Background: A. gambiae transglutaminase AgTG3 cross-links Plugin within seminal fluids.
Results: AgTG3 and Plugin were purified and their structure and activity characterized in vitro.
Conclusion: AgTG3 forms a dimer in solution, analogous to human FXIII. Plugin is nonglobular in solution. AgTG3 is Ca2+-dependent and prefers Plugin as a substrate.
Significance: Inhibition of AgTG3 cross-linking of Plugin is a possible method to chemosterilize male Anopheles mosquitoes.
Keywords: Analytical Ultracentrifugation, Circular Dichroism (CD), Protein Expression, Transglutaminases, X-ray Scattering
Abstract
Male Anopheles mosquitoes coagulate their seminal fluids via cross-linking of a substrate, called Plugin, by the seminal transglutaminase AgTG3. Formation of the “mating plug” by cross-linking Plugin is necessary for efficient sperm storage by females. AgTG3 has a similar degree of sequence identity (∼30%) to both human Factor XIII (FXIII) and tissue transglutaminase 2 (hTG2). Here we report the solution structure and in vitro activity for the cross-linking reaction of AgTG3 and Plugin. AgTG3 is a dimer in solution and exhibits Ca2+-dependent nonproteolytic activation analogous to cytoplasmic FXIII. The C-terminal domain of Plugin is predominantly α-helical with extended tertiary structure and oligomerizes in solution. The specific activity of AgTG3 was measured as 4.25 × 10−2 units mg−1. AgTG3 is less active than hTG2 assayed using the general substrate TVQQEL but has 8–10× higher relative activity when Plugin is the substrate. Mass spectrometric analysis of cross-linked Plugin detects specific peptides including a predicted consensus motif for cross-linking by AgTG3. These results support the development of AgTG3 inhibitors as specific and effective chemosterilants for A. gambiae.
Introduction
Transglutaminases (TGs)2 are ubiquitous enzymes that catalyze the deamidation and transamidation of glutamine and the cross-linking of proteins by formation of ϵ-(γ-glutamyl)-lysine isopeptide bonds (1). Mammals possess multiple TGs that are involved in processes such as blood clotting by Factor XIII (FXIII), regulating cellular responses to stress by tissue transglutaminase (TG2), formation of the epithelial barrier (TG3), and coagulation of seminal plasma (TG4) (2). Similar roles for TGs have been identified in insects yet with a reduced gene set. The single TG gene present in Drosophila is involved in cuticle morphogenesis (3) and coagulation of hemolymph in response to septic injury (4). This TG is conserved in mosquitoes (Culicidae) including the malaria vector Anopheles gambiae (AGAP009100, or AgTG1). A second TG is specific to Culicidae (A. gambiae AGAP009098, or AgTG2), whereas a third TG is specific to Anopheles: A. gambiae AGAP009099, hereafter termed AgTG3.
Although AgTG1 and AgTG2 have yet to be functionally characterized, AgTG3 has recently been found to play a role in male Anopheles fertility (5). In many animals the coagulation of male seminal fluids results in the formation of a “copulatory plug” or “mating plug” that completely occludes the female reproductive tract. A recent study of A. gambiae seminal fluid proteins and male accessory glands showed that the plug is formed by cross-linking of a substrate protein called Plugin (AGAP009368) by AgTG3 (5). Furthermore, dsRNAi-mediated knockdown of AgTG3 inhibited proper sperm storage by females, indicating that correct formation of the mating plug is required for reproductive success of Anopheles males. Within Culicidae, formation of a mating plug is specific to Anopheles (6). TG-mediated cross-linking of seminal fluid proteins occurs in rodents (7), but other invertebrates use different mechanisms, such as secretion of O-glycosylated mucins in Caenorhabditis elegans (8) and homopolymerization of PEBII in Drosophila melanogaster (9).
Formation of A. gambiae mating plug is interesting because it represents not only the independent evolution of a seminal TG in an insect but also a target for control of Anopheles, which are vectors for malaria, the world's most devastating parasitic disease. Targeting fertility of insect populations, the sterile insect technique, is a classic method of pest control first used to control the cattle screwworm fly (10). Field tests with radiation-sterilized, chemosterilized, or genetically modified sterile mosquitoes have demonstrated feasibility of the sterile insect technique for public health purposes (11–13), provided that logistical and regulatory issues are considered (14–16). A potent and specific inhibitor of AgTG3 could serve as a safe and effective chemosterilant for use alone or in combination with radiation and/or genetically modified mosquitoes to advance the application of sterile insect technique for Anopheles mosquitoes.
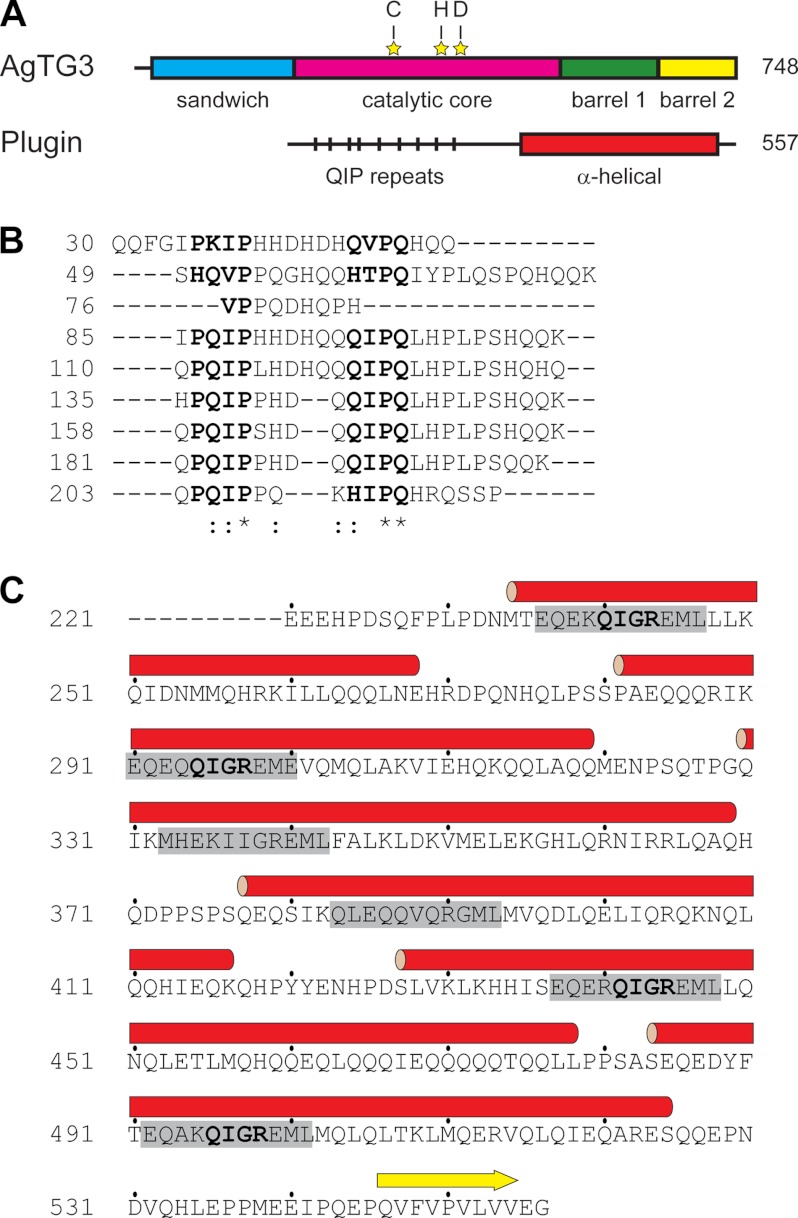
AgTG3 contains four canonical domains of the α/β TG fold and residues of the catalytic triad (Fig. 1A). The substrate Plugin has an unusual sequence with two distinct, repetitive domains. An N-terminal 30-kDa region (Fig. 1B) has a primary sequence dominated by Gln, His, and Pro, with multiple repeats of the consensus sequence PQIP(X)3–6QIPQ(X)8–9K and no predicted secondary structure. In contrast, the C-terminal domain is rich in both Gln and Glu (Fig. 1C), consisting of six α-helices commencing with a consensus sequence EQE(K/R)QIGREML. Thus, the N- and C-terminal domains of Plugin are structurally distinct, yet both are rich in the TG substrate motif QI(P/G) and accompanying Lys residues.
FIGURE 1.
Sequence of AgTG3 and Plugin. A, schematic diagram of AgTG3 and Plugin amino acid sequence with predicted tertiary and secondary structure, respectively. Residues of the catalytic triad indicated by yellow stars. B, alignment of Plugin N-terminal repeats containing QIP motif. C, sequence of Plugin C-terminal domain. The predicted α-helices are shown as red cylinders, with the consensus sequence in each helix highlighted in gray.
AgTG3 is homologous at ∼30% sequence identity to all TGs of known structure: FXIII, hTG2, hTG3, and Pagrus major TG (17–21). FXIII is a dimer in solution whereas other TGs are monomeric. All TGs are activated by calcium (Ca2+), but only one structure, hTG2, bound by a peptidomimetic inhibitor, exhibits the presumed active conformation (21); other structures adopt a conformation in which the catalytic site is occluded by the β-barrel domains even in the presence of Ca2+. Other mechanisms of regulation exist for TGs besides Ca2+. Serum FXIII requires proteolysis by thrombin for activation, although cytoplasmic FXIII can be activated without proteolysis (22). The activation of hTG2 is inhibited by the binding of GTP (23). Hence, some initial questions regarding the mechanism of AgTG3-mediated cross-linking of Plugin are the oligomeric states of both enzyme and substrate in solution, the specific activity of AgTG3, and the requirements for its activity.
Here we report the expression, purification, and biophysical characterization of AgTG3 and its substrate Plugin. The in vitro reconstitution of Plugin cross-linking by AgTG3 was performed, and specific cross-linking sites within Plugin were identified. We demonstrate that AgTG3 can exist as both a monomer and dimer whereas Plugin has an extended or partially unfolded structure and tendency to oligomerize in solution. We measured the specific activity of purified AgTG3 by a fluorescent assay both in solution and in a high throughput plate-based format and confirm that the activity of AgTG3 is dependent on Ca2+ but not GTP. Our results provide the basis for further molecular characterization of the AgTG3-Plugin mechanism of seminal fluid coagulation and screening for specific AgTG3 inhibitors.
EXPERIMENTAL PROCEDURES
Cloning
Full-length clones of Plugin (AGAP009368) and AgTG3 (AGAP009099) were amplified from male accessory gland cDNAs prepared from 4-days old virgin A. gambiae males and cloned into a pBluescript plasmid. Amplification was performed using the following primers: Plugin_FWD, 5′-GAATTCCATATGAAGGCTTTGGTAGCTCTGC-3′ and Plugin_REV, 5′-GAATTCTTCACTGCGGAAGCACTCC-3′; AgTG3 9099_FWD, 5′-CGGCAGCCATATGTCTACCACCTCTACCAACTACCG-3′ and AgTG3 9099_REV, 5′-CCGGAATTCCAGCTTAAGGAATATCAATGGTGAGA-3′(EcoRI and NdeI restriction sites underlined). AgTG3 was subcloned into pET28a with a C-terminal His6 tag. A C-terminal fragment of Plugin encoding amino acid residues 345–557 (Plugin-C) was amplified by PCR with primers 5′-GCTTCCATGGCACTCAAGCTCG-3′ and 5′-GCTTCTCGAGACCTTCGACGACC-3′ (NcoI/XhoI restriction sites underlined). The PCR product was digested with NcoI/XhoI and ligated into the pHis-parallel vector (24), comprising an N-terminal His6 tag and tobacco etch virus cleavage site. All constructs were verified by DNA sequencing.
Purification of AgTG3 and Plugin
Recombinant protein was expressed in Escherichia coli using LB medium, RosettaTM (Novagen) for AgTG3, and BL21(DE3) for Plugin-C. Cells were induced at A600 ∼ 0.5 with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and grown overnight at 18 °C. All purification steps were performed at 4 °C.
For AgTG3 purification, cells were lysed by sonication in 20 mm HEPES, pH 7.5, 500 mm NaCl, 20 mm imidazole, 10% (w/v) glycerol. The lysate was clarified by centrifugation at 40,000 × g at 4 °C for 30 min, loaded onto a HisTrap HP column (GE Healthcare), and eluted with a 0–1 m imidazole gradient. AgTG3 was further purified by Superdex 200 (GE Healthcare) size exclusion chromatography (SEC) in 25 mm HEPES, pH 7.5, 0.1 m NaCl, 10% (w/v) glycerol, 2 mm TCEP.
For Plugin-C purification, cells were lysed by sonication in 20 mm HEPES, pH 7.5, 500 mm NaCl, 20 mm imidazole, and the lysate was clarified by centrifugation. The supernatant was loaded onto a HisTrap HP column, eluted with a 0–600 mm imidazole gradient and dialyzed against a tobacco etch virus cleavage buffer (25 mm Tris-HCl, pH 8.0, 150 mm NaCl). Except in the case of plate-based assays, the His6 tag was removed by treatment with tobacco etch virus protease overnight at room temperature and reapplication to a HisTrap column. Plugin-C was further purified by Superdex 200 SEC in 25 mm HEPES, pH 7.5, 0.1 m NaCl.
Circular Dichroism Spectroscopy (CD)
Both AgTG3 and Plugin-C were concentrated to A280 = 0.5 in 10 mm KH2/K2HPO4, pH 7.4, 1 mm DTT. Spectra were obtained on a Jasco J-810 (average of three scans, 1-nm step) at 25 °C. Data were deconvoluted using K2D2 (25).
Size Exclusion Chromatography with Multiangle Laser Light Scattering (SEC-MALLS)
Plugin-C (0.3 mg/ml) was loaded onto a Superdex 200 (10/300) column (GE Healthcare) equilibrated with 20 mm HEPES, pH 7.5, 100 mm NaCl, 0.5 mm TCEP. Peaks were detected with an in-line UV detector (Jasco UV975) at 280 nm, a light scattering detector (DAWN EOS; Wyatt Technology Corp.) at 690 nm, and a refractive index detector (Optilab; Wyatt Technology Corp.). Protein concentration was based on differential refractive index (dn/dc). The molecular mass was determined from the Debye plot of light scattering intensity versus scattering angle (Astra software; Wyatt Technology Corp.).
Analytical Ultracentrifugation (AUC)
AgTG3 and Plugin-C (0.5 mg/ml) were analyzed by sedimentation velocity in a Beckman XL-1 centrifuge at 20 °C and 42,000 × g (AgTG3), 55,000 × g (Plugin). Data were analyzed with SEDFIT (26) with parameters ¯ν = 0.7337 cm3 g−1, ρ = 1.0096 g cm−3, η = 0.01 P for AgTG3, and ¯ν = 0.7369 cm3 g−1, ρ = 1.0050 g cm−3, η = 0.01 P for Plugin-C.
Small Angle X-ray Scattering (SAXS)
Ca2+-AgTG3 was purified by SEC in 25 mm HEPES, pH 7.5, 100 mm NaCl, 10 mm CaCl2, 1 m glycerol, and prepared at 1.0–5.0 mg/ml. Plugin-C was prepared in TBS at 0.5 mg/ml. SAXS data were collected at NSLS beamline X9 (Brookhaven National Laboratory). A 50-μl sample was loaded by autosampling into a capillary cuvette (1 mm) at 60 μl min−1, 2-s exposures. Data were analyzed using in-house (pyXS) and ATSAS (27) software.
FITC Incorporation Assay
Plugin-C cross-linking was performed in 25 mm HEPES, pH 7.5, 10 mm CaCl2, 10 mm DTT, 0.5 mm FITC-cadaverine (FITC-CAD) at room temperature. After 2 h, the reaction was quenched by heat denaturation of proteins in the presence of Laemmli buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was scanned using Alpha Imager 2200 software.
Specific Activity Assays
All assays are the average of three measurements. For the solution assay, Plugin-C was reductively methylated with dimethylamine borane using standard procedures (28). Assay conditions were 1.25 ml of 150 mm NaCl, 55 mm HEPES, pH 7.5, 10 mm DTT, 100 μm FITC-CAD, 1 mg/ml Plugin-C, 9.6 μg/ml AgTG3, and 5 mm CaCl2. Solutions were prewarmed to 30 °C before addition of CaCl2 (6.25 μl of 1 m stock) to initiate reactions. At the indicated time points, 200-μl aliquots were removed, mixed with 22 μl of 0.5 m iodoacetamide, and incubated for 30 min at room temperature. Aliquots were diluted to 0.5 ml with PBS and loaded onto a PD-10 desalting column (GE Healthcare) followed by 2 ml of PBS, eluted with 1.5 ml, and diluted to 3.5 ml with PBS. Fluorescence (485/515) was measured on a JY Horiba Fluorolog-3 and fluorescence quantified relative to a FITC-CAD standard curve.
ELISAs for specific activity relative to guinea pig TG2 were performed with a standard kit (Sigma CS1070). For Plugin plated-based assay, Plugin-C (25 μg/well) was incubated in black 96-well Ni-coated plates (Pierce 15342) for 2 h at room temperature or overnight at 4 °C. Plates were washed three times with 200 μl of TBS and loaded with 50 μl of AgTG3 (diluted to given concentration with water), then 50 μl of 2× assay buffer (2× TBS, 20 mm CaCl2, 100 μm FITC-CAD, 2 mm DTT). Wells were incubated for 60 min at room temperature, washed three times with 200 μl of TBS, and filled to 100-μl final volume for fluorescence measurement in a microplate reader (Biotek Synergy 2). Inhibition was performed by 30-min preincubation with 50 mm iodoacetamide (Sigma A3221).
Mass Spectrometry
Cross-linked Plugin-C was separated on SDS-PAGE, excised, and subjected to in-gel tryptic digest. Peptides were separated on a Waters nanoACQUITY (75 μm × 250 mm eluted at 300 nl/min) and analyzed on a LTQ Orbitrap mass spectrometer. Results were analyzed using MassMatrix (29). Identities of crosslinked peptides were confirmed by Collision-induced Dissociation tandem mass spectrometry (CID-MS/MS).
RESULTS
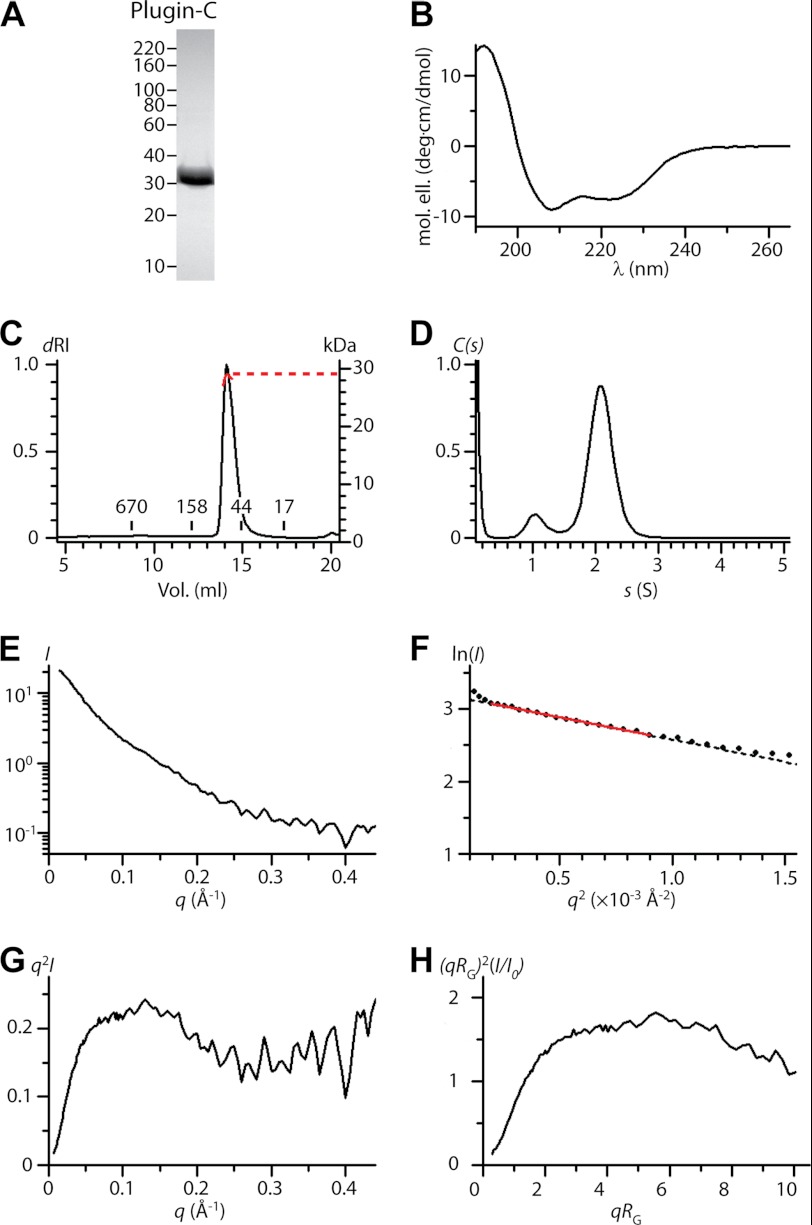
AgTG3 was expressed and purified to homogeneity in E. coli (Fig. 2A). Purification and SDS-PAGE were equivalent in reducing and nonreducing conditions; hence, AgTG3 contains no disulfide bonds. The AgTG3 CD spectrum (Fig. 2B) corresponds to 40–50% β-sheet and 5–15% α-helix. In the presence of 10 mm CaCl2 AgTG3 elutes from Superdex 200 column with an apparent molecular mass of ∼160 kDa suggesting a dimeric species (Fig. 2C). In the presence of 1 mm EDTA AgTG3 appears more heterogeneous, with the appearance of a shoulder peak at the apparent molecular mass of a monomer. AUC sedimentation velocity analysis of Ca2+-AgTG3 (Fig. 2D) gives a molecular mass estimate of ∼170 kDa (s20,w = 7.0, f/f0 = 1.4), consistent with the estimate from SEC.
FIGURE 2.
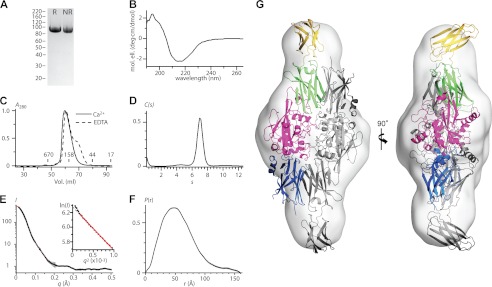
Structure of AgTG3 in solution. A, reducing (R) and nonreducing (NR) SDS-PAGE of purified AgTG3. B, CD spectrum. C, SEC in presence of Ca2+ (solid line) and EDTA (dashed line). Retention volume of molecular mass standards is indicated. D, sedimentation velocity C(s) distribution for Ca2+-AgTG3, s20,w = 7.0 (f/f0 = 1.4). E–H, Ca2+-AgTG3 SAXS analysis. E, I versus q (black line) fit by p(r) distribution (red line) (inset) Guinier plot fit to RG = 42.7 Å (limit qRG = 1.35). F, p(r) distribution, Dmax = 160 Å. G, superposition of ab initio volume with P2 symmetry to Ca2+-AgTG3 homology model. One monomer of the dimer is colored according to protein domains shown in Fig. 1A.
We used SAXS to further analyze the structure of Ca2+-AgTG3 in solution. In 1 m glycerol Ca2+-AgTG3 gave consistent scattering curves in the concentration range 1.0–5.0 mg/ml (Fig. 2E). Guinier analysis (Fig. 2E, inset) at 1 mg/ml determined a radius of gyration RG = 42.7 Å. Scattering at zero angle (I0) was directly proportional to concentration, although a systematic increase in RG suggests some nonideality of the solute. Calibration with a 1.0 mg/ml solution of BSA gave an estimated molecular mass of 173 kDa, consistent with that of a dimer. The radial distribution function p(r) calculated by GNOM (30) requires Dmax = 160 Å (Fig. 2F), almost 4× the RG, implying that the Ca2+-AgTG3 dimer is a prolate ellipsoid. Ab initio shape determination was performed using 20 independent models calculated by DAMMIF (31) with P2 symmetry and normalized spatial discrepancy (32) NSD = 0.67 ± 0.18.
The most probable volume for the Ca2+-AgTG3 dimer (Fig. 2G) suggests a structure similar to that of the Ca2+-bound FXIII dimer (18), yet with an open conformation for each AgTG3 monomer, analogous to that observed for inhibitor-bound hTG2 (21). A homology model for Ca2+-AgTG3 based on hTG2 (Protein Data Bank ID 2Q3Z) was superimposed on the FXIII dimer (Protein Data Bank ID 1GGU) and aligned with the most probable volume; imposition of P2 symmetry gives a good fit to the model.
Full-length Plugin is susceptible to proteolysis and aggregates in solution. Plugin-C (residues 345–557) was purified to homogeneity (Fig. 3A). The CD spectrum (Fig. 3B) has minima at 208 nm and 222 nm consistent with predominant α-helical secondary structure. At ≤0.5 mg/ml Plugin-C exhibits an apparent molecular mass of 90 kDa on SEC, but static light scattering (Fig. 3C) is consistent with a monomeric molecular mass of ∼30 kDa, suggesting that the protein has a nonglobular conformation in solution. AUC sedimentation velocity analysis (Fig. 3D) also yielded a molecular mass estimate of ∼30 kDa for Plugin-C (s20,w = 2.1 S, f/f0 = 1.4).
FIGURE 3.
Structure of Plugin in solution. A, SDS-PAGE of purified Plugin-C. B, CD spectrum. C, SEC-MALLS, concentration (dRI, black line) and calculated molecular mass from static light scattering (red line). Retention volume of molecular mass is standards indicated. D, sedimentation velocity C(s) distribution, s20,w = 2.1, (f/f0 = 1.4). E–H, Plugin-C SAXS analysis. E, I versus q curve. F, Guinier plot fit to RG = 42.8 Å (limit qRG = 1.33). G, absolute and H, dimensionless Kratky plot for Plugin-C.
We analyzed Plugin-C (0.5 mg/ml) by SAXS to confirm its extended structure in solution. The scattering curve (Fig. 3E) resembles that of an unfolded or heterogeneous sample, precluding calculation of a p(r) function. Guinier analysis (Fig. 3F) yields an estimated molecular mass of ∼30 kDa based on I0 (calibrated with BSA) but an unusually large radius of gyration (RG = 43 Å). In such cases the Kratky plot provides a qualitative measure of flexibility in a scattering particle (33, 34). Both absolute (Fig. 3G) and dimensionless (Fig. 3H) Kratky plots for Plugin-C suggest a partly unfolded tertiary structure. At concentrations >0.5 mg/ml Plugin-C forms oligomers or soluble aggregates that interfere with particle analysis.
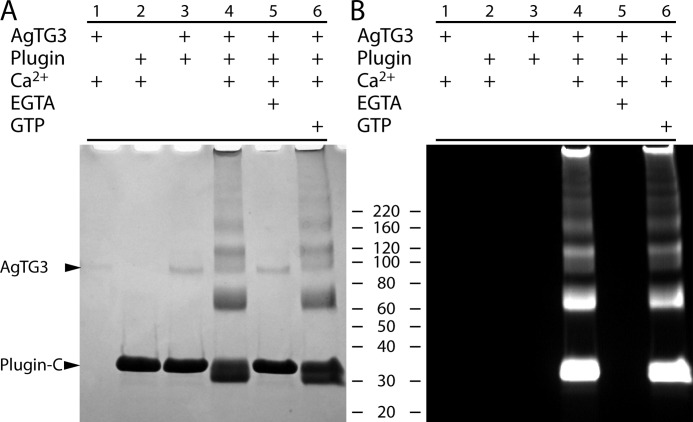
Cross-linking of Plugin by AgTG3 was previously demonstrated using endogenous extracts of male accessory glands, monodansylcadaverine, Ca2+, and GTP (5). We therefore reconstituted AgTG3 cross-linking of Plugin-C in vitro. Combining AgTG3 and Plugin-C in the presence of Ca2+ produced a ladder of cross-linked Plugin-C bands of increasing molecular mass (Fig. 4A). The presence of FITC-CAD led to fluorescent labeling of both the monomer and cross-linked bands (Fig. 4B), confirming that cross-linking was the result of transamidation. The requirement for Ca2+ was confirmed by the inhibitory effect of the chelating agent EGTA. Whereas Ca2+ binding sites are conserved between AgTG3 and other TGs, the GTP binding site found in tissue TG (TG2) is not: only two binding site residues are conservative substitutions (S171A, F174G, R476E, R478K, S482E, R580K). Consistent with this observation, no effect on AgTG3 cross-linking of Plugin-C was detected in the presence of GTP.
FIGURE 4.
Cross-linking of Plugin by AgTG3. A, Coomassie staining and B, fluorescence for SDS-PAGE of cross-linking reactions in presence of FITC-CAD. Lanes: 1, AgTG3 and Ca2+; 2, Plugin and Ca2+; 3, AgTG3 and Plugin-C without Ca2+; 4, AgTG3, Plugin-C, and Ca2+; 5, AgTG3, Plugin-C, Ca2+, and EGTA; 6, AgTG3, Plugin-C, Ca2+, and GTP.
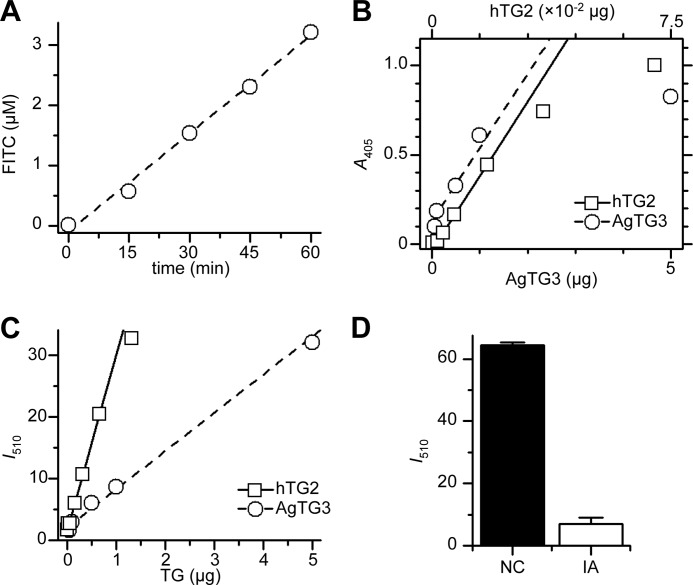
Next, we used the conjugation of FITC-CAD to Plugin-C to determine the specific activity of transamidation for AgTG3. Reductive methylation of Plugin-C prevents the transamidation of lysine residues. Fluorescence was measured after incubation for 0–60 min at 30 °C and buffer exchange (PD-10 column) to remove unbound FITC-CAD (Fig. 5A). In a 1.25-ml total volume, 1 mg/ml (40 μm) Plugin-C, 100 μm FITC-CAD, 1.2 μg of AgTG3 cross-linked FITC-CAD to Plugin-C at 0.408 μm min−1, yielding a specific activity of 4.25 × 10−2 unit mg−1 or 3.5 unit μmol−1 AgTG3 (1 unit = 1 μmol min−1 FITC-CAD).
FIGURE 5.
Specific activity of AgTG3. A, conjugation of FITC-CAD to Plugin-C in solution by AgTG3 (9.6 μg/ml, 30 °C). B, HRP-streptavidin assay (TMB, A405) for conjugation of biotin-TQQVEL to polylysine plates by 1.5 units mg−1 guinea pig TG2 (□) or AgTG3 (○). C, fluorescence (I510) for conjugation of FITC-CAD to Plugin-coated plates by 1.5 units mg−1 guinea pig TG2 (□) or AgTG3 (○). D, inhibition of AgTG3 by iodoacetamide (IA) versus negative control (NC) in Plugin/FITC-CAD assay.
The specific activity of AgTG3 determined above is 100× lower than is typically observed for standard TGs such as guinea pig TG2 (35). To confirm this result we performed a standard ELISA for transamidation of a biotinylated peptide (Btn-TVQQEL-OH) to polylysine-coated plates, with guinea pig TG2 (1.5 unit mg−1) as a standard (Fig. 5B). The relative specific activity of AgTG3 was 0.04 ± 0.01 unit mg−1, significantly less than is measured for human Factor XII or TG2 (1 unit = 1 μmol min−1 hydroxamate from Nα-Z-Gln-Gly, NH2OH, pH 6.0 at 37 °C).
To test whether the specific activity of AgTG3 may be substrate-dependent we repeated our FITC-CAD assay in a plate-based format. His6-tagged Plugin-C was adsorbed to Ni-coated plates and incubated with FITC-CAD and either guinea pig TG2 or AgTG3 for 60 min at room temperature (Fig. 5C). The specific activity of AgTG3 was 0.3–0.4 unit mg−1 relative to guinea pig TG2, an 8–10-fold increase compared with the previous assay. Iodoacetamide irreversibly inhibits transglutaminases by acetylating the active site cysteine. AgTG3 (10 μg) was efficiently inhibited by 30-min preincubation with iodoacetamide in the plate-based Plugin/FITC-CAD assay (Fig. 5D).
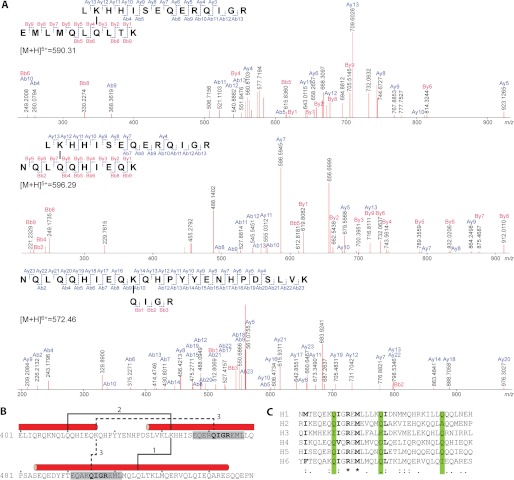
To identify specific AgTG3 cross-linking sites within Plugin-C the high molecular mass band observed on SDS-PAGE (Fig. 4) was excised and subjected to in-gel tryptic digestion and tandem mass spectrometry. Mass spectrometry data were input into the MassMatrix (29) search engine, which evaluates the monoisotopic masses of peptides, looking for a mass shift of −17 Da due to release of ammonia during isopeptide formation. Three specific peaks: m = 2947.5 ([M+H]5+, m/z = 590.5), m = 2977.4 ([M+H]5+, m/z = 596.5), and m = 3429.7 ([M+H]6+, m/z = 572.6) were detected. The first peak corresponds to Plugin peptides 432–445 (m = 1729.9) and 500–509 (m = 1233.7) minus 17 Da resulting in a monoisotopic mass m = 2946.6. The calculated mass is within error of m = 2947.5 (±1 atomic mass unit, accuracy ±0.02%). The identity of cross-linked species was confirmed by y and b ion fragments (Fig. 6A). On the basis of the fragments observed and the sequences of two tryptic peptides, we deduce that Lys-433 in peptide 432–445 and either Gln-504 or Gln-506 in peptide 500–509 are linked by an ϵ-(γ-glutamyl)lysine isopeptide bond. By similar calculations, we found that the second peak (m = 2977.4) corresponds to Plugin peptides 432–445 and 408–417 with the isopeptide bond formed between Lys-433 and either Gln-411 or Gln-412. The third peak (m = 3429.7) corresponds to Plugin peptides 409–431 and either 442–445 or 496–449 with the isopeptide bond formed between Lys-417 and either Gln-442 or Gln-496.
FIGURE 6.
Identification of Plugin cross-linking sites. A, CID-MS/MS analyses and corresponding fragment ion series for three specific peaks identified by mass spectrometric analysis of cross-linked Plugin-C tryptic digest. B, location of cross-linking sites within Plugin-C sequence. C, alignment of α-helices in Plugin C-terminal domain. The positions of known cross-linked Gln residues are highlighted in green, conserved or highly similar residues are shown in bold.
Interestingly, whereas Plugin-C contains 10 lysine residues, two of the three cross-linked peptides detected involve the same lysine residue Lys-433 cross-linked to Gln residues Gln-411 and Gln-506, in the preceding and subsequent α-helices, respectively (Fig. 6B). The third cross-link is between Lys-417 and the sequence QIGR, which occurs twice in Plugin-C within the consensus repeat near the N terminus of each predicted α-helix. The Gln residues involved in cross-linking are present in a majority of the six α-helices within the C-terminal domain of Plugin (Fig. 6C).
DISCUSSION
We have characterized A. gambiae seminal transglutaminase AgTG3 and the cross-linking of its native substrate Plugin in vitro. Intriguingly, we find that AgTG3 forms a dimer in solution, similar to the mammalian blood-clotting enzyme FXIII, yet distinct from other transglutaminases of known structure. AgTG3 has similar (∼30%) sequence identity to all of these enzymes. This raises the question: what features of the TG sequence are responsible for dimerization of AgTG3/FXIII versus tissue-type transglutaminases?
This question can be addressed by mapping the dimer interface of the Ca2+-bound FXIII reported by PISA (36) to the alignment of AgTG3 with TGs of known structure (Fig. 7). For the majority of this interface AgTG3 is no more similar to FXIII than other TGs with two exceptions. First, tissue-type TGs have an extended loop following the second α-helix of the core domain (hTG2 200–206), absent from AgTG3 and FXIII, which would interrupt the FXIII dimer interface. Second, AgTG3/FXIII share a charged residue in the loop prior to the catalytic aspartate within the core domain, the βCD loop, at the center of the dimer interface. Specifically FXIII Arg-408 (AgTG3 Lys-417) is replaced by hTG2 Cys-370, which forms a vicinal disulfide with Cys-371 in the active conformation of hTG2 and is implicated in oxidative inhibition of tissue TGs (37).
FIGURE 7.
Sequence alignment of AgTG3(22–748) with transglutaminases of known structure: FXIII, hTG2, hTG3, and P. major TG. Secondary structure elements colored and labeled by domain. Identical residues are shaded gray, similar residues are boxed in gray. Catalytic residues are shaded yellow, Ca2+-binding residues (based upon hTG3) are shaded pink. FXIII residues at the dimer interface are shaded green. Numbering corresponds to AgTG3 full-length sequence. Figure was generated with ESPRIPT (42).
An important assumption in the analysis above is that both the monomeric structure and dimeric interface of AgTG3 are equivalent to those of FXIII. Further structural studies of AgTG3 are necessary to test this assumption. Hence, a caveat to our analysis is that AgTG3 diverges significantly from all mammalian TGs, including numerous amino acid substitutions and unique extensions, notably in two loops within the β-sandwich domain. A high resolution structure of AgTG3 would be required to address the molecular basis of AgTG3 dimerization.
Whereas AgTG3 adopts a folded tertiary structure, Plugin-C has an extended or partially unfolded tertiary structure despite clear α-helical secondary structure and a propensity for oligomerization or aggregation in solution. Oligomerization of TG substrates is found for both FXIII and seminal TGs. Fibrin αC domains preassemble into polymers which are then reinforced by cross-linking with FXIIIa (38, 39), and seminal vesicle secretion proteins in rodents form disulfide-bridged high molecular mass complexes prior to cross-linking by the seminal transglutaminase TG4 (40). We suggest that oligomerization of Plugin is mediated by coiled-coil interactions between the repetitive helical regions of the C-terminal domain that are subsequently reinforced by AgTG3 cross-linking. The N-terminal domain of Plugin has a high density of putative cross-linking sites and is expected to form a densely cross-linked polymer, so association of the C-terminal domain may have a significant effect on the density and mechanical properties of the Anopheles mating plug.
The specific activity of AgTG3 is 8–10 times lower than that of other well characterized TGs. Although this may arise from the use of E. coli to express AgTG3, we note that both FXIII and hTG2 have been expressed in an active form from this host (37, 41). From studies of other TGs we propose two explanations. First, AgTG3 includes an N-terminal extension of ∼20 amino acids that may bind across the dimer interface, as is observed in FXIII. Activation of serum FXIII includes cleavage of this sequence by thrombin: if a similar mechanism held for AgTG3 the purified enzyme may exhibit autoinhibition resulting in suboptimal activity in vitro. Second, hTG2 has been shown to exhibit redox regulation of activity (37). AgTG3 contains a number of cysteine residues that are not conserved with mammalian TGs; despite the lack of evident disulfides in the purified protein these cysteines may be involved in oxidative inhibition of AgTG3.
AgTG3 has higher relative activity toward Plugin-C/FITC-CAD versus Btn-TVVQEL-OH and polylysine-coated plates. Hence, AgTG3 may have some preference for specific sequence motifs in Plugin. Among the cross-linking sites identified is the motif QIGR present in the majority of α-helices in the C-terminal domain (Fig. 6C). Its similarity to the motif present in the N-terminal domain of Plugin suggests that QI(G/P) is a preferred motif for cross-linking by AgTG3.
AgTG3 activity toward Plugin-C/FITC-CAD, however, may be due to a preference for (i) polypeptide or α-helical versus hexapeptide substrates, or (ii) soluble polyamine versus surface-bound lysine. Further experiments are necessary to discriminate between these hypotheses. Nevertheless, the current signal-to-noise achieved with the nonspecific inhibitor iodacetamide is sufficient for in vitro high throughput screening of prospective AgTG3 inhibitors (Z′ = 0.84).
The mechanism for formation of the copulatory plug in A. gambiae bears intriguing similarities to the mammalian blood-clotting cascade: a dimeric TG activated by calcium to act upon a specific substrate capable of preassembly of a oligomeric/polymeric state. Given their diverse functions, these similarities appear to be an instance of convergent evolution. Future investigation of AgTG3 will provide insight into the evolution of TGs and advance our basic understanding of fertility as well as the development of potential chemosteriliants for male Anopheles.
Acknowledgments
We thank Dr. Ian Berke (Molecular Biophysics & Biochemistry, Yale University) for assistance with AUC data collection and analysis; Drs. Marc Allaire and Lin Yang of NSLS beamline X9 for assistance with SAXS data collection and processing; and the Yale Keck Center Biophysical and Proteomics facilities for SEC-MALLS and MS data collection.
This work was supported, in whole or in part, by National Institutes of Health Career Development Award K22AI085112-01 from the NIAID (to R. H. G. B.). This work was also supported by European Research Council FP7 ERC Starting Grant Project “Anorep” Grant 260897 (to F. C.). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
- TG
- transglutaminase
- AgTG
- Anopheles gambiae TG
- AUC
- analytical ultracentrifugation
- CAD
- cadaverine
- FXIII
- Factor XIII
- h
- human
- MALLS
- multiangle laser light scattering
- RG
- radius of gyration
- SAXS
- small angle x-ray scattering
- SEC
- size exclusion chromatography
- TBS
- Tris-buffered saline
- TCEP
- Tris(2-carboxyethyl)phosphine.
REFERENCES
- 1. Folk J. E. (1983) Mechanism and basis for specificity of transglutaminase-catalyzed ϵ-(γ-glutamyl) lysine bond formation. Adv. Enzymol. Relat. Areas Mol. Biol. 54, 1–56 [DOI] [PubMed] [Google Scholar]
- 2. Lorand L., Graham R. M. (2003) Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 3. Shibata T., Ariki S., Shinzawa N., Miyaji R., Suyama H., Sako M., Inomata N., Koshiba T., Kanuka H., Kawabata S. (2010) Protein crosslinking by transglutaminase controls cuticle morphogenesis in Drosophila. PLoS One 5, e13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Z., Wilhelmsson C., Hyrsl P., Loof T. G., Dobes P., Klupp M., Loseva O., Mörgelin M., Iklé J., Cripps R. M., Herwald H., Theopold U. (2010) Pathogen entrapment by transglutaminase: a conserved early innate immune mechanism. PLoS Pathog. 6, e1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rogers D. W., Baldini F., Battaglia F., Panico M., Dell A., Morris H. R., Catteruccia F. (2009) Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PLoS Biol. 7, e1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuval B. (2006) Mating systems of blood-feeding flies. Annu. Rev. Entomol. 51, 413–440 [DOI] [PubMed] [Google Scholar]
- 7. Williams-Ashman H. G., Beil R. E., Wilson J., Hawkins M., Grayhack J., Zunamon A., Weinstein N. K. (1980) Transglutaminases in mammalian reproductive tissues and fluids: relation to polyamine metabolism and semen coagulation. Adv. Enzyme Regul. 18, 239–258 [DOI] [PubMed] [Google Scholar]
- 8. Palopoli M. F., Rockman M. V., TinMaung A., Ramsay C., Curwen S., Aduna A., Laurita J., Kruglyak L. (2008) Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 454, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lung O., Wolfner M. F. (2001) Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem. Mol. Biol. 31, 543–551 [DOI] [PubMed] [Google Scholar]
- 10. Bushland R. C., Lindquist A. W., Knipling E. F. (1955) Eradication of screw-worms through release of sterilized males. Science 122, 287–288 [DOI] [PubMed] [Google Scholar]
- 11. Asman S. M., McDonald P. T., Prout T. (1981) Field studies of genetic control systems for mosquitoes. Annu. Rev. Entomol. 26, 289–318 [DOI] [PubMed] [Google Scholar]
- 12. Harris A. F., Nimmo D., McKemey A. R., Kelly N., Scaife S., Donnelly C. A., Beech C., Petrie W. D., Alphey L. (2011) Field performance of engineered male mosquitoes. Nat. Biotechnol. 29, 1034–1037 [DOI] [PubMed] [Google Scholar]
- 13. Helinski M. E., Hassan M. M., El-Motasim W. M., Malcolm C. A., Knols B. G., El-Sayed B. (2008) Towards a sterile insect technique field release of Anopheles arabiensis mosquitoes in Sudan: irradiation, transportation, and field cage experimentation. Malar. J. 7, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dame D. A., Curtis C. F., Benedict M. Q., Robinson A. S., Knols B. G. (2009) Historical applications of induced sterilisation in field populations of mosquitoes. Malar. J. 8, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El Sayed B. B., Malcolm C. A., Babiker A., Malik E. M., El Tayeb M. A., Saeed N. S., Nugud A. H., Knols B. G. (2009) Ethical, legal and social aspects of the approach in Sudan. Malar. J. 8, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lehane M. J., Aksoy S. (2012) Control using genetically modified insects poses problems for regulators. PLoS Negl. Trop. Dis. 6, e1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahvazi B., Boeshans K. M., Idler W., Baxa U., Steinert P. M. (2003) Roles of calcium ions in the activation and activity of the transglutaminase 3 enzyme. J. Biol. Chem. 278, 23834–23841 [DOI] [PubMed] [Google Scholar]
- 18. Fox B. A., Yee V. C., Pedersen L. C., Le Trong I., Bishop P. D., Stenkamp R. E., Teller D. C. (1999) Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography. J. Biol. Chem. 274, 4917–4923 [DOI] [PubMed] [Google Scholar]
- 19. Liu S., Cerione R. A., Clardy J. (2002) Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. U.S.A. 99, 2743–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noguchi K., Ishikawa K., Yokoyama K. i., Ohtsuka T., Nio N., Suzuki E. (2001) Crystal structure of red sea bream transglutaminase. J. Biol. Chem. 276, 12055–12059 [DOI] [PubMed] [Google Scholar]
- 21. Pinkas D. M., Strop P., Brunger A. T., Khosla C. (2007) Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol. 5, e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kristiansen G. K., Andersen M. D. (2011) Reversible activation of cellular factor XIII by calcium. J. Biol. Chem. 286, 9833–9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Achyuthan K. E., Greenberg C. S. (1987) Identification of a guanosine triphosphate-binding site on guinea pig liver transglutaminase: role of GTP and calcium ions in modulating activity. J. Biol. Chem. 262, 1901–1906 [PubMed] [Google Scholar]
- 24. Sheffield P., Garrard S., Derewenda Z. (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 25. Perez-Iratxeta C., Andrade-Navarro M. A. (2008) K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konarev P. V., Volkov V. V., Soklova A. V., Koch M. H., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 [Google Scholar]
- 28. Rayment I. (1997) Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 276, 171–179 [PubMed] [Google Scholar]
- 29. Xu H., Freitas M. A. (2009) MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 [Google Scholar]
- 31. Franke D. A., Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozin M. B., Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 [Google Scholar]
- 33. Kratky O. (1982) in Small Angle X-ray Scattering (Glatter O., Kratky O. eds) pp. 361–386, Academic Press, London [Google Scholar]
- 34. Durand D., Vivès C., Cannella D., Pérez J., Pebay-Peyroula E., Vachette P., Fieschi F. (2010) NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 169, 45–53 [DOI] [PubMed] [Google Scholar]
- 35. Leblanc A., Day N., Ménard A., Keillor J. W. (1999) Guinea pig liver transglutaminase: a modified purification procedure affording enzyme with superior activity in greater yield. Protein Expr. Purif. 17, 89–95 [DOI] [PubMed] [Google Scholar]
- 36. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 37. Stamnaes J., Pinkas D. M., Fleckenstein B., Khosla C., Sollid L. M. (2010) Redox regulation of transglutaminase 2 activity. J. Biol. Chem. 285, 25402–25409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsurupa G., Mahid A., Veklich Y., Weisel J. W., Medved L. (2011) Structure, stability, and interaction of fibrin αC-domain polymers. Biochemistry 50, 8028–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsurupa G., Pechik I., Litvinov R. I., Hantgan R. R., Tjandra N., Weisel J. W., Medved L. (2012) On the mechanism of αC polymer formation in fibrin. Biochemistry 51, 2526–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tseng H.-C., Tang J.-B., Gandhi P. S. S., Luo C.-W., Ou C.-M., Tseng C.-J., Lin H.-J., Chen Y.-H. (2012) Mutual adaptation between mouse transglutaminase 4 and its native substrates in the formation of copulatory plug. Amino Acids 42, 951–960 [DOI] [PubMed] [Google Scholar]
- 41. Lai T.-S., Santiago M. A., Achyuthan K. E., Greenberg C. S. (1994) Purification and characterization of recombinant human coagulant factor XIII A-chains expressed in E. coli. Protein Expr. Purif. 5, 125–132 [DOI] [PubMed] [Google Scholar]
- 42. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]