Background: The trithorax protein Ash2l is a core component of the MLL complex essential for H3K4 methylation and mouse embryonic development.
Results: Ash2l depletion promotes differentiation and chromatin compaction.
Conclusion: Ash2l is essential for pluripotency and open chromatin in mouse ES cells.
Significance: This work describes for the first time genome-wide localization of Ash2l and provides insight into how open chromatin is maintained in ES cells.
Keywords: Chromatin, Chromatin Histone Modification, Chromatin Immunoprecipitation (ChIP), Embryonic Stem Cell, Stem Cells
Abstract
Embryonic stem (ES) cells exhibit general characteristics of open chromatin, a state that may be necessary for ES cells to efficiently self-renew while remaining poised for differentiation. Histone H3K4 and H3K9 trimethylation associate as a general rule, with open and silenced chromatin, respectively, for ES cell pluripotency maintenance. However, how histone modifications are regulated to maintain open chromatin in ES cells remains largely unknown. Here, we demonstrate that trithorax protein Ash2l, homologue of the Drosophila Ash2 (absent, small, homeotic-2) protein, is a key regulator of open chromatin in ES cells. Consistent with Ash2l being a core subunit of mixed lineage leukemia methyltransferase complex, RNAi knockdown of Ash2l was sufficient to reduce H3K4 methylation levels and drive ES cells to a silenced chromatin state with high H3K9 trimethylation. Genome-wide ChIP-seq analysis indicated that Ash2l is recruited to target loci through two distinct modes and enriched at a family of genes implicated in open chromatin regulation, including chromatin remodeler Cdh7, transcription factor c-Myc, and H3K9 demethylase Kdm4c. Our results underscore the importance of Ash2l in open chromatin regulation and provide insight into how the open chromatin landscape is maintained in ES cells.
Introduction
Embryonic stem (ES) cells are derived from the inner cell mass of mammalian blastocysts and able to self-renew and differentiate into all lineages in vitro and in vivo (1–3). The mechanisms of how ES cells maintain self-renewal and pluripotency offer insights and further our understanding of mammalian development and are fundamental for developing therapeutics in regenerative medicine. Recent studies established the importance of “open” chromatin, characterized by a predominance of euchromatin over heterochromatin, in maintenance of ES cell pluripotency (4–7). Euchromatin has a less condensed chromosomal architecture and is generally associated with transcriptional activation, whereas the condensed conformation of heterochromatin often signals transcriptional repression. It has been postulated that the ES cell open chromatin represents a global transcriptionally permissive state, contributing to pluripotency and developmental plasticity (8–11). In the presence of differentiation inducing signals, this open conformation affords ES cells the ability to rapidly alter gene expression to profiles appropriate for the desired cell lineage (5, 12, 13).
In all cell types, both histones and DNA can undergo enzymatic modifications, including histone methylation, acetylation, phosphorylation, ubiquitination, and DNA methylation, all of which are intimately linked to transcription and other biological processes (14, 15). For example, methylation of histone H3K4, especially di- and trimethylation, correlates with active gene transcription, whereas H3K9 methylation correlates with gene silencing and heterochromatin formation (16–19). In ES cells, genome-wide surveys of histone marks indicate that ES cell chromatin features higher levels of histone H3K4 trimethylation (H3K4me3) and H3/H4 acetylation, accompanied by lower levels of H3K9 trimethylation (H3K9me3), compared with the chromatin status of differentiated cells (11, 20, 21). Moreover, facultative heterochromatin (marked by H3K27me3) is preferred over constitutive heterochromatin (marked by H3K9me3) to allow more permissive transcription (22). Indeed, bivalent domains, marked by both H3K4me3 and H3K27me3 in ES cells, have been proposed as a poised state for developmentally important genes (23–25), one that allows rapid activation of such genes during embryonic differentiation.
Multiple chromatin modifying enzymes, epigenetic regulators, and transcription factors interact to establish an open chromatin state. Notably, in ES cells, examples include histone methyltransferases, histone demethylases, and histone acetyltransferases, recruited by transcription factors such as canonical stem cell factors (e.g. Oct4 and Nanog) and c-Myc (7, 26–28). For example, Kdm4c, a histone demethylase that catalyzes the removal of the H3K9me3 mark, regulates global H3K9me3 levels and is required for maintained pluripotency in mouse ES cells (27). In addition, chromatin-remodeling proteins such as Chd1 (chromodomain helicase DNA binding protein 1) play an essential role in maintaining open chromatin and pluripotency in ES cells (7). Genome-wide localization analysis of Chd1 shows its strong correlation with enrichment of Pol II and H3K4me3 but not bivalent domains in ES cells (26). Chd7, another Chd family member, co-localizes with Oct4, Nanog, Sox2, and p300 at enhancers of actively transcribed genes, indicating a likely role for Chd7 in promoting open chromatin in ES cells (29). For induced pluripotent stem cell generation, c-Myc appears to promote active transcription and open chromatin by increasing proliferation and opposing differentiation (30–32). Although it is clear that a multitude of chromatin remodeling proteins participate in the dynamic process of histone mark establishment and regulation, the mechanisms that maintain high H3K4me3 and low H3K9me3 in ES cells remain unclear.
In mammals, the highly conserved mixed lineage leukemia (MLL)3 protein complexes add methyl groups to histone H3K4 and contribute to H3K4me1, me2, and me3 marks across the genome (33). Full activation of the MLL complex histone methyltransferase activities depends on its core components such as trithorax proteins Ash2l, Wdr5, Rbbp5, and Dpy-30 (34–36). In fact, the MLL complex cannot trimethylate H3K4 in the absence of Ash2l (37). Ash2l-deficient mice die at gestation, suggesting an essential role of Ash2l in development (38). Recent studies have shown that Ash2l can directly bind to DNA through its winged helix motif and prefers CG-rich sequences (39, 40). The in vivo relevance of these findings has yet to be demonstrated. Moreover, mass spectrometry analysis of affinity-purified H3K4me3 complexes in HeLa cells reveals that the chromodomain of CHD1 can directly bind the H3K4me3 mark and forms a complex with Ash2L, suggesting that CHD1 function may depend on the H3K4me3 mark generated by the Ash2L-containing MLL complex (41). However, a direct role of Ash2l in regulating an open chromatin and ES cell self-renewal remains to be determined.
In this study, we investigated the function of Ash2l in ES cell maintenance. We found that depletion of Ash2l by siRNAs resulted in a loss of pluripotency manifested by changes in morphology and gene expression profiles. In addition, Ash2l RNAi-treated cells displayed reduced global H3K4me3 and increased H3K9me3, suggesting possible coordination between these two histone marks for maintaining open chromatin. Whole-genome ChIP-seq analysis revealed multiple potential targets of Ash2l that may participate in open chromatin regulation. These findings indicate that Ash2l is required for ES cell pluripotency and the global, active chromatin modifications associated with ES cells. Furthermore, our work suggests that a regulatory network of specific epigenetic factors interact and sustain the open chromatin state of ES cells.
EXPERIMENTAL PROCEDURES
ES Cell Line and Antibodies
AB2.2 mouse embryonic stem cells (passage 18, kindly provided by Darwin Core Facility, Baylor College of Medicine, Houston, TX) were grown in ES cell medium with 15% fetal bovine serum (FBS) either on irradiated mouse embryonic fibroblasts as feeder cells or with LIF in the absence of feeder cells.
The following antibodies were used: anti-Ash2l (A300–112A and A300–489A, Bethyl Laboratories; sc-81184, Santa Cruz Biotechnology); anti-Oct4 (Santa Cruz Biotechnology); anti-HP1α (2616, Cell Signaling Technology); anti-histone H3 (ab1791, Abcam), anti-H3K4me2 (07-030, Upstate); anti-H3K4me3 (ab8580, Abcam); and anti-H3K9me3 (ab8898, Abcam).
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as described previously (42). Mouse ES cells (80–90% confluence) were cross-linked in 1% formaldehyde before lysis and sonication. The resulting extract was precleared (50 μl of protein A beads, 10 μl of IgG, 10 μl of 5% BSA, and 5 μg of sheared salmon sperm DNA), and then incubated overnight at 4 °C with 3 μg of the appropriate antibodies plus 30 μl of protein A-agarose beads, 1 μl of BSA (5%), and 25 μg of sheared salmon sperm DNA. Beads were then washed to elute the bound complexes for reverse cross-linking and DNA purification.
RNAi Knockdown and RT-qPCR
ES cells were plated in gelatin-coated 24-well plates (5 × 104 cells/well) in 500 μl of ES cell medium. Approximately 40 pmol of Stealth RNAiTM siRNAs (Invitrogen) were transfected using Lipofectamine (Invitrogen). Total RNAs were isolated using the RNeasy mini kit (Qiagen). Reverse transcription was performed using iScript Select cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using an ABI StepOnePlus real-time PCR system and SYBR Green Master Mix (Invitrogen). The siRNA sequences used were as follows: Ash2l KD1, 5′-CAAGAAGGCCAGAAGUGAUCCUUUA; Ash2l KD2, 5′-UGGAGACCUUUACAGAGCCUGCUUA; and control KD, 5′-UUCCUCUCCACGCGCAGUACAUUUA.
Immunostaining and Fluorescence Microscopy
Cells plated on coverslips were fixed with 4% paraformaldehyde (20 min) and permeabilized with 0.25% Triton X-100 (10 min) at room temperature, followed by blocking with 3% normal goat serum, before primary and secondary antibody incubation. Primary antibodies used are: anti-Ash2l (sc-81184, Santa Cruz Biotechnology), anti-Oct4 (sc-5279, Santa Cruz Biotechnology), anti-H3K9me3 (ab8898, Abcam), and anti-HP1α (2616, Cell Signaling Technology). FITC and Texas Red conjugated secondary antibodies were obtained from Invitrogen. DAPI (4′,6′-diamidino-2-phenylindole) was used to stain DNA. Fluorescence microscopy was performed on a TE200 microscope (Nikon, New York, NY) equipped with a CoolSNAP-fx CCD camera.
Whole-genome ChIP-seq Analysis
Two independent genome-wide Ash2l ChIP-seq experiments were performed essentially as described previously (43). IgG ChIP-seq served as negative control. Alignment of short reads to the mouse genome (mm9) was carried out using SOAP (44), allowing for a maximum of two mismatches. Reads that produced no initial hits were repeatedly remapped with successive removal of the last 10 bp (whenever necessary) until the reads reached <25 bp. All reads that were uniquely aligned were further analyzed (supplemental Table S1).
Peak detection was performed with model-based analysis of ChIP-seq (version 1.40) (45) using default parameters (p = 1 × 10−5). For two-sample ChIP-seq analysis, model-based analysis of ChIP-seq linearly scales the control sample for comparison with Ash2l ChIP-seq samples. Model-based analysis of ChIP-seq first uses high-confidence fold enrichment (default 32) peaks to model shift size and then detects peaks with a significant tag enrichment compared with background (Poisson distribution, p = 1 × 10−5). About 45% binding regions and >68% binding genes were found to overlap in the two independent Ash2l ChIP-Seq experiments. To ensure the quality of binding sites, only peaks detected in both ChIP-Seq experiments were further annotated.
Peak annotation was carried out with mouse genomic sequences (mm9). Gene locus information was extracted based on UCSC exon locus annotation and BioMart (version 61). For exon annotation, the longest transcript was taken as the representative for each gene. Sites of peaks were assigned based on their relative location to gene loci. For example, peaks within −2 kb to +1 kb from transcriptional start sites were defined as promoters. Peaks outside of promoters but within ±10 kb of transcriptional start sites or termination sequences were designated as 5′-UTR or 3′-UTR. Distal regions were peaks >10 kb upstream or downstream of 5′-UTR and 3′-UTR, respectively. All remaining peaks were classified as unknown or not defined. Annotation follows the order of promoters, exons, introns, 5′-UTR and 3′-UTR, and distal regions. Gene Ontology (biological processes) and KEGG pathway analysis were performed using DAVID bioinformatics resources (46). Probabilities were evaluated by Bonferroni correction, and values of ≤0.001 were considered significant. Genomic sequences that were up to 200 nt up- or downstream from the top 200 predicted binding sites (ranked by enrichment) were used for motif search with the multiple expectation maximization for motif elicitation (MEME) and motif alignment and search tool (MAST) software (47). Sources for previously published ChIP-Seq data used in our analysis are listed below (Table 1).
TABLE 1.
Accession nos. for previously published ChIP-seq data
| Data | Accession no. |
|---|---|
| Rbbp5, negative | GSE22934 (PMID 21477851) |
| H3k4me3, RNA polymerase II, WCEa | GSE12241 (PMID 19946276) |
| RA treatment, Nanog siRNA microarray data | GSE4679 (PMID 16767105) |
a WCE, whole cell extract; RA, retinoic acid.
RESULTS
Ash2l Is Important for ES Cell Pluripotency
The global chromatin structure of ES cells is characterized as relatively open and devoid of heterochromatin (8, 11, 48), with discrete regions of trimethylated H3K27 and H3K4 bivalency associated with essential developmental regulatory genes (21, 23–25, 49–51). Given the importance of Ash2l in catalytic methylation of H3K4 by the MLL histone methyltransferase complex and in embryonic development (34, 38), we hypothesized that Ash2l may play an important role in determining the chromatin status of ES cells and in turn maintain ES cell self-renewal. Indeed, Ash2l expression was downregulated during ES cell differentiation (Fig. 1a). To further test our hypothesis, we used two independent siRNA duplexes to transiently knock down Ash2l in ES cells and examined their effect on ES cell biology. The efficacy of Ash2l knockdown was confirmed by RT-qPCR and Western blotting. As shown in Fig. 1, b and c, two siRNA duplexes could efficiently reduce the mRNA and protein levels of Ash2l (>80%). This Ash2l inhibition led to decreased mRNA expression of self-renewal and pluripotency factors such as Nanog, Oct4, Errb, and Rex1 (Fig. 1c). Importantly, RNAi knockdown of Ash2l by both Ash2l siRNA duplexes resulted in a reduction of alkaline phosphatase staining and an increase in the percentage of differentiated cells (Fig. 1d). In addition, we were able to observe up-regulated expression of specific differentiation genes in the mesoderm lineage (Fig. 1e). These findings are in line with the essential role of Ash2l in early embryonic development (38) and support the idea that Ash2l is important for ES cell pluripotency.
FIGURE 1.
Ash2l is critical for maintaining ES cell pluripotency. a, mouse ES cells were treated with retinoic acid (RA) and then harvested at the indicated time points for Western blotting with the indicated antibodies. Tubulin served as loading control. b, mouse ES cells were transfected with control (ctrl) or two different Ash2l siRNA oligonucleotides for knockdown. Extracts were collected after 4 days for Western blotting with the indicated antibodies. GAPDH served as loading control. Relative levels of Ash2l after normalization with loading controls were determined using NIH ImageJ software. c–e, self-renewal was impaired in Ash2l knockdown cells. Ash2l RNAi treated cells from (b) were examined for expression of the indicated self-renewal genes by RT-qPCR (c), alkaline phosphatase activities (d), and expression of the indicated lineage markers by RT-qPCR (e). Error bars indicate S.E. (n = 3).
Ash2l Regulates the Open Chromatin in Mouse ES Cells
We noted that Ash2l RNAi cells remained weakly positive for alkaline phosphatase activity (Fig. 1d), indicating that the Ash2l RNAi cells still retained certain properties of the undifferentiated state. These phenotypes are reminiscent of Chd1 loss-of-function ES cells, which accumulated heterochromatin with Chd1 knockdown (26). Thus, Ash2l may participate in regulation of an open chromatin structure, which is characteristic of ES cells and intimately associated with their pluripotent state. Open chromatin is generally marked by H3K4 trimethylation, whereas silenced chromatin is marked by H3K9 trimethylation (7). If the effects of reduced Ash2l expression are directly linked to the ability of Ash2l to regulate an open chromatin state of ES cells, loss of Ash2l may have marked effects on the level of trimethylation of H3K4 and H3K9 present in ES cell chromatin. tk;2Indeed, transient knockdown of Ash2l in mouse ES cells led to a global decrease in H3K4me3 levels and concomitant increase in global H3K9me3 levels (Fig. 2, a and b, and supplemental Fig. S1).
FIGURE 2.
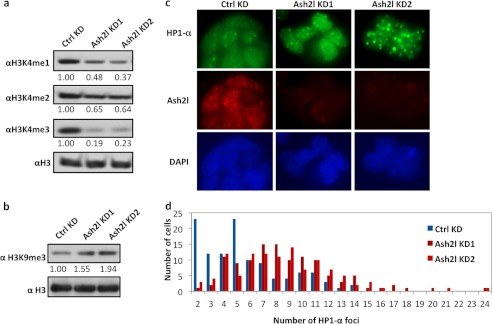
Ash2l regulates histone marks and open chromatin in ES cells. a and b, mouse ES cells were analyzed by Western blotting with the indicated antibodies 2 days after transfection with Ash2l siRNA oligonucleotides as described above. Relative expression was determined using ImageJ software using H3 as control (ctrl). c, mouse ES cells that were transfected twice (on day 0 and 2) with the indicated Ash2l siRNA duplexes and then immunostained with anti-HP1α and Ash2l antibodies on day 4. d, quantification of data from c. Over 100 nuclei were examined for each treatment, and the number of HP1α foci was plotted (p < 1 × 10−8).
To further investigate the effect of Ash2l knockdown on ES cell chromatin, we performed immunostaining studies to detect HP1α, which are enriched at heterochromatin regions (20, 26). In line with our detection of up-regulated global H3K9me3 levels with loss of Ash2l (Fig. 2b), we also observed a marked increase of HP1α foci in Ash2l KD cells (Fig. 2, c and d). Given that Ash2l loss gave rise to more heterochromatin domains prior to differentiation, our data indicate that Ash2l is required for the maintenance of open chromatin in ES cells.
Genome-wide ChIP-seq Analysis of Ash2l Binding Sites in Mouse ES Cells
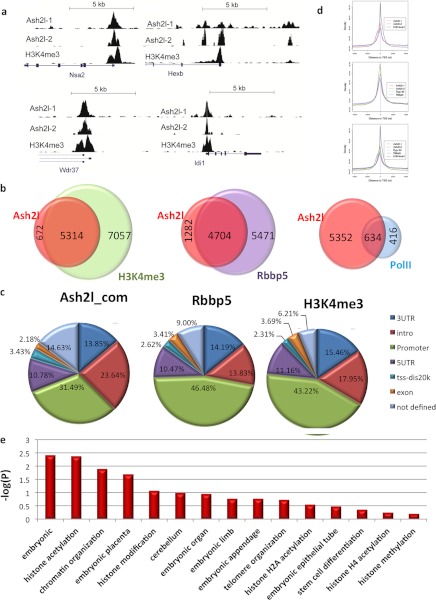
Next, we carried out genome-wide ChIP-seq analysis to identify Ash2l sites of chromatin enrichment in mouse ES cells. Our ChIP-seq experiments yielded >2.4 × 106 uniquely mapped short reads for Ash2l (supplemental Fig. S2). Short reads from IgG were used as controls for peak detection. A total of 9,807 sites were identified for Ash2l, corresponding to 5,986 genes (also see “Experimental Procedures” for details) (supplemental Table S1). Compared with published data sets for H3K4me3 and H3K27me3 (50, 52), nearly 89% of Ash2l targets identified from our ChIP-seq experiments overlap with enrichment of H3K4me3, an indication that Ash2l may indeed positively regulate H3K4me3 (Fig. 3a). Consistent with interactions of Ash2l and Rbbp5 trithorax proteins, as members of a MLL complex (53, 54), ∼79% of the Ash2l binding sites coincide with those of Rbbp5 (Fig. 3b) (55), further validating our ChIP-seq results and supporting chromatin interactions of Ash2l in ES cells as a member of an MLL complex. Significant co-occupancy was also observed for Ash2l and RNA polymerase II (60%) (52), indicating preferential association of Ash2l with transcriptionally active euchromatin. In contrast, Ash2l binding sites poorly overlap with the repressive mark H3K27me3 (33%) (data not shown). Additionally, the distributions of Ash2l, Rbbp5, Dpy-30, and H3K4me3 localization were strikingly similar and co-enriched around transcriptional start sites (Fig. 3, c and d). Notably, gene ontology analysis revealed that genes important for ES cell self-renewal (including embryonic morphogenesis, histone modification, and chromosome organization) were among the top categories for Ash2l-bound genes (Fig. 3e). Taken together, these findings indicate that Ash2l globally associates with euchromatin in ES cells and may regulate the expression of genes that modulate histone modification and chromosome organization.
FIGURE 3.
Whole-genome ChIP-seq analysis of Ash2l. a, representative Ash2l binding profiles at its target genes from whole-genome ChIP-seq analysis. The binding patterns of Ash2l on Nsa2, Hexb, Wdr37, and Idi1 were compared with those of H3K4me3. Peak detection was performed with model-based analysis of ChIP-seq (version 1.40) using default settings (p < 10−5). b, a significant portion of ChIP-seq identified Ash2l target genes overlap with those of H3K4me3, Rbbp5, and PolII. c, distribution of Ash2l ChIP-seq binding regions was presented. For comparison, similar analysis was performed for Dpy-30, Rbbp5, and H3K4me3, based on published data sets. Ash2l_com, data from both ChIP-seq experiments. d, binding site distribution of Ash2l, H3K4me3, Dpy-30, and Rbbp5 relative to the transcriptional start sites of RefSeq genes. e, gene ontology analysis of Ash2l-bound genes identified from our whole-genome ChIP-seq experiments. TSS, transcription start site.
Ash2l may be recruited to target genes through its association with transcription factors (56–58) and MLL subunits such as Rbbp5, Dpy30, and Wdr5 (36). However, Rbbp5 and Wdr5 can also form protein complexes that are distinct from those containing Ash2l (34, 37, 55). Recent structural and biochemical studies have suggested that Ash2l may in fact directly bind target genes in vitro through its winged-helix DNA binding domain (39, 40). To further investigate these two possibilities of Ash2l chromatin recruitment in vivo, we analyzed our Ash2l ChIP-seq data for Ash2l consensus binding motifs. As shown in Fig. 4a, de novo binding motif analysis revealed two distinct motifs (referred to as TTCC and G-rich motifs here), suggesting two different binding modes. Notably, the G-rich motif is consistent with the Ash2l-binding motif identified by in vitro SELEX experiments using the Ash2l winged-helix DNA binding domain (40). When we examined several target regions containing either of the two Ash2l binding motifs, we found Ash2l to be most enriched at sequences containing the respective binding motif (Fig. 4b and supplemental Table S2). These data combined validate our ChIP-seq results, support two different modes through which Ash2l may be recruited to its target genes, and indicate that the G-rich motif may be directly recognized by Ash2l.
FIGURE 4.
Ash2l may be recruited to ES cell chromatin through two distinct mechanisms. a, two consensus binding motifs of Ash2l were derived based on analysis of the top 200 Ash2l binding sites. b, candidate targets for both motifs were selected for validation of Ash2l enrichment, where specific regions containing Ash2l binding motifs (as indicated) were examined by ChIP-qPCR using multiple sets of primers (supplemental Table S2). Motif 1, cog8 and cdc5l; Motif 2, Jmjd5 and Sirt6.
Ash2l Regulates Open Chromatin Status through Its Target Genes
Our results thus far indicate that Ash2l can regulate H3K4 methylation, concomitant with active gene transcription and has broad effects on ES cell maintenance. The enzymatic activity of Ash2l complex at specific gene targets may be further amplified by a network of Ash2l target genes, which promotes more global outcomes for ES cell stability and maintenance. Based on our ChIP-seq data, genes encoding pluripotency factors Nanog and Oct4, transcription factor c-Myc, and chromodomain-containing chromatin remodeling enzymes Chd1 and Chd7 are targets of Ash2l binding (supplemental Fig. S3), which was confirmed by ChIP-qPCR analysis (Fig. 5a). Ash2l directly regulated the expression of c-Myc and Chd7, but not Chd1, as revealed by Ash2l knockdown (Fig. 5b). These results suggest that the effects of Ash2l in controlling open chromatin structure are manifested further by regulation of c-Myc and Chd7 expression.
FIGURE 5.
Ash2l regulates c-Myc, Chd7, and Kdm4c for open chromatin maintenance. a, ChIP assays were performed using mouse ES cells and anti-Ash2l antibodies, the co-precipitated DNA was analyzed by qPCR for the indicated genes. IgG was used as negative control. Error bars indicate S.E. (n = 3). b, Mouse ES cells were transfected with Ash2l siRNA oligonucleotides and analyzed by RT-qPCR for Ash2l expression 2 days after transfection. Error bars indicate S.E. (n = 3). c, RT-qPCR analysis of the expression level of the indicated genes in cells from b. Error bars indicate S.E. (n = 3). Student t test was performed to determine p values (indicated by an asterisk). d, ChIP assays were performed using mouse ES cells and anti-Ash2l antibodies, and the co-precipitated DNA was analyzed by qPCR using two sets of primers specific for Kdm4c (Kdm4c-A and -B). IgG served as control. Error bars indicate S.E. (n = 3).
In addition to chromatin remodeling, establishment of euchromatin also requires the repression of heterochromatin marks such as methylated H3K9, which is maintained by methyltransferases and demethylases for H3K9 (27, 59). In the case of Ash2l-mediated effects on chromatin structure in ES cells, we predict that genes encoding H3K9 demethylases are gene targets of Ash2l, which would tip the balance toward loss of H3K9 methylation. Indeed, several H3K9 demethylases were identified as Ash2l-bound genes by our ChIP-seq analysis (supplemental Fig. S3). Notably, previous reports showed that RNAi-mediated knockdown of the H3K9me3 demethylase Kdm4c led to increased H3K9me3 levels and triggered differentiation of ES cells (27), suggesting Kdm4c as a potential target of Ash2l in ES cells. Gene annotation by BioGPS indicates that Kdm4c but not Kdm4b is highly enriched in ES cells (supplemental Fig. S4, a and b), consistent with a specific role for Kdm4c in ES cells. Using ChIP-qPCR, we confirmed that the Kdm4c gene locus was indeed targeted by Ash2l (Fig. 5d). Importantly, Kdm4c expression is controlled by Ash2l, as Ash2l KD reduced Kdm4c but not Chd1 expression (Fig. 5c). Analysis of publicly available microarray data, quantifying RNA expression in Nanog-depleted and retinoic acid-treated ES cells revealed similarly reduced Kdm4c expression (supplemental Fig. S4c) (60). In addition, our Ash2l ChIP-seq experiments also indicated an enrichment of Ash2l on the Kdm4c locus (supplemental Fig. S5). This pathway convergence underscores the importance of an Ash2l-Kdm4c axis in ES cell maintenance. Furthermore, these data collectively indicate that c-Myc, Chd7, and Kdm4c are targeted for modification by Ash2l, acting as a network of chromatin modifiers and transcription factors that promotes an open chromatin state of self-renewing, pluripotent ES cells.
DISCUSSION
We have demonstrated here that Ash2l is necessary for maintaining open chromatin and self-renewal of ES cells. As a core subunit of MLL complexes, Ash2l controls histone H3K4 methylation. Knocking down Ash2l in ES cells resulted in a decrease in H3K4 methylation while elevating heterochromatin marks. Consequently, the chromatin structure of Ash2l KD cells physically switched from an open state to a more closed conformation. In line with increased heterochromatin marks, we found that knocking down Ash2l resulted in global increased H3K9me3 level in knockdown cells (Fig. 2b). These changes are accompanied by lower alkaline phosphatase activities and elevated expression of specific differentiation genes, driving these ES cells toward mesodermal lineages. These results are consistent with an essential role of Ash2l in early embryogenesis (38). Our findings highlight the importance of Ash2l in the dynamic regulation of euchromatin and heterochromatin and support the hypothesis that this chromatin regulatory process is a crucial component of the ES cell maintenance program (26).
Analysis of our Ash2l ChIP-seq data and previously published studies has revealed genome-wide co-occupancy of Ash2l, Rbbp5, Dpy-30, as well as histone H3K4me3 on ES cell chromatin (55, 61). These observations underscore their importance for establishing and maintaining an open chromatin in ES cells, and are consistent with Ash2l/Rbbp5/Dpy-30 forming a core complex that is required for establishing and maintaining H3K4me3 (37, 61). We identified two in vivo Ash2l binding motifs in this study. Notably, one of the motifs resembles the GC-rich binding pattern that was previously found to bind Ash2l winged-helix domain in vitro (40), suggesting that a portion of Ash2l may directly bind to DNA independent of trithorax complexes in vivo. The two motifs therefore may correspond to the two modes of Ash2l binding to genomic DNA, direct or indirect, and reflect two distinct mechanisms by which MLL complexes may be recruited to their target genes. Different from other MLL subunits, Ash2l can bind DNA and may function as a DNA-binding factor itself to directly recruit MLL complex to modify histones. In fact, Ash2l may control expression of a subset of genes independent of transcription factors. Although Ash2l, Rbbp5, Dpy30, and Wdr5 have been thought as core components for MLL complexes (34), functional differences among these proteins have also been suggested (34, 55, 61). Further studies are needed to tease apart the function of various subcomplexes for these proteins to shed more light on how ES chromatin is maintained and regulated.
Our genome-wide ChIP-seq experiments indicate that Ash2l preferentially occupies genes important for embryonic morphogenesis, histone modification, and chromosome organization. Among Ash2l target genes are several known open chromatin regulators, including Chd1, Chd7, Kdm4b, Kdm4c, and c-Myc (26, 27, 29, 62–64). These factors have been shown to maintain open chromatin through chromatin remodeling, transcriptional control, and H3K9me3 demethylation (26, 27, 29, 62–64). We provide evidence here that Chd7, c-Myc, and Kdm4c likely are transcriptionally regulated by Ash2l and augment Ash2l function in creation of open chromatin (supplemental Fig. S6). Our model does not exclude the possibility that binding of Ash2l physically protects euchromatin or that H3K4me3 marks act as a platform in recruitment of additional factors that regulate the formation of open chromatin. For example, Chd1 can bind H3K4me3 directly through its chromodomain and remodels local chromatin for the open chromatin state (41). Further studies are expected to gain deeper insight into the mechanism by which Ash2l is required for maintaining open chromatin in ES cells.
Given the complexity and dynamic nature of ES cell self-renewal and open chromatin maintenance, alterations in the composition and activity of Ash2l complexes during ES cell differentiation warrant further investigation. One possible approach is to utilize advanced genomic and proteomic methods to systematically dissect Ash2l signaling networks and chromatin dynamics (65, 66). Taken together, our results demonstrate that Ash2l is necessary for ES cell pluripotency and open chromatin regulation and carries out its function through multiple target proteins.
This work was supported by National Basic Research Program (973 Program 2010CB945400 and 2012CB911201); National Natural Science Foundation of China (NSFC 91019020); NIGMS, National Institutes of Health (GM081627), NCI, National Institutes of Health (CA133249 and GM095599); the Welch Foundation (Q-1673), and the Leukemia and Lymphoma Society. This work was also supported by the Dan L. Duncan Cancer Center (P30CA125123) and the Administrative and Genome-wide RNAi Screens Cores (IDDRC P30HD024064).

This article contains supplemental Tables S1 and S2 and Figs. S1–S6.
- MLL
- mixed lineage leukemia
- qPCR
- quantitative PCR
- KD
- knockdown.
REFERENCES
- 1. Rossant J. (2001) Stem cells from the Mammalian blastocyst. Stem Cells 19, 477–482 [DOI] [PubMed] [Google Scholar]
- 2. Loh Y. H., Yang L., Yang J. C., Li H., Collins J. J., Daley G. Q. (2011) Genomic approaches to deconstruct pluripotency. Annu. Rev. Genomics Hum. Genet. 12, 165–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans M. (2011) Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat. Rev. Mol. Cell Biol. 12, 680–686 [DOI] [PubMed] [Google Scholar]
- 4. Park S. H., Park S. H., Kook M. C., Kim E. Y., Park S., Lim J. H. (2004) Ultrastructure of human embryonic stem cells and spontaneous and retinoic acid-induced differentiating cells. Ultrastruct. Pathol. 28, 229–238 [DOI] [PubMed] [Google Scholar]
- 5. Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D. J., Dash C., Bazett-Jones D. P., Le Grice S., McKay R. D., Buetow K. H., Gingeras T. R., Misteli T., Meshorer E. (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahmed K., Dehghani H., Rugg-Gunn P., Fussner E., Rossant J., Bazett-Jones D. P. (2010) Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLOS One 5, e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. (2011) Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niwa H. (2007) Open conformation chromatin and pluripotency. Genes Dev. 21, 2671–2676 [DOI] [PubMed] [Google Scholar]
- 9. Mattout A., Meshorer E. (2010) Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 22, 334–341 [DOI] [PubMed] [Google Scholar]
- 10. Koh F. M., Sachs M., Guzman-Ayala M., Ramalho-Santos M. (2010) Parallel gateways to pluripotency: open chromatin in stem cells and development. Curr. Opin. Genet. Dev. 20, 492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T. (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ying Q. L., Nichols J., Chambers I., Smith A. (2003) BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- 13. Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 14. Wu H., Sun Y. E. (2006) Epigenetic regulation of stem cell differentiation. Pediatr. Res. 59, 21R–25R [DOI] [PubMed] [Google Scholar]
- 15. Kubicek S., Schotta G., Lachner M., Sengupta R., Kohlmaier A., Perez-Burgos L., Linderson Y., Martens J. H., O'Sullivan R. J., Fodor B. D., Yonezawa M., Peters A. H., Jenuwein T. (2006) The role of histone modifications in epigenetic transitions during normal and perturbed development. Ernst Schering Res. Found Workshop, 57, 1–27 [DOI] [PubMed] [Google Scholar]
- 16. Shilatifard A. (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eissenberg J. C., Shilatifard A. (2010) Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 19. Hublitz P., Albert M., Peters A. H. (2009) Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 53, 335–354 [DOI] [PubMed] [Google Scholar]
- 20. Krejcí J., Uhlírová R., Galiová G., Kozubek S., Smigová J., Bártová E. (2009) Genome-wide reduction in H3K9 acetylation during human embryonic stem cell differentiation. J. Cell Physiol. 219, 677–687 [DOI] [PubMed] [Google Scholar]
- 21. Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A. G. (2006) Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538 [DOI] [PubMed] [Google Scholar]
- 22. Ram E. V., Meshorer E. (2009) Transcriptional competence in pluripotency. Genes Dev. 23, 2793–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 24. Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G. A., Stewart R., Thomson J. A. (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell stem cell 1, 299–312 [DOI] [PubMed] [Google Scholar]
- 25. Zhao X. D., Han X., Chew J. L., Liu J., Chiu K. P., Choo A., Orlov Y. L., Sung W. K., Shahab A., Kuznetsov V. A., Bourque G., Oh S., Ruan Y., Ng H. H., Wei C. L. (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1, 286–298 [DOI] [PubMed] [Google Scholar]
- 26. Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M. J., Heidersbach A., Ramalho-Santos J., McManus M. T., Plath K., Meshorer E., Ramalho-Santos M. (2009) Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loh Y. H., Zhang W., Chen X., George J., Ng H. H. (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orkin S. H., Hochedlinger K. (2011) Chromatin connections to pluripotency and cellular reprogramming. Cell 145, 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schnetz M. P., Handoko L., Akhtar-Zaidi B., Bartels C. F., Pereira C. F., Fisher A. G., Adams D. J., Flicek P., Crawford G. E., Laframboise T., Tesar P., Wei C. L., Scacheri P. C. (2010) CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6, e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 31. Smith K. N., Singh A. M., Dalton S. (2010) Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin C. H., Jackson A. L., Guo J., Linsley P. S., Eisenman R. N. (2009) Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 28, 3157–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malik S., Bhaumik S. R. (2010) Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 277, 1805–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., Roeder R. G. (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 [DOI] [PubMed] [Google Scholar]
- 35. Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Y., Cao F., Wan B., Dou Y., Lei M. (2012) Structure of the SPRY domain of human Ash2L and its interactions with RbBP5 and DPY30. Cell Res. 22, 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steward M. M., Lee J. S., O'Donovan A., Wyatt M., Bernstein B. E., Shilatifard A. (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 13, 852–854 [DOI] [PubMed] [Google Scholar]
- 38. Stoller J. Z., Huang L., Tan C. C., Huang F., Zhou D. D., Yang J., Gelb B. D., Epstein J. A. (2010) Ash2l interacts with Tbx1 and is required during early embryogenesis. Exp. Biol. Med. 235, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y., Wan B., Wang K. C., Cao F., Yang Y., Protacio A., Dou Y., Chang H. Y., Lei M. (2011) Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 12, 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sarvan S., Avdic V., Tremblay V., Chaturvedi C. P., Zhang P., Lanouette S., Blais A., Brunzelle J. S., Brand M., Couture J. F. (2011) Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain. Nat. Struct. Mol. Biol. 18, 857–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sims R. J., 3rd, Millhouse S., Chen C. F., Lewis B. A., Erdjument-Bromage H., Tempst P., Manley J. L., Reinberg D. (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28, 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 43. Yang D., Xiong Y., Kim H., He Q., Li Y., Chen R., Songyang Z. (2011) Human telomeric proteins occupy selective interstitial sites. Cell Res. 21, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., Wang J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 47. Bailey T. L., Williams N., Misleh C., Li W. W. (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic. Acids Res. 34, W369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meshorer E., Misteli T. (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 7, 540–546 [DOI] [PubMed] [Google Scholar]
- 49. Vastenhouw N. L., Zhang Y., Woods I. G., Imam F., Regev A., Liu X. S., Rinn J., Schier A. F. (2010) Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chamberlain S. J., Yee D., Magnuson T. (2008) Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26, 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Young M. D., Willson T. A., Wakefield M. J., Trounson E., Hilton D. J., Blewitt M. E., Oshlack A., Majewski I. J. (2011) ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 39, 7415–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cao F., Chen Y., Cierpicki T., Liu Y., Basrur V., Lei M., Dou Y. (2010) An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PloS one 5, e14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Avdic V., Zhang P., Lanouette S., Groulx A., Tremblay V., Brunzelle J., Couture J. F. (2011) Structural and biochemical insights into MLL1 core complex assembly. Structure 19, 101–108 [DOI] [PubMed] [Google Scholar]
- 55. Ang Y. S., Tsai S. Y., Lee D. F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., Wang J., Rendl M., Bernstein E., Schaniel C., Lemischka I. R. (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rampalli S., Li L., Mak E., Ge K., Brand M., Tapscott S. J., Dilworth F. J. (2007) p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 14, 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tan C. C., Sindhu K. V., Li S., Nishio H., Stoller J. Z., Oishi K., Puttreddy S., Lee T. J., Epstein J. A., Walsh M. J., Gelb B. D. (2008) Transcription factor Ap2δ associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proc. Natl. Acad. Sci. U.S.A. 105, 7472–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Demers C., Chaturvedi C. P., Ranish J. A., Juban G., Lai P., Morle F., Aebersold R., Dilworth F. J., Groudine M., Brand M. (2007) Activator-mediated recruitment of the MLL2 methyltransferase complex to the β-globin locus. Mol. Cell 27, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bilodeau S., Kagey M. H., Frampton G. M., Rahl P. B., Young R. A. (2009) SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 23, 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. (2006) Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 [DOI] [PubMed] [Google Scholar]
- 61. Jiang H., Shukla A., Wang X., Chen W. Y., Bernstein B. E., Roeder R. G. (2011) Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144, 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schnetz M. P., Bartels C. F., Shastri K., Balasubramanian D., Zentner G. E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T., Crawford G. E., Scacheri P. C. (2009) Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 19, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin C. H., Lin C., Tanaka H., Fero M. L., Eisenman R. N. (2009) Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PloS One 4, e7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim J., Woo A. J., Chu J., Snow J. W., Fujiwara Y., Kim C. G., Cantor A. B., Orkin S. H. (2010) A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lu R., Markowetz F., Unwin R. D., Leek J. T., Airoldi E. M., MacArthur B. D., Lachmann A., Rozov R., Ma'ayan A., Boyer L. A., Troyanskaya O. G., Whetton A. D., Lemischka I. R. (2009) Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu J. I., Lessard J., Crabtree G. R. (2009) Understanding the words of chromatin regulation. Cell 136, 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]