Background: DNA-protein cross-links (DPCs) are formed by DNA-damaging agents.
Results: DPCs on the translocating strand but not on the nontranslocating strand block hexameric replicative helicases in a size-dependent manner. Stalled helicases dissociate from DNA with a half-life of 15–36 min.
Conclusion: DPCs on the translocating and nontranslocating strands constitute helicase and polymerase blocks, respectively.
Significance: Reversible and irreversible protein roadblocks may have distinct effects on replisomes.
Keywords: DNA Damage, DNA Enzymes, DNA Helicase, DNA Repair, DNA Replication, DNA-Protein Cross-link
Abstract
DNA-protein cross-links (DPCs) are formed when cells are exposed to various DNA-damaging agents. Because DPCs are extremely large, steric hindrance conferred by DPCs is likely to affect many aspects of DNA transactions. In DNA replication, DPCs are first encountered by the replicative helicase that moves at the head of the replisome. However, little is known about how replicative helicases respond to covalently immobilized protein roadblocks. In the present study we elucidated the effect of DPCs on the DNA unwinding reaction of hexameric replicative helicases in vitro using defined DPC substrates. DPCs on the translocating strand but not on the nontranslocating strand impeded the progression of the helicases including the phage T7 gene 4 protein, simian virus 40 large T antigen, Escherichia coli DnaB protein, and human minichromosome maintenance Mcm467 subcomplex. The impediment varied with the size of the cross-linked proteins, with a threshold size for clearance of 5.0–14.1 kDa. These results indicate that the central channel of the dynamically translocating hexameric ring helicases can accommodate only small proteins and that all of the helicases tested use the steric exclusion mechanism to unwind duplex DNA. These results further suggest that DPCs on the translocating and nontranslocating strands constitute helicase and polymerase blocks, respectively. The helicases stalled by DPC had limited stability and dissociated from DNA with a half-life of 15–36 min. The implications of the results are discussed in relation to the distinct stabilities of replisomes that encounter tight but reversible DNA-protein complexes and irreversible DPC roadblocks.
Introduction
DNA is replicated by the replisome, which is a multiprotein complex comprising replicative DNA helicase, DNA polymerase, and other factors (1–3). The replicative helicases unwind the parental double-stranded DNA (dsDNA) into two single strands, and DNA polymerases synthesize leading and lagging strands in continuous and discontinuous modes, respectively, using the separated strands as templates. The mechanism is well conserved from phages and bacteria to higher organisms (1–4). The replisome proceeds through the barrier of DNA-associated proteins such as nucleosomes and site-specific DNA-binding proteins. The replicative helicases disrupt nucleosomes in eukaryotes, probably with the aid of histone modifications and chaperones (3). They can also unwind DNA bound by DNA-binding proteins with variable efficiencies (5, 6) and dislodge proteins from dsDNA (7). However, the proteins associated with DNA are often covalently trapped on DNA to generate DNA-protein cross-links (DPCs)2 by exposure to endogenous and exogenous DNA-damaging agents (8–11). It has been shown that DPCs inhibit the replication of plasmids (12–14) and possibly chromosomal DNA, indicating that the progression of the replisome is impeded by DPCs in vivo. Although DPCs constitute absolute blocks to several DNA polymerases in vitro (15–17), the impediment of the replisome by DPCs is more closely associated with replicative helicases that unwind DNA at the front of the replication fork. However, little is known about how replicative helicases respond to proteins that are covalently immobilized on DNA.
Replicative helicases such as the phage T7 gene 4 protein (T7gp4), simian virus 40 large T antigen (Tag), and Escherichia coli DnaB protein are characterized by their ring-shaped homohexameric structure with a central channel that accommodates DNA (18). The eukaryotic replicative helicase also assembles into a ring-shaped heterohexamer of minichromosome maintenance (Mcm) proteins 2–7 (19, 20). In addition, a subcomplex comprising Mcm4, Mcm6, and Mcm7 (Mcm467) forms a ring-shaped heterohexamer containing two respective subunits and exhibits a helicase activity in vitro (21–24). Coupled with the hydrolysis of NTP (usually ATP), T7gp4 and DnaB helicases translocate along the lagging template strand with 5′–3′ polarity and disrupt the hydrogen bonds between two strands, whereas Tag and Mcm helicases translocate along the leading template strand with 3′–5′ polarity (18, 19). The central channel of Tag, DnaB, and archaeal Mcm is wide enough to accommodate dsDNA (25–27), and DnaB and Mcm467 can pass over it without unwinding (24, 28).
The unwound strand bearing base lesions such as cyclobutane dimer, 6-4 photoproduct, and thymine glycol passes through the central channel of Tag and DnaB helicases (29, 30). Benzo[a]pyrene-DNA adducts also pass through the central channel of T7gp4, but less efficiently (31). Consequently, the base lesions are delivered to DNA polymerase and inhibit dNTP incorporation or subsequent primer elongation, acting as polymerase blocks. Conversely, a large streptavidin tetramer (60 kDa) acts as a helicase block and strongly (but not completely) inhibits the helicase activity of T7gp4 (32), DnaB (24), archaeal Mcm (33), and yeast Mcm467 (24), when it is tethered via biotin to the translocating strand but not to the nontranslocating strand. However, the association between streptavidin and biotin, albeit very tight, can also be disrupted by certain hexameric replicative helicases including phage T4gp41 and archaeal Mcm (34, 35) and by the nonreplicative Dda helicase (36). Thus, streptavidin tethered to DNA mimics a cross-linked protein, but is not completely equivalent to cross-linked proteins due to its latent dissociating property. In addition, chromatins are rich in small proteins such as histones (11.3–15.3 kDa) and nucleoid proteins (mostly 9.2–19 kDa) (37). Their abundance and close association with DNA make them potential targets of DPC formation. However, to our knowledge, whether such small proteins impede the progression of replicative helicases when they are covalently trapped on DNA has yet to be elucidated.
It is thus of interest to clarify how covalent and irreversible association between proteins and DNA affects the translocation of replicative helicases. In the present study we have assessed the critical size of the DPCs that block the progression of hexameric replicative helicases. To this end we constructed model helicase substrates containing DPCs of various sizes and assayed them for the in vitro unwinding activity of DnaB, T7gp4, Mcm467, and Tag. We show here that DPCs on the translocating strand but not on the nontranslocating strand block the progression of the helicases in a size-dependent manner, with a threshold size of cross-linked proteins for clearance by the helicases of 5.0–14.1 kDa. Furthermore, the stalled helicases by a large DPC dissociate from DNA with a half-life of 15–36 min. This finding is discussed in relation to the stability of the roadblocked replisome.
EXPERIMENTAL PROCEDURES
DNA and Proteins
The oligonucleotide containing oxanine was synthesized chemically as reported (38). Other oligonucleotides with normal compositions were obtained from Tsukuba Oligo Service. Proteins used for DPC formation were adrenomedullin (ADR), parathyroid hormone (PTH), gastric inhibitory polypeptide (GIP), midkine (MID), histone H2A (H2A), T4 endonuclease V (TEV), high mobility group 1,2 (HMG), and human NEIL1 DNA glycosylase (NEI) (Fig. 1A). With the exception of NEI, which was purified as reported (39), these proteins were purchased from Peptide Institute or Roche Diagnostics. T7gp4 and E. coli DnaB were obtained from BioHelix and ProFoldin, respectively. Human Mcm467 complex consisting of Mcm6 and histidine-tagged Mcm4 and Mcm7 was purified from coexpressed High5 cells as reported (40, 41). Tag was purified from recombinant baculovirus-infected Sf27 cells as reported (42). Forked DNA substrates containing DPCs for helicase assays (Fig. 1B) were constructed using methods similar to those described previously (13, 17, 43–45). Briefly, a 25-mer oligonucleotide containing oxanine was ligated enzymatically with dT60 or dT40 using a scaffold DNA. The resulting 85-mer and 65-mer were purified by denaturing PAGE and incubated with a protein (ADR, PTH, or GIP) to form a cross-link between oxanine and a protein. DNA containing a DPC was purified by denaturing PAGE and annealed to a dT-tailed complementary strand. The control substrate containing oxanine was prepared similarly without cross-link reactions. Alternatively, the 85-mer and 65-mer containing oxanine prepared as above were annealed to a dT-tailed complementary strand and incubated with a protein (MID, H2A, TEV, HMG, or NEI) to form a cross-link. DNA containing a DPC was loaded onto SDS-PAGE without heat denaturation, separated, and recovered from the gel by electroelution. Analysis of DPC-containing DNA substrates by denaturing PAGE or SDS-PAGE confirmed that they were free of unmodified DNA.
FIGURE 1.
DPC-containing DNA substrates used for helicase reactions. A, cross-linked proteins, including their abbreviations and sizes. B, structures of forked DNA substrates. The substrates consisted of a 25-mer duplex containing DPC, dT60, and dT40 tails for helicase loading, and a 32P label at the indicated 5′ positions. Proteins were tethered to oxanine (O) in leading and lagging template strands via an amide bond (see also Fig. 4C).
Helicase Reactions
Typically, forked DNA substrates (2 fmol, Fig. 1B) were incubated with DnaB (50 ng, approximately 160 fmol as a hexamer), T7gp4 (50 ng, approximately 130 fmol as a hexamer), Mcm467 (50 ng as total protein, approximately 92 fmol as a hexamer), or Tag (15 ng, approximately 31 fmol as a hexamer) in a helicase reaction buffer (20 μl) at 37 °C for up to 15 min. The helicase reaction buffers had the following compositions: 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 20 mm MgCl2, 5 mm dithiothreitol, 3 mm ATP, and 10% glycerol for DnaB; 35 mm Tris acetate (pH 7.5), 11 mm magnesium acetate, 5 mm dithiothreitol, 2 mm dTTP, and 0.01% Triton X-100 for T7gp4; and 50 mm Tris-HCl (pH 7.0), 20 mm mercaptoethanol, 10 mm magnesium acetate, 10 mm ATP, and 0.5 mg/ml BSA for Mcm467 and Tag. The amount of helicases or incubation time was varied when necessary (indicated in figures). The reaction was terminated by the addition of SDS (final concentration 2%). The sample was mixed with an equal volume of 50% sucrose and separated by 10% native PAGE. After drying the gel, the radioactivities of substrates and unwound products were quantified using a Fuji BAS2000 bioimaging analyzer.
Analysis of DNA-Helicase Complexes
A forked DNA substrate containing NEI-DPC on the translocating strand (2 fmol, Fig. 1B) was incubated with DnaB (50 ng), T7gp4 (50 ng), or Mcm467 (100 ng as total protein) as described for helicase reactions except that Tris-HCl was replaced by Hepes-KOH in buffer to avoid the inactivation of the cross-linker (see below). After 20 min of reaction initiation, a cold control DNA substrate without DPC (100 fmol, 50-fold excess over the DPC substrate) was added to the reaction mixture as a competitor to quench the loading of helicases onto the DPC substrate. The complexes were stabilized every 10 or 20 min by 2 mm Sulfo-EGS (Pierce), which is a bifunctional cross-linker that cross-links between the NEI protein and stalled helicases. After incubation with Sulfo-EGS for 30 min, the sample was mixed with SDS (final concentration 2%) followed by an equal volume of 50% sucrose and then separated by 10% native PAGE. Free DNA and DNA-helicase complexes were quantified as described for the helicase reactions. The half-lives of the DNA-helicase complexes based on an exponential decay model were calculated by regression analysis using DeltaGraph version 5 (Red Rock Software). For comparison, helicase reactions without the competitor were also performed, and DNA-helicase complexes were analyzed similarly.
RESULTS
DPCs on the Translocating Strand Block Helicases in a Size-dependent Manner
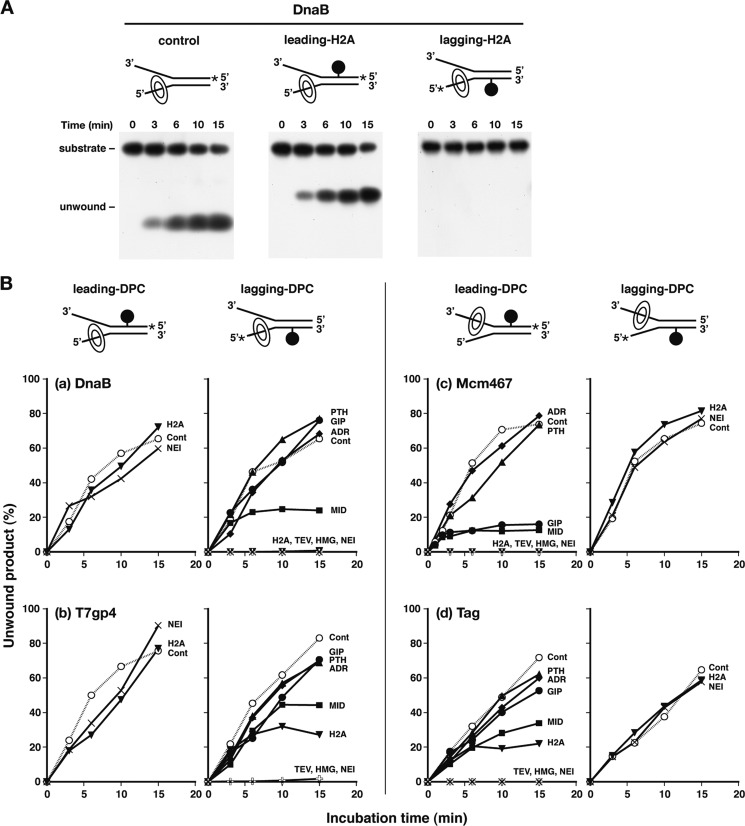
To clarify the effect of DPCs on replicative helicases, we performed unwinding reactions using DPC substrates and DnaB, T7gp4, Mcm467, and Tag. The DNA substrates consisted of a DPC-containing duplex region of 25 bp and two single-stranded dT tails (Fig. 1B). For convenience, the two strands of the helicase substrates were designated leading and lagging template strands according to the polarity of the splayed dT arms. Typical gel data obtained with DnaB and Mcm467 are shown in Fig. 2A and supplemental Fig. 1. The time courses of the formation of unwound products are shown in Fig. 2B. DnaB and T7gp4 helicases unwound the substrate containing NEI, which is the largest cross-linked protein (43.7 kDa) used in this study, at a rate comparable with that for the control substrate (no DPC) when NEI-DPC was placed in the leading template strand (Fig. 2B, a and b, left graphs). This was also the case for H2A-DPC (14.1 kDa). In contrast, Mcm467 and Tag helicases unwound the substrate containing NEI- and H2A-DPCs at a rate comparable with that for the control substrate when NEI- and H2A-DPCs were placed in the lagging template strand (Fig. 2B, c and d, right graphs). Considering the polarities of helicase translocation, these results indicate that DPCs on the nontranslocating strand do not impede the progression of the replicative helicases.
FIGURE 2.
DPCs on the translocating strand but not on the nontranslocating strand block the progression of replicative helicases. A, PAGE analysis of DnaB reaction products. Control and H2A-DPC substrates were incubated with DnaB as described in B, and products were separated by 10% native PAGE. In the substrates shown in the schemes, the black circle, asterisks, and oval rings indicate DPC, a 32P label, and helicase, respectively. B, time courses of DNA unwinding by DnaB (a), T7gp4 (b), Mcm467 (c), and Tag (d) with substrates containing DPCs in the leading template strand (leading-DPC, left graphs) and the lagging template strand (lagging-DPC, right graphs). In a–d, DNA substrates (2 fmol) were incubated with DnaB (50 ng), T7gp4 (50 ng), Mcm467 (50 ng as total protein), or Tag (15 ng) at 37 °C for the indicated times. The percentages of unwound products relative to the total substrates are plotted against the reaction time. The data are the average of two independent experiments. The abbreviations next to the plotted lines indicate cross-linked proteins (see Fig. 1A). The reactions with control templates without DPCs (Cont) are shown by dotted lines and open circles.
Unlike DPCs on the nontranslocating strand, those on the translocating strand blocked the helicases to various extents. With DnaB, smaller cross-linked proteins (ADR, PTH, and GIP; 3.6–5.0 kDa) on the lagging template strand constituted no roadblock, whereas larger ones (H2A, TEV, HMG, and NEI; 14.1–43.7 kDa) were absolute roadblocks (Fig. 2Ba, right graph). Interestingly, the unwinding reaction was biphasic for MID of an intermediate size (6.8 kDa). The reaction for MID initially proceeded at a rate comparable with that of the control and then reached a plateau when approximately 20% of the substrate was unwound. With T7gp4, smaller and larger proteins also constituted no and absolute roadblocks, respectively (Fig. 2Bb, right graph). A biphasic mode of unwinding reactions was observed for MID and H2A, with plateaus of approximately 40 and 30%, respectively. With Mcm467 and Tag, smaller and larger proteins on the leading template strand constituted no and absolute roadblocks, respectively (Fig. 2B, c and d, left graphs). A biphasic mode of unwinding reactions was observed for Mcm467 with GIP and MID and for Tag with MID and H2A.
The correlations between the unwinding efficiency of helicases and the size of cross-linked proteins are shown in Fig. 3. The plots indicate that proteins smaller than 3.7 kDa (ADR and PTH) on the translocating strand do not impede the progression of DnaB, T7gp4, Mcm467, and Tag helicases, whereas those larger than 16.0 kDa (TEV, HMG, and NEI) completely block their progression. Proteins of intermediate sizes (i.e. 5.0–14.1 kDa) differentially block the helicases, probably due to subtle differences in the interaction between the central channel of helicases and cross-linked proteins. According to the data shown in Fig, 3, the critical sizes of DPCs for clearance by helicases are 6.8 kDa for DnaB, 6.8–14.1 kDa for T7gp4, 5.0–6.8 kDa for Mcm467, and 6.8–14.1 kDa for Tag. Although not conclusive, phage and viral helicases (T7gp4 and Tag) appear to have a slightly high clearance limit with respect to the size of DPCs. The yeast Mcm2–7 complex exhibits significant DNA unwinding activity in vitro (46). However, thus far we have observed that the human heterohexameric Mcm2–7 complex exhibits only moderate activity to the control substrate (data not shown), and so it has not been possible to study its activity to DPC substrates.
FIGURE 3.
The critical size of DPCs for clearance by replicative helicases is 5.0–14.1 kDa. The percentages of unwound products relative to the total substrates after a reaction time of 15 min (Fig. 2B) are plotted against the sizes of cross-linked proteins (Fig. 1A). The results with leading- and lagging-DPC substrates are represented by open and closed circles, respectively. The numbers on the graphs indicate the critical sizes of cross-linked proteins (in kDa) for clearance by respective helicases.
The Orientation of Cross-Linked Proteins May Affect the Translocation of Helicases through DPCs
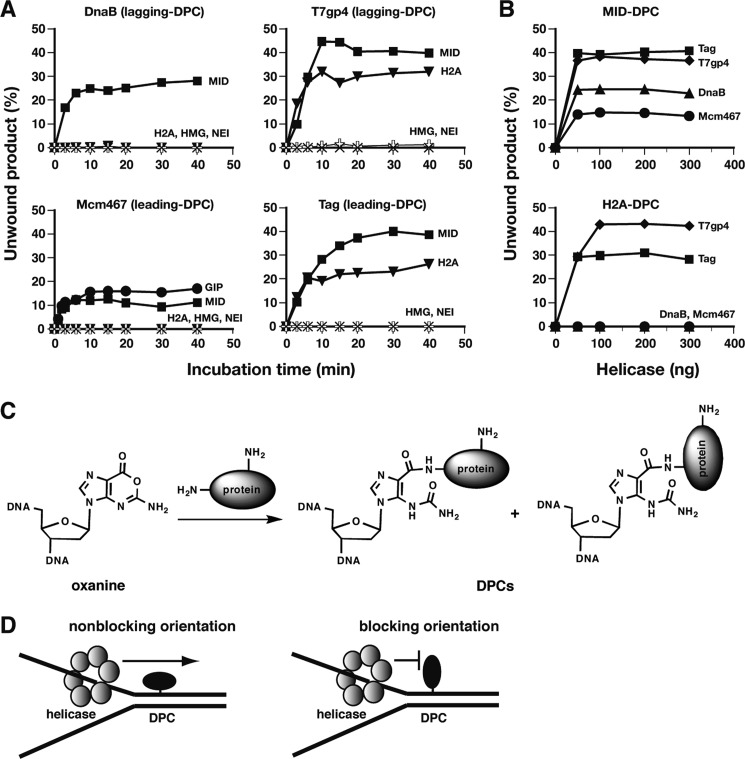
The unwinding reactions occurred in a biphasic mode with a plateau for DPCs of intermediate sizes (Fig. 2B). We investigated whether the plateau of reactions was altered by extending incubation time or by increasing/decreasing the amount of helicases. Extension of the incubation time up to 40 min (originally 15 min in Fig. 2B) had no effect on the plateau level for all helicases (Fig. 4A). The larger cross-linked proteins also remained absolute roadblocks to the helicases. The effect of the amount of helicases was assessed mostly using MID- and H2A-DPCs. The unwinding reactions with MID-DPC under standard reaction conditions were biphasic for all helicases, and those with H2A-DPC were biphasic for T7gp4 and Tag (Fig. 2B). The increase in the amount of helicases up to 300 ng (originally 50 ng for DnaB, T7gp4, and Mcm467, and 15 ng for Tag) did not change the plateau level for MID- and H2A-DPCs (Fig. 4B). Similarly, the decrease in the amount of helicases (1.3–5.4 ng), where the molar ratios of substrate to hexameric helicase were 1:2 for DnaB, T7gp4, and Tag and 1:5 for Mcm467, did not change the plateau level either, although the plateau was reached after prolonged incubation time (30–60 min) due to a decrease in the initial rate of unwinding (supplemental Fig. 2). Taken together, these results indicate that the plateaus of the unwinding reactions were independent of the incubation time and the amount of helicases. The present results further suggest that the DPC that exhibited a biphasic mode of a reaction contains two forms of a cross-linked protein, with one being a block to helicases and the other not being a block. In the present study we used proteins that are rich in lysine (and arginine) to prepare the DPCs. According to a previous study that showed that multiple lysine sites of a protein are modified by the reaction with free 2′-deoxyoxanosine (47), it is possible that the protein in DPC was anchored at different sites in the present study (Fig. 4C). Given that proteins are not completely spherical, one orientation of the anchored protein, but not the other, may result in steric crash to the surface peptide of the helicase central channel (Fig. 4D). We infer from the present results that subtle differences in the orientation of cross-linked proteins may be responsible for the biphasic mode of reactions observed for DPCs of intermediate sizes.
FIGURE 4.
Orientation of cross-linked proteins may affect the translocation of helicases through DPCs. A, effect of prolonged helicase reactions on the yield of unwound products. The helicase reactions were performed as described in Fig. 2B except that the reaction time was extended to 40 min. The percentages of unwound products relative to the total substrates are plotted against the reaction time. The abbreviations next to plotted lines indicate cross-linked proteins (see Fig. 1A). B, effect of the increasing amounts of helicases on the yield of unwound products. The helicase reactions were performed as described in Fig. 2B except that increasing amounts of helicases were used. MID- and H2A-DPCs on the lagging template strand were used for DnaB and T7gp4, whereas those on the leading template strand were used for Mcm467 and Tag. The percentages of unwound products are plotted against the amount of helicases. The data in A and B are based on a single experiment. C, reaction scheme for oxanine and a protein that results in two orientations of cross-linked protein. The amino groups of lysine residues involved in cross-linking are shown by “–NH2.” D, schemes showing that cross-linked proteins in nonblocking and blocking orientations differentially affect the translocation of replicative helicases through DPC.
Stalled Helicases by DPC Dissociate from DNA with a Half-life of 15–36 Min
We analyzed the stability of DNA-helicase complexes when helicases were stalled by DPCs. For this we used NEI-DPC, which was a complete block to helicases when placed on the translocating strand (Fig. 2B). The NEI-DPC substrate was incubated with a helicase, and the loading of helicases onto the DPC substrate was quenched by the addition of a competitor 20 min after initiating the reaction. The remaining DNA-stalled helicase complexes were stabilized by a cross-linker (Sulfo-EGS) and analyzed by native PAGE. Fig. 5A shows the typical gel data for DnaB and Mcm467 without and with the competitor. The DNA-helicase complexes were completely separated from free DNA, although it was not clear whether the trapped complexes contained the complete subunits of helicases. Such complexes were not observed in the present PAGE analysis when the control substrate (no DPC) was used for the reaction or when the complex was not stabilized by cross-linking (data not shown). The quantification of free DNA and the complex for DnaB, T7gp4, and Mcm467 helicases showed that in the absence of the competitor, the amount of DNA-stalled helicase complexes increased with incubation time, indicating the continuous loading of helicases onto free substrates (Fig. 5B). However, adding the competitor DNA at 20 min decreased the amount of DNA-stalled helicase complexes, indicating the dissociation of stalled helicases from DNA. The half-life was evaluated using the data after addition of the competitor by assuming a simple exponential decay of the complex: the half-life was 36 min for DnaB, 15 min for T7gp4, and 33 min for Mcm467 (Fig. 5C). The half-life of DNA-stalled Tag complexes could not be determined due to the nonspecific dsDNA binding activity of Tag (48). With Tag, we observed dT tail-dependent and -independent complex formation when DNA-helicase complexes were trapped by the cross-linker (supplemental Fig. 3). Thus, the trapped complex observed for Tag was probably a mixture of unwinding and nonspecific complexes. Such nonspecific (i.e. dT tail-independent) complex formation was not observed for DnaB, T7gp4, and Mcm467 (supplemental Fig. 3).
FIGURE 5.
Replicative helicases stalled by DPC dissociate from DNA with a half-life of 15–36 min. A, PAGE analysis of DNA-stalled helicase complexes. DNA substrates containing NEI-DPC on the translocating strand (2 fmol) were incubated with DnaB (50 ng) or Mcm467 (100 ng as total protein). For analysis of the dissociation kinetics of stalled helicases, a competitor (100 fmol of cold control DNA substrate without DPC) was added to the reaction mixture at 20 min to quench the loading of helicases onto the DPC substrate. The complexes were stabilized by Sulfo-EGS every 10 or 20 min and analyzed by 10% native PAGE (lower gels for DnaB and Mcm467). Upper gels for DnaB and Mcm467 show PAGE data obtained in the reactions without a competitor. B, changes in the amount of DNA-stalled helicase complexes with incubation time. The percentages of DNA-stalled helicase complexes relative to the total DPC substrates are plotted against the incubation time: filled circles, without a competitor; open circles, with a competitor. The data are means ± S.D. (error bars) based on three or four independent experiments. C, stabilities of DNA-stalled helicase complexes. The half-lives were evaluated from the decay of complexes after addition of the competitor (open circles in B) by assuming exponential decay.
DISCUSSION
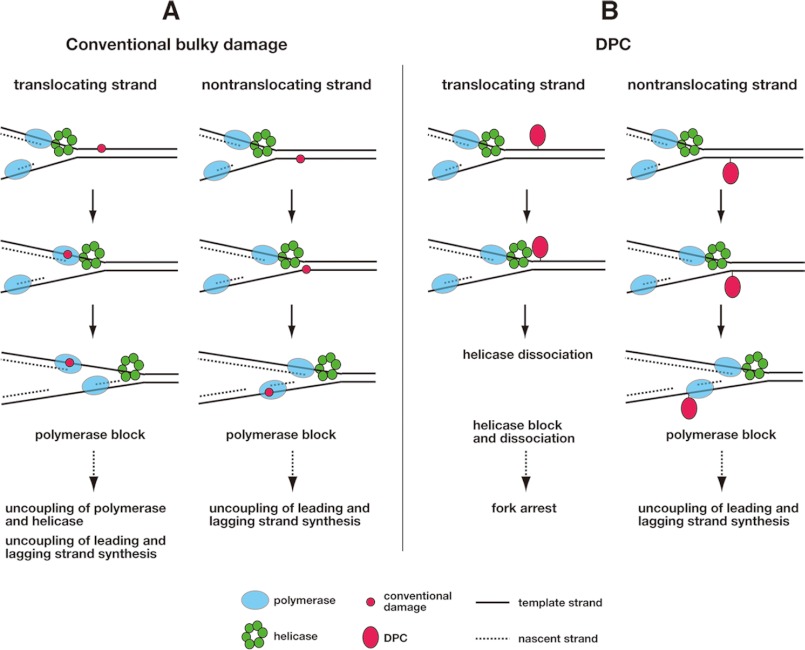
The present study has shown that DPCs on the translocating strand impede the progression of hexameric replicative helicases in a size-dependent manner (Fig. 3). The critical size of cross-linked proteins for clearance by helicases was 5.0–14.1 kDa. These data indicate that the central channel of the dynamically translocating hexameric helicases can accommodate only relatively small proteins. Thus, small chromatin proteins such as histones (11.3–15.3 kDa) and nucleoid proteins (mostly 9.2–19 kDa) will severely block the helicases when they are cross-linked to the translocating strand. Although DPCs constitute strong blocks to several DNA polymerases (15–17), the present results highlight an alternative mechanism of replisome blockage that involves the inhibition of replicative helicases that unwind DNA at the front of the replication fork. In addition, the present results suggest the distinct fates of replisomes upon encountering conventional bulky damage and large DPCs. Conventional bulky damage both on the translocating and nontranslocating strands are cleared by helicases and arrest DNA polymerase (Fig. 6A). This can further lead to functional uncoupling of polymerase and helicase as well as that of leading and lagging polymerases. In eukaryotes functional uncoupling of polymerase and helicase activates a checkpoint kinase ATR that directs the DNA damage response (49). DPCs on the translocating strand block the helicase, immediately halting leading and lagging strand synthesis (Fig. 6B). This will preclude functional uncoupling of polymerase and helicase and of leading and lagging polymerases. In contrast, DPCs on the nontranslocating strand do not block the helicase and act like conventional bulky damage. Accordingly, the mechanism underlying stalled fork processing and the concurrent events of damage signaling may differ significantly for DPCs on the translocating and nontranslocating strands.
FIGURE 6.
Possible fates of replisomes that encounter conventional bulky damage and DPCs. A, fates of the replisome that encounters conventional bulky damage on the translocating (left) and nontranslocating (right) strands of the helicase. B, fates of the replisome that encounters a DPC on the translocating (left) and nontranslocating (right) strands of the helicase. Note that the scheme was drawn for eukaryotic replication, where the replicative helicase translocates on the leading template strand.
Aside from DNA replication, DPCs adversely affect other aspects of DNA transactions such as repair and transcription (10), both of which involve unwinding of dsDNA. It has been shown that DPCs impede the progression of the UvrD helicase involved in nucleotide excision repair only when placed on the translocating strand (14). The progression of T7 RNA polymerase that transiently disrupts dsDNA for transcription is also blocked by DPCs only when DPCs are placed on the transcribed strand (17). It follows that replicative helicases (DnaB, T7gp4, Mcm467, and Tag), UvrD helicase, and T7 RNA polymerase are blocked by DPCs on the translocating or transcribed strand, but not by those on the nontranslocating or nontranscribed strand. Therefore, these enzymes with a DNA unwinding activity will respond similarly upon head-on and lateral collisions with DPCs.
Several models of DNA unwinding have been proposed for hexameric helicases. In the steric exclusion model, the hexameric helicase encircles and translocates along one strand and sterically excludes the complementary strand from the central channel (19, 50). In other models, dsDNA is pumped into the central channel of the helicase, and a pair of hexameric helicases unwinds the dsDNA by either twisting it apart or extruding it through channels in the complex (19, 50). Our results showing the differential effects of DPCs in the translocating and nontranslocating strands (Figs. 2 and 3) are consistent with the steric exclusion model and reinforce the conclusion obtained in the previous work using streptavidin-biotin roadblocks (24, 32, 33). If dsDNA was to be pumped into the central channel of the helicase, DPCs on both strands would impede the progression of the helicase. However, this was not the case for all helicases tested. The eukaryotic replicative helicase contains Cdc45, heterohexameric Mcm2–7, and GINS (CMG complex) (19, 51). It has been recently shown that Xenopus CMG uses the steric exclusion mechanism for DNA unwinding during translocation (52). Thus, yeast Mcm467 (24), human Mcm467 (this study), and Xenopus CMG (52) share the same DNA unwinding mechanism during translocation, although the Mcm467 subcomplex and CMG have significantly different subunit integrities. It has been proposed that Tag uses the dsDNA pump mechanism (53). In this mechanism, Tag forms a double hexamer complex and pumps dsDNA into the central channel where dsDNA is separated. The separated single-stranded DNA is extruded from the side channel. However, the present result with Tag argues against this model and favors the steric exclusion mechanism, because DPCs on the nontranslocating strand did not inhibit the unwinding reaction (Fig. 2Bd, right graph). In this context it is noteworthy that the papillomavirus replicative helicase E1, whose structural and biochemical properties are similar to those of Tag, is thought to use the steric exclusion mechanism (54).
In the present study we determined the in vitro stability of replicative helicases stalled by NEI-DPC. Stalled DnaB, T7gp4, and Mcm467 helicases exhibited limited stability and dissociated from DNA with a half-life of 15–36 min (Fig. 5C). The following contrasting results were reported regarding the stability of E. coli replisomes: the replisome blocked by an array of repressor-operator complexes loses the ability to continue replication with a half-life of 4–6 min in vitro (55), whereas it retains the ability to resume replication upon removal of the block for several hours in vivo (56). The dissociation of stalled DnaB from DNA observed in the present study accounts at least in part for the inactivation of the replisome in vitro, although the in vitro half-lives of stalled replisome (4–6 min) and DnaB (36 min) differ considerably. The inactivation of the replisome due to loss of DnaB also seems to be consistent with the observations that reactivation of stalled replication fork requires reloading of DnaB (or replication machinery) via the PriA helicase in E. coli (57) and that the priA mutant is hypersensitive to DPC-inducing agents (58). However, further studies are necessary to account for the discrepancy between the in vitro and in vivo stabilities of roadblocked E. coli replisomes (see also below). In yeast, it has been shown that the replisome stalled by tight (but reversible) DNA-protein complexes is stable in vivo and that DNA synthesis continues through the barriers after a transient pause (approximately 30 min) (59, 60). Thus, the CMG complex is likely to be retained in the stalled replisome in yeast cells. In contrast, a recent study of in vitro replication of plasmids with Xenopus egg extracts has shown that Mcm7 (a component of CMG complex) dissociates from DNA with an approximate half-life of 10 min when the progression of replisome is blocked by an interstrand cross-link (52). In keeping with this observation (52), we found that human Mcm467 stalled by NEI-DPC dissociates from DNA with a half-life of 33 min. Regarding the apparent difference in the stability of replicative helicases in vivo and in vitro, it should be pointed out that tight but reversible DNA-protein complexes are used in in vivo studies, whereas irreversible roadblocks (interstrand cross-links and DPCs) are used in in vitro studies. Accordingly, the replisome could proceed by gradually disrupting reversible protein roadblocks in cells while retaining the helicase in the replisome. However, this does not occur if the replisome is completely arrested by irreversible roadblocks such as interstrand cross-links and DPCs. Therefore, it is tempting to speculate that replicative helicases (or replisome) exhibit significantly different stabilities to reversible and irreversible roadblocks. It is also possible that eukaryotic cells possess a factor that disrupts reversible protein roadblocks to assist replicative helicases or that actively retains the replicative helicase in the stalled replisome (59, 61).
Acknowledgments
We thank Minako Takuwa for technical assistance and the members of the Ide laboratory for helpful discussions and comments on the manuscript.
This work was supported in part by a grant-in-aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H. I.), a grant-in-aid for Young Scientists (B) from the Japan Society for the Promotion of Science (to T. N.), and a Marine Biomaterials Research Center grant from the Marine Biotechnology Program funded by the Ministry of Land, Transport, and Maritime Affairs, Korea (to S. P. P.).

This article contains supplemental Figs. 1–3.
- DPC
- DNA-protein cross-link
- ADR
- adrenomedullin
- CMG
- replicative helicase containing Cdc45, Mcm4–7, and GINS
- GIP
- gastric inhibitory polypeptide
- H2A
- histone H2A
- HMG
- high mobility group
- Mcm
- minichromosome maintenance
- Mcm467
- Mcm subcomplex containing Mcm4, Mcm6, and Mcm7
- MID
- midkine
- NEI
- NEIL1 DNA glycosylase
- PTH
- parathyroid hormone
- Sulfo-EGS
- ethylene glycol bis(sulfosuccinimidylsuccinate)
- T7gp4
- T7 gene 4 protein
- Tag
- simian virus 40 large T antigen
- TEV
- T4 endonuclease V.
REFERENCES
- 1. Johnson A., O'Donnell M. (2005) Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74, 283–315 [DOI] [PubMed] [Google Scholar]
- 2. Hamdan S. M., Richardson C. C. (2009) Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 3. Alabert C., Groth A. (2012) Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167 [DOI] [PubMed] [Google Scholar]
- 4. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79, 89–130 [DOI] [PubMed] [Google Scholar]
- 5. Yancey-Wrona J. E., Matson S. W. (1992) Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 20, 6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shin J. H., Santangelo T. J., Xie Y., Reeve J. N., Kelman Z. (2007) Archaeal minichromosome maintenance (MCM) helicase can unwind DNA bound by archaeal histones and transcription factors. J. Biol. Chem. 282, 4908–4915 [DOI] [PubMed] [Google Scholar]
- 7. Kaplan D. L., O'Donnell M. (2002) DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell 10, 647–657 [DOI] [PubMed] [Google Scholar]
- 8. Barker S., Weinfeld M., Murray D. (2005) DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 589, 111–135 [DOI] [PubMed] [Google Scholar]
- 9. Pommier Y. (2009) DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 109, 2894–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ide H., Shoulkamy M. I., Nakano T., Miyamoto-Matsubara M., Salem A. M. (2011) Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 711, 113–122 [DOI] [PubMed] [Google Scholar]
- 11. Shoulkamy M. I., Nakano T., Ohshima M., Hirayama R., Uzawa A., Furusawa Y., Ide H. (2012) Detection of DNA-protein crosslinks (DPCs) by novel direct fluorescence labeling methods: distinct stabilities of aldehyde and radiation-induced DPCs. Nucleic Acids Res. 40, e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuo H. K., Griffith J. D., Kreuzer K. N. (2007) 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 67, 8248–8254 [DOI] [PubMed] [Google Scholar]
- 13. Nakano T., Morishita S., Katafuchi A., Matsubara M., Horikawa Y., Terato H., Salem A. M., Izumi S., Pack S. P., Makino K., Ide H. (2007) Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein cross-links. Mol. Cell 28, 147–158 [DOI] [PubMed] [Google Scholar]
- 14. Kumari A., Minko I. G., Smith R. L., Lloyd R. S., McCullough A. K. (2010) Modulation of UvrD helicase activity by covalent DNA-protein cross-links. J. Biol. Chem. 285, 21313–21322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Novakova O., Kasparkova J., Malina J., Natile G., Brabec V. (2003) DNA-protein cross-linking by trans-[PtCl2(E-iminoether)2]: a concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 31, 6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chválová K., Brabec V., Kaspárková J. (2007) Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 35, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakano T., Ouchi R., Kawazoe J., Pack S. P., Makino K., Ide H. (2012) T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error-prone transcription. J. Biol. Chem. 287, 6562–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patel S. S., Picha K. M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 [DOI] [PubMed] [Google Scholar]
- 19. Bochman M. L., Schwacha A. (2009) The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 73, 652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costa A., Onesti S. (2009) Structural biology of MCM helicases. Crit. Rev. Biochem. Mol. Biol. 44, 326–342 [DOI] [PubMed] [Google Scholar]
- 21. Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 272, 24508–24513 [DOI] [PubMed] [Google Scholar]
- 22. Sato M., Gotow T., You Z., Komamura-Kohno Y., Uchiyama Y., Yabuta N., Nojima H., Ishimi Y. (2000) Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol. 300, 421–431 [DOI] [PubMed] [Google Scholar]
- 23. Lee J. K., Hurwitz J. (2000) Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 275, 18871–18878 [DOI] [PubMed] [Google Scholar]
- 24. Kaplan D. L., Davey M. J., O'Donnell M. (2003) Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 278, 49171–49182 [DOI] [PubMed] [Google Scholar]
- 25. Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J. A., Fanning E., Jochimiak A., Szakonyi G., Chen X. S. (2003) Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423, 512–518 [DOI] [PubMed] [Google Scholar]
- 26. Bailey S., Eliason W. K., Steitz T. A. (2007) Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318, 459–463 [DOI] [PubMed] [Google Scholar]
- 27. Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., Chen X. S. (2003) The structure and function of MCM from archaeal M. thermoautotrophicum. Nat. Struct. Biol. 10, 160–167 [DOI] [PubMed] [Google Scholar]
- 28. Kaplan D. L. (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol. 301, 285–299 [DOI] [PubMed] [Google Scholar]
- 29. Veaute X., Mari-Giglia G., Lawrence C. W., Sarasin A. (2000) UV lesions located on the leading strand inhibit DNA replication but do not inhibit SV40 T-antigen helicase activity. Mutat. Res. 459, 19–28 [DOI] [PubMed] [Google Scholar]
- 30. Suhasini A. N., Sommers J. A., Mason A. C., Voloshin O. N., Camerini-Otero R. D., Wold M. S., Brosh R. M., Jr. (2009) FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J. Biol. Chem. 284, 18458–18470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yong Y., Romano L. J. (1996) Benzo[a]pyrene-DNA adducts inhibit the DNA helicase activity of the bacteriophage T7 gene 4 protein. Chem. Res. Toxicol. 9, 179–187 [DOI] [PubMed] [Google Scholar]
- 32. Hacker K. J., Johnson K. A. (1997) A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry 36, 14080–14087 [DOI] [PubMed] [Google Scholar]
- 33. Graham B. W., Schauer G. D., Leuba S. H., Trakselis M. A. (2011) Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 39, 6585–6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris P. D., Raney K. D. (1999) DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry 38, 5164–5171 [DOI] [PubMed] [Google Scholar]
- 35. Shin J. H., Jiang Y., Grabowski B., Hurwitz J., Kelman Z. (2003) Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J. Biol. Chem. 278, 49053–49062 [DOI] [PubMed] [Google Scholar]
- 36. Byrd A. K., Raney K. D. (2004) Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 11, 531–538 [DOI] [PubMed] [Google Scholar]
- 37. Azam T. A., Ishihama A. (1999) Twelve species of the nucleoid-associated protein from Escherichia coli: sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274, 33105–33113 [DOI] [PubMed] [Google Scholar]
- 38. Pack S. P., Nonogawa M., Kodaki T., Makino K. (2005) Chemical synthesis and thermodynamic characterization of oxanine-containing oligodeoxynucleotides. Nucleic Acids Res. 33, 5771–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katafuchi A., Nakano T., Masaoka A., Terato H., Iwai S., Hanaoka F., Ide H. (2004) Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 279, 14464–14471 [DOI] [PubMed] [Google Scholar]
- 40. Numata Y., Ishihara S., Hasegawa N., Nozaki N., Ishimi Y. (2010) Interaction of human MCM2–7 proteins with TIM, TIPIN, and Rb. J. Biochem. 147, 917–927 [DOI] [PubMed] [Google Scholar]
- 41. Ishimi Y., Komamura-Kohno Y., Kwon H. J., Yamada K., Nakanishi M. (2003) Identification of MCM4 as a target of the DNA replication block checkpoint system. J. Biol. Chem. 278, 24644–24650 [DOI] [PubMed] [Google Scholar]
- 42. Ishimi Y., Matsumoto K. (1993) Model system for DNA replication of a plasmid DNA containing the autonomously replicating sequence from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 90, 5399–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakano T., Katafuchi A., Matsubara M., Terato H., Tsuboi T., Masuda T., Tatsumoto T., Pack S. P., Makino K., Croteau D. L., Van Houten B., Iijima K., Tauchi H., Ide H. (2009) Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 284, 27065–27076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakano T., Katafuchi A., Shimizu R., Terato H., Suzuki T., Tauchi H., Makino K., Skorvaga M., Van Houten B., Ide H. (2005) Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 33, 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., Ide H. (2003) DNA-protein cross-link formation mediated by oxanine: a novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 278, 25264–25272 [DOI] [PubMed] [Google Scholar]
- 46. Bochman M. L., Schwacha A. (2008) The Mcm2–7 complex has in vitro helicase activity. Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 47. Chen H. J., Chiu W. L., Lin W. P., Yang S. S. (2008) Investigation of DNA-protein cross-link formation between lysozyme and oxanine by mass spectrometry. ChemBioChem 9, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 48. Jiao J., Simmons D. T. (2003) Nonspecific double-stranded DNA binding activity of simian virus 40 large T antigen is involved in melting and unwinding of the origin. J. Virol. 77, 12720–12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cimprich K. A., Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takahashi T. S., Wigley D. B., Walter J. C. (2005) Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 30, 437–444 [DOI] [PubMed] [Google Scholar]
- 51. Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu Y. V., Yardimci H., Long D. T., Ho T. V., Guainazzi A., Bermudez V. P., Hurwitz J., van Oijen A., Schärer O. D., Walter J. C. (2011) Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S. (2004) Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60 [DOI] [PubMed] [Google Scholar]
- 54. Enemark E. J., Joshua-Tor L. (2006) Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 [DOI] [PubMed] [Google Scholar]
- 55. McGlynn P., Guy C. P. (2008) Replication forks blocked by protein-DNA complexes have limited stability in vitro. J. Mol. Biol. 381, 249–255 [DOI] [PubMed] [Google Scholar]
- 56. Possoz C., Filipe S. R., Grainge I., Sherratt D. J. (2006) Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 25, 2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gabbai C. B., Marians K. J. (2010) Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair 9, 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Salem A. M., Nakano T., Takuwa M., Matoba N., Tsuboi T., Terato H., Yamamoto K., Yamada M., Nohmi T., Ide H. (2009) Genetic analysis of repair and damage tolerance mechanisms for DNA-protein cross-links in Escherichia coli. J. Bacteriol. 191, 5657–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Calzada A., Hodgson B., Kanemaki M., Bueno A., Labib K. (2005) Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19, 1905–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Labib K., Hodgson B. (2007) Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 8, 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., Zakian V. A. (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12, 1525–1536 [DOI] [PubMed] [Google Scholar]