Background: Thymidine kinase 2 (TK2) deficiency causes severe mitochondrial DNA (mtDNA) depletion due to absence of nucleotides for mtDNA synthesis.
Results: Nucleoside kinase from Drosophila melanogaster was able to rescue TK2-deficient mice.
Conclusion: Nucleotide import into mitochondria can compensate the loss of TK2 in differentiated tissues.
Significance: The results highlight mechanisms to be explored for treatment of mtDNA depletion.
Keywords: DNA Synthesis, Enzymes, Gene Therapy, Mitochondrial DNA, Nucleotide, Nucleotide Pools, Nucleotide Transport, Thymidine Kinase 2
Abstract
A strategy to reverse the symptoms of thymidine kinase 2 (TK2) deficiency in a mouse model was investigated. The nucleoside kinase from Drosophila melanogaster (Dm-dNK) was expressed in TK2-deficient mice that have been shown to present with a severe phenotype caused by mitochondrial DNA depletion. The Dm-dNK+/− transgenic mice were shown to be able to rescue the TK2-deficient mice. The Dm-dNK+/−TK2−/− mice were normal as judged by growth and behavior during the observation time of 6 months. The Dm-dNK-expressing mice showed a substantial increase in thymidine-phosphorylating activity in investigated tissues. The Dm-dNK expression also resulted in highly elevated dTTP pools. The dTTP pool alterations did not cause specific mitochondrial DNA mutations or deletions when 6-month-old mice were analyzed. The mitochondrial DNA was also detected at normal levels. In conclusion, the Dm-dNK+/−TK2−/− mouse model illustrates how dTMP synthesized in the cell nucleus can compensate for loss of intramitochondrial dTMP synthesis in differentiated tissue. The data presented open new possibilities to treat the severe symptoms of TK2 deficiency.
Introduction
The synthesis of mitochondrial DNA (mtDNA) is not cell cycle-regulated and requires a constant supply of deoxyribonucleoside triphosphates (dNTPs) for maintenance of the mitochondrial integrity in quiescent cells. dNTPs are synthesized by two pathways: the de novo pathway and the salvage pathway. The mitochondrial dNTP pool is maintained by salvage of deoxynucleosides within the mitochondria and by importing cytosolic deoxyribonucleotides through specific transporters (1, 2). In addition, the presence of a de novo dTMP biosynthesis pathway was recently demonstrated in mammalian mitochondria (3). The p53-inducible ribonucleotide reductase small subunit (p53R2)3 plays an important role in dNTP synthesis in quiescent cells but is not sufficient to maintain proper dNTP levels for mtDNA (4, 5, 9). Therefore, in nonreplicating cells, the mtDNA synthesis heavily depends on the salvage pathway enzymes thymidine kinase 2 (TK2) and deoxyguanosine kinase (DGUOK) (6). Dependence on these enzymes has proven to be deleterious in deficiencies of TK2 (7) and DGUOK (8) and may also contribute to disease where minor alterations of these enzyme activities affect important cell functions.
Mitochondrial DNA depletion syndrome comprises a heterogeneous group of mitochondrial disorders characterized by reduced levels of mtDNA but with no mutations or deletions of the mtDNA (10). Mutations in the nuclear encoded dNKs, DGUOK and TK2, have been associated with hepatocerebral and myopathic forms of mitochondrial DNA depletion syndrome, respectively (8, 11). Other mutations known to cause mitochondrial DNA depletion syndrome are mutations in p53R2, the succinyl-CoA ligase β subunit (SUCLA2), the succinyl-CoA ligase α subunit (SUCLG1), the catalytic subunit of mitochondrial DNA polymerase (pol γ), the twinkle gene (mitochondrial DNA helicase), and the MPV17 protein (12).
To find possible strategies to treat mtDNA deficiency, some basic questions must be addressed. One important question is whether nucleotides delivered in the nuclear or cytosolic compartment can reach mitochondria and support mtDNA synthesis in quiescent cells. This would be of value because it is known from the antiviral field that mononucleotide analogs can reach the cytosol and act as monophosphate prodrugs targeting viral DNA (13, 14). If such monophosphates can be prodrugs of dTMP and dCMP, they could in theory reverse a TK2 deficiency provided they reach the mitochondrial compartment.
The deoxyribonucleoside kinase from D. melanogaster (Dm-dNK) is a multisubstrate nucleoside kinase that has unique properties to recognize all four natural nucleosides and has a very high catalytic rate (15, 16). Dm-dNK can be expressed at high levels with high enzyme activity in mammalian cells (17, 18) and can be used as a suicide gene in cancer cells (19, 20). The aim of this study was to investigate whether Dm-dNK expression could rescue the severe phenotype of TK2-deficient mice (7). Our results demonstrate that the Dm-dNK-expressing mice (Dm-dNK+/− and Dm-dNK+/−TK2−/−) appear as wild-type (wt) mice regarding growth, behavior, and mtDNA levels during the entire observation period of 6 months. The only evident differences between the Dm-dNK-expressing mice and the wt control mice were a highly elevated thymidine (dThd) phosphorylating activity and a 100-fold increase of the dTTP pool level.
EXPERIMENTAL PROCEDURES
Construction of the Dm-dNK Mice
A mouse line expressing Dm-dNK was constructed by PCR amplification and subsequent cloning into the pcDNA3 vector (Invitrogen). The transgene was cut from the vector using BglII and DraIII restriction enzymes. The Dm-dNK transgene was made through the pronuclear injection technique. Genotyping was performed by isolating DNA from tail tissues for PCR analysis of the presence of the Dm-dNK gene. Tail samples collected from mice ∼14 days old were cut ∼0.25 inches from the tip using sterile scissors, and the mice were ear-marked. DNA was isolated from the tail samples using the DNeasy blood and tissue kit (Qiagen), and the DNA was screened for Dm-dNK gene by PCR using specific primers for the Dm-dNK gene (Dm-dNK-F and Dm-dNK-R, supplemental Table 1). The mice were studied for growth rate, organ weights, mortality, and signs of neurological defects throughout the observation time of 6 months. Dm-dNK+/− mice were crossed with TK2+/− mice to generate Dm-dNK+/− TK2+/− mice, which were intercrossed to generate TK2-deficient mice expressing Dm-dNK (Dm-dNK+ TK2−/−) (see Fig. 3A). Genomic DNA of the Dm-dNK+TK2−/− mice was digested using the SpeI restriction enzyme and analyzed by Southern blot to confirm the absence of the wt TK2 allele, and the DNA was screened for the Dm-dNK gene by PCR as mentioned above. All animal experiments were compliant with the guidelines of the local ethical committee (S104-09, S135-11).
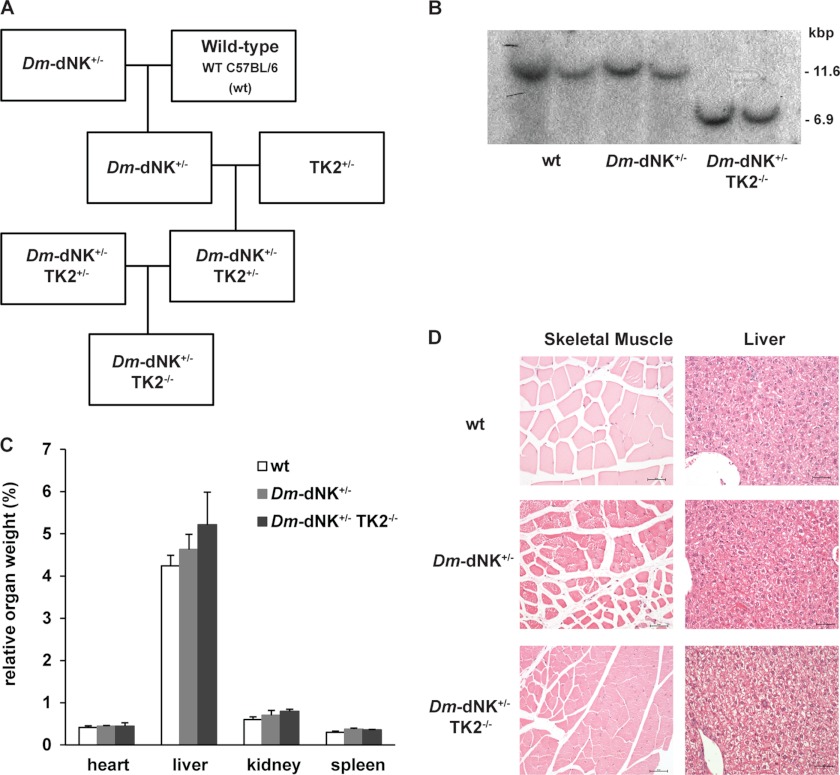
FIGURE 3.
Development and characterization of TK2-deficient mice expressing Dm-dNK (Dm-dNK+/−TK2−/−). A, establishment of the Dm-dNK+/−TK2−/− mice. B, genotyping of wt, Dm-dNK+/−, and Dm-dNK+/−TK2−/− mice (n = 2 each) by Southern blot using SpeI restriction enzyme. The detection of 6.9-kb DNA in lanes 5 and 6 confirms disruption of TK2 gene in the mice. C, relative organ weight (organ weight/body weight) of heart, liver, kidney, and spleen (%) of wt, Dm-dNK+/−, and Dm-dNK+/−TK2−/− mice (n = 3–6, mean ± S.E.). D, histological analysis of anterior straight femoral muscle and liver tissues from 6-month-old wt, Dm-dNK+/−, and Dm-dNK+/−TK2−/− mice (original magnification of each panel: 20×). Scale bar: 50 μm.
Analysis of Protein Expression
Total protein was extracted from tissues using radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P40, 0.05% sodium deoxycholate, 0.1% SDS, and protease inhibitors). Western blot was performed using 4–12% precast Bis-Tris gel (NuPAGE) and Amersham Biosciences Hybond-P membrane (Invitrogen). The presence of Dm-dNK protein was detected using anti-histidine antibody targeted against His tag of the protein (Calbiochem, 1:3000) and anti-mouse IgG linked to horseradish peroxidase (HRP) (GE Healthcare, 1:3000). ECL (GE Healthcare) was used as a substrate for the HRP.
Dm-dNK Subcellular Localization
The subcellular localization of Dm-dNK protein was determined through immunofluorescence. Briefly, HeLa cells were seeded in wells of chamber slide (Thermo Scientific) and cultured overnight at 37 °C, 5% CO2. When the confluence reached 50–80%, the cells were transfected with Dm-dNK construct using FuGENE® 6 reagent (Promega) according to the manufacturer's instructions and incubated for 24 h before mitochondrial staining. Mitochondria were stained with MitoTracker® Red CMXRos (Life Technologies) at the concentration 0.25 ng/μl for 30 min. The cells were then fixed with 10% formaldehyde in PBS for 15 min and treated with cold acetone for 10 min. The subcellular localization of Dm-dNK was detected using anti-histidine primary antibody (Calbiochem) and Alexa Fluor® 488-conjugated secondary antibody (Life Technologies).
Thymidine Phosphorylation Activity
The enzyme assays were carried out as described (18). Briefly, the tissues were homogenized (using tube and pestle (Kimble Chase) or Qiagen TissueRuptor) and suspended in extraction buffer (50 mm Tris-HCl, pH 7.6, 2 mm DTT, 5 mm benzamidine, 0.5 mm phenylmethylsulfonyl fluoride (PMSF), 20% glycerol, and 0.5% Nonidet P40). The suspension was centrifuged at 13,000 rpm for 20 min, and the supernatant was collected and stored at −80 °C. The protein concentrations were determined using Bradford protein assay reagent (Bio-Rad) and BSA as a standard. The enzymatic assays were performed in 50 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 5 mm ATP, 2 mm DTT, 15 mm NaF, 0.5 mg/ml BSA, 40–50 μg of protein, 3 μm [methyl-3H]thymidine (2 Ci/mmol; Moravek), and 7 μm unlabeled thymidine in a volume of 50 μl. For the bromovinyl-2′-deoxyuridine (BVDU) assay, 2.5 μm [5-3H]BVDU (28 Ci/mmol) was used as the substrate. 10 μl of the reaction mixture was spotted on Whatman DE-81 filter discs after incubation at 37 °C at different time points (0, 10, 20, and 30 min). The filters were washed three times in 5 mm ammonium formate, and the filter-bound product was eluted from the filter with 0.1 m KCl and 0.1 m HCl. The radioactivity was quantified by scintillation counting using 3 ml of scintillation buffer.
Quantification of mtDNA by Real-time PCR
The number of mtDNA copies per diploid nucleus in mouse tissues was determined using real-time PCR absolute quantification, using an ABI 7500 Fast system (Applied Biosystems). Total genomic DNA was purified from mouse tissues using the DNeasy blood and tissue kit (Qiagen). 5–10 ng of genomic DNA was used in each reaction. Primers and probe for mouse mt-ND1 gene (mitochondrial encoded NADH dehydrogenase 1; primers, mt-ND1-F and mt-ND1-R; probe, mt-ND1) and for single-copy mouse RPPH1 gene (nuclear encoded ribonuclease P RNA component H1; primers, RPPH1-F and RPPH1-R; probe, RPPH1) were designed for this purpose (supplemental Tables S1 and S2). For each DNA sample, the mitochondrial gene mt-ND1 and the nuclear gene RPPH1 were quantified separately. Standard curves were generated using known numbers of a plasmid containing one copy of each of the two mouse genes referred above. According to the standard curve, the number of copies from each gene was calculated for each sample, and the number of mtDNA copies per diploid nucleus was calculated according to the formula: mtDNA copies per diploid nucleus = 2 × (mt-ND1 gene copies/RPPH1 gene copies).
Quantification of dNTP Pools
dNTP extracts were obtained from skeletal muscle after homogenization (Qiagen TissueRuptor) on ice in 10 volumes (w/v) of cold MTSE buffer (210 mm mannitol, 70 mm sucrose, 10 mm Tris-HCl, pH 7.4, 2 mm EGTA, 0.2 mg/ml BSA) and centrifuged at 1000 × g for 3 min at 4 °C. Supernatants were precipitated with 100% methanol (to a final concentration of 60%), kept for 1–3 h at −20 °C, boiled 3 min, and centrifuged at 20,670 × g for 30 min at 4 °C. Supernatants were evaporated until dry, resuspended in 200 μl of distilled water, and stored at −80 °C until needed. The total dNTP pools were determined as described (21). Briefly, 100-μl reaction volumes were generated by 10 μl of sample or standard with 90 μl of reaction buffer containing 40 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 5 mm DTT, 0.25 mm of specific oligonucleotide template, 0.25 μm [2,8-3H]dATP (15.2 Ci/mmol; for dTTP, dCTP and dGTP determinations; Moravek) or [methyl-3H]dTTP (66.2 Ci/mmol; Moravek) for dATP determination, and 0.2 unit of Klenow DNA polymerase (New England Biolabs) (1 unit catalyzes the incorporation of 10 nmol of deoxynucleotide into acid insoluble product in 30 min at 37 °C). After a 45-min incubation at 37 °C, 10 μl of the reaction mixture was spotted on Whatman DE-81 filter discs. After drying, the filters were washed three times for 10 min in 5% Na2HPO4, once in distilled water, and once in 95% ethanol. The filters are completely dried, and the retained radioactivity was determined by scintillation counting. All experiments were performed in triplicates, and dNTP pools were determined in pmol dNTP/mg of tissue. The preparations of specific oligonucleotides for each dNTPs were as described (22).
Analysis of mRNA Expression
Total RNA was isolated using the RNeasy kit (Qiagen). The cDNA was synthesized using the high capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer's instructions. The expression analysis of all genes was done with specific primers and TaqMan probes (MWG-Biotech), using the endogenous GAPDH gene as a loading control. The PCR reactions were done using TaqMan universal PCR master mix (Applied Biosystems) and run on the Applied Biosystems 7500 Fast equipment. The primers and probes for TK1, TK2, deoxycytidine kinase (dCK), DGUOK, ribonucleoside-diphosphate reductase subunit M2 (RRM2), ribonucleoside-diphosphate reductase subunit M2 B (RRM2B), and GAPDH genes are listed in supplemental Tables S1 and S2.
Analysis of mtDNA Point Mutations
Total DNA was extracted from mice tissue and fragments from mt-Cytb gene (nucleotides 14073–14906) and mitochondrial DNA noncoding region (nucleotides 15357–138) were amplified by high fidelity PCR (PfuUltra high fidelity DNA polymerase, Agilent). The PCR products were cloned into pGEM®-T vector (Promega) after A-tailing the blunt-ended PCR products according to the manufacturer's instructions. Plasmids of multiple clones obtained were sequenced to detect point mutations in those fragments, and mutation rates were calculated.
Histopathology
Selected tissue samples from two mice per genotype were fixed in 4% buffered formaldehyde and transferred to 70% ethanol after 24 h. After routine processing and paraffin embedding, 4-μm-thick sections were mounted on glass slides, stained with hematoxylin and eosin, and viewed under a light microscope.
Statistical Analysis
All experimental data are reported as mean, and error bars in Figs. 3 and 4 indicate S.E. Student's t test was used to analyze differences between the mean values, and a p < 0.05 was considered statistically significant.
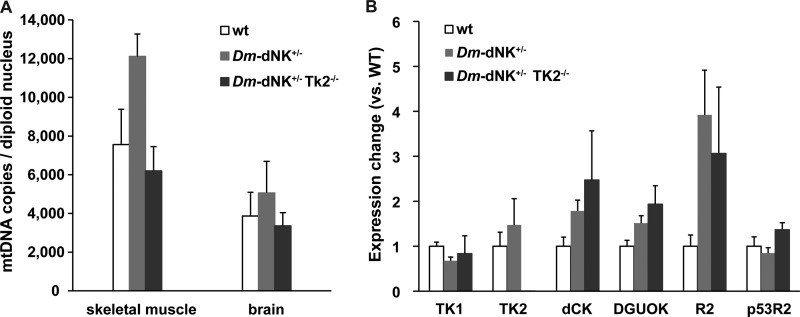
FIGURE 4.
mtDNA copy number and gene expression analysis of Dm-dNK+/−TK2−/− mice. A, mtDNA copies per diploid nucleus in skeletal muscle and brain of 6-month-old wt (n = 6), Dm-dNK+/− (n = 6), and Dm-dNK+/−TK2−/− (n = 3) mice (mean ± S.E.). B, mRNA expression of compensatory enzymes (TK1, TK2, deoxycytidine kinase (dCK), DGUOK, R2, and p53R2) in skeletal muscle of 6-month-old wt, Dm-dNK+/−, and Dm-dNK+/−TK2−/− mice (n = 3 each; mean ± S.E.).
RESULTS
Construction and Characterization of Mice Expressing Dm-dNK
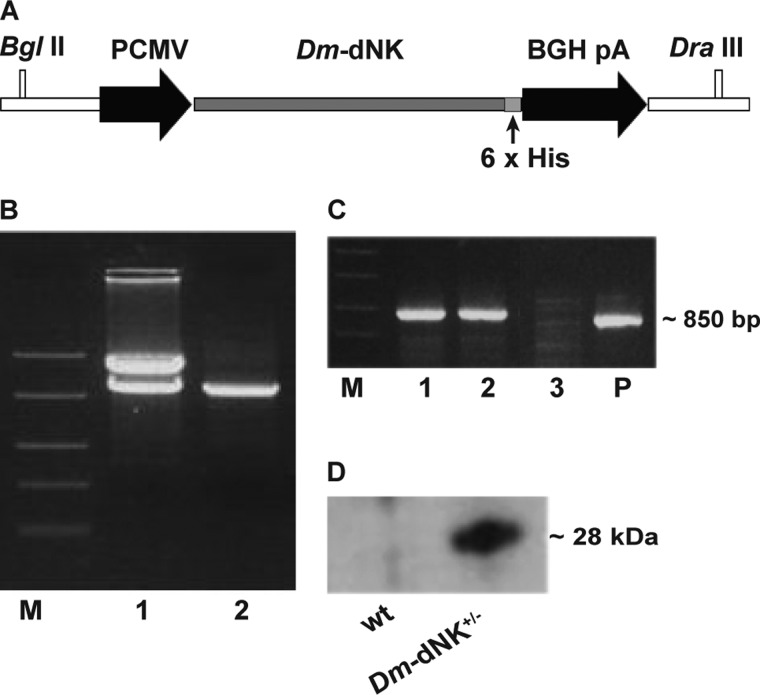
The Dm-dNK transgene was constructed using a pcDNA3 vector (Fig. 1A). Restriction digestion showed the Dm-dNK cDNA present in the construct used for transfection (Fig. 1B). Dm-dNK+/− mice were created by crossing a Dm-dNK+/− mouse with a wt mouse, and the pups (∼14 days) were screened for the presence of the Dm-dNK gene by PCR. The integration of the Dm-dNK transgene in the mouse genome was verified by PCR as shown in lanes 1 and 2 (Fig. 1C). The Dm-dNK protein expression was detected in skeletal muscle of 1-month-old mice by Western blot analysis (Fig. 1D), whereas the control samples of litter mate wt mice did not show Dm-dNK protein expression. Dm-dNK has previously been shown to localize in the cell nucleus when expressed in human cells (20, 23). The Dm-dNK construct used in the present study was also shown to have a nuclear localization when transfected in HeLa cells (supplemental Fig. S1).
FIGURE 1.
Construction and screening of Dm-dNK mice. A, Dm-dNK transgene construct in pcDNA3 vector. BGH pA, bovine growth hormone polyadenylation signal. B, restriction digestion of Dm-dNK using BglII and DraIII enzymes. Lane M, FastRuler middle range DNA ladder (Fermentas); lane 1, pcDNA3-Dm-dNK restriction digested using BglII + DraIII enzymes; lane 2, purified Dm-dNK transgene. C, genotyping of Dm-dNK+/− mice. Lane M, FastRuler middle range DNA ladder (Fermentas); lanes 1–3, DNA samples from three mice; lane P, positive control. D, expression of Dm-dNK protein (28 kDa) in skeletal muscle of wt and Dm-dNK+/− mice.
Thymidine-phosphorylating Activity in Dm-dNK Transgenic Mice
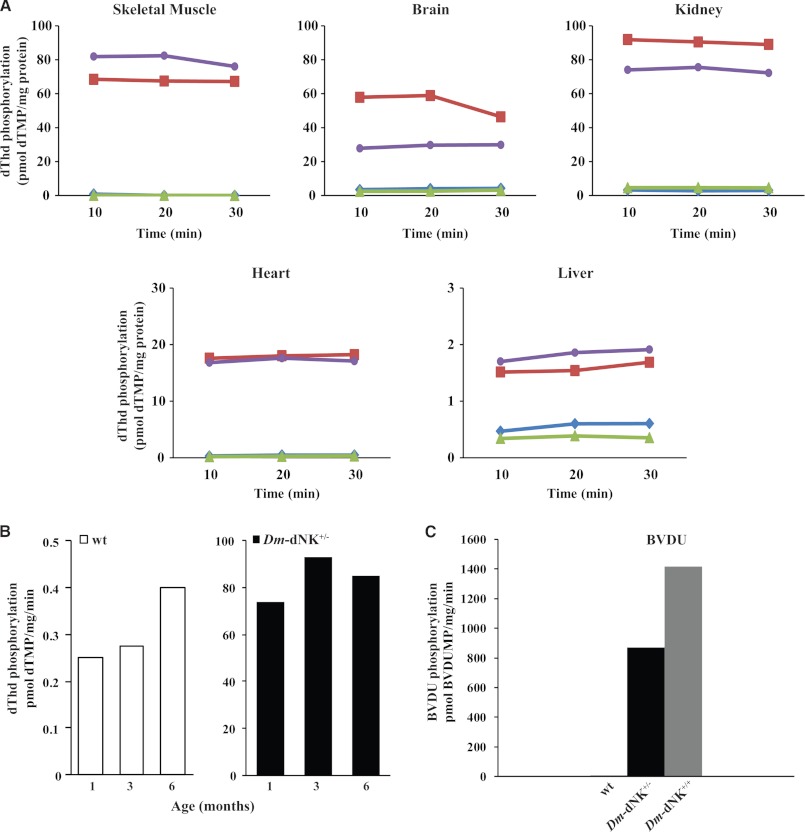
The catalytic activity of Dm-dNK was measured by dThd phosphorylation in tissue extracts from 1-month-old Dm-dNK+/− mice (Fig. 2A). Extracts from brain, skeletal muscle, kidney, heart, and liver all showed increased dThd-phosphorylating activity as compared with corresponding tissues from wt control mice. Highest enzyme activity with ∼80-fold increase was detected in skeletal muscle and kidney, whereas activity in the liver showed a 1.5–2-fold increase. The dThd phosphorylation rates were similar when determined in skeletal muscle of 1-, 3-, and 6-month-old mice (Fig. 2B). In addition, to test for a gene dose effect, phosphorylation of the high affinity substrate BVDU was used to measure enzyme activity in kidneys of 1-month-old Dm-dNK+/− mice, Dm-dNK+/+ mice, and control wt mice. The homozygote Dm-dNK+/+ mice showed a slightly higher enzyme activity (1.5–2-fold) than the heterozygote Dm-dNK+/− mice (Fig. 2C).
FIGURE 2.
Thymidine-phosphorylating activity of Dm-dNK mice. A, enzyme activity determined as [3H]dThd phosphorylation (pmol of dTMP/mg of protein) in extracts of brain, heart, liver, skeletal muscle, and kidney of wt and Dm-dNK+/− mice that were 1-month-old (blue filled diamond, wt 1; green filled triangle, wt 2; red filled square, Dm-dNK+/− 1; purple filled circle, Dm-dNK+/− 2). The enzyme activity was measured over a time period of 30 min. The enzyme activity was higher in the Dm-dNK+/− mice when compared with the wt mice in all the tissues studied, and this increase was statistically significant for all the tissues (p < 0.0001). Data represent average of the three time points at which activity was measured (10, 20, and 30 min). B, estimated activity of Dm-dNK determined as [3H]dThd phosphorylation (pmol of dTMP/mg/min) in extracts of skeletal muscle samples of 1-month-old (n = 2 wt, n = 2 Dm-dNK+/−), 3-month-old (n = 3 wt, n = 2 Dm-dNK+/−), and 6-month-old (n = 3 wt, n = 3 Dm-dNK+/−) mice. No significant difference in enzymatic activity was observed in Dm-dNK+/− samples in 1-, 3-, and 6-month-old mice (p > 0.05). C, difference in enzyme activity determined as phosphorylation of [3H]BVDU (pmol of BVDUMP/mg/min) in kidney of 1-month-old wt (n = 3), Dm-dNK+/− (n = 2), and Dm-dNK+/+ (n = 1) mice.
Development of TK2-deficient Mice Expressing Dm-dNK
TK2-deficient mice expressing Dm-dNK (Dm-dNK+/−TK2−/−) were created as shown in Fig. 3A. Genomic DNA of Dm-dNK+/−TK2−/− mice was analyzed by Southern blot to confirm the absence of the wt TK2 alleles (Fig. 3B). The Dm-dNK+/−TK2−/− mice were fertile, appeared similar in size, and showed normal development in comparison with the wt and Dm-dNK+/− mice over the observation period of 6 months. The Dm-dNK+/−TK2−/− mice showed increased dThd-phosphorylating activity at levels similar to the Dm-dNK+/− mice (supplemental Fig. S2). There was no significant difference in the relative organ weights of heart, liver, kidney, and spleen in these mice (Fig. 3C). There were no major histopathology differences between the mouse lines investigated (Fig. 3D), although a slight increase in tubular protein casts was observed in mice carrying the Dm-dNK transgene (supplemental Fig. S3).
Levels of mtDNA, dNTP Pools, and Expression of Compensatory Enzymes
Quantification of the mtDNA was performed using real-time PCR to determine the number of mtDNA copies per diploid nucleus in brain and skeletal muscle of 6-month-old mice. These tissues were chosen for quantification because the dThd-phosphorylating activity was highest in skeletal muscle, and previous studies show mtDNA depletion in brain of newborn TK2−/− mice (7). No change in mtDNA copy number was observed in tissues of both Dm-dNK+/− and Dm-dNK+/−TK2−/− mice (Fig. 4A). To determine the effect of Dm-dNK activity on intracellular dNTP pools, these were measured in whole cell extracts of skeletal muscle of 1-month-old wt and Dm-dNK+/−TK2−/− mice (Table 1). The results showed that there was a 114-fold increase of the dTTP pool, a 3.7-fold increase of the dCTP pool, and a 1.6-fold increase of the dGTP pool in the Dm-dNK+/−TK2−/− mice. It was not possible to obtain values of the dATP pool, probably because of the presence of a large excess of dTTP in the Dm-dNK+/−TK2−/− extracts that competed with the [3H]dTTP present in the reaction mix.
TABLE 1.
dNTP pool levels in skeletal muscle of 1-month-old WT and Dm-dNK+/−TK2−/− mice
The values for dNTPs are mean ± S.E. of measurements from three independent repeats of three mice each; ND, not determined.
| dNTP pool concentration |
Ratio (Dm-dNK+/−TK2−/−/WT) | ||
|---|---|---|---|
| WT | Dm-dNK+/−TK2−/− | ||
| pmol of dNTP/mg of tissue | |||
| dTTP | 0.32 ± 0.07 | 36 ± 4.0 | 114 |
| dCTP | 0.39 ± 0.02 | 1.4 ± 0.2 | 3.7 |
| dGTP | 0.43 ± 0.03 | 0.67 ± 0.07 | 1.6 |
| dATP | 0.25 ± 0.03 | ND | ND |
The mRNA expression of several deoxyribonucleotide metabolizing enzymes was compared between wt, Dm-dNK+/−, and Dm-dNK+/−TK2−/− mice. The results showed a decrease of the expression of TK2 in the Dm-dNK+/−TK2−/− mice (p < 0.001) and a slight increase in R2 in the Dm-dNK+/− mice (p < 0.05) as compared with the wt mice. No significant change was observed in TK1, deoxycytidine kinase, DGUOK, and p53R2 expression levels (Fig. 4B).
Point mutations in the mtDNA were determined by cloning and sequencing the mt-Cytb gene and mtDNA noncoding control regions in skeletal muscle of 6-month-old wt and Dm-dNK+/− mice (Table 2). The mutation frequencies were not significantly different in the wt and Dm-dNK+/− mice when analyzed in both the noncoding control regions (2.0–3.1 mutations/10 kb) and the mt-Cytb gene (1.7–7.8 mutations/10 kb) (p > 0.05). However, one of the Dm-dNK+/− mice showed a higher mutation frequency (∼14 mutations/10 kb) and indicates the importance of a more extensive mutation analysis in mice of older age.
TABLE 2.
mtDNA point mutations analysis
Skeletal muscle of 6-month-old wild-type and Dm-dNK+/− mice was analyzed.
| Mouse (genotype) |
mt-Cytb gene |
Noncoding control region |
||
|---|---|---|---|---|
| Number of mutations/base pairs sequenced | Mutation frequency (per 10 kb) | Number of mutations/base pairs sequenced | Mutation frequency (per 10 kb) | |
| Wild type 1 | 0/5838 | 0.0 | 2/7567 | 2.6 |
| Wild type 2 | 1/5004 | 2.0 | 3/7567 | 3.9 |
| Wild type 3 | 2/6672 | 3.0 | 2/7567 | 2.6 |
| Dm-dNK+/− 1 | 3/5838 | 5.1 | 2/7567 | 2.6 |
| Dm-dNK+/− 2 | 1/3336 | 3.0 | 2/8648 | 2.3 |
| Dm-dNK+/− 3 | 7/5004 | 14.0 | 1/8648 | 1.2 |
DISCUSSION
The present work was initiated to address questions of importance to develop treatment strategies for specific mtDNA deficiency diseases. The strategy chosen was to express the multisubstrate Dm-dNK in TK2-deficient mice. The Dm-dNK cDNA has previously been expressed in mammalian cells, and the enzyme has been characterized regarding enzyme kinetics and substrate specificity (17, 24). The Dm-dNK enzyme is closely related to the mammalian TK2 enzyme but, in contrast to TK2, Dm-dNK possesses a very high catalytic activity (15). Based on these enzymatic properties, we speculated that the Dm-dNK enzyme would be more efficient, as compared with TK2, to deliver dTTPs for mitochondrial DNA synthesis and thus rescue the severe phenotype of TK2-deficient mice. The alternative would have been to replace the missing TK2 with an identical TK2 transgene. However, TK2 is an enzyme with low catalytic activity and would not be optimal to address the fundamental question regarding cytosolic substitution of dTMP and dCMP as substrates for mtDNA synthesis.
TK1 is only present in dividing cells, whereas TK2 is present both in nondividing and in dividing cells but at much lower levels (25, 26). Accordingly, very low levels of dThd phosphorylation were detected in skeletal muscle, brain, heart, liver, and kidney cells of the wt mice, and the low levels of activity observed probably originated mainly from TK2, although subpopulations of dividing cells may also be present because the tissue preparations contains a mixture of cell types. The dThd-phosphorylating activity observed in the tissue extracts of Dm-dNK-expressing mice (Dm-dNK+/− and Dm-dNK+/−TK2−/−) was a result of the combined activity of the low amount of TK2 enzyme and the Dm-dNK enzyme. The general pattern was a large increase of dThd phosphorylation in all investigated tissues, but there was also a variation depending on the tissue. Because the background TK1 and TK2 activities were very low, the differences most probably were due to a variation in the expression of the Dm-dNK gene. This variation in gene expression between different tissues could be due to differences in epigenetic modifications in different organs expressing Dm-dNK+/− (27). To demonstrate that the increased dThd-phosphorylating activity was mainly due to Dm-dNK activity, and not the induction of TK1, the nucleoside analog substrate BVDU was used in kidney extracts. BVDU is a very efficient substrate for Dm-dNK but a poor substrate for TK1 (28, 29). The large increase of BVDU phosphorylation in kidney extracts of Dm-dNK+/− mice thus supports the conclusion that the Dm-dNK transgene is responsible for the increased dThd-phosphorylating activity.
The effects of a strong and general increase of dThd phosphorylation have not previously been studied in a mammalian system, to our knowledge. This enzymatic reaction is the first step to synthesize dTTP for DNA synthesis. The question of how this dThd-phosphorylating activity affected the dTTP pool was therefore of great interest. dNTP pool imbalances are believed to affect the fidelity of DNA synthesis, and there is substantial evidence for the existence of substrate cycles to fine-tune dTTP levels (30, 31). In the nuclear genome environment, mutagenic dNTP pools are known to activate the DNA damage checkpoint pathways (32). This may only occur if at least one dNTP is present at low levels and thereby limit DNA replication (33). The increased dTTP pools detected in our study are unique to address previous hypotheses regarding consequences of dNTP pool imbalances. We can conclude that the Dm-dNK+/−TK2−/− mice live without gross signs of disturbance with the high dTTP pools during the observation period of 6 months. However, this is a relatively short observation time, and we cannot exclude that the introduced Dm-dNK may affect the mice at an older age. In addition, it must be noted that the dNTP pool measurements are from whole cell extracts and do not reflect intramitochondrial dNTP levels.
Because there was no significant difference in the mtDNA copy number in the Dm-dNK+/− or the Dm-dNK+/−TK2−/− mice, the increase in levels of dTTP may not have affected the fidelity of DNA synthesis. DNA sequencing revealed some mutations in the Dm-dNK+/− mice, but no pattern was found that would account for a high dTTP pool causing this effect, and the increase was not statistically significant. Elevated dTTP pools have also been found in mice deficient in thymidine phosphorylase, which dephosphorylates thymidine to thymine. Mutations in thymidine phosphorylase result in a syndrome called mitochondrial neurogastrointestinal encephalomyopathy presenting with increased dThd and dUrd concentrations (34). Knock-out mice lacking thymidine phosphorylase have been shown to exhibit elevated dTTP pools and late onset mtDNA depletion in brain (35). Further studies of older mice will be necessary to draw conclusions on whether the high dTTP pool causes an increased rate of mutations in the Dm-dNK+/−TK2−/− mice. Studies have shown that limited availability of dCTP is a key factor for mtDNA depletion in mitochondrial neurogastrointestinal encephalomyopathy rather than excess of dTTP (36). Our present study also suggests that thymidine phosphorylase deficiency may cause additional cell alterations and that elevated dTTP pools alone are insufficient to explain the full set of symptoms observed in this disease.
An important result of the present study is the proof that dTMP, synthesized exclusively by an enzyme outside the mitochondria, can compensate for the dTTP required for mtDNA synthesis in quiescent cells. This finding opens possibilities to deliver cytoplasmic monophosphate analogs that can subsequently serve as substrates for dTTP and mtDNA. There are examples of such monophosphate analogs in the antiviral field (13, 14, 37). These compounds bypass the nucleoside kinase step, which is a requirement for treatment of TK2 or DGUOK deficiencies. The clinical success of antiviral nucleotide analog compounds proves that monophosphates can be easily administered to patients and effectively delivered to target cells.
Acknowledgment
We thank Professor Vera Bianchi for discussions and expertise advice regarding measurements of dTTP and dCTP pools in tissue extracts.
This work was supported by the Swedish Cancer Society (Grant CAN 2011/1277), Swedish Research Council (Grant 2010-2828), and Karolinska Institute.
This article was selected as a Paper of the Week.

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- p53R2
- p53-inducible ribonucleotide reductase small subunit
- R2
- ribonucleotide reductase 2
- BVDU
- bromovinyl-2′-deoxyuridine
- DGUOK
- deoxyguanosine kinase
- Dm-dNK
- D. melanogaster-nucleoside kinase
- mt-Cytb
- cytochrome b
- RPPH1
- nuclear encoded ribonuclease P RNA component H1
- TK1
- thymidine kinase 1
- TK2
- thymidine kinase 2
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Bridges E. G., Jiang Z., Cheng Y. C. (1999) Characterization of a dCTP transport activity reconstituted from human mitochondria. J. Biol. Chem. 274, 4620–4625 [DOI] [PubMed] [Google Scholar]
- 2. Ferraro P., Nicolosi L., Bernardi P., Reichard P., Bianchi V. (2006) Mitochondrial deoxynucleotide pool sizes in mouse liver and evidence for a transport mechanism for thymidine monophosphate. Proc. Natl. Acad. Sci. U.S.A. 103, 18586–18591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson D. D., Quintero C. M., Stover P. J. (2011) Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pontarin G., Ferraro P., Håkansson P., Thelander L., Reichard P., Bianchi V. (2007) p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J. Biol. Chem. 282, 16820–16828 [DOI] [PubMed] [Google Scholar]
- 5. Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V. (2012) Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. U.S.A. 109, 13302–13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou X., Johansson M., Solaroli N., Rozell B., Grandien A., Karlsson A. (2010) Hematopoiesis in the thymidine kinase 2 deficient mouse model of mitochondrial DNA depletion syndrome. J. Inherit. Metab. Dis. 33, 231–236 [DOI] [PubMed] [Google Scholar]
- 7. Zhou X., Solaroli N., Bjerke M., Stewart J. B., Rozell B., Johansson M., Karlsson A. (2008) Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum. Mol. Genet. 17, 2329–2335 [DOI] [PubMed] [Google Scholar]
- 8. Mandel H., Szargel R., Labay V., Elpeleg O., Saada A., Shalata A., Anbinder Y., Berkowitz D., Hartman C., Barak M., Eriksson S., Cohen N. (2001) The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat. Genet. 29, 337–441; Correction Nat. Genet. 29, 491 [DOI] [PubMed] [Google Scholar]
- 9. Bourdon A., Minai L., Serre V., Jais J. P., Sarzi E., Aubert S., Chrétien D., de Lonlay P., Paquis-Flucklinger V., Arakawa H., Nakamura Y., Munnich A., Rötig A. (2007) Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 39, 776–780 [DOI] [PubMed] [Google Scholar]
- 10. Suomalainen A., Isohanni P. (2010) Mitochondrial DNA depletion syndromes — Many genes, common mechanisms. Neuromuscul. Disord. 20, 429–437 [DOI] [PubMed] [Google Scholar]
- 11. Saada A., Shaag A., Mandel H., Nevo Y., Eriksson S., Elpeleg O. (2001) Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 29, 342–344 [DOI] [PubMed] [Google Scholar]
- 12. Wang L. (2010) Deoxynucleoside salvage enzymes and tissue specific mitochondrial DNA depletion. Nucleosides Nucleotides Nucleic Acids 29, 370–381 [DOI] [PubMed] [Google Scholar]
- 13. Meier C., Jessen H. J., Balzarini J. (2008) Nucleoside diphosphate prodrugs. Nucleic Acids Symp. Ser. (Oxf.) 83–84 [DOI] [PubMed] [Google Scholar]
- 14. Mehellou Y., Balzarini J., McGuigan C. (2009) Aryloxy phosphoramidate triesters: a technology for delivering monophosphorylated nucleosides and sugars into cells. Chemmedchem 4, 1779–1791 [DOI] [PubMed] [Google Scholar]
- 15. Johansson M., van Rompay A. R., Degrève B., Balzarini J., Karlsson A. (1999) Cloning and characterization of the multisubstrate deoxyribonucleoside kinase of Drosophila melanogaster. J. Biol. Chem. 274, 23814–23819 [DOI] [PubMed] [Google Scholar]
- 16. Munch-Petersen B., Piskur J., Sondergaard L. (1998) Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J. Biol. Chem. 273, 3926–3931 [DOI] [PubMed] [Google Scholar]
- 17. Solaroli N., Johansson M., Balzarini J., Karlsson A. (2007) Enhanced toxicity of purine nucleoside analogs in cells expressing Drosophila melanogaster nucleoside kinase mutants. Gene Ther. 14, 86–92 [DOI] [PubMed] [Google Scholar]
- 18. Solaroli N., Zheng X., Johansson M., Balzarini J., Karlsson A. (2007) Mitochondrial expression of the Drosophila melanogaster multisubstrate deoxyribonucleoside kinase. Mol. Pharmacol. 72, 1593–1598 [DOI] [PubMed] [Google Scholar]
- 19. Zheng X., Johansson M., Karlsson A. (2001) Bystander effects of cancer cell lines transduced with the multisubstrate deoxyribonucleoside kinase of Drosophila melanogaster and synergistic enhancement by hydroxyurea. Mol. Pharmacol. 60, 262–266 [DOI] [PubMed] [Google Scholar]
- 20. Zheng X., Johansson M., Karlsson A. (2000) Retroviral transduction of cancer cell lines with the gene encoding Drosophila melanogaster multisubstrate deoxyribonucleoside kinase. J. Biol. Chem. 275, 39125–39129 [DOI] [PubMed] [Google Scholar]
- 21. Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V. (2010) Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 38, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherman P. A., Fyfe J. A. (1989) Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal. Biochem. 180, 222–226 [DOI] [PubMed] [Google Scholar]
- 23. Bertoli A., Franco M., Balzarini J., Johansson M., Karlsson A. (2005) Altered deoxyribonucleotide pools in T-lymphoblastoid cells expressing the multisubstrate nucleoside kinase of Drosophila melanogaster. FEBS J. 272, 3918–3928 [DOI] [PubMed] [Google Scholar]
- 24. Munch-Petersen B., Knecht W., Lenz C., Søndergaard L., Piskur J. (2000) Functional expression of a multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster and its C-terminal deletion mutants. J. Biol. Chem. 275, 6673–6679 [DOI] [PubMed] [Google Scholar]
- 25. Eriksson S., Arnér E., Spasokoukotskaja T., Wang L., Karlsson A., Brosjö O., Gunvén P., Julusson G., Liliemark J. (1994) Properties and levels of deoxynucleoside kinases in normal and tumor cells; implications for chemotherapy. Adv. Enzyme Regul. 34, 13–25 [DOI] [PubMed] [Google Scholar]
- 26. Wang L., Eriksson S. (2010) Tissue specific distribution of pyrimidine deoxynucleoside salvage enzymes shed light on the mechanism of mitochondrial DNA depletion. Nucleosides Nucleotides Nucleic Acids 29, 400–403 [DOI] [PubMed] [Google Scholar]
- 27. Babinet C. (2000) Transgenic mice: an irreplaceable tool for the study of mammalian development and biology. J. Am. Soc. Nephrol. 11, S88–S94 [PubMed] [Google Scholar]
- 28. Franzolin E., Rampazzo C., Pérez-Pérez M. J., Hernández A. I., Balzarini J., Bianchi V. (2006) Bromovinyl-deoxyuridine: A selective substrate for mitochondrial thymidine kinase in cell extracts. Biochem. Biophys. Res. Commun. 344, 30–36 [DOI] [PubMed] [Google Scholar]
- 29. Wang L., Eriksson S. (2008) 5-Bromovinyl 2′-deoxyuridine phosphorylation by mitochondrial and cytosolic thymidine kinase (TK2 and TK1) and its use in selective measurement of TK2 activity in crude extracts. Nucleosides Nucleotides Nucleic Acids 27, 858–862 [DOI] [PubMed] [Google Scholar]
- 30. Gazziola C., Ferraro P., Moras M., Reichard P., Bianchi V. (2001) Cytosolic high Km 5′-nucleotidase and 5′(3′)-deoxyribonucleotidase in substrate cycles involved in nucleotide metabolism. J. Biol. Chem. 276, 6185–6190 [DOI] [PubMed] [Google Scholar]
- 31. Rampazzo C., Fabris S., Franzolin E., Crovatto K., Frangini M., Bianchi V. (2007) Mitochondrial thymidine kinase and the enzymatic network regulating thymidine triphosphate pools in cultured human cells. J. Biol. Chem. 282, 34758–34769 [DOI] [PubMed] [Google Scholar]
- 32. Zegerman P., Diffley J. F. X. (2009) DNA replication as a target of the DNA damage checkpoint. DNA Repair 8, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 33. Kumar D., Viberg J., Nilsson A. K., Chabes A. (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 38, 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valentino M. L., Martí R., Tadesse S., López L. C., Manes J. L., Lyzak J., Hahn A., Carelli V., Hirano M. (2007) Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). FEBS Lett. 581, 3410–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. López L. C., Akman H. O., García-Cazorla A., Dorado B., Martí R., Nishino I., Tadesse S., Pizzorno G., Shungu D., Bonilla E., Tanji K., Hirano M. (2009) Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum. Mol. Genet. 18, 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. González-Vioque E., Torres-Torronteras J., Andreu A. L., Martí R. (2011) Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). PLoS Genet. 7, e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gisch N., Balzarini J., Meier C. (2008) Studies on enzyme-cleavable dialkoxymethyl-cycloSaligenyl-2′,3′-dideoxy-2′,3′-didehydrothymidinemonophosphates. J. Med. Chem. 51, 6752–6760 [DOI] [PubMed] [Google Scholar]