A is imprinted, and specifically silenced in the brain. Thus, the level of the gene's expression in the nervous system is kept in check through imprinting and is further regulated by neuronal activity.
Mice that carry a deletion in the maternal copy of Ube3A have impairments in learning and in the LTP form of synaptic plasticity90. Experience-dependent maturation of excitatory circuits in the visual cortex is substantially impaired in these same mice. In addition, sensory experience-regulated neocortical plasticity is defective in Ube3A mutant mice91. Specifically, inhibitory drive onto neocortical pyramidal neurons is impaired owing to defective presynaptic vesicle cycling in interneuron populations92. Excitatory drive onto neocortical pyramidal neurons is also diminished, but to a less severe extent92. These studies suggest that Ube3a protein regulates excitatory–inhibitory balance during brain development and that when the level of expression is perturbed ASD may occur. UBE3A is a member of a family of E3 ubiquitin ligases, which catalyse the ubiquitination of substrates, often leading to the degradation of the ubiquitinated substrate by the proteasome. Notably, one of the best-studied targets of UBE3A is the neuronal activity-regulated protein Arc89, suggesting a possible convergence point for UBE3A, FMRP and TSC mutations in the aetiology of ASD. Studies using mice that carry a deletion in the maternally inherited Ube3A revealed that neurons from these mice express increased levels of Arc protein. In addition, UBE3A was found to mediate the ubiquitination and degradation of Arc89. The increased level of Arc expression in the absence of UBE3A results in decreased AMPA receptor cell surface expression and a decrease in the AMPA receptor-mediated currents89. Thus, the dysregulation of Arc levels and AMPA receptor expression at synapses may contribute to the ASD phenotypes observed in Angelman syndrome and possibly ASD that involve CNVs on chromosome 15.
The studies described here on Angelman syndrome, Timothy syndrome and Rett syndromes suggest that the dysregulation of molecular pathways that control activity-dependent gene expression — including those that control the influx of calcium into neurons (CACNA1C in Timothy syndrome), the regulation of chromatin and transcription (MECP2 in Rett syndrome) and the expression of effector genes that act on synapses (Ube3A in Angelman syndrome) — may be crucial to ASD pathogenesis. Thus, it seems likely that further characterization of the signalling network that controls activity-dependent gene expression in neurons will reveal additional genes that are mutated in ASD.
Future directions
The studies reviewed in this manuscript suggest that dysregulation of activity-dependent signalling networks that control synapse development, function and plasticity may contribute significantly to ASD pathogenesis. A variety of studies now provide evidence that mutations that lead to ASD affect how sensory experience modifies synapses, either through local changes in protein phosphorylation at the synapse, the control of mRNA translation or the regulation of gene transcription. However, further experimentation will be required to provide definitive evidence that disruption of activity-dependent programs in neurons is an underlying cause of ASD. In addition, many questions remain to be addressed regarding the molecular mechanisms by which mutations associated with ASD disrupt the activity-dependent pathways that control synapse development, maturation and function.
It will be important to determine if perturbation of activity-dependent gene regulation contributes to ASD pathology. This could be addressed by evaluation of the effects of subtle mutations, such as knock-in mutations in promoters or enhancers, that specifically disrupt activity-dependent gene transcription. Similarly, it will be useful to determine if dysregulation of activity-dependent splicing or translation of mRNAs that are implicated in ASD have a role in the development the disorder, and if disruption of the activity-dependent phosphorylation of specific synaptic proteins leads to ASD phenotypes. For instance, analysis of knock-in mice that carry subtle mutations that disrupt activity-dependent phosphorylation of specific proteins may prove useful in addressing these issues.
To further advance our understanding of ASD, it will also be crucial to continue to investigate the function of ASD candidate genes, particularly those that are part of the activity-dependent signalling network that controls synapse development and function. In this regard, it is important to keep in mind that although not all ASD candidate genes are part of the activity-dependent signalling network, careful examination may reveal additional ASD genes that regulate various aspects of activity-dependent signalling. For example, ASD-associated mutations may disrupt activity-dependent protein synthesis or degradation at or near the synapse. In addition, ASD-associated mutations could disrupt activity-dependent protein localization within the neuron, such as recruitment to the synapse or protein clustering at the synaptic cleft, or neuronal activity-dependent protein phosphorylation events that control channel conductance properties or protein–protein interactions. ASD-associated mutations could also disrupt routes and dynamics of calcium influx into neurons or affect signalling complexes that transmit calcium-dependent signals to control gene transcription. As new ASD loci are identified, it will be important to determine which loci encode proteins that are regulated by neuronal activity, how neuronal activity affects the function of the protein and if the affect on protein function is relevant to the ASD phenotype. One signalling pathway of significant interest in this regard is Ras–MAPK. Mutations in multiple components of this pathway have recently been found to be associated with ASD, including rare mutations in HRAS, MAP2K2 and SYNGAP1, and with syndromes with ASD features (that have mutations in NF1, MAP2K1, MAP2K2)93,94. As the Ras–MAPK signalling pathway is crucial for activity-regulated protein synthesis, activity-dependent gene transcription and mGluR dependent synaptic plasticity23,25,95, mutations in this pathway that are associated with ASD may be disrupting activity-dependent synaptic development and plasticity by altering activity-regulated mRNA translation or gene transcription.
Understanding the molecular basis of ASD will hopefully allow for rational development of therapies for treating these disorders. Several mouse models of ASD, including those of Rett syndrome96,97, Angelman syndrome98, tuberous sclerosis complex71, and Fragile X23 syndrome, and mouse models of rare mutations in SHANK proteins49, have provided convincing evidence that ASD phenotypes can be improved by genetic or pharmacological interventions, providing hope that it will be possible to develop therapeutics for ASD that target the underlying pathophysiology and provide significant clinical benefit. Defining how each type of ASD-associated mutation disrupts synaptic function may be crucial for the development of effective treatments that target the specific molecular defect that occurs in each disorder.
A more general therapeutic approach might be to target a cellular phenotype, such as the imbalance in excitatory–inhibitory neurotransmission that seems to be common to many ASD. The mGluR5 antagonists that are being evaluated for efficacy in Fragile X syndrome are an example of a therapeutic that has been developed to treat a specific disorder, but which might have more general applicability. Pharmacological screens for small molecules that correct deficits in activity-dependent signalling or excitatory–inhibitory balance may also prove useful. For instance, small molecules that target regulators of phosphorylation- or ubiquitin-mediated signalling in neurons may be able to counter the dysregulation of activity-dependent signalling that occurs in certain forms of ASD. Thus, a better understanding of the role of neuronal activity-dependent signalling in the aetiology of ASD may be useful for both advancing our understanding of the molecular basis of ASD and for the development of new therapeutics for treating this cognitive disorder.
Acknowledgements
M.E.G is supported by NIH grant RO1NS048276 and the Rett Syndrome Research Trust. D.H.E is supported by NIH grant K08MH90306.
Footnotes
Author Information: Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Baird G, et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368:210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- 2.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 2012;4:a009886. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nature Neurosci. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 7–10 use exome and entire genome sequencing to demonstrate the importance of rare mutations in ASD and identify new genetic mutations associated with ASD. These studies demonstrate the ability of new sequencing technologies to significantly advance our understanding of the genetic basis of ASD.
- 11.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 12.Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Wiesel TN. Postnatal development of the visual cortex and the influence of environment. Nature. 1982;299:583–591. doi: 10.1038/299583a0. [DOI] [PubMed] [Google Scholar]
- 15.Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu. Rev. Cell Dev. Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Missler M, Sudhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb. Perspect. Biol. 2012;4:a005694. doi: 10.1101/cshperspect.a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naisbitt S, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 18.Sala C, et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- 19.Sheng M, Kim E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect Biol. 2011;3:a005678. doi: 10.1101/cshperspect.a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luscher C, Malenka RC. NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD) Cold Spring Harb. Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardingham GE, Arnold FJ, Bading H. A calcium microdomain near NMDA receptors: on switch for ERK-dependent synapse-to-nucleus communication. Nature Neurosci. 2001;4:565–566. doi: 10.1038/88380. [DOI] [PubMed] [Google Scholar]
- 22.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu. Rev. Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 25.Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- 26.Flavell SW, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrow EM, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides evidence for the hypothesis that genetic mutations associated with autism may disrupt activity-dependent gene expression programs.
- 35.Chahrour MH, et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 2012;8:e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabuchi K, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Etherton MR, Tabuchi K, Sharma M, Ko J, Sudhof TC. An autism-associated point mutation in the neuroligin cytoplasmic tail selectively impairs AMPA receptor-mediated synaptic transmission in hippocampus. EMBO J. 2011;30:2908–2919. doi: 10.1038/emboj.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl Acad. Sci. USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the crucial role for neuroligins in activity-dependent synapse maturation and how this pathway is dysregulated by ASD-associated mutations in neuroligin.
- 42.Kim J, et al. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc. Natl Acad. Sci. USA. 2008;105:9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YB, et al. Neurexin-neuroligin transsynaptic interaction mediates learning-related synaptic remodeling and long-term facilitation in aplysia. Neuron. 2011;70:468–481. doi: 10.1016/j.neuron.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using the Aplysia model system, this study demonstrates that the neurexin-neuroligin interaction regulates activity-dependent synaptic development and plasticity, a process that is dysregulated by an ASD-associated mutation in neuroligin.
- 44.Thyagarajan A, Ting AY. Imaging activity-dependent regulation of neurexin-neuroligin interactions using trans-synaptic enzymatic biotinylation. Cell. 2010;143:456–469. doi: 10.1016/j.cell.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato D, et al. SHANK1 deletions in males with autism spectrum disorder. Am. J. Hum. Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmeisser MJ, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 49.Won H, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]; References 47–49 report the studies of mouse models of Shank mutations to demonstrate the role of Shanks in regulating excitatory neurotransmission.
- 50.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 51.Bingol B, Sheng M. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Hung AY, Sung CC, Brito IL, Sheng M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS ONE. 2010;5:e9842. doi: 10.1371/journal.pone.0009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee SH, et al. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science. 2008;319:1253–1256. doi: 10.1126/science.1150541. [DOI] [PubMed] [Google Scholar]
- 54.Zalfa F, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 55.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl Acad. Sci. USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J. Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dolen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park S, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J. Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou L, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Harlow EG, et al. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron. 2010;65:385–398. doi: 10.1016/j.neuron.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeiffer BE, et al. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barbosa AC, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc. Natl Acad. Sci. USA. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pulipparacharuvil S, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han JM, Sahin M. TSC1/TSC2 signaling in the CNS. FEBS Lett. 2011;585:973–980. doi: 10.1016/j.febslet.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 69.Ehninger D, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nature Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr. Opin. Neurobiol. 2012:873–879. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 71.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that loss of function of either TSC1–TSC2 or FMRP disrupt activity-regulated mRNA translation and synaptic plasticity in opposite directions.
- 72.Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J. Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ceman S, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 74.Narayanan U, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J. Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhat S, et al. CACNA1C (Ca(v)1.2) in the pathophysiology of psychiatric disease. Prog. Neurobiol. 2012;99:1–14. doi: 10.1016/j.pneurobio.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barrett CF, Tsien RW. The Timothy syndrome mutation differentially affects voltage- and calcium-dependent inactivation of CaV1.2 L-type calcium channels. Proc. Natl Acad. Sci. USA. 2008;105:2157–2162. doi: 10.1073/pnas.0710501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pasca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nature Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cohen S, et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates the importance of activity-dependent regulation of MECP2 in the control of synapse development and behaviour.
- 81.Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 82.Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deng JV, et al. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nature Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Zhong X, Chau KF, Williams EC, Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nature Neurosci. 2011;14:1001–1008. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noutel J, Hong YK, Leu B, Kang E, Chen C. Experience-dependent retinogeniculate synapse remodeling is abnormal in MeCP2-deficient mice. Neuron. 2011;70:35–42. doi: 10.1016/j.neuron.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dani VS, et al. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc. Natl Acad. Sci. USA. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moretti P, et al. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Greer PL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides insight into the molecular mechanisms underlying Angelman syndrome and the role of neuronal activity in regulating Ube3A and its targets.
- 90.Jiang YH, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 91.Yashiro K, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nature Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory Imbalance through neuron type-specific synaptic defects. Neuron. 2012;74:793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kelleher RJ, 3rd, et al. High-throughput sequencing of mGluR signaling pathway genes reveals enrichment of rare variants in autism. PLoS ONE. 2012;7:e35003. doi: 10.1371/journal.pone.0035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/s0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 96.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc. Natl Acad. Sci. USA. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Woerden GM, et al. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nature Neurosci. 2007;10:280–282. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]