Background: Carbohydrate binding modules (CBMs) contribute to the enzymatic degradation of complex polysaccharide structures.
Results: New CBMs display specificity for decorated glucans through an extensive hydrophobic platform that interacts with both backbone and side chain structures.
Conclusion: CBMs that bind to complex β-glucans exploit different components of these ligands as specificity determinants.
Significance: CBMs can utilize the side chains of decorated glucans as specificity determinants.
Keywords: Carbohydrate-binding Protein, Isothermal Titration Calorimetry, Plant Cell Wall, Protein Structure, X-ray Crystallography
Abstract
Plant biomass is central to the carbon cycle and to environmentally sustainable industries exemplified by the biofuel sector. Plant cell wall degrading enzymes generally contain noncatalytic carbohydrate binding modules (CBMs) that fulfil a targeting function, which enhances catalysis. CBMs that bind β-glucan chains often display broad specificity recognizing β1,4-glucans (cellulose), β1,3-β1,4-mixed linked glucans and xyloglucan, a β1,4-glucan decorated with α1,6-xylose residues, by targeting structures common to the three polysaccharides. Thus, CBMs that recognize xyloglucan target the β1,4-glucan backbone and only accommodate the xylose decorations. Here we show that two closely related CBMs, CBM65A and CBM65B, derived from EcCel5A, a Eubacterium cellulosolvens endoglucanase, bind to a range of β-glucans but, uniquely, display significant preference for xyloglucan. The structures of the two CBMs reveal a β-sandwich fold. The ligand binding site comprises the β-sheet that forms the concave surface of the proteins. Binding to the backbone chains of β-glucans is mediated primarily by five aromatic residues that also make hydrophobic interactions with the xylose side chains of xyloglucan, conferring the distinctive specificity of the CBMs for the decorated polysaccharide. Significantly, and in contrast to other CBMs that recognize β-glucans, CBM65A utilizes different polar residues to bind cellulose and mixed linked glucans. Thus, Gln106 is central to cellulose recognition, but is not required for binding to mixed linked glucans. This report reveals the mechanism by which β-glucan-specific CBMs can distinguish between linear and mixed linked glucans, and show how these CBMs can exploit an extensive hydrophobic platform to target the side chains of decorated β-glucans.
Introduction
The plant cell wall represents a major nutrient for numerous microbial ecosystems, exemplified by bacterial and fungal communities established in the rumen and large bowel of mammals, where they play an important role in animal nutrition and human health, respectively (1, 2). It is also evident that these composite structures are of increasing industrial significance, particularly in the environmentally relevant bioenergy and bioprocessing sectors (3, 4). The complex physical and chemical structure of the plant cell wall restricts its access to degradative enzymes. Microorganisms that utilize plant biomass as a significant nutrient express extensive repertoires of degradative enzymes, primarily, glycoside hydrolases but also lyases and esterases, which attack the structural polysaccharides of the plant cell wall (5).
A common feature of plant cell wall degrading enzymes is their complex modular architecture in which the catalytic module is appended to one or more noncatalytic carbohydrate binding modules (CBMs)4 (see Ref. 6 for review), which are grouped into sequence-based families on the CAZy database (7). The general function of CBMs is to direct the cognate catalytic modules to their target substrate within the plant cell wall, thereby increasing the efficiency of catalysis (8–10). The majority of CBMs display a β-sandwich fold with the ligand binding site located in either the concave surface presented by one of the β-sheets, a topography that facilitates the targeting of the internal regions of glycan chains (11–13), or in the loops that connect the two sheets (14, 15). This latter binding site can either target the end (15) or, less frequently, the internal regions of glycan chains (14).
The majority of CBMs that target the plant cell wall bind to crystalline cellulose, single chains of β-glucans and xylan (see Ref. 6 for review). Binding to crystalline cellulose by Type A CBMs is mediated by a planar hydrophobic surface, which makes apolar contacts with exposed cellulose chains (16). Ligand recognition in CBMs that bind to the internal regions of single polysaccharide chains can be highly specific, exemplified by the CBM6 from the clostridial xylanase CtXyn10B, which exclusively targets xylan (14), whereas examples of promiscuous specificity include CmCBM6 from the Cellvibrio endoglucanase CmLic5A that binds to both β1,4-glucans and mixed linked β1,3-β1,4-glucans (17), and the CBM62 from the Cellvibrio xylanase CjXyn11A that recognizes β-glucans, xylans, and even β-galactans (15). In both examples plasticity in ligand recognition is achieved through binding to a conserved element of the target glycan, demonstrated by the primary binding site of CmCBM6, which is specific for cellobiose (Glc-β1,4-Glc), a structure found in both cellulose and the mixed linked glucan (17). It is unclear, however, whether a cohort of CBMs exist that recognize diverse glycans by binding to distinct structures in the different target ligands. In addition, all CBMs that recognize β-glucans, also bind to xyloglucan, a β1,4-glucan that is decorated with α1,6-linked xylose residues. It would appear that these proteins accommodate, but do not target the xylose side chains. Indeed, to date no CBMs have been described that display a preference for xyloglucan over other β-glucans (12).
A recent report has identified two modules in a Eubacterium cellulosolvens endoglucanase (EcCel5A), designated hereafter as CBM65A and CBM65B, that bind to both disordered cellulose and mixed linked glucans (18). In this study we have exploited the two CBM65s as a model system to understand the mechanistic basis for the diverse specificities displayed by some CBMs. We show that the CBM65s, uniquely, display a significant preference for xyloglucan. The structure of CBM65B in complex with a xyloglucan-derived oligosaccharide, in combination with mutagenesis studies on CBM65A, revealed the mechanism by which these proteins display a preference for xyloglucan. The ligand binding cleft contains an unusually large number of aromatic residues that are optimized to not only make apolar contacts with the glucan backbone, but also make hydrophobic interactions with the xylose side chains. In addition to the dominant apolar contacts, CBM65A contains two polar residues that play an important role in binding undecorated β-glucans. Gln106 confers specificity for β1,4-glucan (cellulose), whereas Gln110 interacts with both cellulose and mixed linked β1,3-β1,4-glucans.
MATERIALS AND METHODS
Protein Production and Purification
DNA encoding the CBM65A (residues 37–170 of EcCel5A) and CBM65B (residues 581–713 of EcCel5A) were synthesized (NZYTech Ltd., Portugal) with codon usage optimized for expression in Escherichia coli. The synthesized genes contained engineered NheI and XhoI recognition sequences at the 5′ and 3′ ends, respectively, which were used for subsequent subcloning into the E. coli expression vector pET28a (Novagen), generating pCMBAL1 and pCBMAL2, which encode CBM65A and CBM65B, respectively. Both CBMs contain an N-terminal His6 tag. E. coli Tuner DE3 cells harboring pCMBAL1 and pCBMAL2 were cultured in Luria-Bertani broth containing kanamycin (50 μg/ml) at 37 °C to mid-exponential phase (A600 nm = 0.6) and recombinant protein expression was induced by the addition of 0.2 mm isopropyl β-d-1-thiogalactopyranoside and incubation for a further 16 h at 19 °C. The His6-tagged recombinant CBMs, and their respective mutants (see below), were purified from cell-free extracts by immobilized metal-ion affinity chromatography as described previously (19).
For crystallization, CBM65A was further purified by size exclusion chromatography. Following immobilized metal-ion affinity chromatography, fractions containing the purified proteins were buffer exchanged, using PD-10 Sephadex G-25M gel-filtration columns (GE Healthcare), into 50 mm HEPES-Na buffer, pH 7.5, containing 200 mm NaCl and 5 mm CaCl2, and was then subjected to gel filtration using a HiLoad 16/60 Superdex 75 column (GE Healthcare) at a flow rate of 1 ml/min. Purified CBM65A was concentrated using an Amicon 10-kDa molecular mass centrifugal concentrator and washed three times with 1 mm CaCl2. Preparation of E. coli to generate selenomethionine CBM65A was performed as described in Ref. 20 and the protein was purified using the same procedures as employed for the native CBM. Purified CBM65A was concentrated using an Amicon 10-kDa molecular mass centrifugal concentrator and washed three times with 5 mm DTT. SDS-PAGE showed that all the recombinant proteins were more than 95% pure after Coomassie Blue staining.
Site-directed Mutagenesis
Site-directed mutagenesis was carried out employing a PCR-based NZYMutagenesis site-directed mutagenesis kit (NZYTech Ltd.) according to the manufacturer's instructions, using pCBMAL1 as the template. The sequence of the primers used to generate these mutants is displayed in supplemental Table S1. The mutated DNA sequences were sequenced to ensure that only the appropriate mutations had been incorporated into the amplified DNA.
Source of Sugars Used
All soluble polysaccharides and cellooligosacchharides were purchased from Megazyme International (Bray, County Wicklow, Ireland), except apple and citrus pectin, konjak galactomannan, and hydroxyethylcellulose, which were obtained from Sigma, and pustullan, which was obtained from Calbiochem. Catalog numbers of polysaccharides where more than one version exists are: wheat arabinoxylan, P-WAXYM; rye arabinoxylan, P-RAXY; galactomannan, Carob (P-GALML); galactomannan, Guar (P-GGMMV).
Affinity Gel Electrophoresis
Affinity gel electrophoresis was used to screen CBM65A and CBM65B for binding to soluble polysaccharides. The method used was essentially that described by Ref. 17, using the polysaccharide ligands at a concentration of 0.3% (w/v), unless stated otherwise. Electrophoresis was carried out for 4 h at room temperature in native 10% (w/v) polyacrylamide gels. The nonbinding negative control protein was BSA.
Isothermal Titration Calorimetry (ITC)
The thermodynamic parameters of the binding of the CBM65s to soluble polysaccharides and cellooligosaccharides were determined by ITC using a VP-ITC calorimeter (MicroCal, Northampton, MA), as described by Ref. 17. Briefly, titrations were performed at 25 °C by injecting 10-μl aliquots of 5–20 mm oligosaccharide or 10 mg/ml of polysaccharide, in 50 mm Na-HEPES buffer, pH 7.5, into the cell containing 100 μm CBM dialyzed into the Na-HEPES buffer, and the release of heat was recorded. The stoichiometry of binding (n), the association constant Ka, and the binding enthalpy ΔH were evaluated by using MicroCal Origin 7.0 software. The standard Gibbs energy change ΔG0 and the standard entropy change ΔS0 were calculated from ΔG0 = −RT lnKa and ΔG0 = ΔH0 − TΔS0, where R is the gas constant and T the absolute temperature. The polysaccharide at 10 mg/ml was converted into a molarity that gave a stoichiometry of 1 to determine the molar concentration of CBM65 binding sites on the polymer.
Immunofluorescence Cell Wall Imaging
Tobacco stem and Miscanthus x giganteus (Miscanthus) stem sections were prepared, and a 3-stage CBM in situ labeling technique described previously (21, 22) was used to assess the binding of CBM65A. Where appropriate Miscanthus stem sections were incubated, prior to incubation with the CBM65, with a Bacillus subtilis lichenase (Biosupplies Australia) at 20 μg/ml in 0.1 m sodium acetate buffer, pH 5.0, overnight at RT. All tobacco stems sections were pretreated with pectate lyase to remove pectic homogalacturonan as described (23) and where appropriate with a Paenibacillus sp. xyloglucan-specific endo-1,4-β-glucanase (Megazyme International, Ireland) at 20 μg/ml in 0.1 m sodium acetate buffer, pH 5.5, overnight at room temperature. Immunofluorescence microscopy and micrograph capture was carried out as described (23).
Crystallization and Data Collection
The crystals of native apo-CBM65A (∼80 mg/ml) were obtained in 200 mm ammonium sulfate, 100 mm sodium acetate trihydrate, pH 4.6, 22–30% (w/v) PEG 2000. CBM65B (apo and in complex with ligand) was crystallized at 80 mg/ml in 200 mm ammonium acetate, 100 mm tri-sodium citrate, pH 5.6, 30% PEG 4000, and cryo protected in 20% PEG 400 containing ligand where appropriate. Datasets were collected for apo native CBM65A, apo-CBM65B, or CBM65B co-crystallized with 14 mm of the heptasaccharide XXXG (Glc4Xyl3) at beamlines IO2 or IO4 at DIAMOND (Harwell, UK). All data sets were processed using the programs iMosflm (24) or XDS (25) and SCALA (26) from the CCP4 suite (27). The crystal belongs to the hexagonal system, with either the P6122 or P3121 space group for CBM65A and P65 for CBM65B and P43212 for the CBM65B-XXXG complex.
Model Building and Refinement
The structure of native CBM65A was solved using crystals of selenomethionine CBM65A to a resolution of 1.75 Å (Ref. 28; Protein Data Bank 4aek) using PHASER (29). A single solution for the space group P3121 with an LLG score of 700 was obtained. This model was adjusted and refined using REFMAC5 (30) interspersed with model adjustment in COOT (31) to give the final model (PDB 4afm) to a resolution of 1.25 Å. PHASER (29) and the atomic coordinates of apo-CBM65A (PDB 4afm) were used as a search model against the highest resolution data (1.42 Å) obtained for apo-CBM65B. A successful solution was obtained in space group P65 with a TFZ score of 20.8 and LLG of 318. The structure was refined as above. Finally, apo-CBM65B was used as the search model in conjunction with MOLREP (32) to solve the CBM65B-XXXG structure to a resolution of 2.35 Å. The root mean square deviation of the bond lengths, bond angles, and torsion angles and other indicators were continuously monitored using the validation tools in COOT (31) at the end for all the refinements. Data collection and refinement statistics are presented in Table 1.
TABLE 1.
Data collection and structure refinement statistics
| Dataset | CBM65A | CBM65B | CBM65B-XXXG |
|---|---|---|---|
| Source | Soleil–Proxima 1 | Diamond–I02 | Diamond–I04 |
| Detector | Quantum 315r CCD | Pilatus 6M | Quantum 315r CCD |
| Wavelength (Å) | 0.9793 | 0.9795 | 0.9795 |
| Space group | P6122 | P65 | P43 21 2 |
| Unit cell parameters | |||
| a = b (Å) | 48.74 | 83.75 | 57.92 |
| c (Å) | 193.70 | 36.75 | 116.74 |
| Α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution limits (Å) | 38.74–1.25 (1.32–1.25)a | 36.75–1.42 (1.45–1.42) | 58.37–2.35 (2.48–2.35) |
| No. of unique observations | 35,557 | 27,755 | 8,856 |
| Multiplicity | 21.6 (6.2) | 5.3 (5.4) | 6.2 (4.7) |
| Completeness (%) | 91.2 (62.4) | 99.8 (99.9) | 100.0 (100.0) |
| 〈I/σ(I)〉 | 28.20 (4.00) | 11.9 (1.8) | 13.4 (2.0) |
| Rmergeb | 7 (40) | 6 (69) | 7.8 (66.1) |
| Refinement statistics | |||
| Rworkb (%) | 15.80 | 16.98 | 22.44 |
| Rfreeb (%) | 17.30 | 19.73 | 28.03 |
| No. non-H atoms | |||
| No. protein atoms | 1097 | 1060 | 955 |
| No. water molecules | 130 | 166 | 1 |
| No. other | 34 | 6 | NAc |
| No. ligand atoms | NA | NA | 72 |
| Root mean square deviations from ideal values (Å) | |||
| Bond length | 0.031 | 0.025 | 0.012 |
| Angle distance | 2.873 | 2.495 | 1.56 |
| Average B factor (Å2) | |||
| Protein | 11.5 | 21.6 | 50.4 |
| Water | 28.5 | 36.9 | 39.5 |
| Other | 31.4 | 62.3 | NA |
| Ligand | NA | NA | 47.6 |
| Ramachandran plot,c residues in allowed and most favoured regions (%) | 100 | 99.2 | 100 |
| PDB accession code | 4afm | 4ba6 | 2ypj |
a Values in parentheses are for the high resolution shell.
b Rmerge = Σh Σi|I(h,i) − 〈I(h)〉|/Σh Σi I(h,i), where I(h,i) is the intensity of the measurement of reflection; h and 〈I(h)〉 is the mean value of I(h,i) for all i measurements.
c Calculated using MOLPROBITY.
d NA, not applicable.
RESULTS
Quantitative Evaluation of the Binding of CBM65A to Its Ligands
The endoglucanase from E. cellulosolvens, EcCel5A, consists of two GH5 modules and two CBMs designated hereafter as CBM65A and CBM65B, which are located at the N terminus and between the two catalytic modules, respectively (18) (Fig. 1). The two CBMs display 73% sequence identity. Previous qualitative studies showed that both CBM65A and CBM65B bound to acid swollen cellulose, lichenan (β1,3-β1,4 mixed linked glucan), but did not bind to laminarin (β1,3-glucan), Avicel or α-glucans (18). Here we have explored further the specificities of the two protein modules. Recombinant forms of CBM65A and CBM65B, comprising residues 37–170 and 581–713, respectively, of full-length EcCel5A were purified to electrophoretic homogeneity by immobilized metal-ion affinity chromatography. Initially affinity gel electrophoresis was used to screen potential polysaccharide ligands of the two proteins. The data, presented in Table 2, with example gels displayed in Fig. 2A, show that both protein modules, in addition to binding β1,3-β1,4 mixed linked glucans, also bound to highly decorated β1,4-glucans such as xyloglucan and hydroxyethylcellulose, displayed weak affinity for glucomannan, but did not exhibit significant binding to other β1,4-glycans such as xylans, galactomannans, or galactans; no binding to pectin backbone structures or α-glucans were observed. The specificity of the two CBMs appeared to be identical. These data indicate that the CBM65s target β-glucans containing β1,4-linkages.
FIGURE 1.
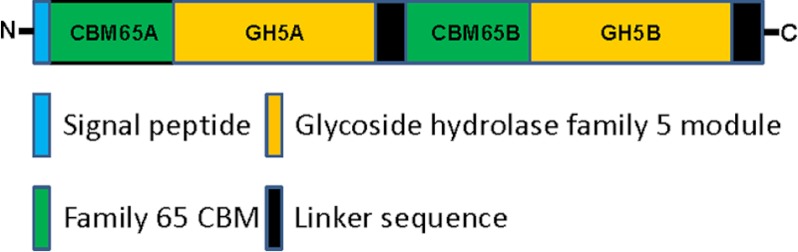
Schematic of EcCel5A.
TABLE 2.
Affinity gel electrophoresis of CBM65A and CBM65B
| Polysaccharide (0.3%) | CBM65Aa | CBM65B |
|---|---|---|
| Celluloses | ||
| HECb | + + + | + + + |
| Lichenan | + + + | + + + |
| Curdlan | − | − |
| CMC | + + | + + |
| Xylans | ||
| Arabinoxylan (rye) | − | − |
| 4-O-Methyl-d-Glucurono-d-xylan | − | − |
| Xylan ( birchwood) | − | − |
| Arabinoxylan (wheat medium viscosity) | + | + |
| Arabinoxylan (wheat; Insoluble) | − | − |
| Other hemicelluloses | ||
| β-Glucan (barley) | + + + | + + + |
| Xyloglucan (Tamarind) | + + + | + + + |
| Mannan (ivory nut) | − | − |
| Galactomannan (locust bean) | + | + |
| Galactomannan (guar gum) | − | − |
| Galactomannan (carob) | − | − |
| Arabinogalactan (larchwood) | − | − |
| Galactan (lupin) | + | − |
| Arabinan (sugar beet) | − | − |
| Konjac glucomannan | + + | + + |
| Pectins | ||
| Rhamnogalacturonan I (soybean) | − | − |
| Rhamnogalacturonan I (potato) | − | − |
| Pectic galactan (lupin) | + | − |
| Pectic galactan (potato) | + | − |
| Polygalacturonic Acid (citrus) | − | − |
| Pectin (apple) | − | − |
| Pectin (citrus) | − | − |
| Other polysaccharides | ||
| Pustulan | − | − |
| Pullulan | − | − |
a Symbols represent: +++, tight binding, ++, significant binding, +, marginal binding, −, no binding.
b HEC, hydroxyethylcellulose.
FIGURE 2.
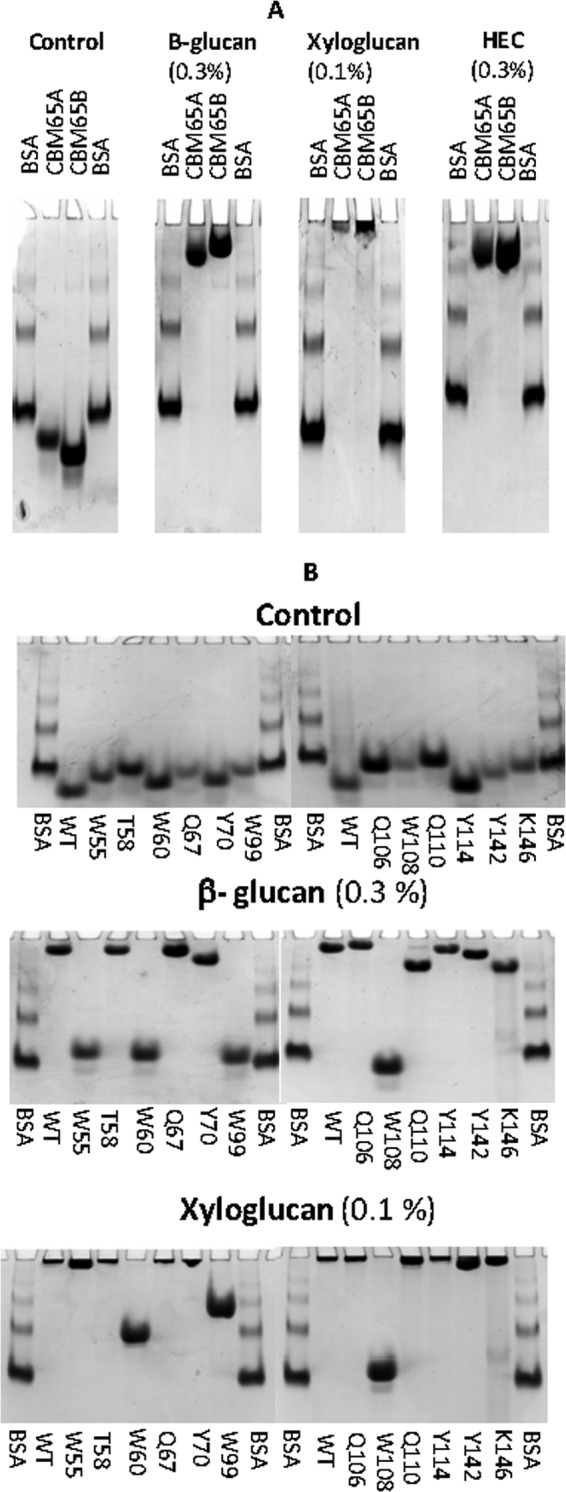
Examples of affinity gel electrophoresis of CBM65A and CBM65B against soluble polysaccharides. Panel A, the two CBM65 proteins were electrophoresed on nondenaturating polyacrylamide gels containing no ligand (control) or 0.3 mg/ml of the target polysaccharide (HEC, hydroxyethylcellulose). BSA was used as a nonpolysaccharide binding control. Panel B, wild type (WT) and mutants of CBM65A were electrophoresed in the presence or absence of the stated polysaccharides.
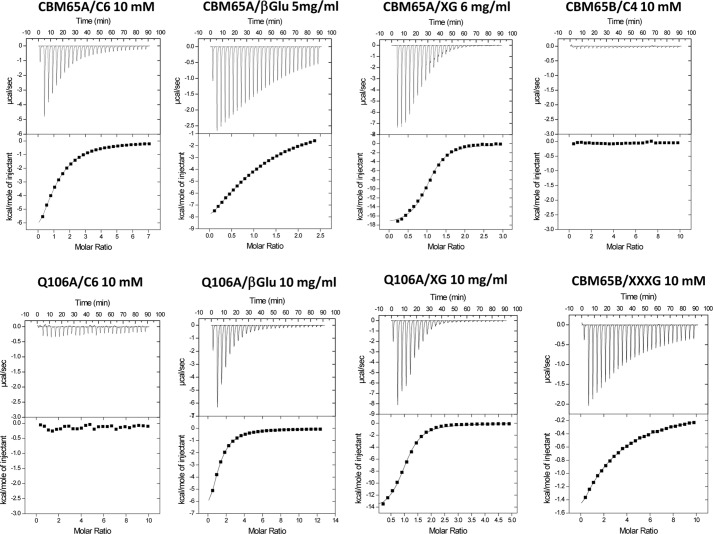
To provide a quantitative assessment of glucan recognition, the thermodynamic parameters of ligand binding were determined by ITC. Example titrations are shown in Fig. 3, and the full data set is displayed in Tables 3 and 4. The CBM65s displayed the highest affinity for xyloglucan, with a Ka of ∼105 m−1, whereas binding to barley β-glucan, a β1,3-β1,4 mixed linked glucan, and hydroxyethylcellulose was ∼10-fold weaker. With respect to oligosaccharides, the CBM65A displayed highest affinity for XXXG (X comprises glucose decorated at O6 with xylose and G corresponds to undecorated glucose), the repeating unit of xyloglucan, with a Ka of 5.6 × 103 m−1, and bound with a similar affinity to cellohexaose (Ka 3.6 × 103 m−1). Although binding to cellopentaose had an estimated Ka value of 1.2 × 103 m−1, no quantifiable binding to cellotetraose or smaller cellulooligosaccharides were observed. Similar to the binding of CBMs to soluble ligands (11, 33, 34) (see Ref. 6 for review), the interaction of the CBM65s with their target polysaccharides and oligosaccharides was driven by enthalpic changes, whereas the decrease in entropy had a negative impact on affinity. The stoichiometry of binding, assuming a single binding site for each CBM protomer, indicated that, at saturation, each protein molecule occupied ∼11 sugar residues arrayed in tandem in the backbone of the various polysaccharides. These data indicate that the two CBM65s binds to the internal regions of β-glucans. The specificity of the CBMs for, predominantly, β1,4- and β1,3-β-1,4 mixed linked glucans is entirely consistent with the activity of the parent enzyme, EcCel5A, which displays much higher activity against lichenan and carboxymethylcellulose than oat spelled xylan (18).
FIGURE 3.
Representative ITC data of CtCBM62 binding to soluble ligands. The ligand (C6, cellohexaose; XG, xyloglucan; β-Glu, barley β-glucan) in the syringe was titrated into CBM65A or Q106A (100 μm) in the cell. The top half of each panel shows the raw ITC heats; the bottom half shows the integrated peak areas fitted using one single binding model by MicroCal Origin software. ITC was carried out in 50 mm Na-HEPES, pH 7.5, at 25 °C.
TABLE 3.
Affinity and thermodynamic parameters of the binding of CBM65A and its variants to polysaccharide and oligosaccharide ligands
The molar concentration of a 1% solution of polysaccharide was iteratively adjusted to give a stoichiometry ∼1. In general each protein covered ∼11 sugar residues at saturation.

a HEC: hydroxyethylcellulose.
b Heptasaccharide derived from xyloglucan in which X is Glc decorated with Xyl, and G is undercorated Glc.
TABLE 4.
Affinity and thermodynamic parameters of the binding of CBM65B and its variant D69A to polysaccharide and oligosaccharide ligands
The molar concentration of a 1% solution of polysaccharide was iteratively adjusted to give a stoichiometry ∼1. In general each protein covered ∼11 sugar residues at saturation.
| CBM65B | Ligand | Ka | ΔG | ΔH | TΔS | n |
|---|---|---|---|---|---|---|
| M−1 | kcal mol−1 | |||||
| Wild type | β-Glucan | 8.2 (±0.2) × 103 | −5.3 | −12.1 ± 0.1 | −6.8 | 1.0 ± 0.0 |
| Wild type | Xyloglucan | 3.3 (±0.1) × 105 | −7.4 | −15.1 ± 0.1 | −7.7 | 1.0 ± 0.0 |
| Wild type | Cellohexaose | 2.3 (±0.2) × 103 | −4.6 | −12.8 ± 1.0 | −8.2 | 1 ± 0.3 |
| Wild type | XXXGa | 1.7 (±0.01) × 103 | −4.4 | −10.2 ± 0.0 | −5.8 | 1.0 ± 0.0 |
| Wild type | HECb | 1.42 (±0.4) × 104 | −5.6 | −7.0 ± 0.1 | −1.4 | 1.0 ± 0.0 |
| D649A | Cellohexaose | 1.5 (±0.05) × 103 | −4.3 | −8.1 ± 0.2 | −3.8 | 1.0 ± 0.2 |
a Heptasaccharide derived from xyloglucan in which X is Glc decorated with Xyl, and G is undecorated Glc.
b HEC, hydroxyethylcellulose.
Structure of CBM65A
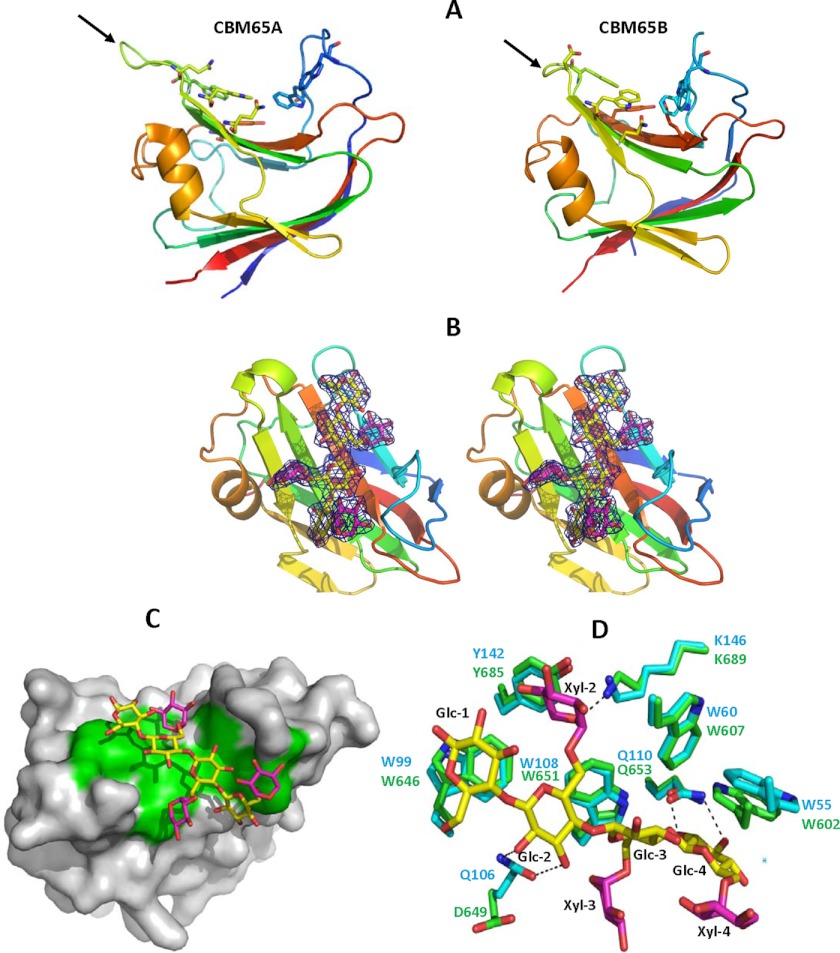
The crystal structure of the apo form of CBM65A was solved previously using the selenomethionine-SAD method (28), whereas the structure of apo-CBM65B was determined by molecular replacement to 1.42-Å resolution, using CBM65A as the search model. The two CBMs adopt a β-sandwich fold in which the β-sheets, comprising the convex (β-sheet 1) and concave surface (β-sheet 2) of the protein, contain five and four anti-parallel β-strands, respectively. The order of the β-strands in β-sheet 1 and β-sheet 2 are β1, β9, β3, β7, β6, and β2, β8, β4, β5, respectively. The β-strands are connected primarily by loops, although there is a small helix extending from Lys125 to Tyr132 in CBM65A and Lys668 to Tyr675 in CBM65B (Fig. 4A). The structures of CBM65B and CBM65A are very similar with an root mean square deviation over the 127 α-carbons of only 0.1 Å. In the majority of CBMs that adopt a β-sandwich fold the structure of these proteins is stabilized by a calcium bound to the loops connecting β3 and β4 (35), however, this conserved metal ion site is absent in both CBM65s.
FIGURE 4.
Structure of CBM65A. Panel A depicts CBM65A and CBM65B as a protein schematic, continuously color ramped from N to C terminus, from blue to red. The ligand binding residues are drawn as sticks. The arrows point to the loop in CBM65A and CBM65B that contain cellohexaose binding residues Gln106 and Asp649, respectively. Panel B shows a stereo representation of the ligand electron density (2Fo − Fc) at 1.5 σ. CBM65B is shown as a schematic representation colored as in panel A. XXXG is shown in stick format with Glc and Xyl carbons colored yellow and magenta, respectively. Panel C shows the solvent-accessible surface of CBM65B with XXXG bound to the surface with the ligand binding aromatic residues shown in green. Panel D shows an overlay of the ligand binding site of CBM65A (carbons of amino acids shown in green) and CBM65B (carbons of amino acids shown in cyan). Dashed lines between atoms show hydrogen bonds. The figure was drawn with PyMOL.
The Ligand Binding Site in CBM65
The ligand binding sites in CBMs that display a β-sandwich fold are, typically, located in the concave surface presented by one of the β-sheets, or at the end of the elliptical protein, within the loops connecting the two β-sheets (see Ref. 6 for review). Inspection of the concave surface of CBM65A and CBM65B reveals a cleft-like structure 20–25 Å long, rich in tryptophan residues. Substituting these aromatic residues with alanine caused a substantial reduction in ligand binding (described in detail below) (Table 3 and Fig. 2B), confirming that the concave surface presented by β-sheet 2 comprises the β-glucan binding site in CBM65A and, by inference, CBM65B.
To explore the mechanism of ligand recognition both CBM65A and CBM65B were co-crystallized with a variety of oligosaccharides. Clear electron density corresponding to XXXG was evident when CBM65B was crystallized in the presence of the xyloglucan-derived oligosaccharide. Despite extensive screening, no crystals of either CBM bound to cellulooligosaccharides, or CBM65A in complex with XXXG, were obtained. The structure of the CBM65B-XXXG complex, at a resolution of 2.35 Å, shows that the backbone of the ligand, comprising β1,4-cellotetraose, makes extensive hydrophobic contacts with the four tryptophans that line the cleft: Glc-1 (reducing end unsubstituted glucose) makes parallel apolar contacts with Trp646, Glc-2 and Glc-3 make extensive hydrophobic interactions with Trp651, whereas Trp602 interacts with Glc-4 through parallel hydrophobic contacts. Perpendicular apolar contacts between Trp607 and Glc-3 completes the interactions between the aromatic residues and the glucan tetrasaccharide backbone. Hydrophobic interactions between the tryptophan residues assist in fixing the orientation of the aromatic residues that bind to the glucan ligand. The topology of the tryptophans imposes a twisted conformation on the cellotetraose between Glc-2 and Glc-4, whereas Glc-2 and Glc-1 are orientated 180° with respect to each other. The only hydrogen bond between the tetrasaccharide backbone of XXXG and CBM65B is between O2 and O3 of Glc-4 with Oϵ1 and Nϵ2 of Gln653 (Fig. 4B).
With respect to the xylose side chains of XXXG, Xyl-3 forms apolar contacts with Trp651 and Xyl-2 makes hydrophobic interactions with Trp607, Trp646, Trp651, and Tyr685. The major polar interactions between XXXG and CBM65B are through O2 and the endocyclic O of Xyl-2, which make hydrogen bonds with the backbone N of Trp607 and the Nζ of Lys689, respectively. The polar and hydrophobic interactions made by the xylose side chains of XXXG make a significant contribution to CBM65 recognition. Indeed the affinity of xyloglucan for the CBM65s is 10-fold greater than undecorated β-glucans, whereas XXXG binds considerably more tightly to CBM65A than cellotetraose (affinity was too low to be quantified). All the residues in CBM65B that interact with XXXG are conserved in CBM65A (Fig. 4D). Thus, the mechanism of ligand recognition is likely to be very similar in the two proteins, although CBM65A may make an additional polar contact with the glucan backbone (see below).
Site-directed Mutagenesis of CBM65A
Site-directed mutagenesis was used to further investigate ligand recognition by CBM65A. The capacity of the mutants to bind to ligands was assessed by ITC (cellohexaose and polysaccharides) and affinity gel electrophoresis (polysaccharides). Examples of the affinity gels are shown in Fig. 2B, with the full dataset reported in Tables 3 and 4. Alanine substitution of Trp55, Trp108, Trp99, or Trp60 in CBM65A, which are structurally equivalent to Trp602, Trp607, Trp646, and Trp651 in CBM65B, abrogated cellohexaose and β-glucan recognition, confirming the importance of these aromatic residues in binding the β-linked glucan backbone in both CBM65A and CBM65B. With respect to polar contacts, mutating Gln110 in CBM65A (Q110A mutant), equivalent to Gln653 in CBM65B, significantly reduced, but did not abrogate, binding to both cellohexaose and β-glucan. It would appear, therefore, that Gln110 and Gln653 contribute to ligand recognition.
Both CBM65A and CBM65B bind to cellohexaose considerably more tightly than cellotetraose (see above), and thus it is possible that the CBM65s make more interactions with the hexasaccharide than the tetrasaccharides. Inspection of the binding cleft downstream of the four tryptophan residues failed to identify obvious ligand binding residues, although it is possible that a tyrosine, at the entrance to the binding cleft (Tyr70 in CBM65A and Tyr617 in CBM65B), and a glutamine (Gln67 in CBM65A and Gln659 in CBM65B) are potential candidates. However, as the Q67A and Y70A mutants of CBM65A displayed similar affinities to the wild type protein (Table 3), it is unlikely that Tyr70/Tyr617 or Gln67/Gln659 contribute to cellulose recognition.
Sequence alignment of CBM65A and CBM65B revealed 73% sequence identity and, as described above, XXXG recognition in CBM65B is conserved in CBM65A (Fig. 4C). A potentially biologically significant difference between the proteins is the loop connecting β4 and β5, which is longer in CBM65A (Trp99 to Gln106) than in CBM65B. Inspection of an overlay of the two proteins indicates that Oϵ1 and Nϵ2 of Gln106 will make hydrogen bonds with O2 and O3 of Glc-2 in the cellotetraose backbone, whereas the equivalent residue in CBM65B, Asp649, will be too distant from the ligand to make a polar contact. To test this hypothesis the specificity of the Q106A mutant of CBM65A and the D649A variant of CBM65B were analyzed. The affinities of the two variants for xyloglucan and barley β-glucan were similar to the corresponding wild type proteins. Although the D649A mutation did not affect the capacity of CBM65B to bind to cellohexaose, the Q106A variant of CBM65A did not display any detectable affinity for cellohexaose, indicating that Gln106 plays a critical role in the recognition of the hexasaccharide, and likely cellulose.
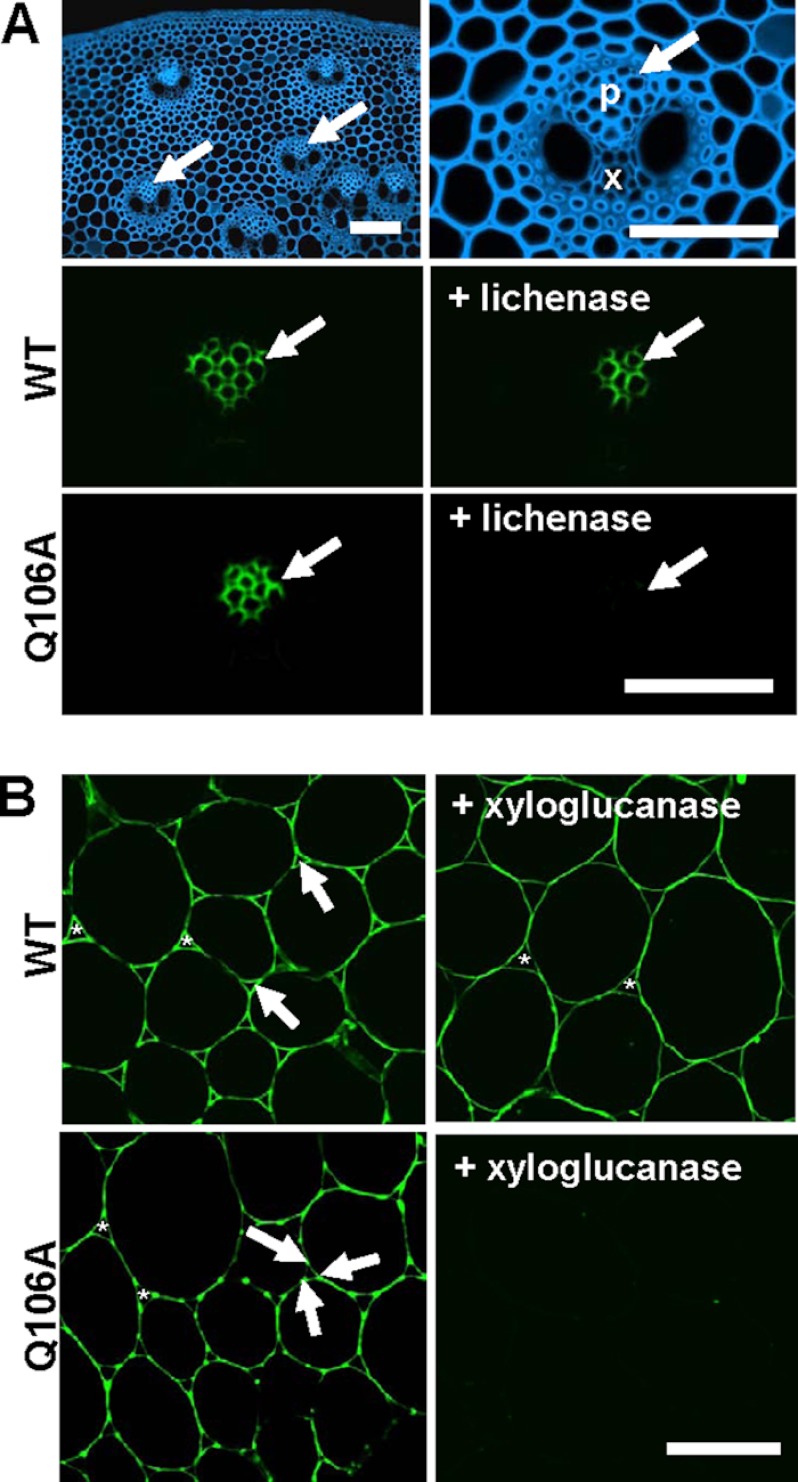
To provide further support for the view that the Q106A mutation has a significant impact on binding to cellulose but not to mixed linked β1,4-β1,3-glucans, the capacity of the mutant to bind to transverse sections of M. x giganteus stem was explored. The data (Fig. 5A) showed that the wild type protein bound specifically to phloem cell walls, before or after treatment with a lichenase, which specifically degrades mixed linked β1,4-β1,3-glucans; the enzyme does not attack β1,4-glucans (36). These data are consistent with the view that CBM65A is able to bind to both cellulose and β1,4-β1,3-glucans, polysaccharides, which are particularly abundant in the phloem cell walls of Miscanthus stems. Although the Q106A mutant bound to phloem cell walls in untreated Miscanthus stems, section treatment with the lichenase abrogated the binding of the CBM65A variant to these cell walls. These data are consistent with the ITC results in showing that the Q106A mutation influences binding to β1,4-glucans, but has no effect on binding to mixed linked β1,4-β1,3-glucans. A comparison of the capacity of the wild type and mutant protein to bind to tobacco stem cell walls, which contain no mixed linked β1,4-β1,3-glucans (Fig. 5B), and in which xyloglucan had been exposed by a prior treatment with pectate lyase, indicated that the wild type protein bound more strongly than Q106A and showed some differential labeling in relationship to cell walls at intercellular spaces (around which xyloglucans are known to be differentially regulated (23)). Section treatment with a xyloglucan-specific endo-1,4-β-glucanase resulted in loss of Q106A binding to cell walls, whereas binding of the wild type protein bound to xyloglucan-depleted cell walls was retained.
FIGURE 5.
Immunofluorescence analysis of CBM65A binding to cell walls in situ. Panel A, transverse sections of M. x giganteus stem. Calcofluor white shows staining of all cell walls (blue) and anatomy of a vascular bundle. In untreated sections both CBMs bind specifically to cell walls of the phloem (p) regions indicated by arrows; x = xylem. After lichenase pre-treatment of the section, before immunofluorescence analysis, wild type CBM65A (WT) still binds to the phloem cell walls but Q106A does not. All fluorescence micrographs have equivalent exposure times. Panel B, transverse sections of tobacco stem showing cell walls in the region of the pith parenchyma after pre-treatment with pectate lyase to remove pectic homogalacturonan. WT and Q106A displayed differential binding to parenchyma cell walls with WT binding strongly to cell walls and particularly to cell wall regions lining intercellular spaces (*) as indicated by arrows (exposure time 25 ms). Q106A bound less strongly to cell walls (exposure time 200 ms) and displayed some preferential binding to adhered cell wall regions at the corners of intercellular spaces; xyloglucan is known to be preferentially located in these regions (23). After a section pre-treatment with a xyloglucan-specific xyloglucanase, WT bound evenly to all cell walls with no differential binding in relationship to intercellular spaces, whereas Q106A did not bind (exposure time for both +xyloglucanase micrographs, 600 ms). Scale bars = 100 μm.
Structural Similarity of CBM65 to Other Proteins
Interrogation of the UNIPROT database revealed only two proteins, an endoglucanase from Cellulosilyticum ruminicola and one from Clostridium lentocellum, which displayed limited sequence similarity with the two CBM65s. The putative C. ruminicola endoglucanase contains two tandem repeated sequences and C. lentocellum a single sequence that displayed ∼30% sequence identity with CBM65A, corresponding to a Z-score of ∼0.085. Of potential significance is the observation that three of the four tryptophans that play a key role in glucan recognition in CBM65A and CBM65B are conserved in these three protein modules (supplemental Fig. S1), suggesting a similar role in β-glucan recognition. We propose, therefore, that CBM65A and CBM65B are the founding members of a new CAZy family, designated CBM65, which includes the two protein modules from the C. ruminicola and one from C. lentocellum putative endoglucanases.
With respect to three-dimensional structural similarity, the structural alignment program DaliLite version 3 revealed that the closest, functionally relevant, structural homolog of CBM65A is CBM30 from Clostridium thermocellum CtCel9D-Cel44A (PDB 2C24), with a Z score of 6.6, root mean square deviation of 3.7 Å over 117 aligned residues out of a possible 120 amino acids, and a total sequence identity of 18%. Several other CBMs showed similar levels of structural similarity with CBM65A, including CBM22 (PDB 1DYO) and CBM15 (PDB 1GNY). Although the overall fold and the location of the ligand binding site are conserved, the β-glucan binding residues in CBM65A and CBM65B are not retained in the other CBMs.
DISCUSSION
This report describes the structure of CBM65A and CBM65B, the founding members of a new CBM family that targets β-glucans. Similar to other CBMs that target β-glucans, the CBM65s display no significant binding to xylan. Such specificity can be achieved by the targeting of O6 groups (productive binding in the case of cellulose binders and through steric clashes in xylan-specific CBMs) that distinguishes gluco- from xylo-configured ligands (11, 14). In the CBM65B-XXXG complex C6 of all the backbone glucose moieties make extensive hydrophobic interactions with the surface tryptophans, and thus are likely to make a significant contribution to overall affinity. Furthermore, the CBM65s are not optimized to bind xylan, which adopts a 3-fold screw axis conformation (37). Xylan-specific CBMs often contain a pair of tryptophans, orientated at 120° with respect to each other, which bind to xylose residues n and n+2 in the polysaccharide (38). Disruption of the orientation of these aromatic residues can convert a xylan binding CBM into a cellulose-specific protein (38). In the CBM65s the ligand binding tryptophans are not optimized to bind to a polysaccharide that has a regular 3-fold helical structure. Indeed Glc-1 and Glc-2 are orientated at 180° to each other, and the hydrogen bond between O3 and O6, which would be absent in a xylan chain, plays a critical role in stabilizing the conformation adopted by these two sugars.
CBM65A and CBM65B display higher affinity for oligosaccharides (e.g. cellohexaose), and particularly polysaccharides (xyloglucan and barley β-glucan), than cellotetraose, which fully occupies the core component of the substrate binding cleft. Although it is formally possible that the two additional residues in cellohexaose provide additional contacts with the protein, mutagenesis of residues in the vicinity of the two glucose moieties in CBM65A did not influence affinity. In cellooligosaccharides with a degree of polymerization >4 the Glc that interacts with Trp99 in CBM65A is internal and thus the pyranose ring is fixed. In cellotetraose the equivalent Glc is at the reducing end of the tetrasaccharides, and hence adopts multiple conformations through mutarotation. Thus, the reduction in entropy upon binding the tetrasaccharide may explain the weak affinity. It is evident, however, that polysaccharides, in which the backbone is either mixed linked or a β1,4-linked polymer binds more tightly than cellohexaose, indicating that fixing the conformation of the terminal sugar is not the sole reason for the tighter binding of ligands with a degree of polymerization >4. There are examples of ligands that extend outside the CBM binding region, which bind more tightly to the protein than smaller glycans that, nevertheless, fully occupy the sugar binding sites (11, 39, 40). It has been suggested that the longer ligands adopt a more fixed conformation, through extensive intra-chain hydrogen bonds, which is optimized to recognize the target CBM (39). An alternative possibility is that the CBMs physically associate, resulting in increased affinity for multivalent ligands through avidity effects (15, 40, 41). However, size exclusion chromatography indicated that CBM65A is a monomer (data not shown), although ligand-induced oligomerization is possible; a phenomenon, which would not be observed by studying the molecular mass of the apoprotein (42).
The observation that the Q106A mutation destroys binding to cellohexaose and cellulose, but not xyloglucan or β1,3-β1,4 mixed linked glucans, is intriguing. These data suggest that CBM65A may display flexibility in ligand recognition, with its binding site capable of recognizing both linear β1,4-glucans and β1,3-β-1,4 mixed linked glucans, and that Gln106 only contributes to cellulose recognition. It is also interesting that CBM65B, despite lacking a functionally equivalent residue to Gln106 displays affinity for cellohexaose, although the mechanism by which the protein module retains this specificity is unclear.
This report provides insights into how a CBM can specifically recognize xyloglucan in preference to other β-glucans. Previously, Najmudin et al. (12) showed that β-glucan binding CBMs can accommodate, but do not display a preference for, xyloglucan. The primary mechanism by which CBM65B binds to the xylose side chains is through apolar interactions with the surface aromatic amino acids. In particular, Tyr685 makes extensive hydrophobic contacts with Xyl-2, although the sugar also makes apolar contacts with Trp607, Trp646, and Trp651. The CBM65s are similar to many CBMs (11, 38, 43) (see Ref. 6 for review), where binding to glycan chains is dominated by hydrophobic interactions with aromatic residues. In typical β-glucan binding CBMs there are three aromatic residues that stack against the sugar rings, or against both faces of the same pyranose. In the CBM65s, however, the binding cleft contains five aromatic residues. Only Glc-4 aligns perfectly with a tryptophan to maximize planar hydrophobic contacts with a tryptophan (Trp602). The side chains of the other aromatic amino acids make apolar contacts with both the backbone Glc and appended xylose residues or, in the case of Tyr685, only with the sugar decoration.
Mutagenesis was used to explore the role of aromatic residues in xyloglucan recognition. Alanine substitution of Trp108 in CBM65A, equivalent to Trp651 in CBM65B, completely abrogated ligand recognition, whereas the mutations W60A and W99A caused a substantial reduction in affinity. This is consistent with the central role Trp651/Trp108 plays in xyloglucan recognition, interacting with Glc-2, Glc-3, Xyl-2, and Xyl-3, whereas also stabilizing the conformation adopted by all the key ligand binding aromatic residues, except Trp602/Trp55. The importance of the central tryptophan in CBM65s has some resonance with studies on CBM2a, where cellulose binding is also dominated by the central aromatic residue (44). Mutation of Trp55 in CBM65A had little influence on affinity for xyloglucan. The equivalent residue in CBM65B, Trp602, although interacting with Glc-4, makes no apolar contact with the xylose side chains, and hence its contribution to xyloglucan recognition is considerably less than the other aromatic residues in the ligand binding cleft. Thus, xyloglucan recognition is dominated by aromatic residues that recognize both the glucan backbone and the xylose side chains.
To summarize, this report describes the biochemical properties of two CBMs that are the founding members of CBM65. The protein modules bind to mixed linked β1,4-β1,3-linear and decorated β1,4-glucans, but displays a preference for the decorated β-glucan, xyloglucan. Specificity for decorated glucans is achieved through an extensive hydrophobic platform that contacts both the glucan backbone and the xylose side chains. Significantly, one of few hydrogen bonds between CBM65A and its ligands confers specificity for cellulose. Thus, this article shows that in CBM65, specificity for diverse β1,4-glucans is not achieved through the targeting of conserved features of these glycans, whereas the work also reveals how the orientation of hydrophobic residues can be optimized to recognize backbone and side chain sugars, providing a model for the recognition of decorated polysaccharides.
This work was supported in part by grants from the Biochemical and Biotechnological Research Council (BB/K001949/1), Wellcome Research Trust (WT097907MA), Fundação para a Ciência e a Tecnologia (PTDC/BIA-PRO/103980/2008 and PTDC/BIA-PRO/69732/2006), and from the European Union Seventh Framework Programme (FP7 2007–2013) under the WallTraC project (Grant Agreement no. 263916). We also acknowledge the Synchrotron Radiation Facilities at SOLEIL (PROXIMA-1) and DIAMOND (beamlines IO2 and IO4).

This article contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (codes 4ACK, 2YPJ, and 4AFM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CBM
- carbohydrate binding module
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank.
REFERENCES
- 1. Flint H. J., Duncan S. H., Scott K. P., Louis P. (2007) Interactions and competition within the microbial community of the human colon. Links between diet and health. Environ. Microbiol. 9, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 2. Mackie R. I., White B. A. (1990) Recent advances in rumen microbial ecology and metabolism. Potential impact on nutrient output. J. Dairy Sci. 73, 2971–2995 [DOI] [PubMed] [Google Scholar]
- 3. Himmel M. E., Bayer E. A. (2009) Lignocellulose conversion to biofuels. Current challenges, global perspectives. Curr. Opin. Biotechnol. 20, 316–317 [DOI] [PubMed] [Google Scholar]
- 4. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance. Engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert H. J. (2010) The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boraston A. B., Kwan E., Chiu P., Warren R. A., Kilburn D. G. (2003) Recognition and hydrolysis of noncrystalline cellulose. J. Biol. Chem. 278, 6120–6127 [DOI] [PubMed] [Google Scholar]
- 9. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., Knox J. P. (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 11. Boraston A. B., Nurizzo D., Notenboom V., Ducros V., Rose D. R., Kilburn D. G., Davies G. J. (2002) Differential oligosaccharide recognition by evolutionarily-related β1,4- and β1,3-glucan-binding modules. J. Mol. Biol. 319, 1143–1156 [DOI] [PubMed] [Google Scholar]
- 12. Najmudin S., Guerreiro C. I., Carvalho A. L., Prates J. A., Correia M. A., Alves V. D., Ferreira L. M., Romão M. J., Gilbert H. J., Bolam D. N., Fontes C. M. (2006) Xyloglucan is recognized by carbohydrate-binding modules that interact with β-glucan chains. J. Biol. Chem. 281, 8815–8828 [DOI] [PubMed] [Google Scholar]
- 13. Simpson P. J., Bolam D. N., Cooper A., Ciruela A., Hazlewood G. P., Gilbert H. J., Williamson M. P. (1999) A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity. Structure 7, 853–864 [DOI] [PubMed] [Google Scholar]
- 14. Czjzek M., Bolam D. N., Mosbah A., Allouch J., Fontes C. M., Ferreira L. M., Bornet O., Zamboni V., Darbon H., Smith N. L., Black G. W., Henrissat B., Gilbert H. J. (2001) The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J. Biol. Chem. 276, 48580–48587 [DOI] [PubMed] [Google Scholar]
- 15. Montanier C. Y., Correia M. A., Flint J. E., Zhu Y., Baslé A., McKee L. S., Prates J. A., Polizzi S. J., Coutinho P. M., Lewis R. J., Henrissat B., Fontes C. M., Gilbert H. J. (2011) A novel, noncatalytic carbohydrate-binding module displays specificity for galactose-containing polysaccharides through calcium-mediated oligomerization. J. Biol. Chem. 286, 22499–22509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Creagh A. L., Ong E., Jervis E., Kilburn D. G., Haynes C. A. (1996) Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. U.S.A. 93, 12229–12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. (2004) The family 6 carbohydrate binding module CmCBM6–2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 18. Yoda K., Toyoda A., Mukoyama Y., Nakamura Y., Minato H. (2005) Cloning, sequencing, and expression of a Eubacterium cellulosolvens 5 gene encoding an endoglucanase (Cel5A) with novel carbohydrate-binding modules, and properties of Cel5A. Appl. Environ. Microbiol. 71, 5787–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Najmudin S., Guerreiro C. I., Ferreira L. M., Romão M. J., Fontes C. M., Prates J. A. (2005) Overexpression, purification and crystallization of the two C-terminal domains of the bifunctional cellulase ctCel9D-Cel44A from Clostridium thermocellum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carvalho A. L., Goyal A., Prates J. A., Bolam D. N., Gilbert H. J., Pires V. M., Ferreira L. M., Planas A., Romão M. J., Fontes C. M. (2004) The family 11 carbohydrate-binding module of Clostridium thermocellum Lic26A-Cel5E accommodates β1,4- and β1,3–1,4-mixed linked glucans at a single binding site. J. Biol. Chem. 279, 34785–34793 [DOI] [PubMed] [Google Scholar]
- 21. McCartney L., Blake A. W., Flint J., Bolam D. N., Boraston A. B., Gilbert H. J., Knox J. P. (2006) Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc. Natl. Acad. Sci. U.S.A. 103, 4765–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCartney L., Gilbert H. J., Bolam D. N., Boraston A. B., Knox J. P. (2004) Glycoside hydrolase carbohydrate-binding modules as molecular probes for the analysis of plant cell wall polymers. Anal. Biochem. 326, 49–54 [DOI] [PubMed] [Google Scholar]
- 23. Marcus S. E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J. J., Farkas V., Pedersen H. L., Willats W. G., Knox J. P. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leslie A. G. W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4/ESF-EAMCB Newsl. Protein Crystallogr. 26, 27–33 [Google Scholar]
- 25. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 27. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luis A. S., Alves V. D., Romao M. J., Prates J. A., Fontes C. M., Najmudin S. (2011) Overproduction, purification, crystallization and preliminary x-ray characterization of a novel carbohydrate-binding module of endoglucanase Cel5A from Eubacterium cellulosolvens. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 491–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Vagin A., Teplyakov A. (1997) MOLREP. An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 33. Bolam D. N., Xie H., Pell G., Hogg D., Galbraith G., Henrissat B., Gilbert H. J. (2004) X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J. Biol. Chem. 279, 22953–22963 [DOI] [PubMed] [Google Scholar]
- 34. Henshaw J., Horne-Bitschy A., van Bueren A. L., Money V. A., Bolam D. N., Czjzek M., Ekborg N. A., Weiner R. M., Hutcheson S. W., Davies G. J., Boraston A. B., Gilbert H. J. (2006) Family 6 carbohydrate binding modules in β-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J. Biol. Chem. 281, 17099–17107 [DOI] [PubMed] [Google Scholar]
- 35. Simpson P. J., Jamieson S. J., Abou-Hachem M., Karlsson E. N., Gilbert H. J., Holst O., Williamson M. P. (2002) The solution structure of the CBM4–2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry 41, 5712–5719 [DOI] [PubMed] [Google Scholar]
- 36. Money V. A., Cartmell A., Guerreiro C. I., Ducros V. M., Fontes C. M., Gilbert H. J., Davies G. J. (2008) Probing the β-1,3:1,4-glucanase, CtLic26A, with a thio-oligosaccharide and enzyme variants. Org. Biomol. Chem. 6, 851–853 [DOI] [PubMed] [Google Scholar]
- 37. Nieduszy I., Marchess R. H. (1972) Structure of β-d-(1–4′)-xylan hydrate. Biopolymers 11, 1335–1344 [Google Scholar]
- 38. Simpson P. J., Xie H., Bolam D. N., Gilbert H. J., Williamson M. P. (2000) The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J. Biol. Chem. 275, 41137–41142 [DOI] [PubMed] [Google Scholar]
- 39. Boraston A. B., Healey M., Klassen J., Ficko-Blean E., Lammerts van Bueren A., Law V. (2006) A structural and functional analysis of α-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J. Biol. Chem. 281, 587–598 [DOI] [PubMed] [Google Scholar]
- 40. Charnock S. J., Bolam D. N., Nurizzo D., Szabó L., McKie V. A., Gilbert H. J., Davies G. J. (2002) Promiscuity in ligand-binding. The three-dimensional structure of a Piromyces carbohydrate-binding module, CBM29–2, in complex with cello- and mannohexaose. Proc. Natl. Acad. Sci. U.S.A. 99, 14077–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bolam D. N., Xie H., White P., Simpson P. J., Hancock S. M., Williamson M. P., Gilbert H. J. (2001) Evidence for synergy between family 2b carbohydrate binding modules in Cellulomonas fimi xylanase 11A. Biochemistry 40, 2468–2477 [DOI] [PubMed] [Google Scholar]
- 42. Flint J., Nurizzo D., Harding S. E., Longman E., Davies G. J., Gilbert H. J., Bolam D. N. (2004) Ligand-mediated dimerization of a carbohydrate-binding molecule reveals a novel mechanism for protein-carbohydrate recognition. J. Mol. Biol. 337, 417–426 [DOI] [PubMed] [Google Scholar]
- 43. Szabo L., Jamal S., Xie H., Charnock S. J., Bolam D. N., Gilbert H. J., Davies G. J. (2001) Structure of a family 15 carbohydrate-binding module in complex with xylopentaose. Evidence that xylan binds in an approximate 3-fold helical conformation. J. Biol. Chem. 276, 49061–49065 [DOI] [PubMed] [Google Scholar]
- 44. McLean B. W., Bray M. R., Boraston A. B., Gilkes N. R., Haynes C. A., Kilburn D. G. (2000) Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose. Specificity and identification of functionally important amino acid residues. Protein Eng. 13, 801–809 [DOI] [PubMed] [Google Scholar]