Background: LRP1 is a scavenger receptor involved in the clearance of apoptotic cells and myelin vesicles.
Results: Novel ligands for LRP1 were discovered in CNS myelin by affinity purification combined with proteomics.
Conclusion: Some ligands are intracellular proteins, suggesting a function for LRP1 in the clearance of necrotic debris.
Significance: LRP1 mediates the removal of cellular waste and could maintain homeostasis.
Keywords: Lipoprotein-like Receptor (LRP), Multiple Sclerosis, Myelin, Necrosis (Necrotic Death), Phagocytosis, Citrullination
Abstract
In the central nervous system (CNS), fast neuronal signals are facilitated by the oligodendrocyte-produced myelin sheath. Oligodendrocyte turnover or injury generates myelin debris that is usually promptly cleared by phagocytic cells. Failure to remove dying oligodendrocytes leads to accumulation of degraded myelin, which, if recognized by the immune system, may contribute to the development of autoimmunity in diseases such as multiple sclerosis. We recently identified low density lipoprotein receptor-related protein-1 (LRP1) as a novel phagocytic receptor for myelin debris. Here, we report characterization of the LRP1 interactome in CNS myelin. Fusion proteins were designed corresponding to the extracellular ligand-binding domains of LRP1. LRP1 partners were isolated by affinity purification and characterized by mass spectrometry. We report that LRP1 binds intracellular proteins via its extracellular domain and functions as a receptor for necrotic cells. Peptidyl arginine deiminase-2 and cyclic nucleotide phosphodiesterase are novel LRP1 ligands identified in our screen, which interact with full-length LRP1. Furthermore, the extracellular domain of LRP1 is a target of peptidyl arginine deiminase-2-mediated deimination in vitro. We propose that LRP1 functions as a receptor for endocytosis of intracellular components released during cellular damage and necrosis.
Introduction
Multiple sclerosis (MS)2 is an autoimmune disease in which CNS myelin is destroyed and the survival of myelin-producing oligodendrocytes is compromised (1). The etiology of MS is poorly understood, but, in some cases, oligodendrocyte apoptosis is detected without the involvement of immune cell infiltrates (2). Thus, the failure to clear apoptotic cells and cellular debris may initiate inflammation and the autoimmune response characteristic for MS (3).
Low density lipoprotein receptor-related protein-1 (LRP1) is a scavenger receptor with key roles in the phagocytosis of myelin debris (4) and apoptotic cells (5, 6). LRP1 is a member of the LDL receptor gene family, first recognized as a receptor for apolipoprotein E and the serum protease inhibitor α2-macroglobulin (7, 8). To date, >40 different ligands for LRP1 have been identified, including proteases, growth factors, heat shock proteins, extracellular matrix proteins, and foreign toxins (9). LRP1 is a two-chain receptor in which the ligand-binding α chain is entirely extracellular and non-covalently associated with the membrane-spanning β chain (9). Upon binding to LRP1 on the cell surface, LRP1 ligands are internalized, dissociate in acidified endosomes, and are delivered to lysosomes, whereas LRP1 is recycled to the cell surface (10). LRP1 also mediates endocytosis of cell surface receptors (11) and, in this manner, participates in the regulation of cell signaling events (12, 13).
In addition to its function as a phagocytic receptor for myelin, LRP1 also participates in myelin-mediated inhibition of axon formation (14). The goal of this study was to characterize the interactome of LRP1 in myelin. We designed fusion proteins corresponding to the second and fourth clusters of complement-like repeats (CCRII and CCRIV), two major extracellular ligand-binding domains in LRP1 (15). CCRII or CCRIV fusion proteins were incubated with myelin protein extracts in the presence or absence of RAP, an LRP1 ligand binding inhibitor (16), and associated partners were identified by tandem mass spectrometry.
Using this approach, we have identified >70 LRP1 ligands in CNS myelin. Validation studies were performed for two myelin-specific LRP1 ligands: cyclic nucleotide phosphodiesterase (CNP, also called CNPase) and peptidyl arginine deiminase-2 (PAD2). CNP is an enzyme highly expressed in oligodendrocytes, with functions in tubulin polymerization and oligodendrocyte process outgrowth (17). CNP is an abundant component of myelin, accounting for up to 4% of the total protein content (18). Although myelination is normal in CNP knock-out mice, they succumb to axonal loss and neurodegeneration due to impaired communication between cells (19). Furthermore, CNP is a potential auto-antigen in MS, as CNP-reactive T cells can be found in MS patients with active disease (20, 21). Another potential auto-antigen in MS is the myelin basic protein (MBP), which we previously identified as a ligand for LRP1 (4). MBP is a substrate for our second chosen target for validation, enzyme peptidyl arginine deiminase 2, or PAD2 (22). PAD2 catalyzes the conversion of protein arginine residues into citrulline, in a reaction called deimination or citrullination (23). Similar to CNP, PAD2 is a component of the myelin sheath and is the only member of the PAD family expressed in the healthy CNS (23, 24). MS patients contain higher levels of MBP citrullination, which is thought to contribute to MBP degradation and loss of myelin sheath integrity (25–28). Similarly, MBP citrullination and PAD2 activity correlate with disease in animal models of MS (29, 30). Our studies demonstrate that both CNP and PAD2 interact with LRP1 CCRs as well as with the full-length LRP1. Finally, we show that LRP1 can be citrullinated by endogenous PAD2 in vitro and that citrullination decreases the endocytic function of LRP1.
Interestingly, many of the newly identified LRP1 ligands were intracellular proteins, indicating a novel role for LRP1 in the clearance of cellular debris, potentially released during necrosis. We further demonstrate for the first time that LRP1 regulates the removal of necrotic cells and cellular debris using model T cells and oligodendrocytes. Necrotic waste is harmful to surrounding cells and is generated during catastrophic events such as trauma and hypoxia or when the apoptotic cell clearance system is saturated (31). Furthermore, necrotic debris has been shown to initiate a proinflammatory response by macrophages (32). During MS, improper clearance of apoptotic cells and necrotic debris would thus generate an inflammatory environment, which would further fuel disease severity. Taken together, the data presented here suggest that, by mediating cellular and myelin-associated debris removal after cell damage, LRP1 may inhibit the immune response generated by excessive cellular debris at sites of inflammation.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Glutathione S-transferase (GST), GST-RAP, human shed LRP1, and rat LRP1 were purified as described previously (33). The following antibodies were used: 8G1 anti-LRP1 (sc-57353, Santa Cruz Biotechnology), anti-LRP1 C-terminal (L2170, Sigma), anti-LRP1 N-terminal (L2420, Sigma), anti-CNP (5664, Cell Signaling Technologies), CD11b-Alexa Fluor 488 (53-0112, eBioscience), and anti-GRP78 (610978, BD Biosciences). Anti-PAD2 serum was obtained by immunizing PAD2−/− mice with GST-PAD2. Anti-citrulline (modified) detection kit was used to detect protein citrullination (17-347, Millipore). BV2, LRP1-positive (PEA10), and LRP1-deficient (MEF2) fibroblasts were obtained from ATCC and cultured in DMEM high glucose, supplemented with 2 mm l-glutamine, 10% FBS, and 1% penicillin/streptomycin. The RAW 264.7 murine macrophage cell line and Jurkat lymphocyte cell line were obtained from ATCC and cultured in RPMI 1640, supplemented with 2 mm l-glutamine, 10% FBS, and 1% penicillin/streptomycin. The N20.1 oligodendrocyte cell line was cultured in DMEM high glucose/F12 at 34 °C, supplemented with 10% FBS, sodium bicarbonate, 100 μg/ml G418, 20 μg/nl gentamycin, and 1% penicillin/streptomycin. For differentiation into oligodendrocytes, cells were switched to DMEM high glucose/F12, supplemented with 1% FBS, 100 μg/ml G418, and 1% penicillin/streptomycin, and cultures were maintained at 39 °C for 3 to 5 days as described (34).
Cloning, Expression, and Purification of CCRII and CCRIV
CCRII and CCRIV from human LRP1 were cloned into pFuse-rFC2 (Invivogen, San Diego, CA) as described in the literature (15) to generate Fc-fusion proteins. CCRII includes EGF repeat 4 and complement repeats 3 to 10 (LRP1 amino acids 806 to 1184). CCRIV spans complement repeats 21 to 31 (LRP1 amino acids 3332 to 3778). CCRII and CCRIV were expressed in CHO-K1 cells and purified from the culture supernatants on protein A-agarose resin (GE Healthcare).
CCR Pulldowns and Proteomics
Myelin vesicles were purified from mouse brains as described previously (35). Myelin-associated proteins were solubilized in radioimmune precipitation assay buffer, containing 100 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS, supplemented with 1 mm CaCl2 and complete protease inhibitor mixture (Roche Applied Science). Protein extracts (2 mg) were incubated with 1 pm of CCRII, CCRIV, or Fc with or without 20 pm of GST-RAP for 16 h at 4 °C. CCRII, CCRIV, and Fc were recovered by adding protein A-agarose beads for 1 h at 20 °C. After extensive washing with radioimmune precipitation assay buffer, proteins were either digested with trypsin or eluted with SDS sample buffer for SDS-PAGE and immunoblot analysis. Trypsin digestion was performed in the presence of ProteaseMAX surfactant as described by the manufacturer (Promega, Madison, WI). Proteins associated with CCRII, CCRIV, and Fc were identified by LC-MS/MS as described previously (36).
Cloning, Expression, and Purification of PAD2-His, GFP-RAP, and GST-CNP
Murine PAD2 open reading frame (ORF) was cloned into pET-30a(+) vector (Novagen). Human RAP ORF was cloned downstream of GFP into pET-30a(+). pET-30A(+) allows bacterial expression and contains a histidine tag for affinity purification. Murine CNP ORF was cloned into pGEX-2T (GE Healthcare) to allow bacterial expression and GST tag-mediated purification. PAD2-His and GST-CNP were purified by affinity chromatography using the Profinia purification kit and chromatography system (Bio-Rad).
Immobilization Binding Studies
Purified BSA, rat LRP1, or fibronectin (25 nm) (Sigma) were diluted in PBS and absorbed on ELISA plates for 12 h at 4 °C. Wells were blocked for 1 h with PBS containing 1% BSA. PAD2-His (25 nm) was added to the wells for 1 h at 20 °C. The wells were washed, and retained PAD2-His was detected using anti-PAD2 serum, in an ELISA format using ABTS as a colorimetric substrate. Alternatively, purified BSA, GST, GST-CNP, or GST-RAP (350 nm) were diluted in PBS and absorbed on ELISA plates for 12 h at 4 °C. Wells were blocked for 1 h with PBS containing 1% BSA. Human shed LRP1 (50 nm) was added to the wells for 1 h at 20 °C. The wells were washed with PBS, and retained LRP1 was detected with antibody 8G1, in an ELISA format using ABTS as a colorimetric substrate.
Citrullination Assay
RAW 264.7 cell extract and brain protein extract were prepared in ice-cold extraction buffer (50 mm HEPES, 150 mm NaCl, 1% Triton X-100, 10% glycerol) containing complete protease inhibitor mixture without EDTA (Roche Applied Science). Equal amounts of protein extracts were incubated with 10 μg of PAD2-His for 3 h at 37 °C in the presence or absence of 10 mm CaCl2. LRP1 was recovered by adding 10 μg of GST-RAP and glutathione-agarose beads (33). Glutathione-agarose beads were washed with the extraction buffer. Proteins were eluted with SDS sample buffer for SDS-PAGE and immunoblot analysis. For cell surface citrullination, LRP1-expressing and -deficient fibroblasts were treated for 6 h at 37 °C with 150 nm recombinant PAD2 in DMEM containing 0.3% BSA. Cell surface proteins were labeled with biotin and purified as described (36).
GFP-RAP Endocytosis
LRP1-positive (PEA10) and LRP1-deficient (MEF2) fibroblasts were treated for 3 h at 37 °C with 150 nm recombinant PAD2 in DMEM containing 0.3% BSA. After washing, cells were treated with 500 nm of GFP-RAP for 3 h at 37 °C. After washing with PBS, cells were detached with trypsin and washed twice with PBS containing 5% FBS. Cells were analyzed by flow cytometry on a BD FACSCalibur (BD Biosciences). Data were plotted using the FlowJo software (Tree Star).
Phagocytosis of Necrotic Cells
LRP1-positive (PEA10) and LRP1-deficient (MEF2) fibroblasts were labeled with CFSE according to the manufacturer's instructions (Invitrogen). To prepare necrotic cells, Jurkat or N20.1 cells were heat shocked at 55 °C for 20 min and then incubated for 4 h at 37 °C. Necrosis was confirmed by microscopy using trypan blue exclusion, or by flow cytometry using annexin V-FITC and 7-amino-actinomycin D staining, according to manufacturers instructions (eBioscience). Necrotic and live cells were labeled with Cypher5 (10 μm, GE Healthcare) for 20 min at 37 °C in DMEM, and excessive dye was quenched by resuspending the cells in DMEM containing 10% FBS. Necrotic and live cells were then added to the CFSE-labeled fibroblasts in DMEM containing 10% FBS for 2 h at 37 °C (10 to 1). After washing with PBS, cells were detached with trypsin and washed twice with PBS containing 5% FBS. In experiments with BV2 cells, necrotic and live cells were added for 30 min at 37 °C (4:1). BV2 cells were distinguished from N20.1 by staining with CD11b-Alexa Fluor 488 antibody. Cells were analyzed by flow cytometry on a BD FACSCalibur (BD Biosciences). Data were plotted using the FlowJo software (Tree Star).
RESULTS
Preparation of CCRII and CCRIV
To identify LRP1 ligands present in CNS myelin, we prepared two fusion proteins corresponding to CCRII and CCRIV of human LRP1 (Fig. 1A). These two CCRs mediate the binding of LRP1 ligands, with the possible exception of α2-macroglobulin (9, 15). CCRII and CCRIV were expressed as soluble Fc-fusion proteins in CHO-K1 cells (Fig. 1A) and tested by examining binding of GST-RAP in a pulldown assay. RAP binds to LRP1 and blocks the binding of most known LRP1 ligands (9). Equivalent amounts of CCRII, CCRIV, or Fc (control) were incubated with GST-RAP, followed by incubation with glutathione-agarose beads. After washing, samples were separated by SDS-PAGE alongside the purified proteins, and the gel was stained with Coomassie Blue. CCRII and CCRIV, but not Fc alone, bound to GST-RAP (Fig. 1B).
FIGURE 1.
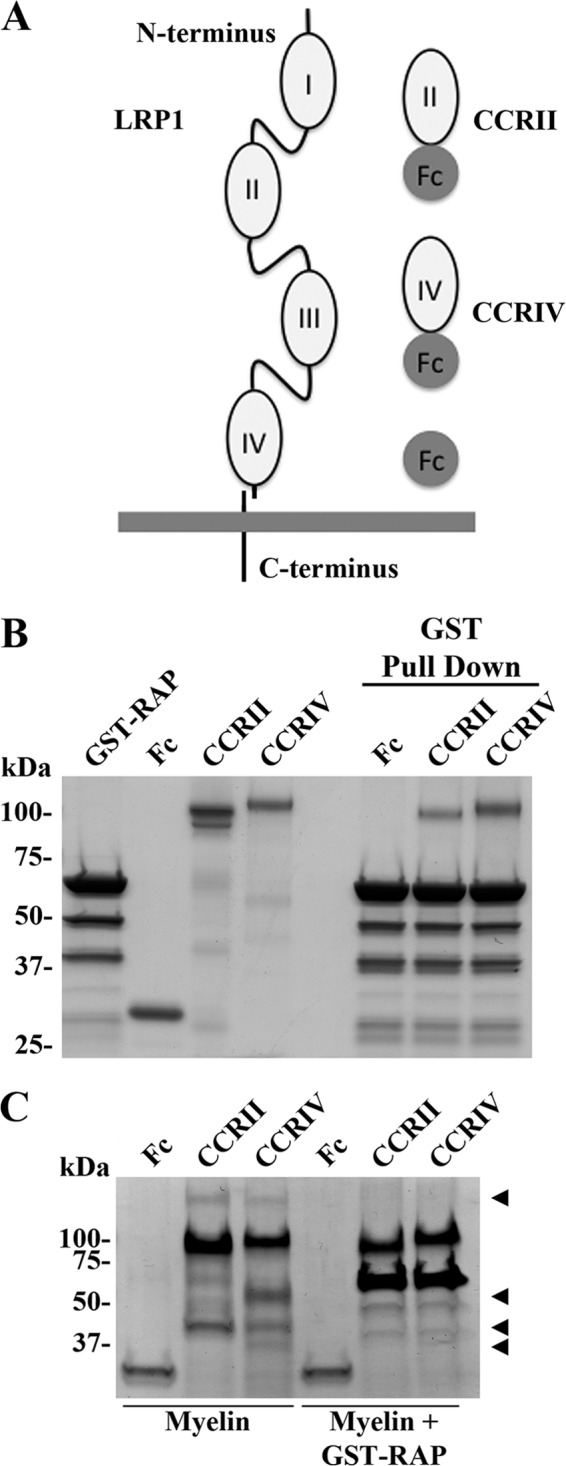
CCRII and CCRIV bind LRP1 ligands. A, schematic representation of LRP1 and CCRII and CCRIV constructs. B, Fc, CCRII, and CCRIV were incubated with GST-RAP as described under “Experimental Procedures,” and GST-RAP was recovered by incubation with glutathione-agarose beads. Samples and purified proteins were analyzed by Coomassie staining. C, Fc, CCRII, and CCRIV were incubated with myelin protein extract in the presence or absence of GST-RAP. Fc and CCRs were recovered by adding protein A-agarose beads. Samples were analyzed by Coomassie staining.
Next, equivalent amounts of CCRII, CCRIV, or Fc were incubated with CNS myelin protein extracts in the presence or absence of excess GST-RAP. Fc fusion proteins were recovered by pull down with protein A-agarose beads. As shown in Fig. 1C, several protein bands are purified from myelin with CCRII and CCRIV, but not with Fc (arrowheads). Treatment with GST-RAP blocks the binding of these proteins to CCRs (Fig. 1C). These results demonstrate that the recombinant Fc-fusion proteins, CCRII and CCRIV, are functional in ligand binding and may be used to identify LRP1 ligands.
Identification of LRP1 Ligands in CNS Myelin by Tandem Mass Spectrometry
CCRII, CCRIV, or Fc were incubated with CNS myelin protein extracts. Associated proteins were recovered by protein A-agarose pulldown as described above. As a control, CCRII, CCRIV, and Fc were preincubated with a 20-fold molar excess of GST-RAP to inhibit ligand binding. Preparations were trypsin-digested, and the resulting peptides were identified by mass spectrometry using an LTQ-Orbitrap. As an additional control, fusion proteins also were analyzed without incubation with myelin extracts to identify ligands derived from CHO-K1 cells, which may co-purify with the CCRs. Using the described method, 72 proteins were identified as ligands for CCRII and CCRIV (supplemental Table 1). As expected, LRP1 peptides were present only when CCRs were analyzed and not Fc. LRP1 peptide abundance was not affected by the presence of myelin protein or by a large excess of GST-RAP, indicating that these reagents did not influence CCR-binding to protein A-agarose. LRP1 partners identified in our screens were separated into two categories: myelin-specific LRP1 ligands (Table 1 and Fig. 2A) and CHO-K1 cell proteins co-purifying with CCRs (Table 2 and Fig. 2B). When CCRII and CCRIV were used to identify myelin-specific LRP1 ligands (Table 1 and Fig. 2A), we confirmed the binding of MBP and myelin-associated glycoprotein, two proteins that we have previously reported as ligands for LRP1 (4, 14). The myelin proteins, CNP and PLP, also were identified as ligands for LRP1 using our screen.
TABLE 1.
Myelin proteins associating with CCRII and CCRIV
The gene products below were identified by tandem mass spectrometry as partners for LRP1 CCRs with (+) or without (−) RAP. Mean spectral counts and S.D. are shown (n = 3).
| Gene name | Fc | CCRII | CCRIV | Fc | CCRII | CCRIV |
|---|---|---|---|---|---|---|
| RAP | − | − | − | + | + | + |
| Cnp | 0 ± 0 | 83.7 ± 0.2 | 73.7 ± 0.3 | 7.6 ± 0.9 | 6.7 ± 1.1 | 11 ± 0.4 |
| Sept7 | 0 ± 0 | 33.7 ± 7 | 34.2 ± 0.37 | 0 ± 0 | 0 ± 0 | 0.7 ± 1.3 |
| Mtap1b | 0 ± 0 | 20.7 ± 0.8 | 27.7 ± 0.7 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Sept2 | 0 ± 0 | 19 ± 0.4 | 17.5 ± 0.2 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Sept8 | 0 ± 0 | 18.2 ± 0.2 | 23.5 ± 0.3 | 0.3 ± 1.7 | 0 ± 0 | 0.2 ± 2 |
| Padi2 | 0 ± 0 | 17.7 ± 0.3 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Mtap1a | 0 ± 0 | 17.2 ± 0.4 | 22.7 ± 0.5 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Plcb1 | 0 ± 0 | 16.7 ± 0.7 | 3.5 ± 1.8 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Sept4 | 0 ± 0 | 13 ± 0.5 | 14.7 ± 0.8 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Glul | 0 ± 0 | 12.2 ± 0.2 | 19 ± 0.2 | 0 ± 0 | 6 ± 0.5 | 5.5 ± 0.2 |
| Pacsin1 | 0 ± 0 | 11 ± 0.1 | 2 ± 0.5 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Rock2 | 0 ± 0 | 10.5 ± 0.8 | 11.5 ± 0.6 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Anln | 0 ± 0 | 8.7 ± 0.6 | 4.5 ± 0.8 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Mbp | 0 ± 0 | 8.5 ± 0.4 | 9.5 ± 0.6 | 1 ± 1.7 | 10 ± 0 | 8.7 ± 0.5 |
| Gnao1 | 0 ± 0 | 8.2 ± 0.3 | 8.2 ± 0.3 | 0 ± 0 | 2 ± 0.5 | 1.25 ± 0.8 |
| Slk | 0 ± 0 | 7.7 ± 0.9 | 11.7 ± 0.6 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Myo18a | 0 ± 0 | 7.5 ± 0.3 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Plp1 | 1.7 ± 0.7 | 7 ± 0.2 | 6 ± 0.2 | 1 ± 0 | 5.7 ± 0.3 | 5 ± 0.3 |
| Ank2 | 0 ± 0 | 6.75 ± 0.3 | 6.2 ± 0.1 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Csrp1 | 0 ± 0 | 6.5 ± 0.7 | 5.5 ± 0.6 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
FIGURE 2.
Identification of LRP1 CCR ligands. A, myelin proteins interacting with LRP1 CCRs were as follows: Cnp, CNP; Sept7, septin-7; Mtap1b, microtubule-associated protein-1b; Sept2, septin-2; Sept8, septin-8; Padi2, PAD2; Mtap1a, microtubule-associated protein-1a; Plcb1, phosphatidylinositol-bisphosphate phosphodiesterase β-1; Sept4, septin-4; Glul, glutamine synthetase. B, endogenous proteins associating with LRP1 CCR were as follows: Glg1, Golgi glycoprotein-1; Lgmn, legumain; Timp2, tissue inhibitor of metalloproteinase-2; Sparc, secreted protein acidic and rich in cysteine; Fhl1, four and a half LIM domains protein 1; Mmp19, matrix metalloproteinase-19; Hpse, heparanase; Mmp9, matrix metalloproteinase-9; Fcho2, FCH domain only protein-2; Col5a2, collagen 5.
TABLE 2.
Proteins co-purifying with CCRII and CCRIV in CHO-K1 cells
The gene products below were identified by tandem mass spectrometry as partners for LRP1 CCRs with (+) or without (−) RAP. Mean spectral counts and S.D. are shown (n = 3).
| Gene name | Fc | CCRII | CCRIV | Fc | CCRII | CCRIV |
|---|---|---|---|---|---|---|
| RAP | − | − | − | + | + | + |
| Lrp1 | 0 ± 0 | 29.2 ± 0.1 | 75 ± 0.2 | 3.3 ± 1.7 | 22.6 ± 0.1 | 58.5 ± 0.3 |
| Glg1 | 0 ± 0 | 68.5 ± 0.1 | 71.7 ± 0.2 | 0.7 ± 1.7 | 3 ± 0.5 | 1 ± 0.8 |
| Lgmn | 0 ± 0 | 0 ± 0 | 15.7 ± 0.3 | 0.7 ± 0.9 | 0 ± 0 | 0 ± 0 |
| Lrpap1 | 0.4 ± 1.4 | 10 ± 0.2 | 15.2 ± 0.1 | 3 ± 1.7 | 71 ± 0.2 | 46.2 ± 0.2 |
| Timp2 | 0 ± 0 | 1 ± 0.8 | 12.2 ± 0.3 | 0.30 ± 1.7 | 0 ± 0 | 0.20 ± 2 |
| Sparc | 0 ± 0 | 16.7 ± 0.1 | 10.2 ± 0.3 | 0 ± 0 | 0.7 ± 0.9 | 0.7 ± 1.3 |
| Fhl1 | 0 ± 0 | 0 ± 0 | 4.7 ± 0.5 | 0 ± 0 | 0 ± 0 | 0.5 ± 2 |
| Mmp19 | 0 ± 0 | 6.7 ± 0.1 | 4.5 ± 0.3 | 0 ± 0 | 1.3 ± 0.4 | 4 ± 0.3 |
| Hpse | 0 ± 0 | 9.7 ± 0.4 | 3 ± 0.3 | 0.3 ± 1.7 | 0.3 ± 1.7 | 0 ± 0 |
| Mmp9 | 0 ± 0 | 3.2 ± 0.5 | 1.2 ± 0.8 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Fcho2 | 0 ± 0 | 2 ± 0 | 1 ± 0.8 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Col5a2 | 0 ± 0 | 5 ± 0.76 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
Surprisingly, myelin-specific CCR partners included many intracellular proteins. We identified members of the septin family (septins 2, 4, 7, and 8) and microtubule-associated proteins, Mtap1a and Mtap1ab, as novel LRP1 ligands (Table 1 and Fig. 2A). As shown in Table 2 and in Fig. 2B, 10 proteins co-purified at significant levels with CCRII and CCRIV when the fusion proteins were expressed in CHO-K1 cells. This list includes known LRP1 ligands such as RAP, matrix metalloproteinase-9, and tissue inhibitor of metalloproteinase 2 (37, 38) together with previously unidentified LRP1 ligands, such as the proteases matrix metalloproteinase-19 and legumain, and the extracellular matrix proteins SPARC and collagen 5 (col5a2). As anticipated, following GST-RAP treatment, the spectral counts for RAP binding to CCRII and CCRIV increased from 10 to 71 and from 15.2 to 46.2, respectively. This result shows that exogenous RAP can efficiently bind to the CCRs.
In both categories of LRP1 ligands, comparable binding to CCRII and CCRIV was most frequently observed. However, we were also able to identify proteins that bind specifically to CCRII or CCRIV, but not both. Collagen 5 (Col5a2), PAD2 or Padi2, and MYO18 (Myo18a) bind only to CCRII, whereas legumain, tissue inhibitor of metalloproteinase 2, and FHL1 (four and a half LIM domains 1) bind only to CCRIV (Fig. 2B). Taken together, our results suggest that the binding sites for a variety of LRP1 ligands reside within a specific domain of LRP1, allowing inhibitor design for targeted disruption of specific LRP1-ligand interactions.
PAD2 and CNP Are Novel LRP1 Ligands in Myelin
To validate the results obtained using proteomics we chose a candidate approach. We selected two myelin-specific proteins, CNP and PAD2, as targets for validation, due to the proposed involvement of these proteins in neurodegeneration and MS. CNP is a major protein component of myelin, an early marker of oligodendrocyte myelination (39), and a potential auto-antigen in MS (20, 21). The proposed function of CNP is to mediate the compaction of myelin by organizing the tubulin network (40). PAD2 is a member of the PAD family of Ca2+-dependent enzymes, which catalyze the post-translational modification of protein arginines, in a process called deimination or citrullination (24). PAD2 expression and the level of citrullination correlate with active disease in both human MS and the animal models of MS (25–30).
We first confirmed binding of PAD2 and CNP to CCRII and CCRIV by immunoblot analysis. As shown in Fig. 3A, PAD2 in whole CNS myelin interacted with CCRII, but not with CCRIV or Fc, confirming the results of our proteomic analysis (Fig. 3A). CNP bound to both CCRII and CCRIV but not the control protein Fc (Fig. 3B). GST-RAP treatment completely inhibited association of both PAD2 and CNP with the CCRs (Fig. 3).
FIGURE 3.

PAD2 and CNP bind to specific CCRs. Purified myelin protein extracts were incubated with Fc, CCRII, and CCRIV in the presence or absence of GST-RAP as described under “Experimental Procedures.” Fc and CCR were recovered with protein A-agarose beads. Samples and myelin extracts were analyzed by immunoblot to detect PAD2 (A) and CNP (B). The vertical line was used to highlight a longer exposure of the same membrane to show expression of PAD2 in myelin extract. Arrows indicate GST-RAP recognized by the anti-PAD2 mouse serum developed against a GST-PAD2 fusion protein.
We next determined whether recombinant PAD2 and CNP bind to full-length LRP1. We expressed PAD2 as a His-tagged fusion protein and CNP as a GST-tagged fusion protein in bacteria, as described under “Experimental Procedures.” Protein preparations were first tested for purity by SDS-PAGE. As shown in Fig. 4, recombinant PAD2 and CNP migrated as single bands with an apparent size of 80 and 60 kDa, respectively, as anticipated (Fig. 4A). Binding of PAD2-His and GST-CNP to LRP1 was then evaluated using an ELISA format. Purified LRP1, fibronectin, and BSA were immobilized in microtiter plates. Recombinant PAD2-His bound to LRP1, but did not bind to purified fibronectin or BSA under the same conditions (Fig. 4B).
FIGURE 4.

PAD2 and CNP interact with full-length LRP1. A, PAD2-His and GST-CNP were expressed as fusion proteins in bacteria and subjected to SDS-PAGE with Coomassie staining. B, BSA, FN (Fibronectin), and LRP1 were adsorbed onto plastic wells and incubated with purified PAD2-His. PAD2 binding to the immobilized phase was detected in an ELISA format, using anti-PAD2 serum, as described under “Experimental Procedures.” ***, p < 0.001. C, BSA, GST, GST-CNP, and GST-RAP were adsorbed onto plastic wells as described under “Experimental Procedures” and incubated with soluble human LRP1. LRP1 binding to the immobilized phase was detected in an ELISA format, using LRP1 α-chain-specific antibody 8G1. ***, p < 0.001.
To study binding of LRP1 to CNP, BSA, GST, GST-CNP, and GST-RAP were immobilized in microtiter plates. Shed LRP1, which was purified from human plasma, was then incubated with the immobilized proteins. Binding of shed LRP1 to GST-CNP wells was significantly increased compared with binding to BSA or GST alone (Fig. 4C). Shed LRP-1 binding to immobilized GST-RAP was used as a positive control.
LRP1 Is Citrullinated by PAD2
Protein citrullination converts positively charged arginine residues into the neutral residue, citrulline, which can substantially alter protein structure and function (23). To test whether LRP1 is a target for PAD2-mediated citrullination, we performed in vitro citrullination assays. RAW 264.7 cell protein extracts were incubated with or without PAD2 in the presence or absence of Ca2+. LRP1 was then recovered by affinity precipitation with GST-RAP, and samples were subjected to immunoblot analysis. As is shown in Fig. 5A, when cell extracts were incubated with PAD2 in the presence of Ca2+, LRP1 citrullination was readily detected. As expected, LRP1 was precipitated under all conditions. Other proteins also were citrullinated by PAD2, as determined by immunoblotting for citrullinated proteins in the RAW 264.7 cell extracts. As a control, we determined that levels of LRP1 and Grp78 were present in equal quantity in the initial cell extracts incubated with PAD2.
FIGURE 5.

LRP1 is a substrate for PAD2-mediated citrullination. A, RAW264.7 cell extracts were incubated with or without purified PAD2 and CaCl2, as described under “Experimental Procedures.” LRP1 was then purified using GST-RAP pulldown. Samples and input were analyzed by immunoblotting (IB) using LRP1 α-chain-specific antibody or with an antibody specific for citrullinated proteins. Grp78 immunoblot was used as a control of protein load. B, brain protein extract was incubated with or without CaCl2, or with CaCl2 and purified PAD2, as described under “Experimental Procedures.” LRP1 was then purified using GST-RAP pulldown. Samples and input were analyzed by immunoblotting using LRP1 α-chain-specific antibody or with an antibody specific for citrullinated proteins.
Next, we examined whether LRP1 can be citrullinated in whole brain extracts (Fig. 5B). Protein extracts from mouse brain were prepared and supplemented with Ca2+ alone, or with recombinant PAD2 and Ca2+. LRP1 was recovered by affinity precipitation with GST-RAP and protein citrullination was examined. As is shown in Fig. 5B, in the absence of added Ca2+, we did not detect citrullination of LRP1. However, when we added Ca2+ to the brain extracts, to activate endogenous PAD, LRP1 citrullination was observed. Because PAD2 is the only member of the PAD family expressed in the healthy CNS, it is reasonable to conclude that endogenous PAD2 citrullinates LRP1 (24). Adding recombinant PAD2 to mouse brain extracts, in the presence of Ca2+, induced robust LRP1 citrullination. (Fig. 5B).
In the CNS extracts, LRP1 was one of many proteins that are citrullinated by PAD2, as determined by immunoblot analysis for citrullinated proteins in protein extracts (Fig. 5B). Next, we sought to determine whether extracellular PAD2 can citrullinate cell surface proteins. LRP1-positive and -deficient fibroblasts were treated with recombinant PAD2 for 6 h at 37 °C. Cell surface proteins were labeled with membrane impermeable biotin and recovered by streptavidin affinity precipitation. Affinity precipitates were then analyzed by immunoblot for the presence of citrullination and LRP1. As is shown in Fig. 6A, PAD2 induces a robust citrullination of cell surface proteins. Furthermore, cell surface citrullination is increased in LRP1 positive cells (Fig. 6A). LRP1 was only detected in LRP1 positive cells, as expected.
FIGURE 6.

Cell surface protein citrullination blocks GFP-RAP endocytosis. A, LRP1-positive or -negative fibroblasts were incubated with or without purified PAD2, as described under “Experimental Procedures.” Cell surface proteins were labeled with membrane impermeable biotin and recovered by streptavidin affinity purification. Samples and input were analyzed by immunoblotting using LRP1 β-chain-specific antibody or with an antibody specific for citrullinated proteins. Tubulin immunoblot (IB) was used as a control of protein load. B, LRP1-positive or -negative fibroblasts were pretreated with or without purified PAD2 and then treated with GFP-RAP for 3 h. After washing, cell-associated GFP fluorescence was analyzed by flow cytometry. Results are normalized to the difference of mean GFP-RAP fluorescence between treated LRP1-positive cells and untreated LRP1-positive cells (n = 4; ***, p < 0.001). ns, not significant.
We further tested whether PAD2-mediated citrullination of cell surface proteins affects the endocytic function of LRP1. LRP1-positive and -deficient fibroblasts, previously treated with recombinant PAD2 for 3 h at 37 °C, were incubated with GFP-RAP for 3 h (Fig. 6B). After extensive washing, GFP fluorescence was assessed by flow cytometry as a measure of LRP1-mediated endocytosis. No fluorescence was detected with LRP1-deficient cells, as expected (Fig. 6B). PAD2 treatment induces a small but significant decrease (12 ± 2%, n = 4) in GFP-RAP association with LRP1-positive cells. Therefore, our data suggests that citrullination of cell surface proteins by PAD2 can decrease the endocytic function of LRP1.
LRP1 Is a Phagocytic Receptor for Necrotic Cells
Multiple LRP1-specific ligands identified in our proteomic screen are myelin-enriched cytoplasmic proteins, including components of the cytoskeleton. We thus hypothesized that LRP1 could be a novel receptor involved in the clearance of necrotic cells and cellular debris. To test this hypothesis, we prepared two types of necrotic cells by heat shock treatment: the T cell-like Jurkat cell line and the oligodendrocyte cell line N20.1 (34). Four hours post induction of necrosis, >95% of the cells are trypan blue-positive, indicating the loss of plasma membrane integrity (data not shown). Furthermore, necrotic Jurkat cells were 95% positive for both annexin-5 and 7-amino-actinomycin D when analyzed by flow cytometry (Fig. 7A). Finally, necrotic N20.1 cells showed ruffling of the plasma membrane when compared with live cells by phase contrast microscopy (Fig. 7B).
FIGURE 7.
LRP1 is a phagocytic receptor for necrotic cells. A, annexin 5/7-amino-actinomycin D (7AAD) staining of live and necrotic Jurkat cells. B, phase contrast microscopy of live and necrotic N20.1 cells. C, CFSE-labeled LRP1 positive or negative fibroblasts were incubated with live or necrotic CypHer-labeled Jurkat cells for 2 h. After washing, fibroblast-associated CypHer fluorescence was analyzed by flow cytometry. D, results are normalized to the difference of CypHer median fluorescence between LRP1-positive cells incubated with necrotic cells and untreated LRP1-positive cells (a representative of two independent experiments is shown; ****, p < 0.0001). E, BV2 cells were incubated with live or necrotic CypHer-labeled N20.1 cells for 30 min. After washing, BV2 were stained with CD11b-Alexa Fluor 488 antibody and BV2-associated CypHer fluorescence was analyzed by flow cytometry. F, results are normalized to the difference of CypHer median fluorescence between GST treated cells incubated with necrotic cells and untreated BV2 cells (a representative of six independent experiments is shown). ***, p < 0.001. ns, not significant.
Necrotic Jurkat cells were labeled with CypHer5, a pH-sensitive fluorescent dye (41), and added to CFSE-labeled LRP1 positive and deficient fibroblasts. After 2 h of incubation at 37 °C, engulfment of necrotic cells was analyzed by flow cytometry. As shown in Fig. 7, LRP1-positive cells are phagocytosing necrotic Jurkat cells more efficiently than LRP1-deficient cells (2.7-fold increase, p < 0.0001, n = 3; Fig. 7, C and D). On the other hand, engulfment of live Jurkat cells was not significantly different between LRP1-positive and -deficient fibroblasts. In a complimentary approach, when LRP1-positive fibroblasts were pretreated with an LRP1 antagonist (GST-RAP), engulfment of necrotic cells was inhibited to the levels observed in LRP1-deficient cells (supplemental Fig. 1A). To next examine the function of LRP1 in the phagocytosis of CNS myelin debris, we repeated necrotic cell internalization experiments using the microglial cell line BV2 as model phagocytes and the oligodendrocyte cell line N20.1 as model necrotic cells (34, 42). When BV2 were pretreated with GST-RAP, phagocytosis of necrotic oligodendrocytes was inhibited by 35% (p < 0.001, n = 6; Fig. 7, E and F), whereas no significant difference was detected with live oligodendrocytes following GST or GST-RAP treatment. Finally, endocytosis of necrotic oligodendrocytes was also inhibited in LRP1-deficient fibroblasts (supplemental Fig. 1B).
Together, our results demonstrate that LRP1 binds intracellular proteins present in myelin. We also describe a new function for LRP1 as a phagocytic receptor for necrotic cells and cellular debris that could be present in the CNS.
DISCUSSION
Proper clearance of dead cells is critical for tissue homeostasis. Defects in apoptotic cell clearance can lead to secondary necrosis and release of cellular components into the extracellular environment, generating an inflammatory stimulus that could lead to the development of autoimmune diseases, including MS (3, 43).
LRP1 was recently reported to function as a phagocytic receptor for apoptotic cells and myelin debris (4–6). Although LRP1 was originally described as a scavenger receptor for extracellular proteins (9), it is now clear that LRP1 function is more complex than just mediating endocytosis (9). LRP1 can regulate cell signaling events and functions in inflammation and neurogenesis (12, 44).
The goal of the present study was to characterize LRP1 ligands present in CNS myelin. The LRP1 ligand-binding domains CCRII and CCRIV were prepared as fusion proteins and used in an affinity-based proteomics screen. Using this approach, we identified >70 ligands for LRP1. These ligands could be separated into two categories based on their cellular localization: extracellular and intracellular proteins. Some of the proteins were identified because of their ability to co-purify with the CCRs during expression and purification (Table 2 and Fig. 2B). This category included known ligands for LRP1, such as RAP, matrix metalloproteinase-9, and tissue inhibitor of metalloproteinase 2 (16, 37, 38). The most abundant LRP1 ligand discovered in the extracellular protein category is a member of the FGF receptor family: Golgi glycoprotein 1 (GLG1)/cysteine-rich fibroblast growth factor receptor/E-selectin ligand 1. GLG1 function remains elusive but studies using GLG1 knock-out mice suggest a role during development as an FGF18 receptor (45). Furthermore, GLG1 is an E-selectin ligand involved in leukocyte extravasation during inflammation (46). By mediating the removal of GLG1 from extracellular spaces, LRP1 may modulate leukocyte migration.
We also identified membrane bound myelin-specific proteins during our screen. We confirmed the interaction of MBP and myelin-associated glycoprotein with LRP1 (Table 1 and supplemental Table 1) (4, 14) and also detected PLP1 as LRP1 ligands (Table 1). The most surprising result we obtained by proteomics screening was the identification of numerous intracellular LRP1 ligands in myelin (Table 1). These included cytoskeleton proteins such as septin 2, 4, 7, and 8; MTAP1a and 1b; and ROCK. MTAP and septins are known components of myelin (47–49). As myelin wraps multiple times around the axon, the formation and maintenance of the myelin sheath is dependent on the cytoskeleton, probably explaining the abundance of cytoskeletal proteins in myelin preparations (50).
We selected two intracellular proteins as candidates for validation as LRP1 binding partners: CNP and PAD2. CNP is a major component of myelin, with a potential role in myelin formation and compaction (40). Clearance of CNP by LRP1 may be critical because it has been identified as a potential auto-antigen in MS (21), and auto-reactive populations of CNP-specific T cells were identified in MS patients with active disease (20). Similarly, PAD2 is a common component of the CNS myelin (24), and its up-regulation is thought to be an early marker of demyelinating disease due to the reduced stability of the citrullinated protein components of the myelin sheath (25). We demonstrate here that LRP1 is a potential regulator of demyelination via interaction with CNP and PAD2. Furthermore, LRP1 can itself be post-translationally modified, at least in vitro, by endogenous PAD2 present in brain extract. We also demonstrate that LRP1 function in endocytosis is regulated by extracellular PAD2-mediated citrullination. Whether PAD2-mediated citrullination affects endocytosis of only a subset, or all LRP1 ligands, remains to be determined.
Finally, we demonstrate here for the first time a new function for LRP1 in clearing necrotic debris. LRP1 has been previously described as a phagocytic receptor for apoptotic cells (5, 6). In this study, we demonstrate that LRP1 function was required for optimal engulfment by both non-professional (fibroblasts) and CNS professional (microglia) phagocytes of necrotic T cells and necrotic oligodendrocytes. Removal of dead and dying oligodendrocytes and infiltrating or resident activated lymphocytes could be of critical importance in neuroinflammatory states such as MS, as a means of removal of potential auto-antigens and danger signals.
In conclusion, our study demonstrates that LRP1 specifically associates with numerous proteins in myelin and is a novel phagocytic receptor for necrotic cells. To our knowledge, we are the first to describe that LRP1 can interact with a variety of cytoplasmic proteins through the extracellular domain. Our results strongly indicate that the function of LRP1 extends beyond regulation of extracellular matrix and cell surface proteins and is consistent with the proposed function of LRP1 in mediating the removal of apoptotic cells and necrotic debris during inflammatory diseases of the CNS, including MS. Further elucidation of the mechanisms by which LRP1 may regulate inflammatory diseases could pave the way for the development of therapeutic treatments, perhaps based on CCRII and CCRIV, which would be utilized to inhibit inflammation and the autoimmune response.
This work was supported by National Institutes of Health Grants R21 NS071347 (to A. G.), R01 HL-060551 (to S. L. G.), and R01 GM085117 and R01 AI067460 (to K. A. M.). This work was also supported by the Philip S. Magaram, Esq., Research Award from the Arthritis Foundation (to S. A.).

This article contains supplemental Table 1 and Fig. 1.
- MS
- multiple sclerosis
- LRP1
- low density lipoprotein receptor related-1
- CNP
- cyclic nucleotide phosphodiesterase
- CCR
- cluster of complement repeats
- PAD2
- peptidyl arginine deiminase 2
- ABTS
- 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid)
- GLG1
- golgi glycoprotein 1
- MBP
- myelin basic protein
- PLP1
- proteolipid protein 1
- CNP
- 2′,3′-cyclic nucleotide 3′ phosphodiesterase
- CFSE
- carboxyfluorescein diacetate succinimidyl ester
- RAP
- receptor associated protein.
REFERENCES
- 1. Compston A., Coles A. (2008) Multiple sclerosis. Lancet 372, 1502–1517 [DOI] [PubMed] [Google Scholar]
- 2. Barnett M. H., Prineas J. W. (2004) Relapsing and remitting multiple sclerosis: pathology of the newly forming lesion. Ann. Neurol. 55, 458–468 [DOI] [PubMed] [Google Scholar]
- 3. Botto M., Dell'Agnola C., Bygrave A. E., Thompson E. M., Cook H. T., Petry F., Loos M., Pandolfi P. P., Walport M. J. (1998) Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 19, 56–59 [DOI] [PubMed] [Google Scholar]
- 4. Gaultier A., Wu X., Le Moan N., Takimoto S., Mukandala G., Akassoglou K., Campana W. M., Gonias S. L. (2009) Low-density lipoprotein receptor-related protein 1 is an essential receptor for myelin phagocytosis. J. Cell Sci. 122, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gardai S. J., McPhillips K. A., Frasch S. C., Janssen W. J., Starefeldt A., Murphy-Ullrich J. E., Bratton D. L., Oldenborg P. A., Michalak M., Henson P. M. (2005) Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123, 321–334 [DOI] [PubMed] [Google Scholar]
- 6. Ogden C. A., deCathelineau A., Hoffmann P. R., Bratton D., Ghebrehiwet B., Fadok V. A., Henson P. M. (2001) C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194, 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., Stanley K. K. (1988) Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 7, 4119–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strickland D. K., Ashcom J. D., Williams S., Burgess W. H., Migliorini M., Argraves W. S. (1990) Sequence identity between the α 2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J. Biol. Chem. 265, 17401–17404 [PubMed] [Google Scholar]
- 9. Lillis A. P., Van Duyn L. B., Murphy-Ullrich J. E., Strickland D. K. (2008) LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol. Rev. 88, 887–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gonias S. L., Wu L., Salicioni A. M. (2004) Low density lipoprotein receptor-related protein: regulation of the plasma membrane proteome. Thromb. Haemost. 91, 1056–1064 [DOI] [PubMed] [Google Scholar]
- 11. Gonias S. L., Gaultier A., Jo M. (2011) Regulation of the urokinase receptor (uPAR) by LDL receptor-related protein-1 (LRP1). Curr. Pharm. Des. 17, 1962–1969 [DOI] [PubMed] [Google Scholar]
- 12. Gaultier A., Arandjelovic S., Niessen S., Overton C. D., Linton M. F., Fazio S., Campana W. M., Cravatt B. F., 3rd, Gonias S. L. (2008) Regulation of tumor necrosis factor receptor-1 and the IKK-NF-κB pathway by LDL receptor-related protein explains the antiinflammatory activity of this receptor. Blood 111, 5316–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zurhove K., Nakajima C., Herz J., Bock H. H., May P. (2008) γ-Secretase limits the inflammatory response through the processing of LRP1. Sci. Signal 1, ra15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stiles T. L., Dickendesher T. L., Gaultier A., Fernandez-Castaneda A., Mantuano E., Giger R. J., Gonias S. L. (2012) LDL Receptor-related Protein-1 is a sialic acid-independent receptor for myelin-associated glycoprotein (MAG) that functions in neurite outgrowth inhibition by MAG and CNS myelin. J. Cell Sci. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neels J. G., van Den Berg B. M., Lookene A., Olivecrona G., Pannekoek H., van Zonneveld A. J. (1999) The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem. 274, 31305–31311 [DOI] [PubMed] [Google Scholar]
- 16. Herz J., Goldstein J. L., Strickland D. K., Ho Y. K., Brown M. S. (1991) 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/α 2-macroglobulin receptor. J. Biol. Chem. 266, 21232–21238 [PubMed] [Google Scholar]
- 17. Lee J., Gravel M., Zhang R., Thibault P., Braun P. E. (2005) Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J. Cell Biol, 170, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozlov G., Lee J., Elias D., Gravel M., Gutierrez P., Ekiel I., Braun P. E., Gehring K. (2003) Structural evidence that brain cyclic nucleotide phosphodiesterase is a member of the 2H phosphodiesterase superfamily. J. Biol. Chem. 278, 46021–46028 [DOI] [PubMed] [Google Scholar]
- 19. Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P. E., Griffiths I. R., Nave K. A. (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 33, 366–374 [DOI] [PubMed] [Google Scholar]
- 20. Muraro P. A., Kalbus M., Afshar G., McFarland H. F., Martin R. (2002) T cell response to 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) in multiple sclerosis patients. J. Neuroimmunol. 130, 233–242 [DOI] [PubMed] [Google Scholar]
- 21. Rösener M., Muraro P. A., Riethmüller A., Kalbus M., Sappler G., Thompson R. J., Lichtenfels R., Sommer N., McFarland H. F., Martin R. (1997) 2′,3′-cyclic nucleotide 3′-phosphodiesterase: a novel candidate autoantigen in demyelinating diseases. J. Neuroimmunol. 75, 28–34 [DOI] [PubMed] [Google Scholar]
- 22. Lamensa J. W., Moscarello M. A. (1993) Deimination of human myelin basic protein by a peptidylarginine deiminase from bovine brain. J. Neurochem. 61, 987–996 [DOI] [PubMed] [Google Scholar]
- 23. Vossenaar E. R., Zendman A. J., van Venrooij W. J., Pruijn G. J. (2003) PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays 25, 1106–1118 [DOI] [PubMed] [Google Scholar]
- 24. Wood D. D., Ackerley C. A., Brand B. v., Zhang L., Raijmakers R., Mastronardi F. G., Moscarello M. A. (2008) Myelin localization of peptidylarginine deiminases 2 and 4: comparison of PAD2 and PAD4 activities. Lab. Invest. 88, 354–364 [DOI] [PubMed] [Google Scholar]
- 25. Moscarello M. A., Wood D. D., Ackerley C., Boulias C. (1994) Myelin in multiple sclerosis is developmentally immature. J. Clin. Invest. 94, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood D. D., Bilbao J. M., O'Connors P., Moscarello M. A. (1996) Acute multiple sclerosis (Marburg type) is associated with developmentally immature myelin basic protein. Ann. Neurol. 40, 18–24 [DOI] [PubMed] [Google Scholar]
- 27. Kim J. K., Mastronardi F. G., Wood D. D., Lubman D. M., Zand R., Moscarello M. A. (2003) Multiple sclerosis: an important role for post-translational modifications of myelin basic protein in pathogenesis. Mol. Cell Proteomics 2, 453–462 [DOI] [PubMed] [Google Scholar]
- 28. Musse A. A., Harauz G. (2007) Molecular “negativity” may underlie multiple sclerosis: Role of the myelin basic protein family in the pathogenesis of MS. Neurobiology of Multiple Sclerosis 79, 149–172 [DOI] [PubMed] [Google Scholar]
- 29. Mastronardi F. G., Noor A., Wood D. D., Paton T., Moscarello M. A. (2007) Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J. Neurosci. Res. 85, 2006–2016 [DOI] [PubMed] [Google Scholar]
- 30. Musse A. A., Li Z., Ackerley C. A., Bienzle D., Lei H., Poma R., Harauz G., Moscarello M. A., Mastronardi F. G. (2008) Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis. Model Mech. 1, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Savill J., Fadok V. (2000) Corpse clearance defines the meaning of cell death. Nature 407, 784–788 [DOI] [PubMed] [Google Scholar]
- 32. Stern M., Savill J., Haslett C. (1996) Human monocyte-derived macrophage phagocytosis of senescent eosinophils undergoing apoptosis. Mediation by α v β 3/CD36/thrombospondin recognition mechanism and lack of phlogistic response. Am. J. Pathol. 149, 911–921 [PMC free article] [PubMed] [Google Scholar]
- 33. Gorovoy M., Gaultier A., Campana W. M., Firestein G. S., Gonias S. L. (2010) Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J. Leukoc. Biol. 88, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Studzinski D. M., Benjamins J. A. (2003) Regulation of CNS glial phenotypes in N20.1 cells. J. Neurosci. Res. 73, 31–41 [DOI] [PubMed] [Google Scholar]
- 35. Norton W. T., Poduslo S. E. (1973) Myelination in rat brain: method of myelin isolation. J. Neurochem. 21, 749–757 [DOI] [PubMed] [Google Scholar]
- 36. Gaultier A., Simon G., Niessen S., Dix M., Takimoto S., Cravatt B. F., 3rd, Gonias S. L. (2010) LDL receptor-related protein 1 regulates the abundance of diverse cell-signaling proteins in the plasma membrane proteome. J. Proteome Res. 9, 6689–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emonard H., Bellon G., Troeberg L., Berton A., Robinet A., Henriet P., Marbaix E., Kirkegaard K., Patthy L., Eeckhout Y., Nagase H., Hornebeck W., Courtoy P. J. (2004) Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2.TIMP-2 complex through a thrombospondin-independent mechanism. J. Biol. Chem. 279, 54944–54951 [DOI] [PubMed] [Google Scholar]
- 38. Hahn-Dantona E., Ruiz J. F., Bornstein P., Strickland D. K. (2001) The low density lipoprotein receptor-related protein modulates levels of matrix metalloproteinase 9 (MMP-9) by mediating its cellular catabolism. J. Biol. Chem. 276, 15498–15503 [DOI] [PubMed] [Google Scholar]
- 39. Gravel M., Peterson J., Yong V. W., Kottis V., Trapp B., Braun P. E. (1996) Overexpression of 2′,3′-cyclic nucleotide 3′-phosphodiesterase in transgenic mice alters oligodendrocyte development and produces aberrant myelination. Mol. Cell Neurosci. 7, 453–466 [DOI] [PubMed] [Google Scholar]
- 40. Bifulco M., Laezza C., Stingo S., Wolff J. (2002) 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc. Natl. Acad. Sci. U.S.A. 99, 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adie E. J., Francis M. J., Davies J., Smith L., Marenghi A., Hather C., Hadingham K., Michael N. P., Milligan G., Game S. (2003) CypHer 5: a generic approach for measuring the activation and trafficking of G protein-coupled receptors in live cells. Assay Drug Dev. Technol. 1, 251–259 [DOI] [PubMed] [Google Scholar]
- 42. Henn A., Lund S., Hedtjam M., Porzgen P., Leist M. (2009) The suitability of BV2 cells as alternative model system for primary microglia cultures or animal experiments of brain inflammation. ALTEX 26, 83–94 [DOI] [PubMed] [Google Scholar]
- 43. Elliott M. R., Ravichandran K. S. (2010) Clearance of apoptotic cells: implications in health and disease. J. Cell Biol. 189, 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mantuano E., Inoue G., Li X., Takahashi K., Gaultier A., Gonias S. L., Campana W. M. (2008) The hemopexin domain of matrix metalloproteinase-9 activates cell signaling and promotes migration of schwann cells by binding to low-density lipoprotein receptor-related protein. J. Neurosci. 28, 11571–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyaoka Y., Tanaka M., Imamura T., Takada S., Miyajima A. (2010) A novel regulatory mechanism for Fgf18 signaling involving cysteine-rich FGF receptor (Cfr) and δ-like protein (Dlk). Development 137, 159–167 [DOI] [PubMed] [Google Scholar]
- 46. Hidalgo A., Peired A. J., Wild M. K., Vestweber D., Frenette P. S. (2007) Complete identification of E-selectin ligands on neutrophils reveals distinct functions of PSGL-1, ESL-1, and CD44. Immunity 26, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garner C. C., Garner A., Huber G., Kozak C., Matus A. (1990) Molecular cloning of microtubule-associated protein 1 (MAP1A) and microtubule-associated protein 5 (MAP1B): identification of distinct genes and their differential expression in developing brain. J. Neurochem. 55, 146–154 [DOI] [PubMed] [Google Scholar]
- 48. Buser A. M., Erne B., Werner H. B., Nave K. A., Schaeren-Wiemers N. (2009) The septin cytoskeleton in myelinating glia. Mol. Cell Neurosci. 40, 156–166 [DOI] [PubMed] [Google Scholar]
- 49. Hall P. A., Russell S. E. (2012) Mammalian septins: dynamic heteromers with roles in cellular morphogenesis and compartmentalization. The Journal of pathology 226, 287–299 [DOI] [PubMed] [Google Scholar]
- 50. Bauer N. G., Richter-Landsberg C., Ffrench-Constant C. (2009) Role of the oligodendroglial cytoskeleton in differentiation and myelination. Glia 57, 1691–1705 [DOI] [PubMed] [Google Scholar]




