Background: Oct-3/4 and Sox2 form a complex to regulate gene expression and maintain pluripotency.
Results: Sall4 directly interacts with Oct-3/4 or Sox2. Sall4 and Sox2 occupy the same promoter regions of genes active in ES cells.
Conclusion: Sall4 is involved in transcriptional networks retaining pluripotency, in combination with Oct-3/4 or Sox2.
Significance: This study provides a molecular understanding of how ES cells maintain pluripotency.
Keywords: Cell Differentiation, Chromatin Immunoprecipitation (ChIP), Microarray, Stem Cells, Transcription Factors, Embryonic Stem Cell, Pluripotency, Transcriptional Networks
Abstract
A small number of transcription factors, including Oct-3/4 and Sox2, constitute the transcriptional network that maintains pluripotency in embryonic stem (ES) cells. Previous reports suggested that some of these factors form a complex that binds the Oct-Sox element, a composite sequence consisting of closely juxtaposed Oct-3/4 binding and Sox2 binding sites. However, little is known regarding the components of the complex. In this study we show that Sall4, a member of the Spalt-like family of proteins, directly interacts with Sox2 and Oct-3/4. Sall4 in combination with Sox2 or Oct-3/4 simultaneously occupies the Oct-Sox elements in mouse ES cells. Overexpression of Sall4 in ES cells increased reporter activities in a luciferase assay when the Pou5f1- or Nanog-derived Oct-Sox element was included in the reporter. Microarray analyses revealed that Sall4 and Sox2 bound to the same genes in ES cells significantly more frequently than expected from random coincidence. These factors appeared to bind the promoter regions of a subset of the Sall4 and Sox2 double-positive genes in precisely similar distribution patterns along the promoter regions, suggesting that Sall4 and Sox2 associate with such Sall4/Sox2-overlapping genes as a complex. Importantly, gene ontology analyses indicated that the Sall4/Sox2-overlapping gene set is enriched for genes involved in maintaining pluripotency. Sall4/Sox2/Oct-3/4 triple-positive genes identified by referring to a previous study identifying Oct-3/4-bound genes in ES cells were further enriched for pluripotency genes than Sall4/Sox2 double-positive genes. These results demonstrate that Sall4 contributes to the transcriptional network operating in pluripotent cells together with Oct-3/4 and Sox2.
Introduction
Embryonic stem (ES)2 cells are characterized by two unique potentials, self-renewal and pluripotency. A small number of transcription factors including Oct-3/4 and Sox2 play a key role in maintaining these properties. Pou5f1 (encoding Oct-3/4) and Sox2 show similar expression profiles during early development. At the blastocyst stage Pou5f1 and Sox2 are expressed in the inner cell mass, a group of cells retaining pluripotency, but not in trophectodermal cells. In vitro, Pou5f1 and Sox2 are expressed in undifferentiated ES cells and become repressed once ES cells are induced to differentiate. The level of Pou5f1 and Sox2 expression is precisely controlled to maintain pluripotency (1–3). Oct-3/4 and Sox2 are among the factors required for reprogramming of somatic cells into induced pluripotent stem cells, underscoring the critical roles of these transcription factors in establishing pluripotency (4).
Expression of Pou5f1 and Sox2 is regulated by enhancers containing a closely juxtaposed Oct-3/4-binding site and a Sox2-binding site (5–7). These bipartite Oct-3/4- and Sox2- binding elements are called the Oct-Sox element. It is suggested that Oct-3/4 and Sox2 form a complex that cooperatively recognizes the Oct-Sox element to regulate their expression, thereby maintaining the pluripotency in ES cells (5, 6, 8, 9). Five other genes, Fgf4, Fbxo15, Nanog, Utf1, and Lefty1, apparently possess Oct-Sox elements and are proposed to be similarly regulated by the Oct-3/4-Sox2 complex (10–14).
ES cells establish and maintain a transcriptional network to achieve the precise control of the key transcription factors required for pluripotency. Many target genes of their transcription factors have been identified in ChIP-on-chip or ChIP-seq analyses in mouse ES cells (8, 9). These analyses led to the identification of two noteworthy features of the network. First, some transcription factors bind to the regulatory elements of members of the network, including themselves, thereby forming positive feedback loops among the network members. Second, members of the network physically interact to form a complex to establish another layer of feedback within the network. Global targets of transcription factors interacting with Oct-3/4 or Nanog have been determined. It is suggested that promoters occupied by multiple transcription factors are active in pluripotent cells, and the consensus motif recognized by these factors is similar to the Oct-Sox element (9). Therefore, it is likely that some of these factors form a complex and function at the Oct-Sox element, but it remains to be determined which proteins comprise the complex.
Sall4 (Sal-like 4) is a member of the Spalt (Sal)-like (Sall) family of proteins containing multiple C2H2 zinc finger motifs. Spalt was first isolated as a homeotic gene required for the development of the head and tail regions in Drosophila (15, 16). Sal-like family proteins have been identified in various species ranging from Drosophila to humans. Sall4-null mice die during peri-implantation, and heterozygous mutant mice exhibit anorectal anomalies and exencephaly (17–19). Sall4 knockdown in ES cells leads to differentiation into the trophoblast lineage (20), and Sall4-deleted ES cells proliferate inefficiently (21). It was suggested that this effect was caused by a decrease of Pou5f1 expression that is regulated by Sall4. However, some reports showed that Sall4-null ES cells maintain the expression of Pou5f1 (17, 18). Sakaki-Yumoto et al. (17) report that Sall4-null ES cells maintain pluripotency, but Tsubooka et al. (18) show that Sall4-null ES cells tend to differentiate under leukemia inhibitory factor conditions. Moreover, it has been shown that Sall4 positively regulates the efficiency of induced pluripotent stem cell generation from mouse somatic cells. Sall4 occupies the promoter regions of genes occupied by Oct-3/4, Sox2, and Nanog (22). As such, it is believed that Sall4 is among the key regulators in the transcriptional network in ES cells. Efforts for systematic isolation of Oct-3/4- and Sox2-interacting factors identified Sall4 as a binding protein (23, 24).
In this study we addressed the question of which proteins form a complex at the Oct-Sox element in mouse ES cells. We identified proteins associated with the Oct-Sox elements by purifying proteins that bind to the elements in vitro. We found that Sall4 directly binds with Sox2 and Oct-3/4 and that Sall4 is associated with the Oct-Sox elements in vivo. Furthermore, we have revealed that Sall4 and Sox2 occupy the same regions of the promoters or enhancers, including the Oct-Sox elements, in a set of genes that is enriched for those active in maintaining pluripotency in ES cells. We furthermore have shown that Sall4, Sox2, and Oct-3/4 are all present at the promoter regions of genes involved in maintaining pluripotency. These results thus demonstrate that Sall4 is involved in the transcriptional regulatory network maintaining pluripotency along with Oct-3/4 and Sox2.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
E14 mouse ES cells, cultured in feeder-free conditions, were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen), with 15% fetal bovine serum (FBS, Hyclone), 2 mm l-glutamine (Invitrogen), 0.1 mm nonessential amino acids (Invitrogen), 1 mm sodium pyruvate (Invitrogen), 100 μm 2-mercaptoethanol (Sigma), and leukemia inhibitory factor. Transfection of plasmids into mouse ES cells was performed using Lipofectamine 2000 (Invitrogen). COS-7 cells were cultured in DMEM with 10% FBS. Transfection of plasmids into COS-7 cells was performed using FuGENE HD (Roche Applied Science).
Immunoprecipitation-Immunoblotting
Anti-Sall4 antibody was raised against a recombinant N-terminal 533-residue peptide expressed in Escherichia coli. Other primary antibodies were as follows: anti-Sox2 antibody from Santa Cruz (Y-17X) and Abcam (ab15830); anti-Oct-3/4 antibody from Santa Cruz (H-134 and N-19X); anti-HA mouse monoclonal antibody clone 16B12 from Berkeley Antibody Co., Inc.; anti-HA rat monoclonal antibody clone 3F10 from Roche Applied Science; anti-FLAG M2 mouse monoclonal antibody from Sigma (F3165). Extracts of mouse ES cells or COS-7 cells were diluted and incubated with antibodies for 2 h at 4 °C. The antibody complexes were pulled down with Dynabeads Protein G (Invitrogen). For FLAG-Sall4, COS-7 extracts were diluted and incubated with Anti-FLAG M2 affinity gel (Sigma). For immunoblotting, proteins were loaded onto SDS-PAGE gels, transferred onto polyvinylidene fluoride membranes (Millipore), and detected with ECL, ECL plus, or ECL advance immunoblotting detection reagents (GE Healthcare).
GST Pulldown Assay
Bacterial GST-Sall4-N and GST-Sall4-C fusion proteins were purified using glutathione-Sepharose beads (Amersham Biosciences). For in vitro expression of Oct-3/4 or Sox2, TNT Quick Coupled Transcription/Translation System (Promega) was used in the presence of [35S]methionine. The protein complexes were pulled down using glutathione beads.
RNA Interference and RT-Quantitative PCR
For RNAi design, 19-base pair gene-specific regions were selected using siDirect. Oligonucleotides were cloned into pSuper.retro.puro (Oligoengine). Transfection of shRNA plasmids into mouse ES cells was performed using Lipofectamine 2000 (Invitrogen). Puromycin selection was performed starting 1 day after transfection and continued for 3 days. RNA was isolated with the RNeasy Plus mini kit (Qiagen) according to the manufacturer's protocol. RNAs were reverse-transcribed using an RNA PCR kit (TaKaRa) and oligo-dT primers. Quantitative PCR was then performed using StepOnePlus Real-time PCR System (Applied Biosystems).
Colony Formation Assay
One day after transfection with shRNA plasmids, ES cells were plated at a clonal density in 6-well plates and incubated for 4 days under puromycin selection. Cells were stained with alkaline phosphatase using an alkaline phosphatase kit (Sigma) to count the alkaline phosphatase-positive colonies.
Reporter Assay
Reporter plasmids and phRL-TK control plasmid were transfected using Lipofectamine 2000 (Invitrogen). Two days after the transfection, luciferase activities were measured with the dual luciferase assay system (Promega).
ChIP, Sequential ChIP (Re-ChIP), and ChIP-on-chip
ChIP assays were carried out as previously described (25). Sequential ChIP (Re-ChIP) assays were performed as previously described, with modifications (26). Extracted chromatin was prepared as previously described (25). Protein complexes were eluted by incubating the immunoprecipitated beads for 30 min at 37 °C in 25 μl 10 mm dithiothreitol. After the beads were pelleted by centrifugation, the supernatant was diluted 50 times and subjected to another round of immunoprecipitation. The immunoprecipitated beads were washed once with buffer A (20 mm Tris-Cl (pH 8), 1 mm EDTA, 1% Triton X-100, and 150 mm NaCl, 1 mm PMSF), twice with buffer B (20 mm Tris-Cl (pH 8), 2 mm EDTA, 1% Triton X-100, 150 mm NaCl, 1 mm PMSF), once with buffer C (20 mm Tris-Cl (pH 8), 2 mm EDTA, 1% Triton X-100, 0.1% SDS, 500 mm NaCl, 1 mm PMSF), and once with buffer D (10 mm Tris-Cl (pH 8), 1 mm EDTA, 0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate). For ChIP and sequential ChIP analyses, quantitative PCR was performed using the 7300 real-time PCR system or StepOnePlus Real-Time PCR System (Applied Biosystems). For ChIP-on-chip analysis, the immunoprecipitated DNA was amplified by in vitro transcription, labeled with biotin, and hybridized to GeneChip Mouse Promoter 1.0R Arrays (Affymetrix). Hybridization and scanning were performed according to protocols from Affymetrix. Using Tiling Analysis Software (Affymetrix), the probe signals were calculated as the ratio relative to input (bandwidth = 250). Integrated Genome Browser (Affymetrix) was used to visualize the data and gene annotations. To identify the protein binding regions, the following conditions were used: threshold = 0.27, Max Gap = 100, Min Run = 350. Genes between 5.5 kb upstream and 2.5 kb downstream of transcription start site were annotated using genome annotation (mm8) from NCBI. Among the regions positive for both Sall4 and Sox2, Sall4-positive regions overlapping with Sox2-positive regions were termed “overlapping,” and the rest of the Sall4-positive regions were “non-overlapping.” The promoter array data have been deposited in the Gene Expression Omnibus (GEO) database with accession number GSE40072.
Data Analyses
To detect the fold differences of gene expression in ES cells relative to differentiated ES cells, we obtained microarray data from a previous study (27). Genes were annotated using Na32 from Affymetrix. The ratios of CCE_RA_d0/CCE_RA_d6 were calculated and plotted. The ratios in each group were compared using the Mann-Whitney U test.
Gene Ontology (GO) Analyses
Total 18781 genes were annotated with GO terms. 12350 GO terms were examined. Fisher's exact test was used to calculate p values for each GO term.
RESULTS
Sall4 Interacts with Sox2
It has been shown that the ternary complex consisting of Oct1, which is closely related to Oct-3/4, Sox2, and a target DNA, induces structural changes in each of the three components, thereby establishing cooperative binding (28). Anticipating that additional proteins modulate such a complex, we sought to isolate proteins specifically bound with the ternary complex of Oct-3/4, Sox2, and target DNA. To identify such proteins, nuclear extracts derived from mouse E14 ES cells (29) were subjected to affinity precipitation using biotin-tagged oligonucleotides containing the Oct-Sox element (supplemental Fig. S1). First, nuclear extracts were preincubated with a biotin-tagged double-stranded (ds) oligonucleotide containing a mutated Oct-Sox element (mut-OS oligo, TTGTTCCT-TCCCATT; the underlines indicate the substituted nucleotides for the consensus Oct-Sox element sequence) that does not bind to the Oct-3/4-Sox2 complex. These same nucleotide substitutions diminished the promoter activity of the Nanog gene (10). The Oct-3/4-Sox2-mut-OS oligonucleotide complex did not form in an EMSA experiment (supplemental Fig. S1B). After the preincubation, avidin-conjugated magnetic beads were added, and mut-OS bound protein fractions were recovered by magnetic separation. Next, the mut-OS unbound protein fraction (supernatant) was incubated with a biotin-tagged ds-oligonucleotide containing the wild-type Oct-Sox element (wt-OS oligo, ATTAGCAT-AACAATG). The wt-OS nucleotide consensus sequence was deduced from the Oct-Sox elements found in the Fgf4, Fbxo15, Nanog, Sox2, and Utf1 genes (7, 10, 12–14). EMSA experiments confirmed that the Oct-3/4-Sox2 complex formed with the wt-OS oligonucleotide (supplemental Fig. S1B). After the incubation, avidin-conjugated magnetic beads were added, and the wt-OS-bound protein fractions were collected by magnetic separation. Wt-OS-bound and mut-OS-bound protein fractions were analyzed by SDS-PAGE followed by silver staining (supplemental Fig. S1C). All proteins present in the mut-OS-bound and wt-OS-bound fractions were analyzed by mass spectrometry. Proteins present in the wt-OS-bound fraction but not in the mut-OS-bound fraction were listed. By setting a cutoff of proteins whose score was more than 420 or less than 79, a total of 74 proteins were detected (supplemental Table S1). We found Sall4 protein (Gene ID 30424972; score, 257, marked with an asterisk in supplemental Table S1) was included in this collection. This suggested to us that Sall4 associates specifically with the Oct-3/4-Sox2-DNA ternary complex. Because it was reported that Sall4 knockdown induced differentiation in mouse ES cells and that expression of Pou5f1 is regulated by Sall4 (20), we further analyzed Sall4 in this study. Other candidates will be investigated in the future.
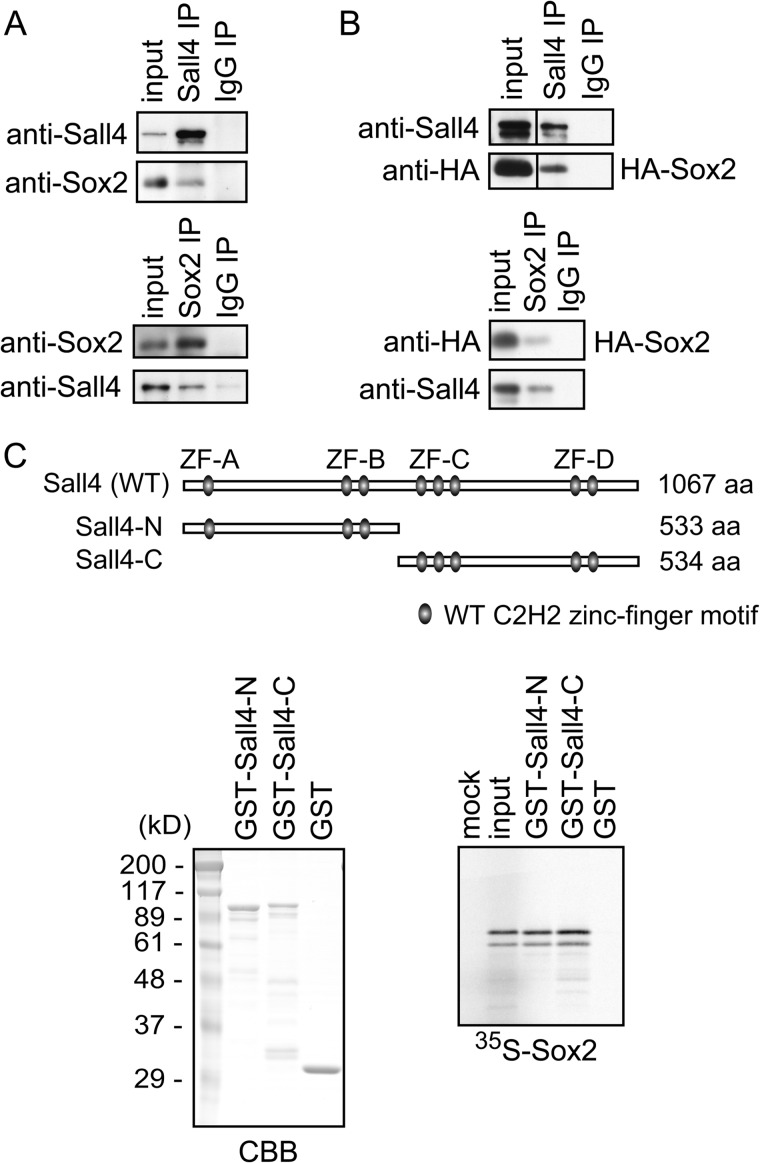
To determine whether Sall4 interacts with Sox2, we performed an immunoprecipitation (IP) and immunoblotting (IB) assay (IP-IB assay) using mouse ES cell extracts. Endogenous Sox2 was co-immunoprecipitated with endogenous Sall4 using an anti-mouse Sall4 antibody. Conversely, endogenous Sall4 was detected in immunoprecipitates obtained with anti-mouse Sox2 antibody, as previously reported (Fig. 1A) (24). Collectively, these results suggest that endogenous Sall4 and Sox2 form a protein complex in ES cells. The protein interaction between Sall4 and Sox2 was also detected when COS-7 cells overexpressing mouse Sall4 and HA-tagged mouse Sox2 were similarly analyzed (Fig. 1B), suggesting that ES-specific factors are not required for the interaction between Sall4 and Sox2.
FIGURE 1.
Sall4 interacts with Sox2. A, IP-IB assays of endogenous Sall4 and Sox2 using ES cell extracts are shown. Proteins were immunoprecipitated and detected by immunoblotting using indicated antibodies. B, IP-IB assays of transiently overexpressed mouse Sall4 and HA-tagged mouse Sox2 in COS-7 cells are shown. The two panels in the top figure were obtained from the same chemiluminescence image. C, shown is a GST pulldown assay. Top, shown is a schematic diagram of Sall4 and Sall4 mutants. Gray ovals show C2H2 zinc finger motifs. These motifs are clustered in four regions, ZF-A, -B, -C, and -D. Bottom, recombinant GST, GST-Sall4-N, and GST-Sall4-C proteins expressed in E. coli were purified using glutathione Sepharose beads (left). In vitro translated and 35S-labeled Sox2 was incubated with the glutathione-Sepharose beads, and bound protein was eluted and detected by autoradiography (right). CBB, Coomassie Brilliant Blue.
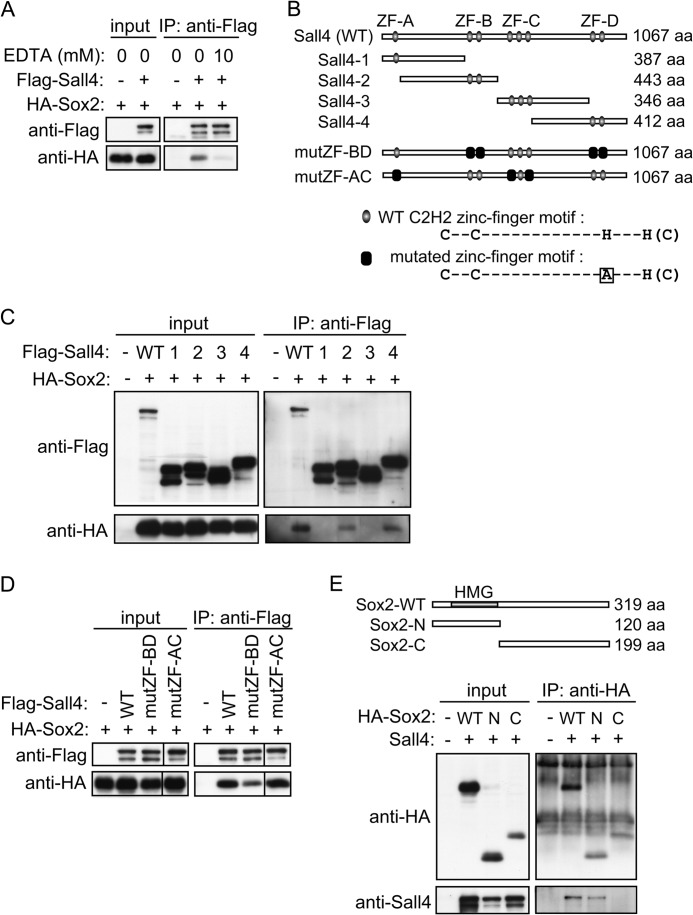
To examine whether Sall4 and Sox2 directly interact, we performed GST pulldown assays. Sall4 contains eight putative C2H2 zinc finger motifs distributed in four zinc finger (ZF) clusters A to D (hereinafter called ZF-A through ZF-D, Fig. 1C). We generated two mutually exclusive deletion mutants corresponding to the N-terminal half (Sall4-N) and the C-terminal half (Sall4-C) of Sall4 (Fig. 1C). GST-Sall4-N and GST-Sall4-C fusion proteins were purified from E. coli using glutathione Sepharose beads. In vitro translated, 35S-labeled Sox2 was incubated with GST alone, GST-Sall4-N, or GST-Sall4-C. We found that both GST-Sall4-N and GST-Sall4-C interacted with recombinant Sox2, but GST alone did not (Fig. 1C). These results suggest that Sall4 and Sox2 physically interact directly and that the N-terminal and C-terminal halves of Sall4 are redundantly responsible for the interaction with Sox2. To examine whether the zinc finger motifs of Sall4 were responsible for the Sox2-Sall4 interaction, cell extracts from COS-7 cells simultaneously overexpressing FLAG-Sall4 and HA-Sox2 were treated with 10 mm EDTA to remove divalent cations before IP-IB experiments. IP-IB assays using anti-FLAG and anti-HA antibodies demonstrated that the interaction between HA-Sox and FLAG-Sall4 was largely disrupted in the presence of EDTA, suggesting that protein folding of zinc finger motifs is important for the interaction (Fig. 2A) (30, 31).
FIGURE 2.
Zinc finger motif clusters of Sall4 play a major role in the interaction with Sox2. A, IP-IB assays of FLAG-Sall4 and HA-Sox2 are shown. FLAG-Sall4 and HA-Sox2 were transiently expressed in COS-7 cells as indicated, and IP-IB assays were performed with or without treatment of extracts with 10 mm EDTA before IP-IB procedures as indicated. B, shown is a schematic diagram of Sall4 and Sall4 mutants. Gray ovals show WT C2H2 zinc finger motifs. Black boxes represent mutated zinc finger motifs containing histidine-to-alanine amino acid substitutions, as shown at the bottom. aa, amino acids. C, IP-IB assays of FLAG-tagged Sall4 deletion mutants and HA-Sox2 are shown. FLAG-tagged wild-type (WT) Sall4, FLAG-Sall4-1, -2, -3, or -4 deletion mutants (indicated as 1, 2, 3, and 4, respectively), and HA-Sox2 were transiently co-expressed in COS-7 cells as indicated, and IP-IB assays were performed. Structures of FLAG- Sall4-1, -2, -3 and -4 are indicated in B. D, IP-IB assays of FLAG-tagged Sall4 point mutants and HA-Sox2 are shown. FLAG-tagged Sall4-WT, mutZF-BD, or mutZF-AC mutants and HA-Sox2 were transiently co-expressed in COS-7 cells as indicated, and IP-IB assays were performed. Structures of mutZF-BD and mutZF-AC are indicated in B. Two panels displayed as connected were obtained from the same chemiluminescence image. E, IP-IB assays of HA-tagged Sox2 deletion mutants and Sall4 are shown. HA-tagged Sox2-WT, Sox2-N (N), or Sox2-C (C) and Sall4 were transiently co-expressed in COS-7 cells as indicated, and IP-IB assays were performed.
To further pinpoint the Sox2-interacting regions in Sall4, we constructed FLAG-tagged Sall4 deletion mutants Sall4-1 through Sall4-4 as well as wild-type Sall4 (Sall4-WT) (Fig. 2B). Each individual mutant was co-expressed with HA-Sox2 in COS-7 cells. We detected co-immunoprecipitated HA-Sox2 in Sall4-WT, -2, and -4 immunoprecipitates (Fig. 2C), suggesting that C2H2 zinc finger motif clusters B and D can independently interact with Sox2.
We generated full-length mutants containing amino acid substitutions in the C2H2 zinc finger motifs of Sall4 by substituting an alanine residue for one of two histidine residues that presumably coordinate a zinc ion. The constructs Sall4-mutZF-BD and Sall4-mutZF-AC contained these substitutions in the C2H2 zinc finger motif in clusters B and D and clusters A and C, respectively (Fig. 2B). We detected a significant decrease in the amount of HA-Sox2 in Sall4-mutZF-BD immunoprecipitates (Fig. 2D) but not in Sall4-mutZF-AC immunoprecipitates compared with Sall4-WT immunoprecipitates, indicating that C2H2 zinc finger motif clusters B and D play a major role in the interaction with Sox2. It is known that the mouse Sall4 gene produces two isoforms, Sall4a and Sall4b, through alternative splicing (32). Sall4a corresponds to the full-length Sall4 designated in this study, whereas Sall4b lacks the region containing C2H2 zinc finger motif clusters B and C. The aforementioned result that Sall4-4 (lacking clusters A, B, and C) bound with Sox2 in vitro suggests that both Sall4a and Sall4b potentially associate with Sox2 in vivo.
Sox2 contains a high mobility group domain in its N-terminal region (Fig. 2E). We constructed HA-tagged Sox2 deletion mutants, Sox2-N and Sox2-C, which retained the high mobility group domain and the C-terminal region, respectively. Wild-type Sox2 (Sox2-WT) or each mutant was individually co-expressed with Sall4 in COS-7 cells. Sall4 was detected in Sox2-WT and Sox2-N immunoprecipitates but not in Sox2-C immunoprecipitates, indicating that the N-terminal 120 amino acids of Sox2 are required for the interaction with Sall4 and suggesting the high mobility group domain of Sox2 is responsible for the interaction (Fig. 2E).
Sall4 Interacts with Oct-3/4
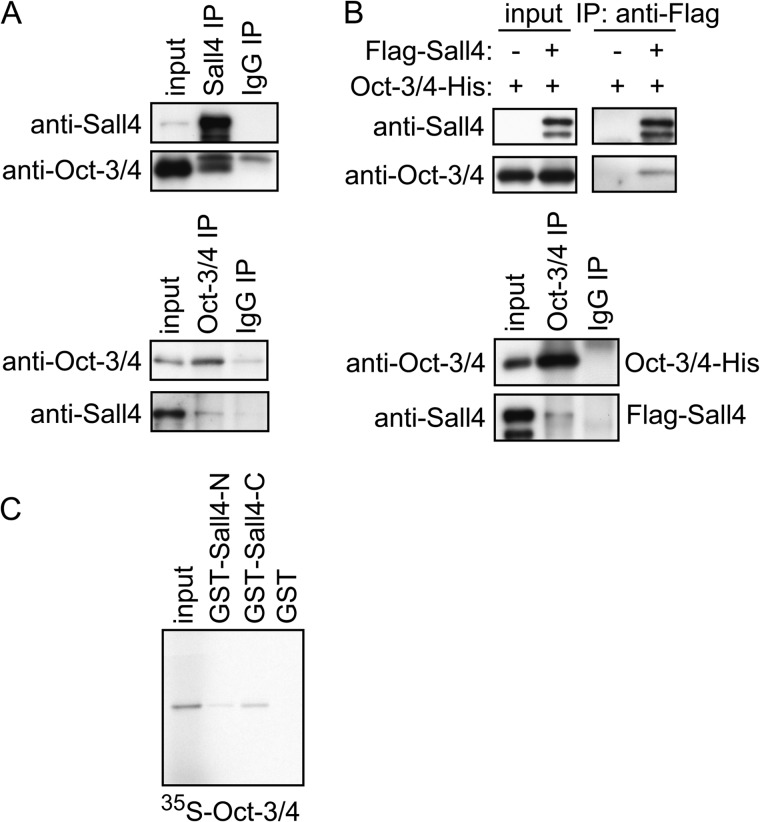
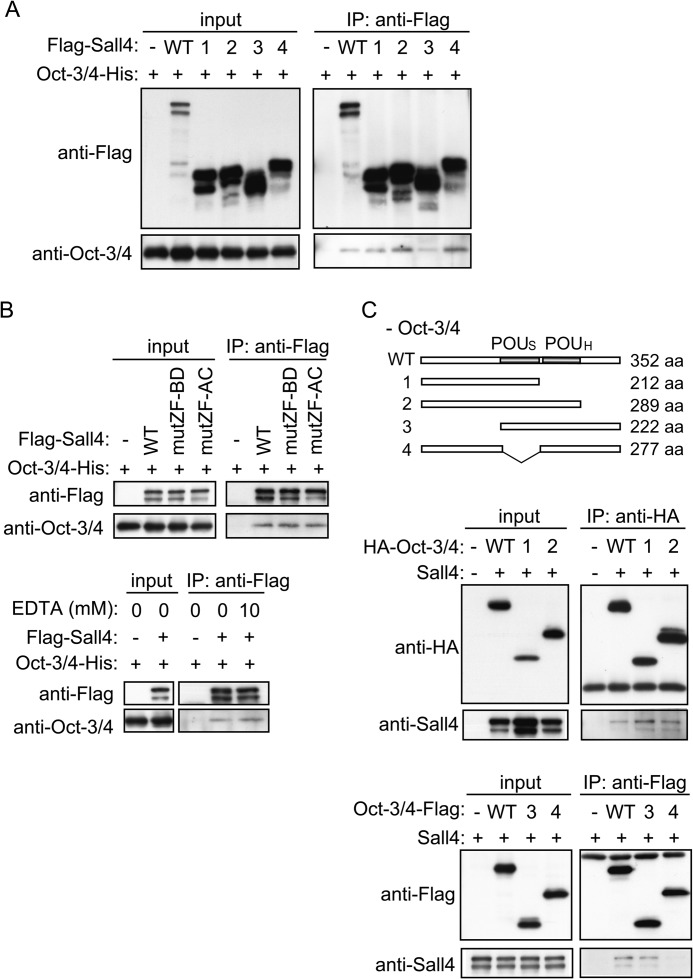
We next examined whether Sall4 interacts with Oct-3/4. We found that endogenous Oct-3/4 and Sall4 were detected in anti-Sall4 and anti-Oct-3/4 immunoprecipitates from mouse ES cell extracts, respectively, as previously reported (Fig. 3A) (23, 33). A similar IP-IB assay suggests that a complex forms between transiently overexpressed FLAG-Sall4 and Oct-3/4-His in COS-7 cells (Fig. 3B). A GST pulldown assay showed that both Sall4-N and Sall4-C recombinant proteins interact with recombinant Oct-3/4, although the interaction between Sall4-N and Oct-3/4 was weak (Fig. 3C). These results suggest that Sall4 directly interacts with Oct-3/4 and that the N-terminal and C-terminal halves of Sall4 are redundantly responsible for this interaction. Sall4 deletion mutants FLAG-Sall4-1 through FLAG-Sall4-4 were individually co-expressed with Oct-3/4-His in COS-7 cells. We found that Oct-3/4-His was co-immunoprecipitated with all FLAG-Sall4 mutants as well as FLAG-Sall4-WT (Fig. 4A). These results suggest that both Sall4a and Sall4b isoforms potentially associate with Oct-3/4 in vivo. Similar amounts of Oct-3/4 were detected in anti-FLAG immunoprecipitates of FLAG-Sall4-WT, -mutZF-BD, and -mutZF-AC (Fig. 4B, top). Moreover, the interaction between Sall4 and Oct-3/4 was not affected by the presence of EDTA (Fig. 4B, bottom). Taken together, these results suggest that zinc finger motifs coordinated with zinc ions are not required for the interaction between Sall4 and Oct-3/4.
FIGURE 3.
Sall4 interacts with Oct-3/4. A, IP-IB assays of endogenous Sall4 and Oct-3/4 in mouse ES cell extracts are shown. Proteins were immunoprecipitated and detected by immunoblotting using indicated antibodies. B, IP-IB assays of FLAG-Sall4 and Oct-3/4-His are shown. FLAG-tagged Sall4 and Oct-3/4-His were transiently expressed in COS-7 cells as indicated, and IP-IB assays were performed. C, shown is a GST pulldown assay. In vitro translated and 35S-labeled Oct-3/4 was analyzed as in Fig. 1C.
FIGURE 4.
Zinc finger motifs of Sall4 are not required for the interaction with Oct-3/4. A, shown are IP-IB assays of FLAG-tagged Sall4 deletion mutants and Oct-3/4-His. FLAG-tagged Sall4-WT, FLAG-Sall4-1, -2, -3, or -4 deletion mutants (indicated as 1, 2, 3, and 4, respectively), and Oct-3/4-His were transiently co-expressed in COS-7 cells as indicated, and IP-IB assays were performed. Structures of FLAG-Sall4-1, -2, -3, and -4 are indicated in Fig. 2B. B, shown are IP-IB assays of FLAG-tagged Sall4 and Oct-3/4-His. FLAG-tagged Sall4-WT, mutZF-BD or mutZF-AC mutants, and Oct-3/4-His were transiently co-expressed in COS-7 cells as indicated, and IP-IB assays were performed (top). Extracts were prepared and treated with or without 10 mm EDTA before IP (bottom). Structures of mutZF-BD and mutZF-AC are indicated in Fig. 2B. C, shown are IP-IB assays of Oct-3/4 deletion mutants and Sall4. Top, shown is a schematic diagram of Oct-3/4 and Oct-3/4–1 through -4 mutants. Middle and bottom, HA- or FLAG-tagged Oct-3/4-WT or Oct-3/4 deletion mutants 1–4 (indicated as 1–4, respectively) and Sall4 were transiently co-expressed in COS-7 cells as indicated, and IP-IB assays were performed. aa, amino acids.
Oct-3/4 contains the POU domain, which is divided into two subdomains, the POU-specific domain (POUS) and the POU-homeodomain (POUH) (34–36) (Fig. 4C, top). We constructed Oct-3/4 deletion mutants Oct-3/4-1 through Oct-3/4-4 as shown in Fig. 4C. We transiently over-expressed Sall4 and either HA-Oct-3/4-WT, -1, or -2 in COS-7 cells. We detected Sall4 in anti-HA immunoprecipitates in all cases, suggesting that POUH and the C-terminal region of Oct-3/4 are dispensable for the interaction between Oct-3/4 and Sall4 (Fig. 4C, middle). We performed similar experiments in which Sall4 and either Oct-3/4-WT-FLAG, -3-FLAG, or -4-FLAG were transiently overexpressed in COS-7 cells. Sall4 was detected in anti-FLAG immunoprecipitates of Oct-3/4-WT-FLAG or -3-FLAG, but not in -4-FLAG immunoprecipitates, suggesting that the POUS domain in Oct-3/4 is required for the interaction with Sall4 (Fig. 4C, bottom).
We analyzed the cellular localization of the overexpressed mutant proteins with immunofluorescence experiments using anti-tag antibodies. FLAG-Sall4-mutZF-BD was detected exclusively in nuclei (supplemental Fig. S2A), ruling out the possibility that mislocalization of FLAG-Sall4-mutZF-BD outside of the nucleus is responsible for its deficient interactions with Sox2. FLAG-Sall4–3 and Oct-3/4–4-FLAG were detected in nuclei at least in a fraction of the transfected cells (supplemental Fig. S2, A and C). HA-Sox2-C was exclusively in nuclei, but appeared as foci, in contrast to overexpressed HA-Sox2-WT that appeared homogeneously in nuclei (supplemental Fig. S2B). It remains to be determined whether these mutant proteins failed to interact with their partner proteins due to protein mislocalization.
Oct-Sox Elements Are Concurrently Occupied by Sall4 and Sox2 or by Sall4 and Oct-3/4
Using a ChIP assay, we examined whether Sall4 interacts with Oct-Sox elements. Mouse ES cells were cross-linked and sonicated. Anti-Sall4 immunoprecipitates were prepared from the extracted chromatin, and the presence of Oct-Sox element DNAs derived from various genes was quantified by real-time PCR using individual primer sets specific to the Oct-Sox elements of seven loci (Pou5f1, Sox2, Nanog, Fgf4, Fbxo15, Utf1, and Lefty1), and two control regions (the first intron of Pou5f1 and the Atbf1 promoter) (Fig. 5A). We found significant enrichments of the Oct-Sox elements derived from the seven loci in anti-Sall4 immunoprecipitates compared with the control regions (Figs. 5B). Similar ChIP experiments were performed using anti-Oct-3/4 or anti-Sox2 antibodies, revealing that the Oct-Sox elements were significantly enriched in anti-Oct-3/4 and anti-Sox2 immunoprecipitates in all cases (Fig. 5B). These results suggest that Sall4, Oct-3/4, and Sox2 occupy the Oct-Sox elements of these genes in mouse ES cells. We asked whether Sall4 and Sox2 concurrently occupy the Oct-Sox elements using the sequential ChIP (Re-ChIP) assay. Cross-linked cell extracts were first immunoprecipitated with anti-Sox2 antibody. Immunoprecipitates were eluted and were subsequently immunoprecipitated with anti-Sall4 antibody or control normal rabbit IgG. We applied the sequential ChIP assay to the Oct-Sox elements derived from the seven loci. We found statistically significant enrichments of the Oct-Sox elements derived from six loci except Utf1 with the anti-Sox2 and anti-Sall4 antibody pair and a tendency toward enrichment, albeit statistically not significant, of the Oct-Sox element of Utf1 (Fig. 5C, top). Similar experiments using anti-Oct-3/4 and anti-Sall4 antibodies revealed that the seven Oct-Sox elements were enriched in anti-Oct-3/4 and anti-Sall4 sequential immunoprecipitates at statistically significant levels (Fig. 5C, bottom). These results strongly suggest that Sall4 in combination with Sox2 or Oct-3/4 simultaneously associates with the Oct-Sox elements in ES cells.
FIGURE 5.
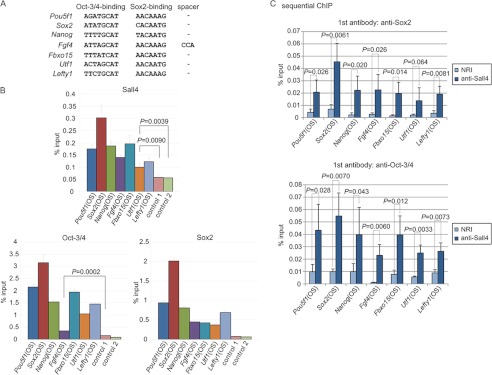
Sall4 co-occupies the Oct-Sox elements concurrently in combination with Oct-3/4 or Sox2. A, shown are the Oct-Sox elements of the indicated genes. Nucleotide sequences of Oct-Sox elements reported with the seven indicated loci are shown (5–7, 10–14). Spacer refers to a sequence intervening between the Oct-3/4 binding and Sox2 binding sequences. B, ChIP analyses show that Sall4, Sox2, and Oct-3/4 occupy the Oct-Sox elements. Control 1 is within the first intron of Pou5f1, and control 2 is within the Atbf1 promoter. The error bars show S.D. OS shows the Oct-Sox element. p values are based on a one-tailed Student's t test (n = 3). C, sequential ChIP (Re-ChIP) analyses showed that Sall4 and Sox2 as well as Sall4 and Oct-3/4 simultaneously occupy the Oct-Sox elements. The error bars show S.D. p values are based on a one-tailed Student's t test (n = 3).
Sall4 Is Required for Maintaining Pluripotency
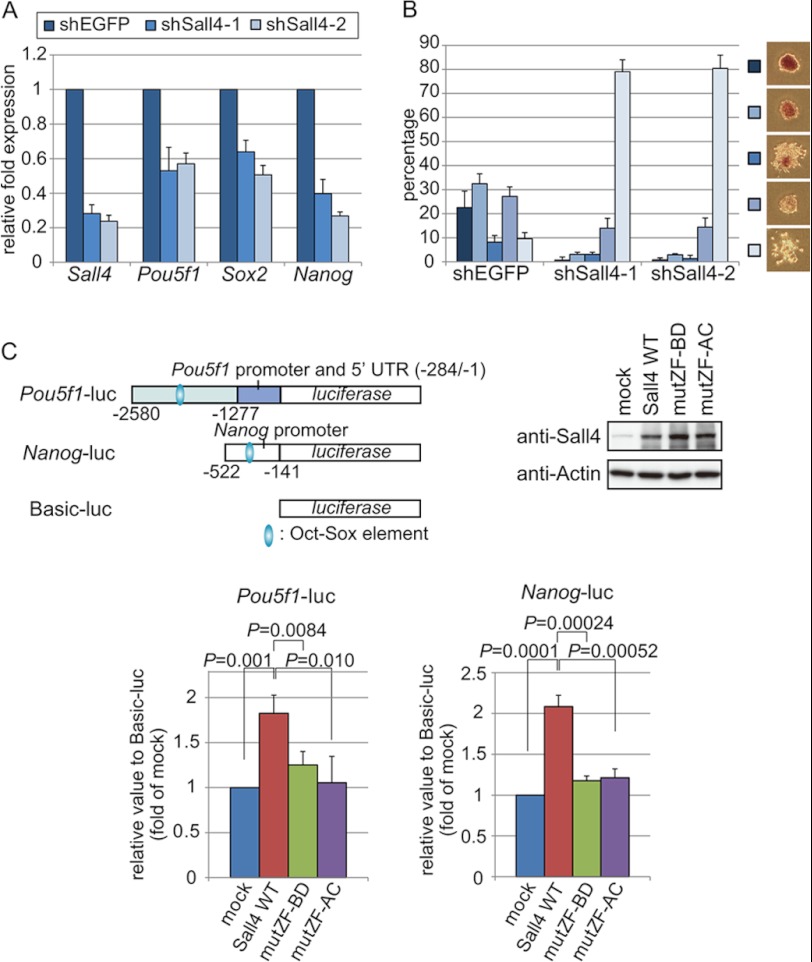
Given that Sall4 associates with the Oct-Sox elements of the Pou5f1, Sox2, and Nanog genes, we explored the biological roles of Sall4 in regulating the expression of these genes. ES cells were transfected with a plasmid expressing either an shRNA targeting Sall4 or a control shRNA targeting EGFP and were selected with puromycin for 3 days. The expression of Sall4 was reduced by ∼70∼80% for two independent Sall4-specific shRNA constructs, as determined by real-time RT-PCR (Fig. 6A). ES cells transfected with shRNAs were plated at a clonal density and stained with alkaline phosphatase after 4 days. Compared with control cells, Sall4 knockdown cells were largely alkaline phosphatase-negative. Each colony was classified into one of five groups according to the relative alkaline phosphatase activity and the shape of the colony, as indicated in Fig. 6B. Sall4-knockdown cells consistently gave rise to significantly increased proportions of colonies with less alkaline phosphatase activity and more scattered morphology (lighter blue color in Fig. 6B), suggesting the loss of pluripotency in Sall4 knockdown cells as reported previously (20). Furthermore, real-time RT-PCR showed that the expression levels of Pou5f1, Sox2, and Nanog were down-regulated in Sall4 knockdown cells, to ∼30∼60% of the respective levels in the control cells (Fig. 6A). We conclude that Sall4 is required for maintaining pluripotency in mouse ES cells.
FIGURE 6.
Sall4 is required for maintaining pluripotency in ES cells. A, expression levels of Pou5f1, Sox2, and Nanog were down-regulated in Sall4 knockdown ES cells. Two types of Sall4-knockdown cells (shSall4-1 and shSall4-2) were established using two independent shRNA expression plasmids. shEGFP was a control of ES cells transfected with an shRNA expression plasmid targeting the EGFP gene. mRNA levels were quantified with RT-quantitative PCR for each of the indicated genes and normalized against that of Actb mRNA. The error bars show S.D. (n = 3). B, Sall4 knockdown results in loss of pluripotency-related phenotypes in mouse ES cells. ES cells were plated at clonal density in 6-well plates 1 day after transfection with shRNA expression plasmids and cultured for 4 days. Cells were stained for alkaline phosphatase. Colonies were classified into five groups as indicated at the right, according to the relative alkaline phosphatase activity and the shape of the colony. Percentages indicate numbers of colonies assigned to each respective group relative to the total number of colonies in that well. Colony types indicated by lighter blue colors are supposed to maintain less pluripotency because they had less alkaline phosphatase activity and showed less demarcated colony shapes. Sall4 knockdown cells showed significantly increased fractions of the least ES-like colonies. The error bars show S.D. (n = 3). C, Sall4 point mutations led to reduced expression of Oct-Sox element-containing luciferase reporters. Structures of the three reporter constructs, Pou5f1-luc, Nanog-luc, and Basic-luc, are shown (top, left). ES cells were co-transfected with individual reporter constructs and one of the three Sall4 constructs expressing Sall4-WT, Sall4-mutZF-BD, or -mutZF-AC. Expression of Sall4 proteins in ES cells was confirmed in an immunoblot experiment (top, right). The luciferase activities expressed by Pou5f1-luc and Nanog-luc were quantified for ES cells expressing mock construct, Sall4-WT, Sall4-mutZF-BD, or -mutZF-AC (bottom). The error bars show S.D. p values are based on a one-tailed Student's t test (n = 3).
We then carried out a luciferase reporter assay using three reporter constructs, Pou5f1-luc, Nanog-luc, and Basic-luc as a control (Fig. 6C). In Pou5f1-luc, a region containing the Oct-Sox element of the Pou5f1 distal enhancer (−2580 to −1277 nucleotides, relative to the translational start site) plus the Pou5f1 promoter and the 5′-untranslated region (−284 to −1 nucleotides) were fused with the luciferase gene. In Nanog-luc, the Nanog promoter (−522 to −141 nucleotides) containing the Oct-Sox element was fused with the luciferase gene. Basic-luc does not contain a eukaryotic promoter or an enhancer. Individual reporter constructs along with one of the Sall4 vectors expressing Sall4-WT, -mutZF-BD, -mutZF-AC, or an empty expression vector were co-transfected into mouse ES cells. Expression of the respective Sall4 proteins was detected by immunoblotting (Fig. 6C). The luciferase activity levels observed in Pou5f1-luc- and Nanog-luc-transfected ES cells were significantly higher in cells co-expressing Sall4-WT compared with the control mock-expressing cells. The increase in the luciferase activity appeared marginally higher in Sall4-mutZF-BD- and -mutZF-AC-expressing cells than the mock-expressing cells (p < 0.05 for Nanog-luc in Sall4-mutZF-BD- and -mutZF-AC-expressing cells and for Pou5f1-luc in Sall4-mutZF-BD-expressing cells; p > 0.05 for Pou5f1-luc in Sall4-mutZF-AC-expressing cells) but markedly lower than in Sall4-WT-expressing cells (Fig. 6C). These results suggest that the four C2H2 zinc finger motif clusters of Sall4 are required to activate the expression of Pou5f1 and Nanog.
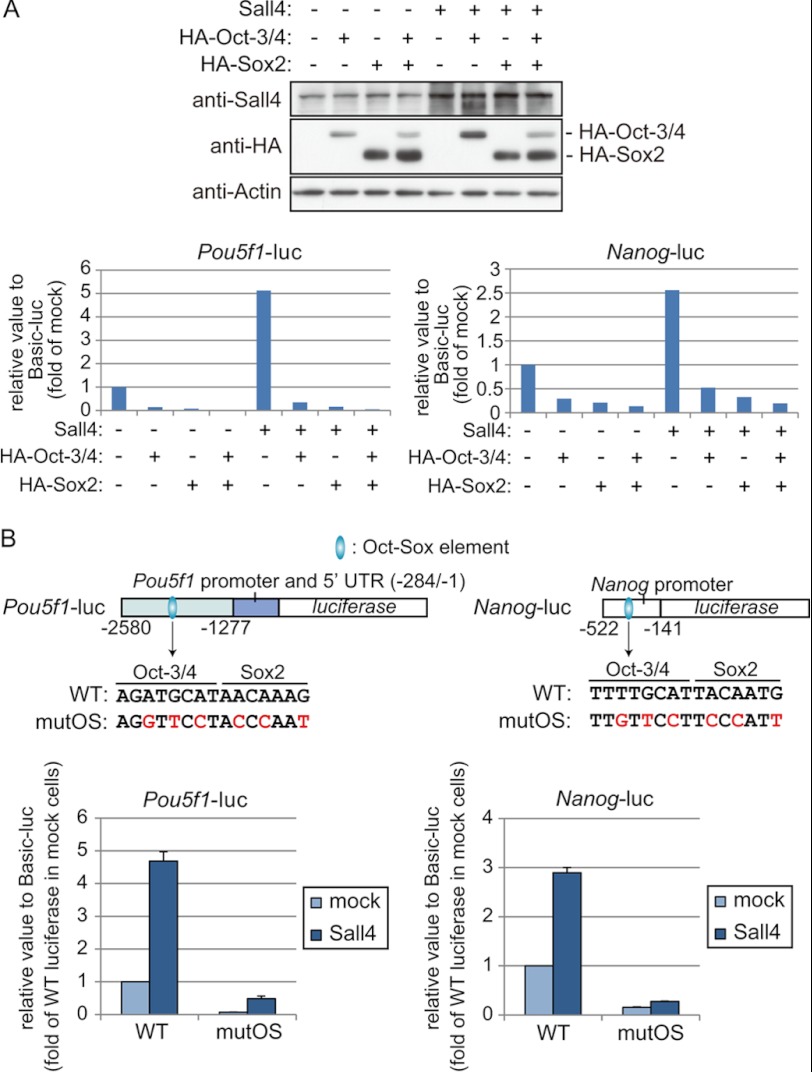
Given that Sall4 in combination with Sox2 or Oct-3/4 simultaneously occupies the Oct-Sox elements in ES cells, we predicted that Sall4, Oct-3/4, and Sox2 would cooperatively activate gene expression of Pou5f1 and Nanog. We examined whether transcriptional activation of Pou5f1-luc and Nanog-luc by Sall4 overexpression is affected by co-transfection of Oct-3/4 or Sox2 expression vectors in ES cells. Surprisingly, in the absence of Sall4 overexpression, the luciferase activity levels of Pou5f1-luc and Nanog-luc were significantly lower in cells overexpressing HA-Oct-3/4 and/or HA-Sox2 compared with the control mock-expressing cells (Fig. 7A). The higher luciferase activity of Pou5f1-luc and Nanog-luc in Sall4-overexpressing cells compared with the control mock-expressing cells was abolished when the Oct-Sox elements in the reporter constructs were mutated (Fig. 7B). These results suggest that Sall4 activates the reporter constructs cooperatively with Oct-3/4 and Sox2 in ES cells. However, the reporter activities in cells co-transfected with Sall4 in combination with HA-Oct-3/4 and/or HA-Sox2 were lower than those in cells transfected with Sall4 alone or the mock empty vector (Fig. 7A). It is not known how overexpression of Oct-3/4 and/or Sox2 diminished the reporter activities in the context of mock- or Sall4-transfected ES cells. It is possible that the experimental conditions did not produce the three proteins with a stoichiometry suitable for the formation of the functional Sall4-Oct-3/4-Sox2 trimeric complex. In this case the protein generated in excess may sequester other proteins required for the trimeric complex to activate the reporter constructs. Alternatively, the overexpression of Oct-3/4 and/or Sox2 may have induced the ES cells to differentiate, as reported previously (1, 3). The effect of the three factors overexpressed in such differentiated cells would not faithfully reflect the effect on the reporter constructs in undifferentiated ES cells.
FIGURE 7.
Mutation of the Oct-Sox element diminishes the luciferase activity of Pou5f1-luc and Nanog-luc. A, three reporter constructs, Pou5f1-luc, Nanog-luc, and Basic-luc, shown in Fig. 6C were used. ES cells were co-transfected with individual reporter constructs and combinations of HA-Oct-3/4, HA-Sox2, and Sall4 expression vectors. Expression of HA-Oct-3/4, HA-Sox2, and Sall4 proteins in ES cells was confirmed in an immunoblot experiment (top). The luciferase activities expressed by Pou5f1-luc and Nanog-luc were quantified for ES cells expressing mock construct, HA-Oct-3/4, HA-Sox2, or Sall4 with the indicated combinations (bottom). B, two reporter constructs, Pou5f1-mutOS-luc and Nanog-mutOS-luc, contain nucleotide substitutions in the Oct-Sox elements of Pou5f1-luc and Nanog-luc, respectively (top). Individual reporter constructs along with the construct expressing Sall4 (WT) were co-transfected into ES cells. The luciferase activities expressed by individual reporter constructs were quantified for ES cells expressing a mock construct or Sall4 (bottom). The error bars show S.D. (n = 3).
ES Cell-specific Genes Are Enriched in Genes Occupied by Sall4, Oct-3/4, and Sox2
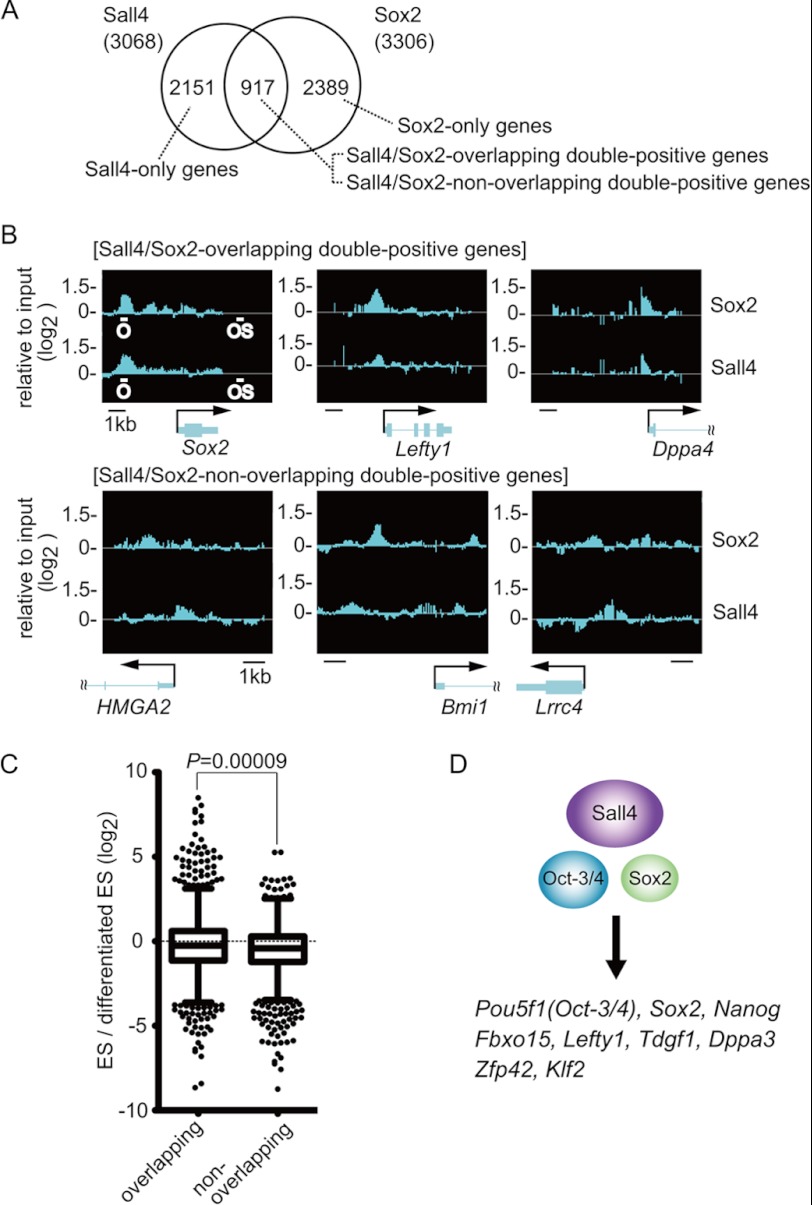
The aforementioned results suggest that Sall4 cooperates with Oct-3/4 and Sox2 to regulate Pou5f1 and Nanog gene expression. Because Pou5f1 and Nanog play a key role in maintaining pluripotency in ES cells in vivo, we were interested in identifying genes that are potentially regulated by Oct-3/4, Sox2, and Sall4 through genome-wide ChIP experiments. Cross-linked, sonicated chromatin extracted from mouse ES cells was immunoprecipitated individually with anti-Sall4 or anti-Sox2 antibody and was hybridized to a DNA microarray (Affymetrix, GeneChip Mouse Promoter 1.0R Array). The microarray contained oligonucleotides that cover ∼6 kb upstream through 2.5 kb downstream of the transcription start site of each gene. We found that Sall4 and Sox2 occupied such defined promoter regions in 3068 and 3306 genes, respectively (hereafter called Sall4-positive and Sox2-positive genes, Fig. 8A), among a total of 20,244 genes that were analyzed with the microarray. 917 genes were positive for both Sall4 and Sox2 (Sall4/Sox2-double-positive genes, DP-genes), and this number of DP-genes was significantly higher than the number expected on the assumption that Sall4 and Sox2 bind independently of each other to individual genes (i.e. 3068 × 3306/202442, p < 2.2 × 10−16, p value was calculated using Fisher's exact test). The promoter regions of Fbxo15, Lefty1, and Nanog were positive for both Sall4 and Sox2 in the microarray, consistent with the results obtained in Fig. 5, supporting the validity of the analysis. These analyses imply that there is a significant tendency for Sall4 and Sox2 to co-occupy target genes.
FIGURE 8.
Sall4 and Sox2 occupancies revealed by genome-wide ChIP analyses. A, a Venn diagram of genes occupied by Sall4 and/or Sox2 is shown. B, shown are chromosomal locations and enrichment ratios of Sall4 and Sox2 for the promoter regions of indicated genes. O and OS in the Sox2 gene map indicate the Oct-3/4 binding motif in the upstream enhancer and the Oct-Sox element in the downstream enhancer, respectively. Sox2 and Sall4 signals at the downstream enhancer were not examined because the promoter array used in this study does not contain oligonucleotides that cover the region. C, box plots of the relative expression levels in undifferentiated ES cells versus differentiated ES cells are shown for the overlapping Sall4/Sox2-double-positive genes (DP-genes) and the non-overlapping DP-genes. p value is scored using a Mann-Whitney U test. D, shown are examples of genes occupied by Sall4, Oct-3/4, and Sox2. The common genes were compiled by comparing the list of Sall4/Sox2-DP genes with that of Oct-3/4 target genes derived from previous ChIP-on-chip analysis. The full lists of genes in the overlapping or non-overlapping DP-genes and Sall4/Sox2/Oct-3/4 triple-positive genes are available upon request.
Interestingly, side-by-side comparisons of Sall4- and Sox2-positive ChIP regions for individual promoters revealed that Sall4 and Sox2 were distributed very similarly along the promoter regions in some genes, such as Sox2, Lefty1, and Dppa4 (Fig. 8B, top). Hereafter, genes in which Sall4 and Sox2 occupied the same regions will be called Sall4/Sox2-overlapping DP-genes. The similar distributions of Sall4 and Sox2 at the nucleotide level along the promoter regions suggest that Sall4 and Sox2 most likely bind to the promoters and/or enhancers of the overlapping DP-genes as a complex. It was reported that the promoter region of Dppa4 potentially contains an Oct-Sox element and the Dppa4 promoter activity depends on the putative Oct-3/4-binding region (37). In contrast to the overlapping DP-genes, Sall4 and Sox2 distributed with distinct localization patterns along the promoters of other DP-genes (examples of HMGA2, Bmi1, and Lrrc4 are shown in Fig. 8B, bottom), which we call Sall4/Sox2-non-overlapping DP-genes.
A previous study reported microarray analyses examining the relative change of expression levels of individual genes during the retinoic acid-induced differentiation of mouse ES cells (27). We accessed the published data from that study and obtained the fold difference of gene expression levels in undifferentiated ES cells (day 0) relative to those in differentiated ES cells that were treated for 6 days with retinoic acid in the absence of leukemia inhibitory factor (day 6). Fold differences in expression of the overlapping DP-genes were significantly larger than those of the non-overlapping DP-genes, suggesting that ES-specific genes are more enriched in the overlapping DP-genes than in the non-overlapping DP-genes (Fig. 8C).
We next subjected the overlapping and non-overlapping DP-genes to a GO analysis. GO terms that showed p values less than 4 × 10−6 were judged as positively enriched. GO terms related to transcriptional activation, such as “regulation of transcription, DNA-dependent” were among the most significantly enriched in both the overlapping and non-overlapping DP-genes (supplemental Table S2). However, the GO term “stem cell maintenance” was enriched only in the overlapping DP-genes, suggesting that genes involved in maintaining pluripotency are significantly enriched in the overlapping DP-genes but not in the non-overlapping DP-genes. Similar analyses of the genes occupied by Sall4 but not by Sox2 (Sall4-only genes) and the genes occupied by Sox2 but not by Sall4 (Sox2-only genes) were performed. The fold difference in expression of Sall4-only genes between undifferentiated and differentiated ES cells was significantly smaller than those of the overlapping DP-genes and was not statistically different from those of the non-overlapping DP-genes (supplemental Fig. S4, left and middle). The fold difference in expression of Sox2-only genes was not statistically different from those of the overlapping DP-genes (supplemental Fig. S4, right). However, the GO term stem cell maintenance was enriched only in the overlapping DP-genes and not in the Sox2-only genes (supplemental Tables S2 and S3). GO terms related to transcriptional activation were highly enriched in the overlapping DP-genes but were not enriched at all in the Sox2-only genes, suggesting that Sox2-only genes were quite different from the overlapping DP-genes. These results suggest that genes involved in maintaining pluripotency, which are expressed at a higher level in ES cells, are likely targeted by a Sall4-Sox2 complex.
Previous studies suggested that Oct-3/4 and Sox2 associate with chromatin in close proximity to synergistically control gene expression (8). We deduced a set of genes predicted to be occupied by three factors, Sall4, Oct-3/4, and Sox2, by comparing the list of Sall4/Sox2-DP-genes revealed in this study with the list of Oct-3/4 target genes identified in the previous ChIP-on-chip analysis (9). A total of 156 genes were identified (Sall4/Sox2/Oct-3/4 triple-positive genes, TP-genes). The number of TP-genes was significantly higher than the number expected by random binding (p < 2.2 × 10−16, p value was calculated using Fisher's exact test), suggesting that the three factors show a tendency to associate with a common set of target genes. The TP-genes include Pou5f1, Sox2, and Nanog, which are known to play primary roles in maintaining or inducing pluripotency in ES cells (Fig. 8D). Fold differences of expression levels in undifferentiated ES cells relative to those in differentiated ES cells were significantly larger for the TP-genes than for the Sall4/Sox2-DP-genes (supplemental Fig. S5). GO analysis of the TP- or DP-genes showed a smaller p value for stem cell differentiation for the TP-genes than for the DP-genes (this term indicates an involvement of genes in the development of stem cells, not the differentiation of stem cells; the group of genes belonging to this term includes Pou5f1, Sox2, and Nanog). In addition, the GO term stem cell maintenance was found only in the TP-genes and not in the DP-genes (supplemental Table S4). These results suggest that co-occupancy by Sall4, Sox2, and Oct-3/4 further enriches ES-specific genes over co-occupancy by Sall4 and Sox2.
DISCUSSION
We have demonstrated that Sall4 directly interacts with Oct-3/4 and Sox2. ChIP-on-chip analyses revealed the presence of genes that associated with both Sall4 and Sox2 (DP-genes). The DP-genes are divided into two sets of genes. In one set (overlapping DP-genes), Sall4-positive and Sox2-positive regions identified by the Affymetrix, GeneChip Mouse Promoter 1.0R Array distributed very similarly, suggesting the two proteins bind to the same promoter and/or enhancer regions as a complex. In the other set (non-overlapping DP-genes), Sall4 and Sox2 were distributed with distinct localization patterns, suggesting that the two proteins bind with the genes independently of each other. Importantly, GO analysis revealed that the overlapping DP-genes are enriched for GO terms “regulation of transcription, DNA-dependent” and “stem cell maintenance,” implying that this set of genes frequently plays a role in maintenance of pluripotency as transcription factors. Most genes targeted by multiple factors, including Oct-3/4, Sox2, and Nanog, are highly expressed in ES cells, whereas genes bound by relatively fewer factors are either expressed or repressed in pluripotent cells (9). Consistent with this, the set of Sall4/Sox2/Oct-3/4-TP genes, which are targeted by Sall4, Sox2, and Oct-3/4, was more highly enriched for genes expressed preferentially in pluripotent ES cells than the set of Sall4/Sox2-DP-genes (supplemental Fig. S5). These results suggest that Sall4, Oct-3/4, and Sox2 likely form a complex on the promoters and/or enhancers of the TP-genes, thereby activating the cognate genes and inducing or maintaining pluripotency in ES cells.
The Sall4 gene produces two isoform proteins, Sall4a and Sall4b, through alternative splicing. Sall4a corresponds to the full-length wild-type protein depicted in Fig. 1C, and Sall4b lacks the zinc finger clusters ZF-B and ZF-C. It was demonstrated that Sall4a is more abundant than Sall4b in mouse undifferentiated ES cells, and neither isoform is produced in differentiated ES cells (32). Sall4a and Sall4b form homo- and hetero-dimers. Using mouse ES cells that expressed biotin-tagged Sall4a or Sall4b, it was found that the Sall4-binding sites were classified into three types; those bound by Sall4a alone (Sall4-alone), by Sall4b alone (Sall4b-alone), and by both Sall4a and Sall4b (Sall4a/Sall4b) (32). Interestingly, in a GO analysis, the gene set of Sall4b-alone target genes was enriched for the term “transcriptional regulation,” but the gene set of Sall4a-alone targets was not. When consensus DNA binding sequences were deduced, Sall4b-alone and Sall4a/Sall4b target sequences were similar and overlapped with the multifactor binding motif targeted by pluripotency-maintaining factors (including Oct-3/4 and Sox2) but the Sall4a-alone target sequences were not (9, 32). Such overlaps between Sall4b-alone and Sall4a/Sall4b target, and Oct-3/4 and Sox2 targets suggest that the Sall4/Sox2-DP and Sall4/Sox2/Oct3/4-TP target genes identified in this study are occupied by Sall4b alone or both Sall4a and Sall4b. Interestingly, our analyses mapping the Sox2 and Oct-3/4 binding domains of the Sall4 protein suggest that both Sall4a and Sall4b potentially associate with Sox2 and Oct-3/4. Future study is required to test whether Sall4a-alone, Sall4b-alone, and Sall4a/Sall4b target genes are indeed bound with Sox2 or Oct-3/4 in vivo or not.
It is known that C2H2 zinc finger motifs can mediate protein-protein interactions. Sall4 contains eight putative C2H2 zinc finger motifs distributed in four clusters A to D (Fig. 1C). We found that C2H2 zinc finger motif clusters B and D in Sall4 are involved in the interaction between Sall4 and Sox2. Expression of Sall4-mutZF-BD, which interacted with Oct-3/4 but impaired the interaction with Sox2, resulted in reduced transcriptional activity compared with expression of Sall4-WT (Fig. 6C), suggesting that the stable formation of an Oct-3/4-Sox2-Sall4-DNA complex is important for transcriptional activation. Expression of Sall4-mutZF-AC, which efficiently interacted with Oct-3/4 and Sox2, also decreased transcriptional activity (Fig. 6C). In this case, we speculate that other unknown proteins that interact with Sall4 through its ZF-A or ZF-C are required for the activation of Pou5f1 and Nanog. Dax1 or Nac1 has been reported to interact with Sall4 and to assemble on the Oct-Sox element, suggesting that candidates of such proteins will likely include Dax1 or Nac1 (9, 23). Future studies are necessary to identify the proteins associated with Sall4 through C2H2 zinc finger motif clusters A or C.
Although it is well established that Sall4 binds to chromatin containing specific DNA sequences, it is not clear whether Sall4 associates with chromatin through its direct binding to specific DNA sequences or by recruitment to chromatin via protein-protein interactions with other DNA-binding proteins. We found that the purified recombinant Sall4 protein did not bind to a ds-oligonucleotide containing the Nanog-derived Oct-Sox element in an EMSA experiment (supplemental Fig. S3), suggesting that Sall4 is recruited to the Oct-Sox element through the protein-protein interactions with the bona fide DNA binding transcription factors Oct-3/4 and/or Sox2. We have found that Sox2 co-occupies the promoter and/or enhancer regions of a set of Sall4 target genes (917 genes in Fig. 8A) but not of a distinct set of Sall4 target genes (2151 Sall4-only genes in Fig. 8A). It is plausible that the presence of Sox2 with Sall4 at Sall4 target genes results in formation of Sall4-DNA complexes with distinct conformations (it is known that Sox2 bends DNA for example (38)). Such a differential conformation of Sall4 complexes may lead to recruitment of different sets of associated proteins, achieving distinct biological functions, such as maintaining pluripotency versus inducing differentiation. Future studies are required to test this hypothesis.
Sall4, Oct-3/4, and Sox2 are associated with Oct-Sox elements (Fig. 5B), whereas these three proteins also occupy regions that do not contain the Oct-Sox element. In ES cells, the Sox2 gene is regulated by two enhancers, the upstream and downstream enhancers. The downstream enhancer contains the Oct-Sox element. In contrast, the upstream enhancer possesses an Oct-3/4 binding motif but not the Oct-Sox element (Fig. 8B) (39). Mutation of the Oct-3/4 binding motif in the upstream enhancer diminished the enhancer activity. Our ChIP-on-chip analysis revealed that both Sall4 and Sox2 occupy the upstream enhancer of Sox2 with very similar distribution patterns (Fig. 8B), and a previous report showed that Oct-3/4 also occupies this enhancer (9). These results suggest that Sox2, Sall4, and Oct-3/4 associate most likely as a complex, with the Oct-3/4 binding motif present in the upstream enhancer of the Sox2 gene despite the fact that the enhancer lacks a Sox2 binding motif. We propose that Oct-3/4 and Sox2 form a ternary complex with target DNAs at both the Oct-3/4 binding motif alone and the Oct-Sox element (Fig. 5B), present in the upstream and downstream Sox2 enhancers, respectively. However, Sox2 does not directly bind to the upstream enhancer (lacking Sox2-element) but binds to the downstream enhancer (possessing both Oct-3/4 and Sox2 elements), suggesting that the structural conformations of the ternary complexes are different at the two enhancers. It is likely that Sall4 is included in the Oct-3/4-Sox2-DNA complexes present at the two enhancers. Sall4 may bind to these complexes in accord with their different conformations, although both complexes consist of Oct-3/4, Sox2, and DNA. We envision that Oct-3/4-Sox2-Sall4-DNA quaternary complexes with or without Sox2 binding to DNA may represent another layer regulating the biological functions of the complex. We have shown that multiple sites of Sall4 redundantly interact with Oct-3/4 or Sox2 (Figs. 1C and 3C). This redundancy may lead to Sall4 binding to Oct-3/4 and Sox2 in Oct-3/4-Sox2-DNA complexes with distinct conformations. It is known that transcription factors operating in ES cells contribute to different tasks by configuring distinct sets of transcriptional networks (9). We suggest that a single factor contributes to different transcription networks by binding to different partner proteins through distinct protein-protein interaction modes. Such seemingly redundant roles of a factor may be enabled through multiple protein-interacting domains as revealed with Sall4 in this study.
Acknowledgments
We are grateful to Dr. Ryuichi Nishinakamura (Kumamoto University) for Sall4 cDNA, to Dr. Takashi Tada (Kyoto University) for Nanog-luc and Nanog-mutOS-luc, and to Dr. Yoichi Shinkai (RIKEN) for E14 mouse ES cells. We thank M. Tamura for technical assistance and A. Katayama, M. Sakamoto, K. Fujimaki, M. Sasaki, F. Maekawa, A. Shirabuchi, and E. Yamazaki for excellent secretarial work.
This work was supported by a grant-in-aid for Cancer Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to F. I.).

This article contains supplemental Tables S1—S7 and Figs. S1—S5.
The promoter array data have been deposited in the Gene Expression Omnibus (GEO) database with accession number GSE40072.
- ES
- embryonic stem
- GO
- gene ontology
- ds
- double-stranded
- mut
- mutated
- OS
- Oct-Sox element
- IP
- immunoprecipitation
- IB
- immunoblotting
- ZF
- zinc finger
- POU
- Pit, Oct, Unc
- luc
- luciferase
- DP
- double-positive
- TP
- triple-positive.
REFERENCES
- 1. Niwa H., Miyazaki J., Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 2. Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., Niwa H. (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- 3. Kopp J. L., Ormsbee B. D., Desler M., Rizzino A. (2008) Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26, 903–911 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 5. Okumura-Nakanishi S., Saito M., Niwa H., Ishikawa F. (2005) Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 280, 5307–5317 [DOI] [PubMed] [Google Scholar]
- 6. Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A. (2002) Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30, 3202–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 9. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakatake Y., Fukui N., Iwamatsu Y., Masui S., Takahashi K., Yagi R., Yagi K., Miyazaki J., Matoba R., Ko M. S., Niwa H. (2006) Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol. Cell. Biol. 26, 7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishimoto M., Fukushima A., Okuda A., Muramatsu M. (1999) The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19, 5453–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tokuzawa Y., Kaiho E., Maruyama M., Takahashi K., Mitsui K., Maeda M., Niwa H., Yamanaka S. (2003) Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23, 2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan H., Corbi N., Basilico C., Dailey L. (1995) Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9, 2635–2645 [DOI] [PubMed] [Google Scholar]
- 15. Kühnlein R. P., Frommer G., Friedrich M., Gonzalez-Gaitan M., Weber A., Wagner-Bernholz J. F., Gehring W. J., Jäckle H., Schuh R. (1994) spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 13, 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jürgens G. (1988) Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 7, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakaki-Yumoto M., Kobayashi C., Sato A., Fujimura S., Matsumoto Y., Takasato M., Kodama T., Aburatani H., Asashima M., Yoshida N., Nishinakamura R. (2006) The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation and cooperates with Sall1 in anorectal, heart, brain, and kidney development. Development 133, 3005–3013 [DOI] [PubMed] [Google Scholar]
- 18. Tsubooka N., Ichisaka T., Okita K., Takahashi K., Nakagawa M., Yamanaka S. (2009) Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 14, 683–694 [DOI] [PubMed] [Google Scholar]
- 19. Warren M., Wang W., Spiden S., Chen-Murchie D., Tannahill D., Steel K. P., Bradley A. (2007) A Sall4 mutant mouse model useful for studying the role of Sall4 in early embryonic development and organogenesis. Genesis 45, 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 21. Yuri S., Fujimura S., Nimura K., Takeda N., Toyooka Y., Fujimura Y., Aburatani H., Ura K., Koseki H., Niwa H., Nishinakamura R. (2009) Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells 27, 796–805 [DOI] [PubMed] [Google Scholar]
- 22. Lim C. Y., Tam W. L., Zhang J., Ang H. S., Jia H., Lipovich L., Ng H. H., Wei C. L., Sung W. K., Robson P., Yang H., Lim B. (2008) Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 3, 543–554 [DOI] [PubMed] [Google Scholar]
- 23. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mallanna S. K., Ormsbee B. D., Iacovino M., Gilmore J. M., Cox J. L., Kyba M., Washburn M. P., Rizzino A. (2010) Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells 28, 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 27. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. (2006) Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 [DOI] [PubMed] [Google Scholar]
- 28. Reményi A., Lins K., Nissen L. J., Reinbold R., Schöler H. R., Wilmanns M. (2003) Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hooper M., Hardy K., Handyside A., Hunter S., Monk M. (1987) HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326, 292–295 [DOI] [PubMed] [Google Scholar]
- 30. Brayer K. J., Kulshreshtha S., Segal D. J. (2008) The protein binding potential of C2H2 zinc finger domains. Cell Biochem. Biophys. 51, 9–19 [DOI] [PubMed] [Google Scholar]
- 31. Tsai R. Y., Reed R. R. (1998) Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 18, 6447–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rao S., Zhen S., Roumiantsev S., McDonald L. T., Yuan G. C., Orkin S. H. (2010) Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol. Cell. Biol. 30, 5364–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010) An expanded Oct4 interaction network. Implications for stem cell biology, development, and disease. Cell Stem Cell 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., Hamada H. (1990) A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472 [DOI] [PubMed] [Google Scholar]
- 35. Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W., Staudt L. M. (1990) A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686–692 [DOI] [PubMed] [Google Scholar]
- 36. Schöler H. R., Ruppert S., Suzuki N., Chowdhury K., Gruss P. (1990) New type of POU domain in germ line-specific protein Oct-4. Nature 344, 435–439 [DOI] [PubMed] [Google Scholar]
- 37. Chakravarthy H., Boer B., Desler M., Mallanna S. K., McKeithan T. W., Rizzino A. (2008) Identification of DPPA4 and other genes as putative Sox2:Oct-3/4 target genes using a combination of in silico analysis and transcription-based assays. J. Cell Physiol. 216, 651–662 [DOI] [PubMed] [Google Scholar]
- 38. Scaffidi P., Bianchi M. E. (2001) Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem. 276, 47296–47302 [DOI] [PubMed] [Google Scholar]
- 39. Iwafuchi-Doi M., Yoshida Y., Onichtchouk D., Leichsenring M., Driever W., Takemoto T., Uchikawa M., Kamachi Y., Kondoh H. (2011) The Pou5f1/Pou3f-dependent but SoxB-independent regulation of conserved enhancer N2 initiates Sox2 expression during epiblast to neural plate stages in vertebrates. Dev. Biol. 352, 354–366 [DOI] [PubMed] [Google Scholar]