Background: The m-AAA protease dislocates transmembrane segments from the mitochondrial inner membrane.
Results: The presence of the m-AAA protease increases the hydrophobicity required for a transmembrane segment to remain in the membrane.
Conclusion: The hydrophobicity thresholds for transmembrane segment retention in the mitochondrial inner membrane differ with or without the m-AAA protease.
Significance: Retention of a transmembrane domain in the inner membrane depends on recognition by the m-AAA protease.
Keywords: Membrane Proteins, Membrane Transport, Membrane Turnover, mitochondria, Yeast, Ccp1, TIM23 Complex, m-AAA Protease
Abstract
Sorting of mitochondrial inner membrane proteins is a complex process in which translocons and proteases function in a concerted way. Many inner membrane proteins insert into the membrane via the TIM23 translocon, and some are then further acted upon by the mitochondrial m-AAA protease, a molecular motor capable of dislocating proteins from the inner membrane. This raises the possibility that the threshold hydrophobicity for the retention of transmembrane segments in the inner membrane is different depending on whether they belong to membrane proteins that are m-AAA protease substrates or not. Here, using model transmembrane segments engineered into m-AAA protease-dependent proteins, we show that the threshold hydrophobicity for membrane retention measured in yeast cells in the absence of a functional m-AAA protease is markedly lower than that measured in its presence. Whether a given hydrophobic segment in a mitochondrial inner membrane protein will ultimately form a transmembrane helix may therefore depend on whether or not it will be exposed to the pulling force exerted by the m-AAA protease during biogenesis.
Introduction
Nuclearly encoded mitochondrial inner membrane proteins are imported into the organelle through the TOM (transporter outer membrane) complex in the outer mitochondrial membrane and engage either the TIM23 or the related TIM22 translocon in the inner membrane. Some TIM23-dependent inner membrane proteins follow a “conservative sorting” pathway, in which they are first fully translocated into the matrix and then inserted into the inner membrane from the matrix side in a process that depends on the Oxa1 translocon (1, 2). Other proteins use a “stop transfer” mechanism in which the transmembrane segments exit laterally from the TIM23 translocon and integrate into the lipid bilayer (2). Finally, the mitochondrial m-AAA protease can dislocate substrate proteins from the inner membrane in an ATP-dependent manner.
Given these different pathways for the insertion and retention of transmembrane segments in the inner membrane, a key goal is to understand the sequence characteristics that distinguish between transmembrane segments that do and those that do not insert into the membrane during passage through the TIM23 translocon and, furthermore, to determine to what extent subsequent dislocation by the m-AAA protease affects membrane retention of different transmembrane segments.
In a recent study (3), we replaced a hydrophobic transmembrane segment in the model yeast inner membrane protein Mgm1 with a series of 19-residue model segments (referred to as H-segments) composed of n Leu and 19 − n Ala residues and determined the number of Leu residues required for 50% retention of the H segment in the inner membrane. The threshold was found to be unexpectedly high (n = 5–6), considerably higher than what we found previously for insertion of H-segments into the endoplasmic reticulum membrane in yeast and mammalian cells (4–6).
Because Mgm1 is not a substrate of the m-AAA protease (7), we looked for another model protein that would allow the study of membrane retention of transmembrane segments in the context of m-AAA protease dislocation activity. Mitochondrial cytochrome c peroxidase (Ccp1) is a heme-binding protein localized in the intermembrane space. During import into the mitochondrion, the N-terminal, positively charged, matrix-targeting sequence in precursor Ccp1 (p-Ccp1)4 is first translocated across the outer and inner membranes, and the following hydrophobic segment is integrated into the inner membrane via the TIM23 complex (see Fig. 1A) (8, 9). The membrane-integrated form is then dislocated from the membrane and concomitantly cleaved at Ala29 in the middle of the hydrophobic segment by the inner membrane m-AAA protease complex, generating intermediate Ccp1 (i-Ccp1). Finally, i-Ccp1 is cleaved between Ser67 and Thr68 by the inner membrane rhomboid protease Pcp1, generating mature Ccp1 (m-Ccp1). In Δyta10 cells, which lack the critical Yta10 subunit of the m-AAA protease, p-Ccp1 is not cleaved and remains anchored in the inner membrane by the hydrophobic transmembrane segment. Interestingly, certain mutations that reduce the hydrophobicity of the Ccp1 transmembrane domain abolish the dependence on the m-AAA protease for membrane dislocation and subsequent cleavage by Pcp1. If, on the other hand, the whole transmembrane segment is deleted, p-Ccp1 becomes fully imported into the matrix and is not cleaved by the Pcp1 protease (8).
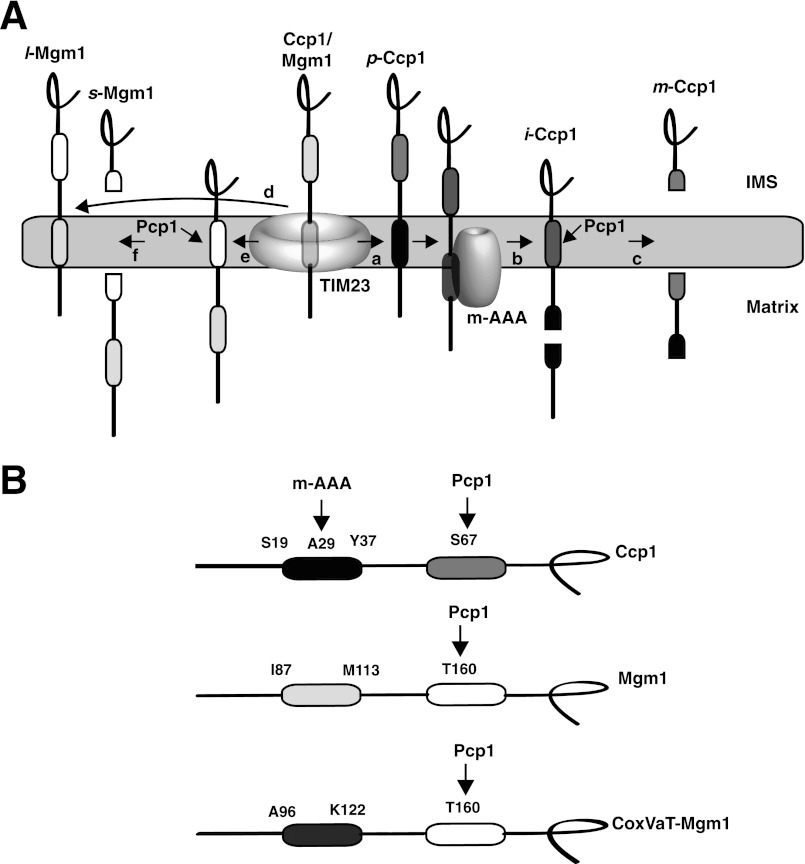
FIGURE 1.
A, maturation pathways for Ccp1 (right) and Mgm1 (left). The first hydrophobic segment (black) in p-Ccp1 triggers insertion from the TIM23 translocon into the inner membrane (arrow a). The membrane-integrated form is dislocated from the membrane and concomitantly cleaved in the middle of the hydrophobic segment by the inner membrane m-AAA protease complex, generating i-Ccp1 (arrow b). Finally, the rhomboid cleavage region (dark gray) in i-Ccp1 is cleaved by the rhomboid protease Pcp1 (arrow c), releasing m-Ccp1 in the intermembrane space (IMS) (8, 9). In the alternative topogenesis model for import of Mgm1 (11), the first hydrophobic segment integrates into the membrane in 30–40% of the molecules (arrow d), resulting in membrane-anchored l-Mgm1. For the remaining molecules, the first hydrophobic segment translocates into the matrix (arrow e), and the second hydrophobic segment is processed by Pcp1 (arrow f), generating s-Mgm1 in the intermembrane space (11). B, the model proteins Ccp1, Mgm1, and CoxVaT-Mgm1. The first hydrophobic segments of Ccp1 (Ser19–Tyr37), Mgm1 (Ile87–Met113), and CoxVaT-Mgm1 (Ala96–Lys122) were replaced with 19-residue H-segments with the composition GGPG(Leun/Ala19−n)GPGG. Pcp1 and m-AAA protease cleavage sites are indicated. In CoxVaT-Mgm1, residues 1–128 of CoxVa (shown in dark gray) are fused to residues 117–902 of Mgm1. Mgm1 H-segments and CoxVaT-Mgm1 H-segments carry a 3×HA tag in the C terminus.
Like Ccp1, Mgm1 has an N-terminal matrix-targeting signal, followed by a mildly hydrophobic segment. During its passage through the TIM23 translocon, the hydrophobic segment is inserted laterally into the inner membrane, yielding the long form of Mgm1 (l-Mgm1), but only in ∼30–40% of the molecules. In the remaining molecules, the hydrophobic segment is translocated into the matrix, and the protein is then cleaved by Pcp1 in a downstream, second hydrophobic segment, giving rise to a shorter form of the protein (s-Mgm1) (10, 11). Translocation of the first hydrophobic segment across the inner membrane is mediated by the matrix-localized mitochondrial import motor (3, 11).
To assess the possible influence of the m-AAA protease on membrane retention of transmembrane segments, we determined the threshold hydrophobicity for retention in the inner membrane of Leun/Ala19−n H-segments engineered into Ccp1 in both the absence and presence of a functional m-AAA protease. Strikingly, we found that in the presence of the m-AAA protease, the membrane retention threshold for H-segments in Ccp1 is n = 5–6, but that in the absence of a functional m-AAA protease, the threshold is n = 1–2. The threshold hydrophobicity for membrane retention is thus significantly higher in the presence than in the absence of a functional m-AAA protease.
The similarity between the threshold hydrophobicity of H-segment retention found for Ccp1 in the presence of the m-AAA protease and that found previously for Mgm1 (3) prompted us to further measure the retention of Mgm1 H-segments in the mitochondrial inner membrane in the absence of a functional m-AAA protease. Contrary to the situation for wild-type Mgm1 (7), we found that the balance between the long and short isoforms in Mgm1 H-segment constructs is strongly affected by the m-AAA protease: in a strain with a nonfunctional m-AAA protease, the threshold for 50% retention in the inner membrane of Mgm1 H-segments is n = 0–1. Replacing the wild-type hydrophobic segment with Leun/Ala19−n H-segments thus converts Mgm1 into an m-AAA protease substrate, explaining the unexpectedly high threshold hydrophobicity observed in our previous study.
These and our previous results (3) suggest a model in which the threshold hydrophobicity for TIM23-mediated insertion in the inner membrane is n ≈ 1–2, with a considerably higher threshold of n ≈ 5–6 required to withstand the force exerted by the m-AAA protease when it extracts protein segments from the membrane.
EXPERIMENTAL PROCEDURES
Plasmid Construction
To facilitate subcloning of an H-segment into the first hydrophobic domain of CCP1, pYX142CCP1HSmaI was prepared by overlap PCR (12) using pYX142CCP1 (8) as a template with the following primer pairs: 5′-GTATCCGAGAGAATTGTGTGA-3′ and 5′-ACCGCCCATAAGAGGGGTGGTCCTGGAGCAGCTGCTGCCGCCGCACTGTTATTGCTTCCCGGGCTG-3′; and 5′-AATGCGGGCTTGCAGAATGGCTTC-3′ and 5′-CTGTTATTGCTTCCCGGGCTGTTAGCTGCCGCAGCAGCAGGACCTGGTGGGTCGCAATCCCACAAG-3′. Various 19-amino acid long H-segments were amplified by PCR as described (3) using pHP84MGM1HA H-segment plasmids as templates (3) and primers 5′-ACCGCCCATAAGAGGGGTGGTCCTGGAGCAGCT-3′ and 5′-GCAGCAGGACCTGGTGGGTCGCAATCCCACAAG-3′ (the underlined sequences are complementary to the upstream and downstream sequences of the SmaI restriction enzyme site in pYX142CCP1HSmaI, and the non-underlined sequences are complementary to the H-segment sequence). For construction of plasmids carrying the H-segments in COXVaT-MGM1, a set of H-segments were amplified by PCR using COXVa H-segment constructs (3) as templates and primers 5′-TGCAAGCTTGATATCGAAATGTTACGTAACACTTTT-3′ and 5′-ACCATGAATAAGGAGTGGAGCTCTTTTACTAAGGAC-3′ (the underlined sequences complement the COXVaT H-segment). H-segment variants were subcloned into SmaI-digested pYX142CCP1HSmaI or pJK110 (3) by homologous recombination as described previously (3, 13).
Western Blot Analysis of Mgm1 and CoxVaT-Mgm1 H-segment Constructs
Yeast transformants of W303-1a (MATa, ade2, can1, his3, leu2, trp1, ura3), Δyta10 (MATa, ade2-1, his3-11,15, yta10::HIS3MX6, trp1-1, leu2,112, ura3-52) (14), or W303-1a rho− carrying a set of MGM1HA (3) or COXVaT-MGM1HA H-segment plasmids were grown overnight in 5 ml of −Leu medium at 30 °C. Whole cell lysates were extracted by TCA precipitation in which 1 A600 unit of yeast cells was harvested; resuspended in 500 μl of Milli-Q water, 75 μl of alkaline mixture (for 1 ml, 641 μl of Milli-Q water, 185 μl of 10 n NaOH, 100 μl of 0.1 m PMSF, and 74 μl of β-mercaptoethanol), and 575 μl of 25% (w/v) TCA; and incubated for 30 min on ice. Samples were washed with 1 ml of 100% (w/v) acetone, and the dried pellet was resuspended in 40 μl of SDS-PAGE sample buffer and incubated for 5 min at 95 °C before loading onto the SDS gels, followed by Western blotting with anti-HA antibody. The ratio of the long and short forms of the protein was quantified on a Fuji LAS-1000 phosphorimaging system using Image Lab v3.0 and Multi Gauge v3.0 software.
Pulse-Chase Analysis of Ccp1 H-segment Constructs
The m-AAA deletion strain (MATα, can1-100, ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-1, Δyta10::HIS3MX6, Δccp1::kanMX4) and the isogenic wild-type strain (MATα, can1-100, ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-1, Δccp1::kanMX4) (8) carrying CCP1 H-segment constructs were grown in −Leu medium at 30 °C. Cells (1.5 A600 units) were harvested and resuspended in synthetic defined medium without ammonium sulfate and methionine. Cells were starved at 30 °C for 20 min, and 30 μCi (per A600 unit of cells) of [35S]Met was added to the culture for 5 min. After the addition of 30 μl of 200 mm nonradioactive methionine (per A600 unit of cells) for 45 min, radiolabeled proteins were chased. The reaction was terminated by adding TCA to a final concentration of 10% (w/v). Yeast cells were disrupted with glass beads (0.5 mm), and the TCA-precipitated homogenate was spun down, resuspended in 50 μl of resuspension solution (3% SDS, 100 mm Tris base (pH 11.0), and 3 mm DTT) per A600 unit of cells, and heated at 100 °C for 10 min. Insoluble debris were pelleted down, and the supernatant was transferred to 700 μl of immunoprecipitation mixture (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, 1 mm PMSF, and 4 μl of 50× yeast protease inhibitor mixture (Roche Applied Science)). A preclearing reaction was performed by the addition of 10 μl of Pansorbin (Calbiochem) in immunoprecipitation mixture and incubation with rotation for 30 min at 4 °C. After centrifugation at 20,000 × g for 1 min, the supernatant was transferred to a new tube and incubated with 2 μl of anti-Ccp1 antibody (8) for 1 h at 4 °C. Finally, 8 μl of Pansorbin was added to immunoprecipitation mixture, and the immunoprecipitation reaction was incubated with rotation at 4 °C for 2 h or longer. Samples were centrifuged at 20,000 × g for 1 min, the supernatant was discarded, and the pellet was washed three times with immunoprecipitation buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, and 0.2% SDS) and once with 10 mm Tris-HCl (pH 7.5). 30 μl of SDS-PAGE sample buffer was added to the pellet, followed by incubation for 5 min at 95 °C before loading onto the SDS gels. Protein bands were visualized in a Fuji FLA-3000 phosphorimaging system.
Growth Rescue of Δccp1 cells with Ccp1 H-segment Constructs
Δccp1 cells (MATα, can1-100, ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-1, Δccp1::kanMX4) (8) carrying CCP1 H-segment constructs were grown in −Leu medium at 30 °C until the A600 reached between 1 and 2 A600 units, resuspended in water, and homogeneously dispersed in a −Leu medium-containing Petri dish (8.5-cm diameter). Filter paper with a diameter of 1 cm and soaked with 5 μl of 10% hydrogen peroxide was placed on the solid medium. Plates were incubated for 2–3 days at 30 °C. The diameter (d) of the region without visible cell growth was measured. Growth rescue relative to Δccp1 cells expressing wild-type Ccp1 was calculated as follows: rescue = 100*(〈db〉 − 〈d〉)/(〈db〉 − 〈dWT〉), where 〈db〉 is the average diameter measured for Δccp1Δyta10 cells expressing wild-type Ccp1, 〈dWT〉 is the average diameter measured for Δccp1 cells expressing wild-type Ccp1, and 〈d〉 is the average diameter measured for Δccp1 cells expressing the Ccp1 H-segment construct in question.
RESULTS
Experimental Setup
In our previous studies on the TIM23-mediated integration of different H-segments into the mitochondrial inner membrane (3, 15), we took advantage of the unique “alternative topogenesis” of Mgm1 (Fig. 1A) to measure the degree of membrane retention of H-segments. The first hydrophobic segment of Mgm1 was replaced with Leun/Ala19−n H-segments, and the degree of membrane retention was quantitated by measuring the relative levels of l- and s-Mgm1.
We applied the same approach to study the dislocation of H-segments from the mitochondrial inner membrane mediated by the m-AAA protease using Ccp1 as a test protein (Fig. 1, A and B). By replacing the Ccp1 hydrophobic domain with H-segments of the composition GGPG(Ln/A19−n)GPGG where n = 0–8 (see supplemental Table S1 for sequences), we determined the dislocation activity of the m-AAA protease on the H-segments using the degree of cleavage of Ccp1 by the rhomboid protease Pcp1 as a measure of membrane retention. m-Ccp1 originating from Pcp1 cleavage is formed only if the H-segment first partitions into the inner membrane and is then dislocated by the m-AAA protease. Ccp1 constructs were expressed from a low-copy plasmid in Δccp1 yeast strains containing either a functional or nonfunctional (Δccp1, Δyta10) m-AAA protease (8). Formation of dislocated, cleaved m-Ccp1 or non-dislocated, full-length p-Ccp1 was assessed by [35S]Met pulse-chase experiments, followed by immunoprecipitation with an antiserum directed against Ccp1 and SDS-PAGE analysis.
m-AAA Protease Affects the Threshold Hydrophobicity for Membrane Retention of Ccp1 H-segments
Consistent with previously published results (8, 9), virtually all wild-type p-Ccp1 was converted to m-Ccp1 in the presence of the m-AAA protease (Yta10), whereas in the absence of Yta10, wild-type p-Ccp1 remained in the membrane-anchored precursor form (Fig. 2A).
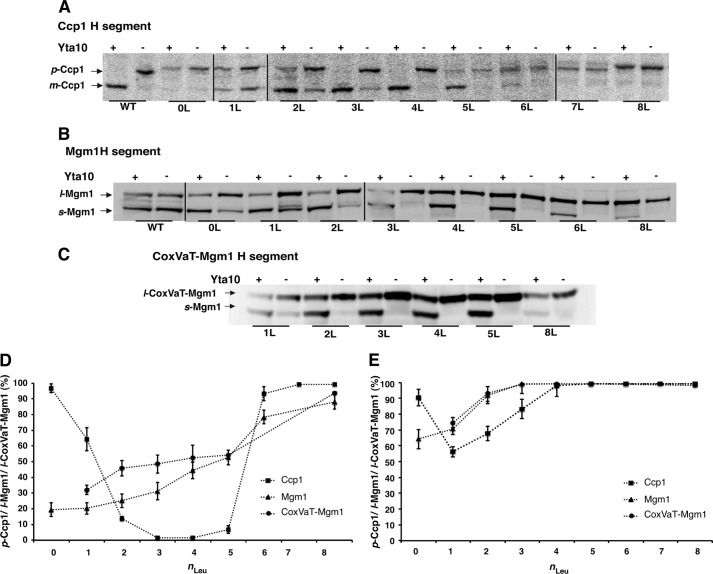
FIGURE 2.
The threshold hydrophobicity for membrane retention is different in the presence and absence of a functional m-AAA protease (Yta10). A, maturation of Ccp1 H-segment constructs expressed in the presence or absence of Yta10 (Δccp1 or Δccp1Δyta10 strain) was assessed by radiolabeling with 35[S]Met, followed by a 45-min chase. p- and m-Ccp1 are indicated. 0L, zero Leu residues. B, maturation of Mgm1 H-segment constructs expressed in the presence or absence of Yta10 (W303-1a or Δyta10 strain) was assessed by Western blotting of whole cell lysates. l- and s-Mgm1 are indicated. C, maturation of CoxVaT-Mgm1 H-segment constructs expressed in the presence or absence of Yta10 (W303-1a or Δyta10 strain) was assessed by Western blotting of whole cell lysates. s-CoxVaT-Mgm1 and s-Mgm1 are indicated. D, relative amounts calculated from A–C of p-Ccp1, l-Mgm1, and l-CoxVaT-Mgm1 expressed in the presence of a functional m-AAA protease. E, same as in D but in the absence of a functional m-AAA protease (Δyta10 strain). Means ± S.E. from at least three independent experiments are shown in D and E (error bars).
Interestingly, in the presence of a functional m-AAA protease, Ccp1 variants with H-segments containing n = 2–5 leucines gave rise mostly to m-Ccp1 (Fig. 2, A and D), whereas in the absence of Yta10, the p-Ccp1 form predominated (Fig. 2, A and E). H-segments with n = 2–5 thus behaved similarly to the natural hydrophobic segment in Ccp1, partitioning into the inner membrane and from there being efficiently dislocated by the m-AAA protease, allowing Pcp1 cleavage and formation of m-Ccp1. In a functional complementation experiment, these Ccp1 variants were expressed in the Δccp1 strain and exposed to hydrogen peroxide (8). Ccp1 in its mature folded form (m-Ccp1) functions as a reactive oxygen species-binding scavenger in the mitochondrial intermembrane space, conferring resistance to oxidative conditions (16). Indeed, Ccp1 with an H-segment containing n = 2–4 leucines fully restored cell growth in the presence of hydrogen peroxide to levels similar to those restored by wild-type Ccp1 (Fig. 3).
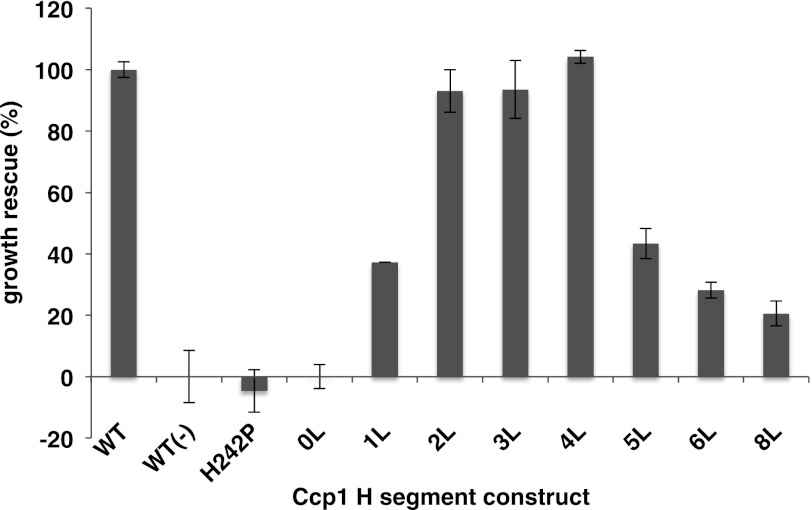
FIGURE 3.
Functional complementation of Δccp1 with Ccp1 H-segment constructs. Ccp1 activity was assessed by growth of a cell suspension on solid medium with filter paper (1-cm diameter) soaked with 5 μl of 10% (v/v) hydrogen peroxide on the center of the plate. The diameter (d) of the circular region around the filter paper with no visible growth after 2–3 days of incubation was measured. Percent growth rescue relative to Δccp1 cells expressing wild-type Ccp1 was calculated as follows: 100*(〈db〉 − 〈d〉)/(〈db〉 − 〈dWT〉), where 〈db〉 is the average diameter measured for Δccp1Δyta10 cells expressing wild-type Ccp1 (WT(−)), 〈dWT〉 is the average diameter measured for Δccp1 cells expressing wild-type Ccp1, and 〈d〉 is the average diameter measured for Δccp1 cells expressing the Ccp1 H-segment construct in question. Δccp1 cells expressing nonfunctional Ccp1 with a mutation (H242P) in the heme-binding domain (8) were included as a negative control. Means ± S.E. from at least three independent experiments are shown (error bars). 0L, zero Leu residues.
It is notable that despite producing similar levels of m-Ccp1 compared with p-Ccp1 in the presence of the m-AAA protease, compared with the construct with n = 2 (Fig. 2D), the Ccp1 construct with n = 5 could only partially restore growth of the Δccp1 strain under oxidative conditions. We have not analyzed this construct further; however, it is possible that the slightly lower amount of m-Ccp1 produced in the Ccp1 construct with n = 5 (Fig. 2A) might be insufficient to fully restore Ccp1 function.
In both the presence and absence of the m-AAA protease, the H-segment with n = 0 yielded almost exclusively p-Ccp1, whereas for n = 1, there was a mixture of p and m-Ccp1 (Fig. 2, A, D, and E). As shown by Tatsuta et al. (8), reducing the hydrophobicity of the hydrophobic domain in wild-type Ccp1 facilitates dislocation from the lipid bilayer and allows Ccp1 maturation by Pcp1 in an m-AAA protease-independent way. Additionally, the same authors found that complete deletion of the hydrophobic domain causes mistargeting of Ccp1 to the matrix, bypassing cleavage by Pcp1. It appears that the n = 0 construct and ∼50% of the molecules in the n = 1 construct are mislocalized to the matrix and therefore not cleaved by Pcp1. The remaining ∼50% of the molecules in the n = 1 construct are found as the m-Ccp1 form regardless of the presence or absence of the m-AAA protease (Fig. 2A), suggesting that the m-AAA protease-independent “facilitated dislocation” mechanism mentioned above may give rise to the observed m-Ccp1 form. Although none of these Ccp1 variants could fully rescue cell growth in the presence of hydrogen peroxide, yeast cells expressing the n = 1 construct grew at a comparable rate to cells expressing the n = 5 construct (Fig. 3).
Finally, Ccp1 carrying H-segments with n = 6–8 produced almost exclusively p-Ccp1 in both the presence and absence of a functional m-AAA protease (Fig. 2, A, D, and E). In light of the Ccp1 maturation mechanism (8), we infer that these very hydrophobic segments partition efficiently into the inner membrane and that the m-AAA protease cannot dislocate them into the matrix.
We conclude that, in the context of Ccp1, the threshold hydrophobicity required for 50% retention of an H-segment in the inner membrane varies depending on the m-AAA protease activity. In the absence of a functional m-AAA protease, the threshold is n = 1–2 (Fig. 2E). In contrast, in the presence of a fully functional m-AAA protease, the threshold is considerably higher at n = 5–6 (Fig. 2D).
m-AAA Protease Affects the Threshold Hydrophobicity for Membrane Retention of Mgm1 H-segments
In our previous study (3), we used Mgm1 as a model protein to study TIM23-mediated insertion of H-segments into the mitochondrial inner membrane. Membrane-anchored l-Mgm1 is generated when the H-segment is retained in the inner membrane, whereas soluble s-Mgm1 is produced when the H-segment is translocated into the matrix and the following hydrophobic segment is cleaved by Pcp1 (Fig. 1A). In the Mgm1 system, the threshold hydrophobicity for 50% retention of the H-segment was found to be n ≈ 5, similar to the threshold obtained for Ccp1 in the presence of the m-AAA protease.
Although maturation of wild-type Mgm1 is independent of the m-AAA protease (10, 11), this unexpected correspondence between the Ccp1 and Mgm1 results prompted us to examine the involvement of the m-AAA protease in the processing of Mgm1 constructs carrying Leun/Ala19−n H-segments. The Mgm1 constructs were expressed in yeast strains with a functional or nonfunctional (Δyta10) m-AAA protease (14). Indeed, in the absence of the m-AAA protease, Mgm1 with an n = 0 H-segment gave ∼65% l-Mgm1, reaching 100% l-Mgm1 for n = 3 (Fig. 2, B and E). These results parallel those seen for Ccp1 in Δyta10 cells, strongly suggesting that Mgm1 constructs with engineered H-segments are substrates of the m-AAA protease, in contrast to wild-type Mgm1.
Finally, CoxVaT-Mgm1, a fusion between the N-terminal part of the inner membrane protein CoxVa (residues 1–128) and the C-terminal part of Mgm1 (residues 117–902) in which the CoxVa transmembrane segment (residues 96–122) has been replaced with Leun/Ala19−n H-segments (Fig. 1B), was tested in both wild-type W303-1a and Δyta10 cells (14). As shown in Fig. 2C, for this series of constructs, we found the threshold hydrophobicity for 50% retention in the inner membrane at n = 0–1 in the absence of m-AAA (Fig. 2E) and at n ≈ 5 in the presence of m-AAA (Fig. 2D), very similar to the results for the Mgm1 constructs.
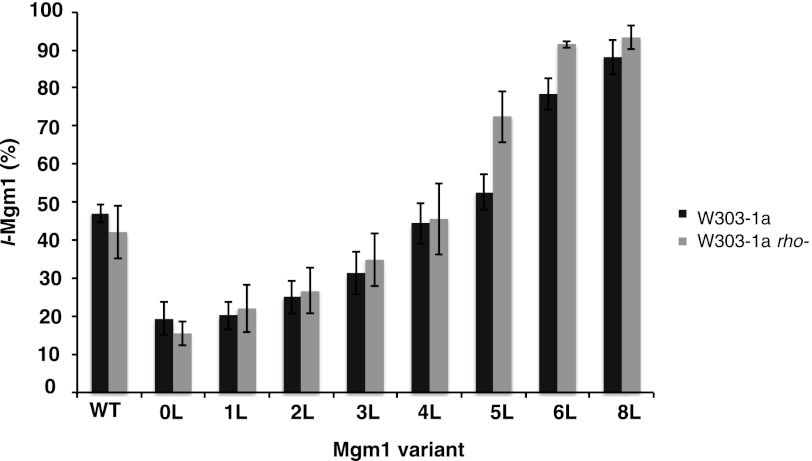
In addition to its role in the maturation of Ccp1 (8, 9) and the mitochondrial ribosomal subunit Mrpl32 (17, 18), the m-AAA protease is a central component of the mitochondrial protein quality control system, being able to extract and degrade misfolded polypeptides from the inner membrane (19, 20). In the latter case, the proteolytic activity of m-AAA is unspecific, and it is unclear how the m-AAA protease differentiates between substrates that are to be degraded from those that are specifically cleaved at a single site (21). Cells lacking a functional m-AAA protease are respiratory-deficient (22) due to impaired translation and assembly of mitochondrially encoded respiratory chain complex subunits (14, 17). We therefore considered the possibility that the variation in the ratio of l- and s-Mgm1 in the Δyta10 strain may not be a direct consequence of m-AAA dislocation activity but rather results from a general mitochondrial malfunction. To this end, we expressed the Mgm1 constructs in the respiratory-deficient W303-1a rho− strain, which carries truncations in the mtDNA (23) and shows a defect in translation of mitochondrially encoded subunits of respiratory chain complexes, as does the m-AAA mutant (14, 17). No variation in the amount of l- and s-Mgm1 compared with the wild-type W303-1a strain was found (Fig. 4). We conclude that the differences in threshold hydrophobicity seen in the presence or absence of the m-AAA protease are due to a direct involvement of the m-AAA protease in the dislocation reaction.
FIGURE 4.
Comparison of membrane retention of Mgm1 H-segment constructs in W303-1a and W303-1a rho− strains. Whole cell lysates were analyzed by SDS-PAGE and Western blotting, and the relative amounts of l-Mgm1 were calculated. Means ± S.E. from at least three independent experiments are shown (error bars).
DISCUSSION
We have analyzed the role of the m-AAA protease in membrane dislocation of model hydrophobic Leun/Ala19−n segments (H-segments) in the context of three different proteins: Ccp1, Mgm1, and a CoxVa-Mgm1 fusion protein. Our results show that in the presence of a functional m-AAA protease, the threshold for membrane retention of H-segments is n = 5–6, whereas in the absence of the m-AAA protease, the threshold is n = 1–2. These data suggest that if the hydrophobicity of a transmembrane domain in an inner membrane protein is higher than n ≈ 5–6, the m-AAA protease will not be able to extract it from the membrane. The unexpectedly high threshold for membrane insertion of Mgm1 H-segment constructs via the TIM23 translocon that we found previously (3) can now be explained as resulting from membrane dislocation of these Mgm1 variants by the m-AAA protease. Possibly, model proteins such as Mgm1 and CoxVa containing engineered H-segments cannot fold and/or oligomerize properly and therefore become substrates of the m-AAA protease (24).
Generally, the hydrophobicity of mitochondrial inner membrane proteins is rather low compared with bacterial inner membrane or eukaryotic plasma membrane proteins (25). Thus, our data suggest that most transmembrane segments would be membrane-extractable by the m-AAA protease, which is important in the processes of protein quality control and turnover in the mitochondrial inner membrane but poses a danger to dislocate a newly synthesized membrane protein. We speculate that this may be why newly made inner membrane proteins often form a complex during early stages of membrane insertion: to evade dislocation by the m-AAA protease.
Acknowledgments
We thank Professor Thomas Langer (Institute for Genetics, University of Cologne) for kindly providing pYX142CCP1, Δccp1, Δyta10, and Δccp1Δyta10 strains and anti-Ccp1 antibody. We thank Dr. Martin Ott (Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden) for the W303-1a rho− strain and for helpful discussion of this work.
This work was supported in part by grants from the Swedish Research Council, the Swedish Cancer Foundation, and the Swedish Foundation for Strategic Research and European Research Council Grant ERC-2008-AdG 232648 (to G. v. H.) and by National Research Foundation of Korea Grants NRF0409-20100093 and GRN-C00048 (to H. K.).

This article contains supplemental Table S1.
- p-Ccp1
- precursor Ccp1
- i-Ccp1
- intermediate Ccp1
- m-Ccp1
- mature Ccp1
- l-Mgm1
- long form of Mgm1
- s-Mgm1
- short form of Mgm1
- m-AAA
- matrix ATPase associated with diverse cellular activities.
REFERENCES
- 1. Hartl F. U., Ostermann J., Guiard B., Neupert W. (1987) Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell 51, 1027–1037 [DOI] [PubMed] [Google Scholar]
- 2. van Loon A. P., Brändli A. W., Schatz G. (1986) The presequences of two imported mitochondrial proteins contain information for intracellular and intramitochondrial sorting. Cell 44, 801–812 [DOI] [PubMed] [Google Scholar]
- 3. Botelho S. C., Osterberg M., Reichert A. S., Yamano K., Björkholm P., Endo T., von Heijne G., Kim H. (2011) TIM23-mediated insertion of transmembrane α-helices into the mitochondrial inner membrane. EMBO J. 30, 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hessa T., Reithinger J. H., von Heijne G., Kim H. (2009) Analysis of transmembrane helix integration in the endoplasmic reticulum in S. cerevisiae. J. Mol. Biol. 386, 1222–1228 [DOI] [PubMed] [Google Scholar]
- 5. Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 [DOI] [PubMed] [Google Scholar]
- 6. Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 [DOI] [PubMed] [Google Scholar]
- 7. Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 8. Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007) m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esser K., Tursun B., Ingenhoven M., Michaelis G., Pratje E. (2002) A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 323, 835–843 [DOI] [PubMed] [Google Scholar]
- 10. Schäfer A., Zick M., Kief J., Steger M., Heide H., Duvezin-Caubet S., Neupert W., Reichert A. S. (2010) Intramembrane proteolysis of Mgm1 by the mitochondrial rhomboid protease is highly promiscuous regarding the sequence of the cleaved hydrophobic segment. J. Mol. Biol. 401, 182–193 [DOI] [PubMed] [Google Scholar]
- 11. Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spee J. H., de Vos W. M., Kuipers O. P. (1993) Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21, 777–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oldenburg K. R., Vo K. T., Michaelis S., Paddon C. (1997) Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arlt H., Steglich G., Perryman R., Guiard B., Neupert W., Langer T. (1998) The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 17, 4837–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osterberg M., Calado Botelho S., von Heijne G., Kim H. (2011) Charged flanking residues control the efficiency of membrane insertion of the first transmembrane segment in yeast mitochondrial Mgm1p. FEBS Lett. 585, 1238–1242 [DOI] [PubMed] [Google Scholar]
- 16. Jiang H., English A. M. (2006) Phenotypic analysis of the ccp1Δ and ccp1Δ-ccp1W191F mutant strains of Saccharomyces cerevisiae indicates that cytochrome c peroxidase functions in oxidative-stress signaling. J. Inorg. Biochem. 100, 1996–2008 [DOI] [PubMed] [Google Scholar]
- 17. Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289 [DOI] [PubMed] [Google Scholar]
- 18. Bonn F., Tatsuta T., Petrungaro C., Riemer J., Langer T. (2011) Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J. 30, 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korbel D., Wurth S., Käser M., Langer T. (2004) Membrane protein turnover by the m-AAA protease in mitochondria depends on the transmembrane domains of its subunits. EMBO Rep. 5, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. (2000) Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5, 629–638 [DOI] [PubMed] [Google Scholar]
- 21. Tatsuta T., Langer T. (2009) AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res. Microbiol. 160, 711–717 [DOI] [PubMed] [Google Scholar]
- 22. Tauer R., Mannhaupt G., Schnall R., Pajic A., Langer T., Feldmann H. (1994) Yta10p, a member of a novel ATPase family in yeast, is essential for mitochondrial function. FEBS Lett. 353, 197–200 [DOI] [PubMed] [Google Scholar]
- 23. Fukuhara H., Moustacchi E., Wesolowski M. (1978) Preferential deletion of a specific region of mitochondrial DNA in Saccharomyces cerevisiae by ethidium bromide and 3-carbethoxy-psoralen: directional retention of DNA sequence. Mol. Gen. Genet. 162, 191–201 [DOI] [PubMed] [Google Scholar]
- 24. Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. (1996) The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85, 875–885 [DOI] [PubMed] [Google Scholar]
- 25. von Heijne G. (1986) Why mitochondria need a genome. FEBS Lett. 198, 1–4 [DOI] [PubMed] [Google Scholar]