Background: Apolipoproteins are lipid emulsifiers with links to additional protective roles.
Results: Small-angle x-ray scattering afforded structural information for full-length apoA-IV.
Conclusion: In the head-to-tail dimer, the N/C-terminal globular domains modulate the twist and curvature of a central helical bundle.
Significance: The lipid affinity of apoA-IV is regulated by opening and closing a molecular clasp.
Keywords: Apolipoproteins, Biophysics, HDL, Lipoprotein, Structural Biology, Small-angle X-ray Scattering
Abstract
ApoA-IV is an amphipathic protein that can emulsify lipids and has been linked to protective roles against cardiovascular disease and obesity. We previously reported an x-ray crystal structure of apoA-IV that was truncated at its N and C termini. Here, we have extended this work by demonstrating that self-associated states of apoA-IV are stable and can be structurally studied using small-angle x-ray scattering. Both the full-length monomeric and dimeric forms of apoA-IV were examined, with the dimer showing an elongated rod core with two nodes at opposing ends. The monomer is roughly half the length of the dimer with a single node. Small-angle x-ray scattering visualization of several deletion mutants revealed that removal of both termini can have substantial conformational effects throughout the molecule. Additionally, the F334A point mutation, which we previously showed increases apoA-IV lipid binding, also exhibited large conformational effects on the entire dimer. Merging this study's low-resolution structural information with the crystal structure provides insight on the conformation of apoA-IV as a monomer and as a dimer and further defines that a clasp mechanism may control lipid binding and, ultimately, protein function.
Introduction
The A class apolipoproteins (apoA-I, apoA-II, apoA-IV, and apoA-V) act as detergents that emulsify lipids to form or associate with lipoprotein particles, particularly HDL. They are considered exchangeable due to their ability to transition between a lipid-free and lipid-bound state, allowing for movement between particles in the plasma. These lipoprotein particles are the body's main mechanism for transporting and delivering lipids. They have distinct functional roles, acting as structural scaffolds and activators of lipoprotein-remodeling factors and interacting with cell surface proteins that modulate lipoprotein metabolism (1, 2).
Because of their importance in lipid biology, the structural characterization of apolipoproteins has been a high priority. Unfortunately, many apolipoproteins undergo rapid transitions between a constellation of self-associated states (dimers, trimers, tetramers, etc.), thus making them difficult to characterize structurally (3). Although some success has been obtained using site-directed mutagenesis to stabilize particular oligomeric forms of apoA-I and apoE, this approach has the drawback of potentially altering protein conformation along with self-association state (4, 5). We recently demonstrated that lipid-free human apoA-IV distributes predominantly between monomers and dimers and that their interconversion is slow enough that they can be isolated and studied individually (6). Thus, we have focused on apoA-IV as a model to gain insight into the structure and self-association of the exchangeable apolipoproteins.
ApoA-IV is the largest member of the exchangeable apolipoprotein family, with a molecular mass of 43 kDa. It is the third most prominent protein found associated with HDL (3, 7). In addition to HDL particle formation, apoA-IV has been shown to have a variety of unique biological roles not shared by other apolipoproteins; for example, in response to lipid absorption, apoA-IV is highly expressed in the intestine and is associated with apoB secretion (8, 9). Furthermore, apoA-IV functions as a satiety signal (10, 11), an antioxidant (12), and an anti-inflammatory agent (13). Recently, apoA-IV has also been shown to regulate insulin secretion, specifically during elevated glucose levels (14). Intriguingly, there is an 8-fold increase of apoA-IV in patients after Roux-en-Y gastric bypass surgery who showed improvements in obesity-related comorbidities (15). Despite such a diversity of protective functions, little is known about the apoA-IV mechanism of action. ApoA-IV can exist in solution (and serum) without lipid in self-associated forms, predominantly as a monomer or dimer (16), and the lipid-bound form remains poorly characterized.
Structurally, apoA-IV has a large hydrophobic core similar to other strong lipid-binding proteins such as apoA-I and apoE3 (4–6). However, it is a relatively poor lipid binder (17). Previous work from our laboratory strongly suggests that an interaction between the N and C termini of apoA-IV holds the protein in a conformation that binds lipids poorly (17–19). Disruption of this “clasp” by introducing the point mutation F334A significantly increased the lipid affinity of apoA-IV (19). Unfortunately, our structure of N/C-terminally truncated apoA-IV did not provide high-resolution structural details pertaining to the clasp mechanism.
To understand the conformational states adopted by full-length apoA-IV, we took advantage of its oligomeric stability and performed small-angle x-ray scattering (SAXS).2 Although of lower resolution than x-ray crystallography, SAXS allowed us to visualize the molecular envelope of full-length apoA-IV, along with truncation variants and the F334A point mutant. Combining this with the partial x-ray structure, we defined how the individual termini affect the global conformation of apoA-IV and further supported the clasp mechanism.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant human apoA-IVWT, apoA-IV64–335, apoA-IV1–335, apoA-IV63–376, and apoA-IVF334A were produced and purified in Escherichia coli as described previously (6). Size-exclusion chromatography (SEC) using a HiLoad 16/60 Superdex 200 column (GE Healthcare) was performed to isolated oligomeric species for the various constructs. The isolated oligomeric species were concentrated at 4 °C using Amicon Ultra-15 centrifuge filter units. To maintain oligomeric homogeneity, concentration was performed in 15-min intervals at low speed (1000 × g). At the end of each interval, the concentrated sample was mixed with the total pool of the less concentrated sample; this minimized the concentration gradient during centrifugation and prevented shifting between oligomeric species. To confirm this, samples were reanalyzed by SEC (Superdex 200 HR 10/300, Amersham Biosciences) using 8–25% gradient native gels (PhastGel system, GE Healthcare).
Sedimentation Velocity
Analytical ultracentrifugation experiments were performed using a Beckman XL-I ultracentrifuge with absorbance optics and a four-hole rotor. Sedimentation velocity was performed in a two-channel carbon-filled Epon centerpiece at 36,000 rpm and 10 °C with protein (0.15 mg/ml) that had been dialyzed into 20 mm NaPO4 (pH 7.4) and 100 mm NaF. Protein was monitored with UV absorbance at 230 nm, and data were analyzed using Sedfit (20).
SEC
To determine the apoA-IV dimer/monomer ratio over time, a HiLoad 16/60 Superdex 200 column on an ÄKTA Explorer equipped with a UV-900 detector and a P-960 sample pump at 4 °C was used. The purified apoA-IV dimer was diluted to 0.15 mg/ml in 50 ml of PBS with a single cOmplete EDTA-free protease inhibitor mixture tablet (catalog number 05056489001, Roche Applied Science), 1 mm sodium azide, and 0.05 mm PMSF. The samples were incubated at 4 °C, and at different time points, a sample was collected and examined using a HiLoad 16/60 Superdex 200 gel filtration column. The elution process was monitored at 215, 235, and 280 nm. Peak integration was performed with the UV absorbance traces. The percent dimer was determined by dividing the area of the dimer peak by the total area of both monomer and dimer peaks.
1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) Clearance Assay
Purified protein was tested for its ability to emulsify suspended DMPC (Avanti Polar Lipids). The assay was performed as described previously (18, 19).
SAXS
SAXS data were collected at beamline 12-ID-B at the Advanced Photon Source at Argonne National Laboratory (Table 1). The beamline was equipped with a flow cell to reduce radiation damage. A total of 120 μl of sample was loaded into the flow cell and oscillated during data collection. Data were collected at room temperature, with an exposure time of 0.5 s and a total of 30 images taken for each sample. Small-angle diffraction images were captured using a PILATUS 2M detector.
TABLE 1.
Data collection
| Data collection parameters | |
| Instrument | 12-ID-B |
| Monochromator type | Side bounce |
| Wavelength (Å) | 1.53 |
| q range (Å−1) | 0.009–0.250 |
| Exposure time (s) | 0.5 |
| Temperature (K) | 293 |
| Software employed | |
| Primary data reduction | PRIMUS |
| Data processing | GNOM |
| Ab initio analysis | GASBOR |
| Validation and averaging | DAMAVER |
| Structure refinement | DireX |
| Computation of model intensities | CRYSOL |
Ab Initio Model Reconstruction
The data collected were converted from two- to one-dimensional using the Irena package for IGOR software (21), and outliers within each set of 30 images were removed and averaged using PRIMUS (22). The averaged data were subtracted from the appropriate buffer blank also in PRIMUS. Subsequently, AutoRG and GNOM were used to determine Rg, Dmax, and I(0) (Table 1). Indirect transformation was performed with GNOM (23); ab initio reconstruction was completed using GASBOR (24). Depending on the data set, monomer (P1) or dimer (P2) symmetry was used as a constraint in GASBOR. Ten ab initio reconstructions for each construct of apoA-IV were performed and averaged using the DAMAVER package (25). The apoA-IV64–335 crystal structure was compared with the scattering data of both apoA-IV64–335 and apoA-IVWT using CRYSOL (26).
DireX
Fitting of the crystal structure into the SAXS envelope, which had additional curvature, was achieved by geometry-based conformational sampling performed by the program DireX (27). An average weighted electron density map was first created from the ab initio reconstruction. Conformational sampling was dictated by the CONCOORD algorithm in 100 total steps (nsteps) with Tirion enhancement. A dynamic elastic network was used for further refinement and to prevent overfitting. The dynamic elastic network used a γ value of 0.0 (den_gamma) and a κ value of 0.2 (den_kappa). A total of 8000 restraints were chosen per iteration (den_no) with a dynamic elastic network strength of 0.4 (den_strength). The structure was not energy-minimized after DireX fitting.
RESULTS
Stability of ApoA-IV
Lipid-free human apoA-IV was originally documented to exist as a mixture of a monomer and dimer, with a slow interconversion between the forms (16). We recently showed that both dimeric and monomeric forms of apoA-IV can be isolated by SEC (6). This is in sharp contrast to other apolipoproteins, such as apoE (28) and apoA-I (29, 30), indicating that apoA-IV oligomerization is uniquely stable.
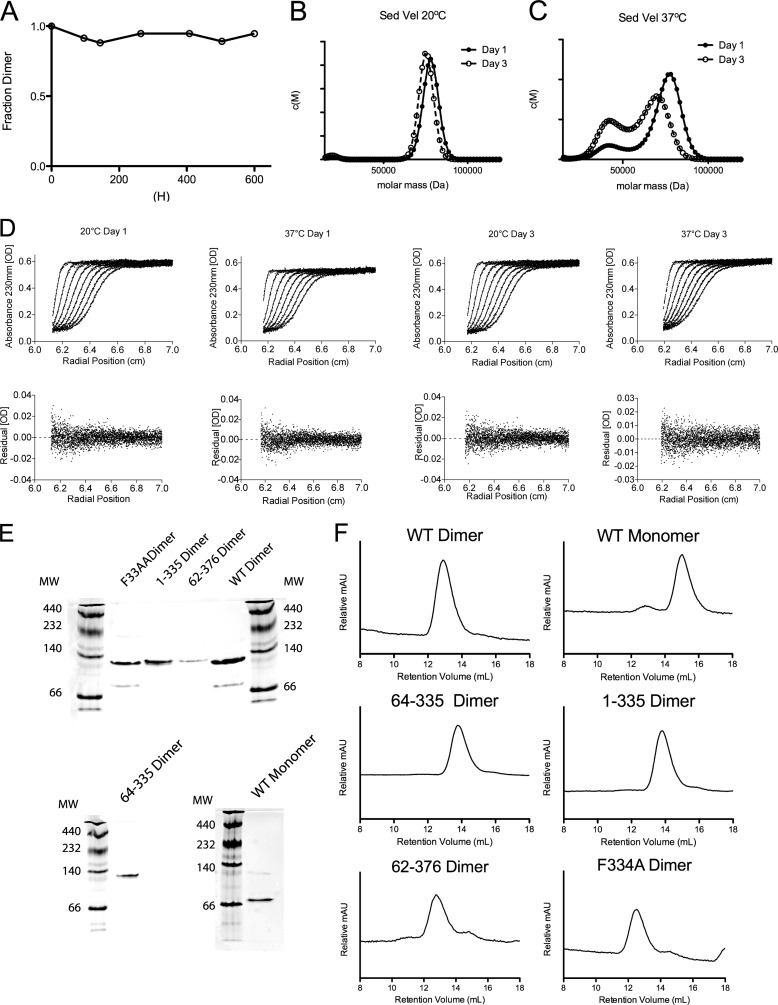
SAXS analysis requires samples that remain molecularly homogeneous for the entirety of the analysis. We first looked at how well apoA-IV maintains the dimeric state at 4 °C. At this temperature, the dimer did not dissociate, even after 600 h (Fig. 1A). As SAXS experiments are carried out at room temperature, the apoA-IV dimer was incubated at 20 °C, and samples were collected and subjected to sedimentation velocity on days 1 and 3. At 20 °C, apoA-IV remained entirely dimeric during the course of 3 days (Fig. 1, B and D). This indicated that, in the time required to collect SAXS data (∼5 min), dimeric apoA-IV would maintain its oligomeric form. As a control to show that apoA-IV can freely distribute, the same experiment was performed at 37 °C. Dissociation of the dimer to the monomer was readily observable on day 1 and to a much greater extent on day 3 (Fig. 1, C and D). Thus, at 20 °C, SAXS analyses were justified. Furthermore, the different truncations and mutations of apoA-IV used in this study did not affect its oligomeric stability. Each variant was isolated and maintained its self-associated state as demonstrated by native gel electrophoresis (Fig. 1E). Additionally, after the SAXS data collection, each sample was analyzed by SEC, which showed that they still maintained their oligomeric states (Fig. 1F).
FIGURE 1.
Quantifying the stability of the apoA-IV monomer and dimer. A, the apoA-IV dimer (0.15 mg/ml) was incubated at 4 °C, and samples were taken at various time points and analyzed by SEC. B and C, sedimentation velocity (Sed Vel) was performed to determine the self-associated state with isolated apoA-IV dimers incubated at 20 °C (B) or 37 °C (C) for 1 or 3 days. After fitting for the frictional ratio (f/f0), the c(s) distribution was transformed into a c(M) distribution of the molecular masses. D, sedimentation velocity data (dots) and corresponding fits (lines). Shown are 8 of the 32 scans used for fitting and their residuals for the sedimentation velocity experiments. E, native gel of apoA-IV and its variants. F, SEC traces of apoA-IV and variants. A retention volume of 12–14 ml is consistent with dimeric size, and 14–16 ml is consistent with a monomer based on molecular size standards.
Dimeric ApoA-IV64–335
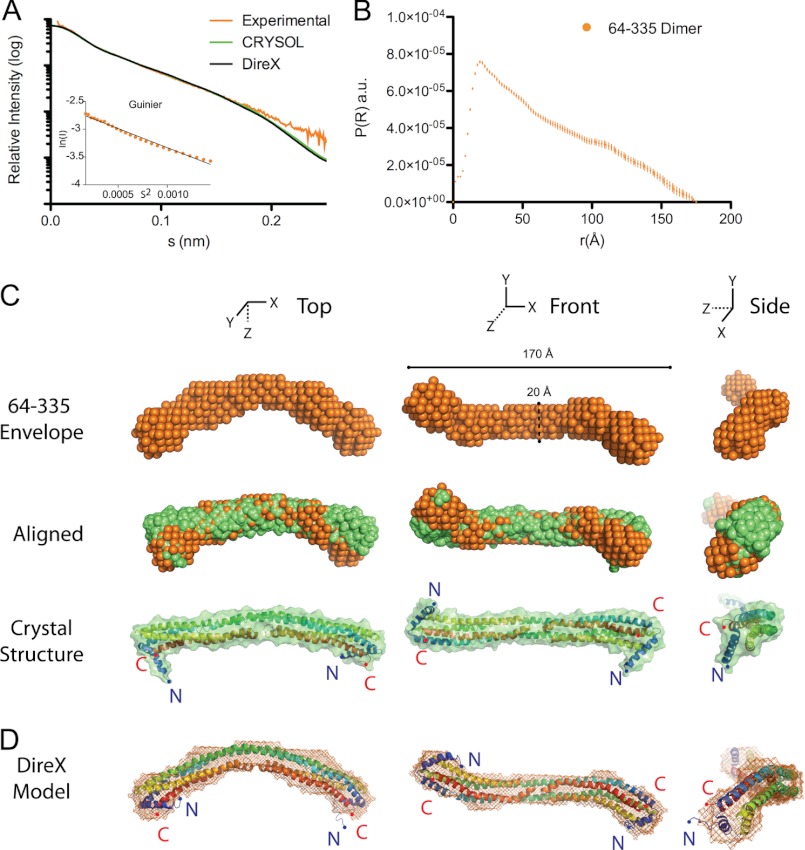
Because apolipoproteins undergo a variety of molecular transitions, we first determined if the observed crystal structure of the terminus-free core domain of apoA-IV (apoA-IV64–335) provides a good model for the protein in solution. We expressed apoA-IV64–335 and isolated its dimer (Fig. 1, E and F), which remained stably dimeric like the full-length form. We collected SAXS data on apoA-IV64–335 in solution at four concentrations ranging from 0.5 to 4 mg/ml (Fig. 2A and Table 2). In all cases, the solution remained free of significant aggregation or repulsion as determined by a linear Guinier plot (Fig. 2A), and I(0) remained consistent over the concentration range examined (Table 2). Although the overall shape of the scattering profile was similar at all protein concentrations analyzed, the data obtained at 4 mg/ml yielded the strongest signal-to-noise intensity and was therefore used for subsequent analysis. The shape of the scattering and pairwise distance distribution function profiles indicated a rod-like structure (Fig. 2B). The diameter of the maximal particle size (Dmax) was determined to be 175 Å using GNOM (Fig. 2, A and B).
FIGURE 2.
Solution state of apoA-IV64–335. A, scattering function and Guinier plot (inset) of dimeric apoA-IV64–335 and simulated scattering function generated with CRYSOL using the apoA-IV64–335 crystal structure (Protein Data Bank code 3S84) and the DireX refined structure. B, pairwise distance distribution function of apoA-IV64–335. a.u., arbitrary units. C, three orthogonal views: top, averaged ab initio reconstructions of apoA-IV64–335 (orange spheres); middle, superposition of the apoA-IV64–335 crystal structure envelope (green spheres) with the SAXS envelope (orange spheres); and bottom, ribbon and surface representation of the apoA-IV64–335 crystal structure. D, the crystal structure was refined using the SAXS envelope with the program DireX. The refined structure of apoA-IV64–335 is shown as a ribbon, and the ab initio reconstruction of apoA-IV64–335 is shown as mesh.
TABLE 2.
Parameters derived from data collection and scattering curves
| Construct | Rg (Guinier) | I(0) (Guinier) | Dmax | Volume | DAMAVER (NSD)a |
|---|---|---|---|---|---|
| Å | cm−1 | Å | Å3 | ||
| ApoA-IV64–335 dimer | |||||
| 4 mg/ml | 48.73 ± 1.41 | 0.074 ± 0.002 | 175 | 1.19 × 105 | 1.49 |
| 2 mg/ml | 49.37 ± 4.53 | 0.035 ± 0.003 | |||
| 1 mg/ml | 44.74 ± 8.16 | 0.016 ± 0.003 | |||
| 0.5 mg/ml | 43.17 ± 17.80 | 0.007 ± 0.002 | |||
| ApoA-IVWT dimer | |||||
| 4 mg/ml | 51.70 ± 0.89 | 0.143 ± 0.003 | 175 | 1.51 × 105 | 1.93 |
| 2 mg/ml | 51.43 ± 1.37 | 0.076 ± 0.002 | |||
| 1 mg/ml | 50.94 ± 2.43 | 0.037 ± 0.002 | |||
| 0.5 mg/ml | 52.11 ± 4.10 | 0.021 ± 0.002 | |||
| ApoA-IVWT monomer | |||||
| 4 mg/ml | 40.04 ± 1.49 | 0.081 ± 0.002 | 90 | 8.81 × 104 | 1.40 |
| 2 mg/ml | 40.06 ± 2.52 | 0.040 ± 0.002 | |||
| 1 mg/ml | 41.34 ± 4.22 | 0.022 ± 0.002 | |||
| 0.5 mg/ml | 44.78 ± 7.04 | 0.012 ± 0.002 | |||
| ApoA-IV1–335 dimer | |||||
| 2 mg/ml | 51.18 ± 1.07 | 0.067 ± 0.000 | 185 | 1.45 × 105 | 1.31 |
| 1 mg/ml | 49.75 ± 2.71 | 0.032 ± 0.000 | |||
| 0.5 mg/ml | 50.66 ± 5.20 | 0.016 ± 0.002 | |||
| ApoA-IV62–376 dimer | |||||
| 2 mg/ml | 54.23 ± 1.12 | 0.051 ± 0.001 | 180 | 1.33 × 105 | 2.29 |
| 1 mg/ml | 51.29 ± 3.32 | 0.026 ± 0.002 | |||
| 0.5 mg/ml | 51.05 ± 6.20 | 0.013 ± 0.002 | |||
| ApoA-IVF334A dimer | |||||
| 2 mg/ml | 52.26 ± 2.17 | 0.037 ± 0.002 | 195 | 1.62 × 105 | 2.49 |
| 1 mg/ml | 53.23 ± 4.23 | 0.020 ± 0.002 | |||
| 0.5 mg/ml | 59.17 ± 6.58 | 0.013 ± 0.002 | |||
| CRYSOL apoA-IV64–335 structure (PDB code 3S84) | 53.11 | 174 | 1.07 × 105 | ||
a NSD, normalized spatial discrepancy; PDB, Protein Data Bank.
From these data, a low-resolution envelope of the protein was generated via ab initio reconstructions. Ten independent reconstructions were performed and then averaged with DAMAVER. Overall, the reconstructions were in agreement, as the mean normalized spatial discrepancy was only 1.49 ± 0.91. The envelope of apoA-IV64–335 reveals a linear elongated rod with a slight curvature extending 170 Å in length and 20 Å in width (Fig. 2C). These dimensions match exceptionally well with our recent crystal structure of apoA-IV64–335, in which residues 75–312 were resolved. (Residues 64–74 and 313–335 were present but were not resolved in the crystal structure.) Additionally, when looking down the long axis, a right-handed twist of ∼90° is observed, similar to the crystal structure. Moreover, the simulated scattering data from the crystal structure fit the experimental scattering data well, with a χ value of 1.16 as determined by CRYSOL, and the experimental radius of gyration (Rg) is consistent with the crystal structure (Fig. 2A and Table 2). These comparisons show that the SAXS and x-ray crystallography data are highly consistent, further validating the dimeric apoA-IV64–335 model.
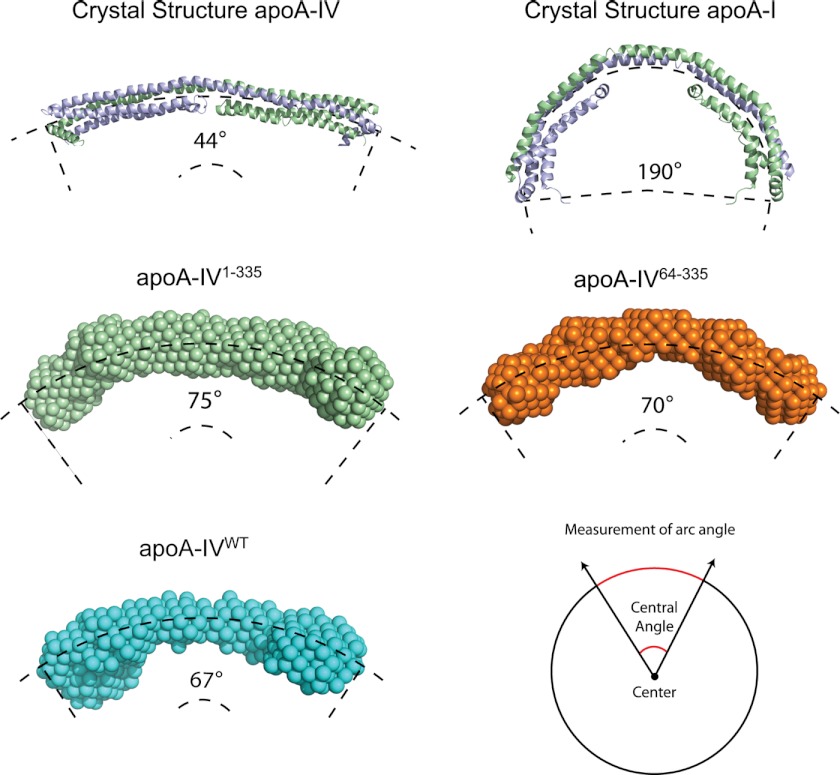
Despite this close agreement, there is a rod curvature difference between the crystal structure and SAXS reconstruction (Fig. 2C). The crystal structure is more linear, whereas the SAXS reconstruction is more curved. When describing the length of the protein as an arc segment, the central angle of the crystal structure measures 44°, whereas the solution structure measures 70° (Fig. 3). This difference in curvature may be attributed to crystallization (31), whereas SAXS is performed in solution. Nevertheless, apoA-IV is still significantly less curved than the recent crystal structure of apoA-I1–182 (4).
FIGURE 3.
Central arc measurements for the crystal structures of apoA-IV (Protein Data Bank code 3S84) and apoA-I (code 3R2P) and ab initio reconstructions of apoA-IV64–335, apoA-IV1–335, and apoA-IVWT.
To better represent the solution state of apoA-IV64–335, we refined the crystal structure against the SAXS envelop with the program DireX (Fig. 2D). Success of the refinement was measured by the correlation coefficient, a numerical value comparing the electron density of the simulation model with the experimental map (32). During the refinement, the correlation coefficient increased from 0.84 to 0.98, indicating that DireX generated a model that was a better fit to the envelope than the crystal structure itself. A low-resolution comparison of the two models indicates that the curvature is the result of a distortion to the helical segments caused by a series of repeating proline residues. In the crystal structure, these specific proline residues align vertically from opposing chains and are thought to provide segmentation and the flexibility needed during lipid binding (6). In our previous description of the crystal structure, we noted that the amount of distortion caused by the introduction of a proline in a helix or kink angle was much lower than in previously reported structures. After modeling with DireX, we measured, as described previously (33), the same proline residues and found an increased average kink of 6° (Table 3). Consequently, the kink angle of the solution structure of apoA-IV64–335 dimer is more in line with the average proline kink angle of 26° seen in other structures (6, 34).
TABLE 3.
Proline kink angles
| Proline | Crystal structure | DireX | Δ |
|---|---|---|---|
| Chain A | |||
| Pro-117 | 21.5° | 32.8° | 11.3° |
| Pro-139 | 14.8° | 16.3° | 1.5° |
| Pro-161 | 16.2° | 23.5° | 7.3° |
| Pro-183 | 13.1° | 16.3° | 3.2° |
| Average | 16.4° | 22.2° | 5.8° |
| Chain B | |||
| Pro-117 | 23.2° | 36.0° | 12.8° |
| Pro-139 | 10.3° | 8.5° | −1.8° |
| Pro-161 | 18.7° | 30.6° | 11.9° |
| Pro-183 | 10.4° | 12.4° | 2° |
| Average | 15.7° | 21.9° | 6.2° |
Overall, the SAXS data of apoA-IV64–335 match exceptionally well with the crystal structure and provided constraints to better model the solution state. This also gave us confidence in the implementation of SAXS to study apoA-IVWT and different truncation constructs.
Dimeric ApoA-IVWT
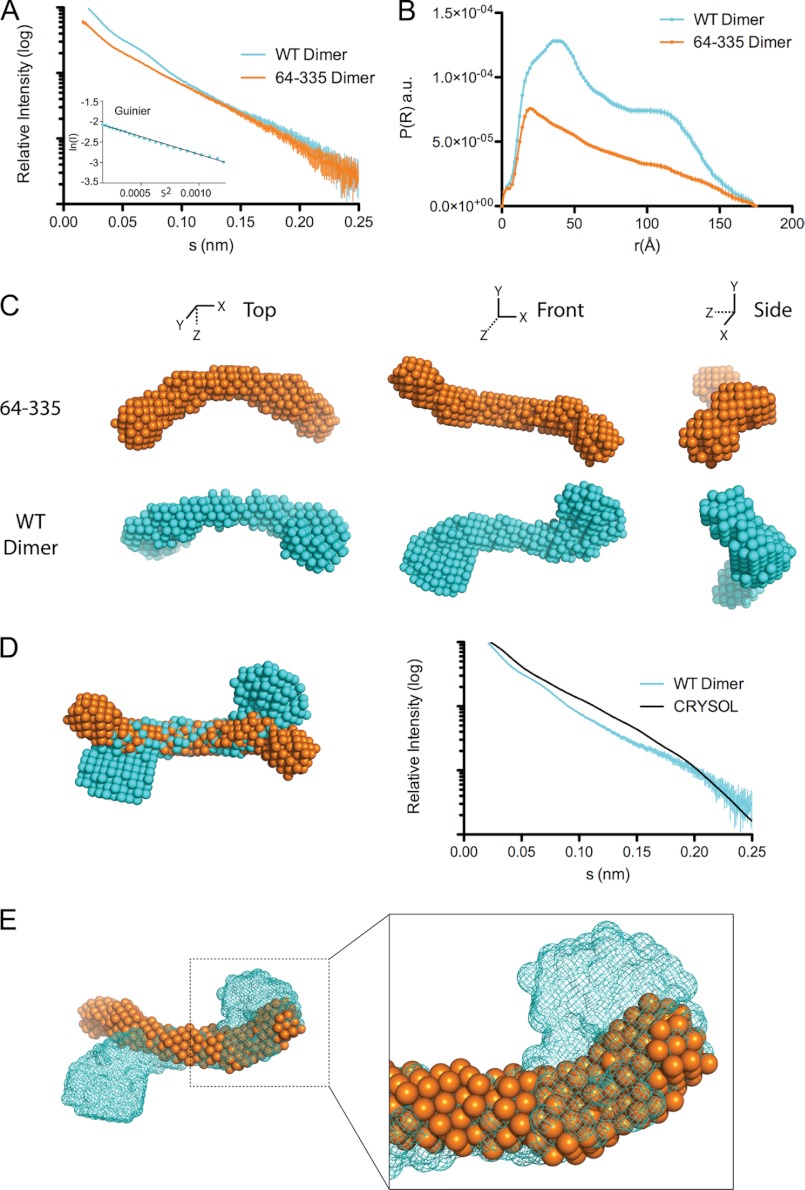
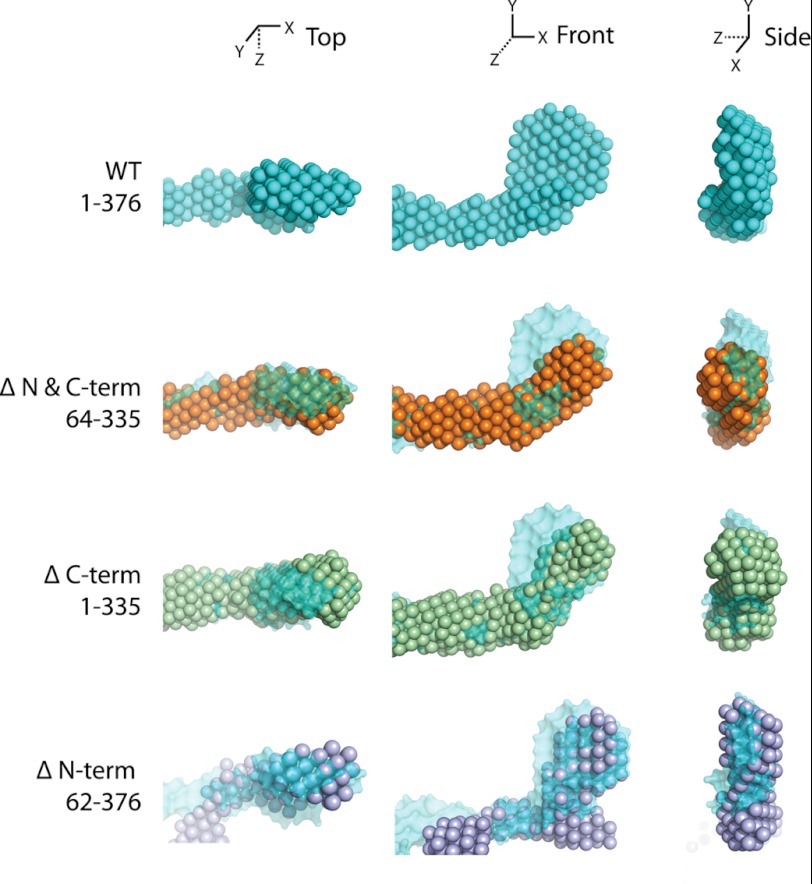
We next performed a SAXS experiment on the full-length apoA-IV dimer for comparison with apoA-IV64–335 (Fig. 4 and Table 2). We first correlated the simulated scattering data from the crystal structure of apoA-IV64–335 with the scattering data from apoA-IVWT. Analysis with the program CRYSOL resulted in a large χ value of 3.99 (Fig. 4D) compared with the value of 1.16 for apoA-IV64–335. This indicates that there are significant structural differences between the two samples. Interestingly, although the pairwise distance distribution function of the full-length apoA-IV dimer again pointed to an elongated structure with a similar Dmax, it depicted a bimodal distribution, indicating a deviation from a purely rod-like structure (Fig. 4B). Aligning the full-length and truncated envelopes shows significant differences in the ends of the rods. Compared with apoA-IV64–335, the ends of the apoA-IVWT rod are significantly more globular, with a substantial increase (∼27%) in the volume of the envelope (Fig. 4E and Table 2). This is consistent with a 35% increase in the molecular mass of the apoA-IVWT dimer (86.8 kDa) versus the apoA-IV64–335 dimer (62.9 kDa). Moreover, aligning the envelopes of apoA-IV64–335 and apoA-IVWT at just one end reveals additional density in the apoA-IVWT envelope. This density spatially corresponds to the visible N- and C-terminal ends in the crystal structure (Fig. 2C). This difference in the reconstructions strongly supports the possibility that the N and C termini interact with each other at both ends of the rod.
FIGURE 4.
Solution state of apoA-IVWT and comparison with apoA-IV64–335. A, scattering function and Guinier plot (inset) of dimeric apoA-IVWT and apoA-IV64–335. B, pairwise distance distribution function of apoA-IVWT and apoA-IV64–335. a.u., arbitrary units. C, three orthogonal views of the averaged ab initio reconstructions of apoA-IV64–335 (top) and apoA-IVWT (bottom). D, alignment of apoA-IVWT and apoA-IV64–335 reconstructions and comparison of apoA-IVWT SAXS scattering function with the simulated scattering function of the apoA-IV64–335 crystal structure generated with CRYSOL. E, superposition of one-half of apoA-IVWT (mesh) and apoA-IV64–335 (spheres) depicting additional density associated with apoA-IVWT.
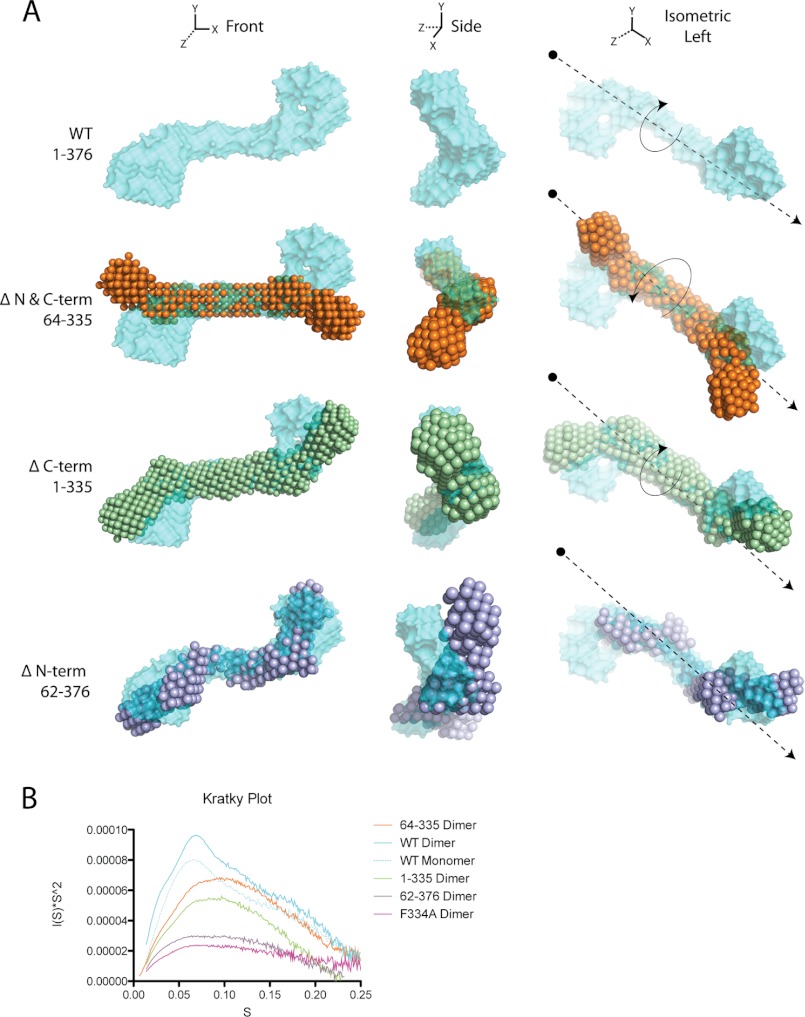
In addition to the increase in mass identified at the ends of the rod, we also observed a difference in the overall twist of the molecule. More specifically, although both apoA-IVWT and apoA-IV64–335 are slightly curved, with a similar central arc of ∼70° (Fig. 3), there is a difference in the direction of the twist (Fig. 4, C and D; see Fig. 8). Whereas apoA-IV64–335 has a left-handed twist when looking down the long axis of the rod, apoA-IVWT has a right-handed twist of ∼120°. This, along with the curvature, creates a chiral mismatch between the two reconstructions (Fig. 4, D and E; see Fig. 8). Although both apoA-IV64–335 and apoA-IVWT have a rod-like core, there are distinctions in volume at the termini and a reversal of the twist along the long axis.
FIGURE 8.
Conformational affects of N/C termini. A, comparison of the overall averaged envelopes of apoA-IV64–335, apoA-IV1–335, and apoA-IV62–376 with apoA-IVWT. The envelopes were superimposed by aligning the central helical bundle. N & C-term, N and C termini. B, Kratky plot for each set of SAXS data that was used for ab initio reconstructions.
Monomeric ApoA-IV
Previously, we proposed a pocketknife model in which the extended helix, which is swapped in the dimer, folds back into itself to form a four-helix bundle in the monomer (6). The model predicts that the atomic contacts are the same between the monomer and dimer. This was supported experimentally, as we previously observed no significant difference in their cross-linking patterns, thermal stability, or secondary structure (6).
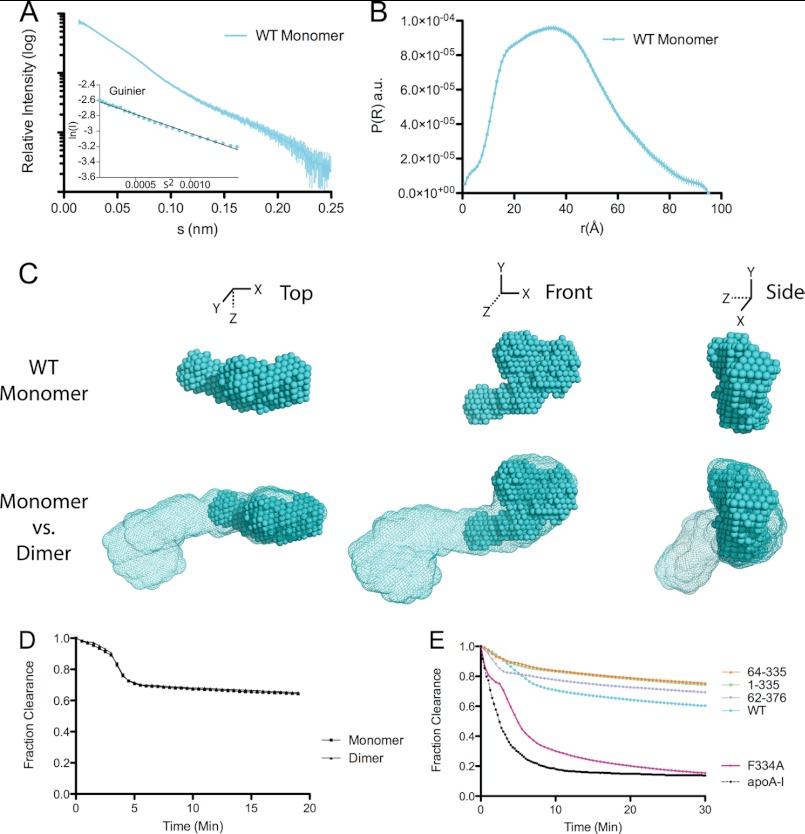
To further evaluate our model, we utilized SAXS to determine a low-resolution structure of monomeric apoA-IVWT (Fig. 5). Again, minimal aggregation was observed as determined by a linear Guinier analysis (Fig. 5A), and apoA-IVWT remained monomeric during the experiment (Fig. 1, E and F). The bell shape of the pairwise distance distribution profile indicated a much more globular structure than the dimer, with a Dmax significantly smaller than that of dimeric apoA-IVWT (Fig. 5B). The envelope generated after ab initio reconstruction shows a compressed structure that was significantly more globular than the rod-like dimer. In fact, the monomer reconstruction fitted exceptionally well into one-half of the dimer reconstruction (Fig. 5C). The envelope of the apoA-IV monomer is roughly half the length (88 Å) of the dimer, but with a similar width (20 Å), which is roughly the width of a four-helix bundle (Fig. 5C). Furthermore, the envelope comparison demonstrates that the additional density observed in dimeric apoA-IVWT compared with the truncated version is also apparent in the monomer envelope (Fig. 5C). This indicates that the N and C termini form an interaction similar to that observed in the dimer, although the interaction would be intramolecular. Additionally, this suggests that the clasp mechanism regulates lipid binding in both the monomeric and dimeric forms. This finding is further supported by DMPC clearance experiments, in which the lipid binding rates of the isolated monomer and dimer were indistinguishable (Fig. 5D). Overall, this is the first structural information on the monomeric form of apoA-IVWT, and the model is supported by its resemblance to exactly half of the dimer.
FIGURE 5.
SAXS of monomeric apoA-IV. A, scattering function and Guinier plot (inset). B, pairwise distance distribution function. a.u., arbitrary units. C, averaged ab initio reconstruction of monomeric apoA-IV (spheres) and superimposed onto the apoA-IVWT dimer reconstruction (mesh). D and E, DMPC clearance assay comparing isolated monomeric and dimeric forms of apoA-IV (D) and comparing the dimeric form of various mutants with apoA-IVWT (E).
Location and Significance of N and C Termini
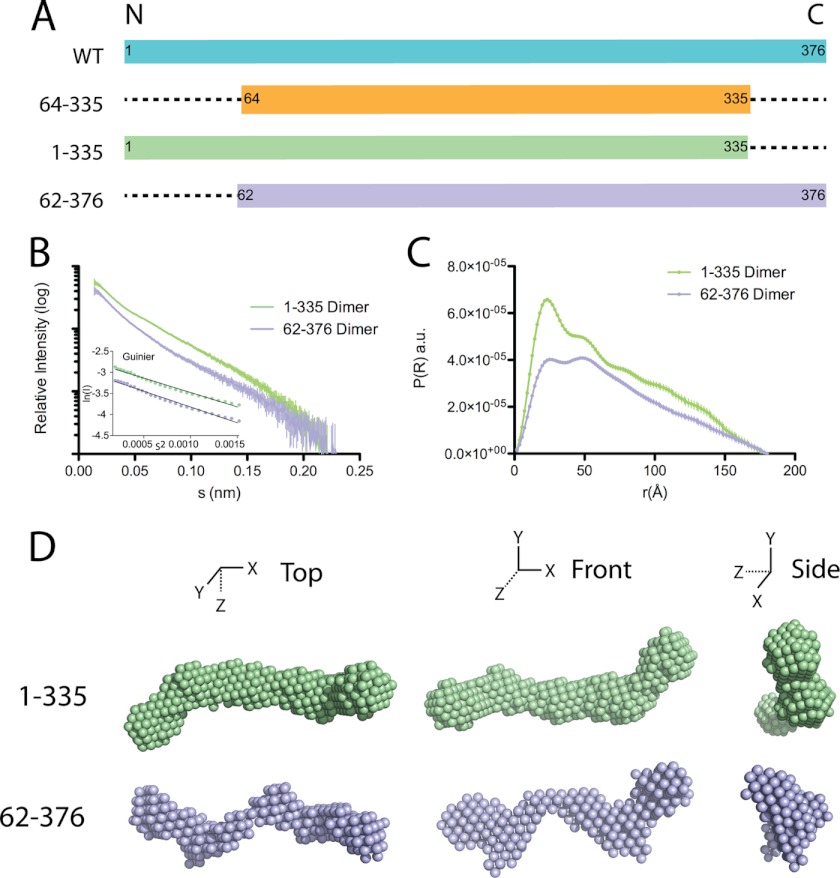
To determine whether the additional volume on the ends of the rod identified in the apoA-IVWT dimer is due to the termini missing from the crystal structure, we analyzed the SAXS envelopes of deletion variants with individual N- and C-terminal truncations. ApoA-IV1–335 was used to identify the additional volume associated with the N terminus, whereas apoA-IV62–376 was used for the C terminus. Each variant was expressed and purified as described above, and the dimer was isolated by SEC. Similar to apoA-IVWT and apoA-IV64–335, the isolated dimers were exceptionally stable (Fig. 1, E and F). ApoA-IV62–376, which lacks residues involved in lipid binding, was shown to be a poor lipid binder (Fig. 5E). Similarly, apoA-IV1–335 also bound lipid poorly. This has been suggested to be a result of an intact clasp, which is highly dependent on interaction of Phe-334 with the N terminus (Fig. 5E) (19). Consistent with previously published results (19), we observed a similar slow lipid binding profile in the DMPC clearance assay when using only the isolated dimer compared with the mixture of monomers and dimers (Fig. 5E).
SAXS data were collected on each mutant, and a summary of the structural statistics is presented in Table 2. Inspection of the pairwise distance distribution revealed that each construct has an extended rod structure that more resembles the profile from apoA-IV64–335 than from apoA-IVWT (Fig. 6C). However, there are obvious differences in the profile of the two mutants that result in distinct molecular envelopes (Fig. 6D). For apoA-IV1–335, the ends of the molecule are less globular compared with apoA-IVWT and with less volume (Fig. 7), suggesting that the missing volume is a result of the C-terminal truncation. Furthermore, comparing apoA-IV1–335 with apoA-IV64–335 shows additional volume that corresponds to the mass of the N terminus (Table 2). The envelope of apoA-IV1–335 also shows a similar curvature of the rod compared with apoA-IVWT and apoA-IV64–335, with a central arc of 75° (Fig. 3). Interestingly, apoA-IV1–335 exhibits a right-handed twist comparable with that of apoA-IVWT (Fig. 8A). This suggests that the addition of the N terminus reverses the left-handed twist of apoA-IV64–335 to a right-handed twist, as seen in apoA-IVWT.
FIGURE 6.
Solution state of apoA-IV1–335 and apoA-IV62–376. A, schematic of the various truncation constructs used for SAXS experiments. B, scattering function and Guinier plot (inset) of dimeric apoA-IV1–335 and apoA-IV62–376. C, pairwise distance distribution function of apoA-IV1–335 and apoA-IV62–376. a.u., arbitrary units. D, three orthogonal views of the averaged ab initio reconstruction of dimeric apoA-IV1–335 and apoA-IV62–376.
FIGURE 7.
Comparison of N and C termini. Shown is the apoA-IVWT averaged reconstruction with various truncation averaged reconstructions aligned at one end for comparison of the N and C termini (N & C-term).
This is in sharp contrast to the envelope of apoA-IV62–376, which is significantly less ordered than the previously examined constructs. This difference is also apparent upon examination of the Kratky plot (Fig. 8B), which yields information about the flexibility and folded state of the molecule (35). Folded and ordered proteins have a bell-shaped profile, which is significantly flattened for apoA-IV62–376 compared with other apoA-IV constructs. Surprisingly, the central rod appears to be kinked at four locations along the length of the core bundle rod (Fig. 6D), thus making it difficult to determine the twist (Fig. 8A). In addition, the ends are less compact, again making it difficult to define the location of the C terminus relative to apoA-IVWT and the other variants (Fig. 7). Taken together, these results imply that alterations to the termini, specifically removing the N terminus, can have dramatic effects on the structure of the core helical bundle.
Structure of Dimeric ApoA-IVF334A
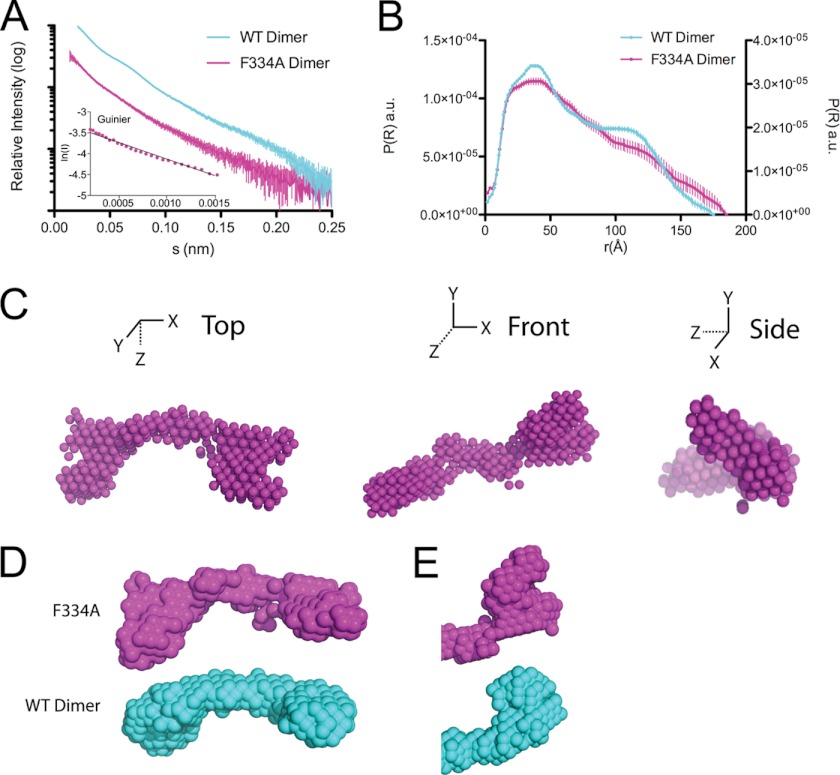
We have shown that the single amino acid mutation F334A could switch apoA-IV from a poor lipid binder to an avid one, on par with the lipid binding potency of apoA-I (Fig. 5E). Therefore, we analyzed dimeric apoA-IVF334A by SAXS to investigate the impact of the F334A mutation on the overall structure (Fig. 9 and Table 2). Again, the purified apoA-IVF334A dimer was stable and exhibited robust lipid binding, as shown in Fig. 5E.
FIGURE 9.
Solution state of apoA-IVF334A. A, scattering function and Guinier plot (inset) of dimeric apoA-IVF334A and apoA-IVWT. B, pairwise distance distribution function of apoA-IVF334A and apoA-IVWT. The y ordinates are for apoA-IVF334A (left) and apoA-IVWT (right). a.u., arbitrary units. C, averaged ab initio reconstructions of apoA-IVF334A. D and E, comparison of the overall structures (D) and terminal bundles (E) of apoA-IVF334A and apoA-IVWT.
Analysis by SAXS revealed that the pairwise distance distribution profile of apoA-IVF334A indicated a structure that was rod-like, but more similar to apoA-IVWT than apoA-IV64–335 (Fig. 9B). Although the bimodal distribution observed in apoA-IVWT is less evident in apoA-IVF334A, the peak of the pairwise distance distribution is more rounded like apoA-IVWT, whereas in apoA-IV64–335, the pairwise distance distribution exhibits a sharp peak. Although we expected the envelope of the single point mutation apoA-IVF334A to closely resemble that of apoA-IVWT, ab initio reconstruction resulted in an average envelope that displays a markedly segmented and less ordered rod, similar to apoA-IV62–376 (Fig. 9, C and D). A more flexible structure was also supported by a flattened Kratky plot (Fig. 8B). Furthermore, significant differences are observed at the ends of the rods despite each occupying a comparable volume (Fig. 9E). In apoA-IVF334A, the end globular domains point away from the core bundle instead of looping back to interact, as seen in apoA-IVWT. This indicates that a single amino acid change can disrupt the ends of the molecule and also significantly affect the global conformation of apoA-IV.
DISCUSSION
Recent structural studies have illuminated how apolipoproteins transition between monomers and dimers, revealing a novel helix-swapping mechanism (4, 6). However, by necessity, both structures were derived from truncated proteins, thus raising questions about what effect the missing components have on the overall structure and whether they are good structural models for apolipoproteins in solution. This is certainly the case for apoA-I, which was solved in two completely different configurations depending on the truncation and crystallization conditions (4, 36). To resolve these issues, we structurally characterized the solution state of a full-length unmodified apolipoprotein, apoA-IV, using SAXS. This information will be important for understanding the mechanisms that occur as apolipoproteins form HDL particles.
We have shown that SAXS is a convenient method for resolving the structures of multiple apoA-IV variants. However, SAXS is a low-resolution, albeit solution-based technique. Therefore, we first had to compare the crystal structure of apoA-IV64–335 with its SAXS envelope to provide orientation and bearing. This showed that the bundle architecture and domain swapping described in the apoA-IV64–335 crystal structure also occur in solution and in the full-length protein. This is important because the concentrations of protein required for crystallographic studies have been suggested to drive domain-swapping interactions (37). In this study, the rod-like structure of apoA-IV was clearly evident at all concentrations examined, supporting that domain swapping was not a crystallization artifact.
From the comparisons of apoA-IV64–335 and apoA-IVWT, it is clear that the N and C termini interact to form a globular domain. Because the missing N and C termini are expected to be highly helical (38), it is likely that this domain is a set of helices forming a compact helical bundle (Fig. 10). This is consistent with circular dichroism measurements (6) and also a comparison with other apolipoproteins that have a core bundle domain flanked by helical termini, such as the C terminus of apoA-I and both N and C termini of apoE3 (4, 5). However, it does not appear that the termini of apoA-I and apoE3 form an additional domain that regulates lipid affinity. Furthermore, the molecular envelops of the monomeric and dimeric forms show that the N and C termini interact in both molecular states. Thus, a structural distinction of apolipoproteins might be how the N and C termini interact with the core bundle to regulate their function. For apoA-IV, our data support that the N and C termini have long-range conformational affects on the core helical bundle.
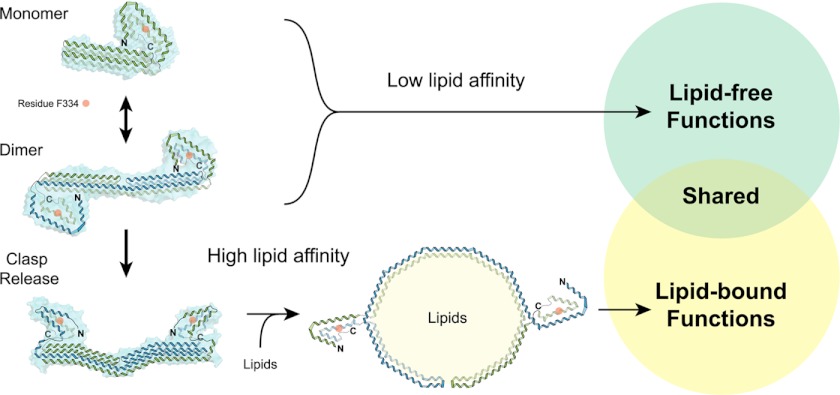
FIGURE 10.
Current model of apoA-IV structure and possible implications for its various functions. ApoA-IV exists predominantly as a monomer and dimer, and both have low lipid affinity. Low lipid affinity is regulated through interactions with the N and C termini, which also interact with the main bundle. Disrupting these interactions increases the lipid affinity of apoA-IV. It is possible that different functions of apoA-IV are associated with a lipid- or lipid-free state, which is mediated through a clasp mechanism.
Previous biochemical experiments suggested that interaction of the apoA-IV N and C termini hinders lipid binding (19). Breaking this configuration through site-directed mutagenesis increased the ability of apoA-IV to emulsify lipids. In this study, our SAXS data support that the N and C termini interact not only with each other but also with the core bundle. Disrupting the clasp, as observed with the F334A mutation, alters the structure of the terminal bundle, implying that significant conformational changes are occurring in this region. Furthermore, the terminal bundle dissociates from the core. One possibility is that Phe-334 interacts directly with the bundle, and its mutation leads to a dissociation of the two domains. Alternatively, Phe-334 could be necessary for the termini to coalesce into a globular structure. Our results show that the F334A mutation not only disrupts the N/C-terminal interaction but also alters how the N/C-terminal bundle interacts with the rod. Without this interaction, the core bundle is not as stable and becomes segmented. This implies that an increase in lipid binding might be associated with a more fluid core bundle. A fluid core bundle was also observed in the N-terminal truncation apoA-IV62–376, but this form remains a poor lipid binder due to deletion of important lipid binding determinants (18). Taken together, these results fundamentally redefine the clasp mechanism as an interaction between the termini and the main bundle instead of just the termini and provide a structural explanation for how the N and C termini can impact lipid binding by altering the overall bundle architecture.
Because apoA-IV exhibits a number of unique biological functions that are not observed with other apolipoproteins, it is intriguing to speculate that these are related to the clasp mechanism. One possibility is that certain biological activities of apoA-IV require a lipid-free state, and therefore, a mechanism is needed to prevent constitutive lipid binding. This could lead to lipid-free functions of apoA-IV (Fig. 10). Disruption of the clasp, possibly through protein or ligand interactions, could convert apoA-IV into an avid lipid binder and facilitate lipid-bound functions of apoA-IV (Fig. 10). Future work will need to associate the functions of apoA-IV with its lipid state.
In summary, we have further defined the conformational starting point of apoA-IV, before it transitions into an HDL, by providing the low-resolution envelops of solution-state apoA-IV in its dimeric and monomeric forms. Combining these envelops with our previously published high-resolution crystal structure has allowed us to better model the solution state of full-length apoA-IV and to determine that the N and C termini can form a globular domain that can interact and impact the overall structure of the bundle. Future studies will be aimed at developing detailed models that fit the molecular envelopes determined here, validating these with additional biophysical techniques, and then performing molecular dynamics studies to determine their plausibility.
Acknowledgments
We thank Drs. Yun-Xing Wang and Xianyang Fan (Protein-Nucleic Acid Interaction Section, Structural Biophysical Laboratory, NCI) for providing the beam time to perform the SAXS experiments and help with data collection. We also thank the X-ray Science Division at Argonne National Laboratory for technical support at beamline 12-ID-B.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM098458 (to T. B. T. and W. S. D.), Grant HL67093 (to W. S. D.), and Training Grant T32 HL007382 (to X. D.).
- SAXS
- small-angle x-ray scattering
- SEC
- size-exclusion chromatography
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine.
REFERENCES
- 1. Lund-Katz S., Phillips M. C. (2010) High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 51, 183–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sorci-Thomas M. G., Thomas M. J. (2012) High density lipoprotein biogenesis, cholesterol efflux, and immune cell function. Arterioscler. Thromb. Vasc. Biol. 32, 2561–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davidson W. S., Hazlett T., Mantulin W. W., Jonas A. (1996) The role of apolipoprotein A-I domains in lipid binding. Proc. Natl. Acad. Sci. U.S.A. 93, 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mei X., Atkinson D. (2011) Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286, 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J., Li Q., Wang J. (2011) Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. U.S.A. 108, 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng X., Morris J., Dressmen J., Tubb M. R., Tso P., Jerome W. G., Davidson W. S., Thompson T. B. (2012) The structure of dimeric apolipoprotein A-IV and its mechanism of self-association. Structure 20, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tabet F., Rye K.-A. (2009) High-density lipoproteins, inflammation and oxidative stress. Clinical Science 116, 87–98 [DOI] [PubMed] [Google Scholar]
- 8. Tso P., Sun W., Liu M. (2004) Gastrointestinal satiety signals. IV. Apolipoprotein A-IV. Am. J. Physiol. Gastrointest. Liver Physiol. 286, G885–G890 [DOI] [PubMed] [Google Scholar]
- 9. Weinberg R. B., Gallagher J. W., Fabritius M. A., Shelness G. S. (2012) ApoA-IV modulates the secretory trafficking of apoB and the size of triglyceride-rich lipoproteins. J. Lipid Res. 53, 736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tso P., Liu M. (2004) Apolipoprotein A-IV, food intake, and obesity. Physiol. Behav. 83, 631–643 [DOI] [PubMed] [Google Scholar]
- 11. Tso P., Liu M., Kalogeris T. (1999) The role of apolipoprotein A-IV in food intake regulation. J. Nutr. 129, 1503–1506 [DOI] [PubMed] [Google Scholar]
- 12. Qin X., Swertfeger D. K., Zheng S., Hui D. Y., Tso P. (1998) Apolipoprotein A-IV: a potent endogenous inhibitor of lipid oxidation. Am. J. Physiol. 274, H1836–H1840 [DOI] [PubMed] [Google Scholar]
- 13. Vowinkel T., Mori M., Krieglstein C. F., Russell J., Saijo F., Bharwani S., Turnage R. H., Davidson W. S., Tso P., Granger D. N., Kalogeris T. J. (2004) Apolipoprotein A-IV inhibits experimental colitis. J. Clin. Invest. 114, 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang F., Kohan A. B., Kindel T. L., Corbin K. L., Nunemaker C. S., Obici S., Woods S. C., Davidson W. S., Tso P. (2012) Apolipoprotein A-IV improves glucose homeostasis by enhancing insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 109, 9641–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Culnan D. M., Cooney R. N., Stanley B., Lynch C. J. (2009) Apolipoprotein A-IV, a putative satiety/antiatherogenic factor, rises after gastric bypass. Obesity 17, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinberg R. B., Spector M. S. (1985) The self-association of human apolipoprotein A-IV. Evidence for an in vivo circulating dimeric form. J. Biol. Chem. 260, 14279–14286 [PubMed] [Google Scholar]
- 17. Pearson K., Saito H., Woods S. C., Lund-Katz S., Tso P., Phillips M. C., Davidson W. S. (2004) Structure of human apolipoprotein A-IV: a distinct domain architecture among exchangeable apolipoproteins with potential functional implications. Biochemistry 43, 10719–10729 [DOI] [PubMed] [Google Scholar]
- 18. Pearson K., Tubb M. R., Tanaka M., Zhang X. Q., Tso P., Weinberg R. B., Davidson W. S. (2005) Specific sequences in the N and C termini of apolipoprotein A-IV modulate its conformation and lipid association. J. Biol. Chem. 280, 38576–38582 [DOI] [PubMed] [Google Scholar]
- 19. Tubb M. R., Silva R. A., Pearson K. J., Tso P., Liu M., Davidson W. S. (2007) Modulation of apolipoprotein A-IV lipid binding by an interaction between the N and C termini. J. Biol. Chem. 282, 28385–28394 [DOI] [PubMed] [Google Scholar]
- 20. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ilavsky J., Jemian P. R. (2009) Irena: tool suite for modeling and analysis of small-angle scattering. J. Appl. Crystallogr. 42, 347–353 [Google Scholar]
- 22. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 23. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 24. Svergun D. I., Petoukhov M. V., Koch M. H. J. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Svergun D., Barberato C., Koch M. H. J. (1995) CRYSOL–a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 27. Wang Z., Schröder G. F. (2012) Real-space refinement with DireX: from global fitting to side-chain improvements. Biopolymers 97, 687–697 [DOI] [PubMed] [Google Scholar]
- 28. Garai K., Frieden C. (2010) The association-dissociation behavior of the ApoE proteins: kinetic and equilibrium studies. Biochemistry 49, 9533–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vitello L. B., Scanu A. M. (1976) Studies on human serum high density lipoproteins. Self-association of apolipoprotein A-I in aqueous solutions. J. Biol. Chem. 251, 1131–1136 [PubMed] [Google Scholar]
- 30. Wong Y. Q., Binger K. J., Howlett G. J., Griffin M. D. W. (2010) Methionine oxidation induces amyloid fibril formation by full-length apolipoprotein A-I. Proc. Natl. Acad. Sci. U.S.A. 107, 1977–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deng X., Davidson W. S., Thompson T. B. (2012) Improving the diffraction of apoA-IV crystals through extreme dehydration. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schröder G. F., Brunger A. T., Levitt M. (2007) Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure 15, 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Visiers I., Braunheim B. B., Weinstein H. (2000) Prokink: a protocol for numerical evaluation of helix distortions by proline. Protein Eng. 13, 603–606 [DOI] [PubMed] [Google Scholar]
- 34. Barlow D. J., Thornton J. M. (1988) Helix geometry in proteins. J. Mol. Biol. 201, 601–619 [DOI] [PubMed] [Google Scholar]
- 35. Mertens H. D. T., Svergun D. I. (2010) Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J. Struct. Biol. 172, 128–141 [DOI] [PubMed] [Google Scholar]
- 36. Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. (1997) Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. U.S.A. 94, 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y., Eisenberg D. (2002) 3D domain swapping: as domains continue to swap. Protein Sci. 11, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adamczak R., Porollo A., Meller J. (2005) Combining prediction of secondary structure and solvent accessibility in proteins. Proteins 59, 467–475 [DOI] [PubMed] [Google Scholar]