Background: PTPMT1 is the mitochondrial phosphatidylglycerophosphate (PGP) phosphatase essential for the biosynthesis of cardiolipin, an integral component of mitochondrial and bacterial membranes.
Results: A PTPMT1-like phosphatase in the bacterium Rhodopirellula baltica has similar PGP phosphatase activity as its mammalian ortholog.
Conclusion: PTPMT1 is evolutionarily conserved.
Significance: The existence of a mammalian-type PGP phosphatase in bacteria provides new insight into the evolution of cardiolipin metabolism.
Keywords: Bacteria, Cardiolipin, Mitochondria, Phosphatase, Tyrosine Protein Phosphatase (Tyrosine Phosphatase), Phosphatidylglycerophosphate
Abstract
Cardiolipin is a glycerophospholipid found predominantly in the mitochondrial membranes of eukaryotes and in bacterial membranes. Cardiolipin interacts with protein complexes and plays pivotal roles in cellular energy metabolism, membrane dynamics, and stress responses. We recently identified the mitochondrial phosphatase, PTPMT1, as the enzyme that converts phosphatidylglycerolphosphate (PGP) to phosphatidylglycerol, a critical step in the de novo biosynthesis of cardiolipin. Upon examination of PTPMT1 evolutionary distribution, we found a PTPMT1-like phosphatase in the bacterium Rhodopirellula baltica. The purified recombinant enzyme dephosphorylated PGP in vitro. Moreover, its expression restored cardiolipin deficiency and reversed growth impairment in a Saccharomyces cerevisiae mutant lacking the yeast PGP phosphatase, suggesting that it is a bona fide PTPMT1 ortholog. When ectopically expressed, this bacterial PGP phosphatase was localized in the mitochondria of yeast and mammalian cells. Together, our results demonstrate the conservation of function between bacterial and mammalian PTPMT1 orthologs.
Introduction
Reversible phosphorylation, a dynamic process carried out by kinases and phosphatases, is vital for the regulation of many cellular signaling events (1, 2). More than 100 human genes encode proteins of the same fold as the classic protein tyrosine phosphatases (PTPs)3 and feature the highly conserved Cys-X5-Arg (CX5R) motif in their active site. These phosphatases were once thought to exclusively dephosphorylate phosphotyrosine. More recent studies, however, have identified several PTPs that can hydrolyze phosphate from phosphoserine/threonine-containing proteins or even nonprotein substrates (3, 4). For instance, the tumor suppressor PTEN and the carbohydrate binding Laforin have been shown to dephosphorylate phosphoinositides and glycogen respectively, both in vitro and in vivo (5–7). The primary amino acid sequences of these phosphatases vary from the classical tyrosine-specific PTPs; therefore, they are categorized into a subfamily called dual specificity phosphatases (DSPs) (2).
We recently characterized a new DSP called PTPMT1 (PTP localized to the mitochondrion 1). Interest in this protein arose due to its strong active site similarity to PTEN. PTPMT1 is the first PTP localized exclusively to the mitochondrion; specifically, it resides in the inner membrane facing the matrix (8). Our recent results identify PTPMT1 as the mammalian phosphatase that mediates the conversion of phosphatidylglycerophosphate (PGP) into phosphatidylglycerol (PG), a step necessary for the de novo biosynthesis of cardiolipin (CL) (Fig. 1A) (9).
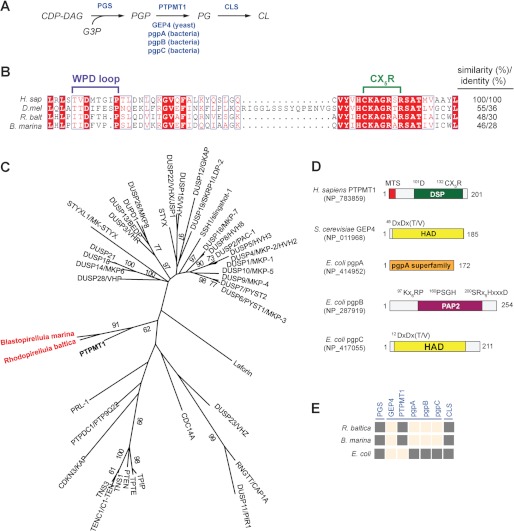
FIGURE 1.
Identification of a putative PTPMT1 ortholog in R. baltica. A, schematic diagram of the CL biosynthesis pathway. Enzymes are highlighted in blue. CDP-DAG, cytidine diphosphate diacylglycerol; G3P, glycerol-3-phosphate. B, active site primary sequence alignment of PTPMT1 orthologs using PROMALS3D and illustrated by ESPript (49, 50). H. sap (H. sapiens), NP_783859; D. mel (D. melanogaster), NP_732901; and R. balt (R. baltica), NP_865112. C, a phylogenetic tree of human and bacterial DSPs constructed using maximum likelihood (ML) method in PHYML. Each branch was tested by 100 bootstrap replicates, and only branches with bootstrap values above 50% were shown. Human PTPMT1 is bolded and bacterial PTPMT1 orthologs are highlighted in red. D, domain architecture of PGP phosphatases. All domains are presented based on analyses using PFAM, CDD, and PROSITE. Catalytic active site sequences are indicated above. MTS, mitochondrial targeting sequence; DSP, dual specificity phosphatase; HAD, haloacid dehalogenase; PAP2, phosphatidic acid phosphatase 2. E, presence and absence matrix for CL de novo synthesis enzymes in Rhodeopirellula. Dark gray squares indicate presence of CL enzyme (column) in target species (row). Peach squares mean no ortholog has been identified, i.e. blast search failed to identify sequences that meet the bidirectional-best-hit criterion (cut-off E-values of 10−10 or less). Sequences of E. coli pgsA, S. cerevisiae GEP4, H. sapiens PTPMT1, E. coli pgpA, pgpB, pgpC, and E. coli cls1 were used as queries for the blast searches.
CL is an anionic phospholipid found in most bacteria and eukaryotes4 (10). It is an essential component of the inner mitochondrial membrane in eukaryotes and regulates mitochondrial ATP production, protein import, and apoptosis (10). In bacteria, CL is one of the major anionic phospholipids and functionally interacts with membrane proteins that are involved in energy metabolism and cellular division (11). Eukaryotes and bacteria utilize a similar strategy to synthesize CL, except in the step of producing CL from PG (12). Therefore, we thought it would be interesting to search for a PTPMT1-like phosphatase in bacteria.
In this study, we have characterized a putative PTPMT1 ortholog in the bacterium Rhodopirellula baltica. We show that this protein functions as a PGP phosphatase and is localized to the mitochondrion when ectopically expressed in yeast or mammalian cells, suggesting that the function of PTPMT1 is evolutionarily conserved. Together, our results contribute to the understanding of the evolution of CL metabolism.
EXPERIMENTAL PROCEDURES
Sequences and Phylogenetic Analysis
Bacterial PTPMT1-like sequences were first collected by searching human PTPMT1 against NCBI non-redundant (NR) database and were then inspected for containing conserved motifs found in eukaryotic PTPMT1s. The phylogenetic tree of selected human and bacterial DSPs was built using only the phosphatase domains. Sequences were aligned by PROMALS3D (13) and adjusted manually. Phylogenetic trees were inferred by using maximum likelihood (ML) and neighbor joining (NJ) methods in MEGA 5.0 (14). Different models of Poisson Correction, Dayhoff matrix, Jones-Taylor-Thornton (JTT) matrix, and JTT + frequency (F) were used, and they are consistent with each other on reliable branches. Support for each interior branch was tested with 100 and 1,000 bootstrap replicates for ML and NJ analyses, respectively. The domain structures of PGP phosphatases were analyzed using PFAM, CDD, and PROSITE (15–17).
Protein Expression and Purification
Mus musculus, Drosophila melanogaster, and R. baltica PTPMT1 orthologs were expressed as fusion proteins with N-terminal GST tag in Escherichia coli BL21 (DE3) CodonPlus RIPL cells (Stratagene) (18). The expression plasmids were constructed by ligating the PCR products of PTPMT1 orthologs into the vector. Cultures were grown at 37 °C in LB medium containing chloramphenicol (34 μg/ml) and kanamycin (50 μg/ml) to an A600 of 0.6. Cultures were then induced with isopropyl-1-thio-β-d-galactose pyranoside (IPTG) to a final concentration of 0.4 mm and allowed to grow overnight at 25 °C. Cells were pelleted down, resuspended in 25 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5 mm TCEP [Tris(2-carboxyethly)phosphine], 1 mm Pefabloc SC, 1 mm bezamidine-HCl, 1 mm PMSF (phenylmethylsulphonyl fluoride), and lysed using a high pressure homogenizer (Avestatin EmulsiFlex C5). Next, the lysed samples were centrifuged at 19,000 rpm for 30 min to remove insoluble material. The resulting supernatants were purified by affinity chromatography using GST-bind resin (Novagen). Finally, fusion proteins were eluted with 50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 10% glycerol, and 15 mm reduced glutathione. Catalytically inactive mutants (CS) were generated using site-directed mutagenesis and similarly purified.
Phosphatase Assays
Phosphatase assays using para-nitrophenylphosphate (pNPP) or [14C]PGP were carried out in assay buffer containing 50 mm sodium acetate, 25 mm bis-Tris, 25 mm Tris, and 2 mm dithiothreitol. The pH of the assay buffer was set at 5.5 for M. musculus PTPMT1, 7.0 for D. melanogaster, and 8.5 for R. baltica protein. pNPP assays were carried out at 37 °C for 10 min as previously described (19). For PGP assays, the 14C-labeled PGP was synthesized as previously described (20). Briefly, the reaction was carried out in the presence of 0.1 m Tris-HCl, pH 8.0, 0.1% Triton X-100, 0.2 mm CDP-DAG (Avanti Polar Lipids), 0.5 mm glycerol-3-phosphate (Sigma), and 13 μm sn-[U-14C]glycerol-3-phosphate (2–4 μCi/μmol, American Radiolabeled Chemicals). The reaction was initiated by the addition of recombinant E. coli PGP synthase and 0.1 m MgCl2 (18). After incubation at 37 °C for 2 h, 0.5 ml of 0.1 N HCl in methanol, 1.5 ml of chloroform, and 3.0 ml of 1 m MgCl2 were added to stop the reaction. The organic phase was then extracted and dried under vacuum. The resulting 14C-PGP was resuspended in 10 mm Tris pH 7.4 via sonication for 5 min. 1.25 × 104cpm of this radiolabeled PGP and 2 μg of recombinant enzymes were then added to the assay buffer. Reactions were incubated at 37 °C for 10 min and quenched by the addition of 0.5 ml of 0.1 N HCl in methanol, 1.5 ml of chloroform, and 3.0 ml of 1 m MgCl2. After extraction, the resulting lipid products were resuspended in chloroform. A Whatman silica gel 60 plate was activated by baking for 1 h at 180 0F under vacuum conditions. The TLC plate was then spotted with the resuspended lipids, dried for 10 min in a fume hood, and developed in a chamber containing chlorofom/methanol/glacial acetic acid (65:25:8). The plate was exposed to a storage phosphor screen and analyzed using a Typhoon 9410 phosphorimager (GE Healthcare).
A malachite green assay was used to quantitatively measure the phosphatase activity (21). The enzyme reactions were carried out in 20 μl of 50 mm sodium acetate, 25 mm bis-Tris, 25 mm Tris, 2 mm dithiothreitol, and indicated amounts of PGP. The pH of assay buffer for each enzyme was described above. Reactions were initiated by the addition of enzyme followed by incubation at 30 °C. Reactions were subsequently terminated by the addition of 20 μl of 0.1 m N-ethylmaleimide and spun down to sediment the lipid aggregates. Twenty-five microliters of the supernatant was added to 50 μl of malachite green reagents and vortexed. Absorbance was measured at 620 nm. The recombinant proteins were free of inorganic phosphate. The nonenzymatic hydrolysis of the substrate was corrected by measuring the increase in the absorbance without the addition of the enzymes.
32P-labeling of Yeast Cells and Lipid Extraction
The plasmids for complementation assays were assembled by inserting the PCR products of the PTPMT1 orthologs with a C-terminal FLAG tag into the pRS415-GPD vector (ATCC). GEP4 knock-out yeast strains are derivatives of S288C obtained from OpenBiosystems (22). After verification by gene-specific PCR, gep4Δ cells were transformed with PTPMT1-encoding plasmids. Cells were grown in YPD at 30 °C until mid-log phase followed by the addition of radiolabeled 32P to the concentration of 10 μCi/ml for 12 h. Lipids were isolated as previously described (23). Briefly, cells were resuspended in solution containing 0.5 m NaCl in 0.1 N HCl and vortexed with glass beads. Lipids were extracted, dried under vacuum, and separated via TLC. For visualizing cardiolipin, a Whatman silical gel 60 plate was used and prepared as described above. The separating solution used was chloroform/methanol/ammonium hydroxide/water (130:75:2:6). For visualizing PGP, a HPTLC silica gel 60 plate was used and the separating solution used was ethyl acetate/isopropanol/ethanol/ammonium hydroxide/water (12:36:12:7.2:28.8) (24). Individual species were identified by co-migration of nonradiolabeled standards. The relative abundance of PGP and CL were determined by using the Typhoon 9410 Variable Mode Imager and ImageQuant TL software, and were statistically analyzed using a Student's t test.
Yeast Complementation Assay
The gep4Δ cells were transformed and grown at 30 °C to mid-log phase as described above. Cells were counted and serially diluted 5-fold. Each dilution was spotted onto either YPD plates or Synthetic Completed Medium with Dextrose (SCD) in the presence of 25 μg/ml ethidium bromide. Plates were incubated for 2 days at the indicated temperatures. For Western blot analysis, 5 A600 of cells were pelleted. Proteins were precipitated with 50% trichloroacetic acid and resuspended in SDS/PAGE loading buffer. Monoclonal anti-Flag antibody was used for analysis (Sigma).
Mitochondrial Fractionation
500 ml cultures of gep4Δ cells transformed with plasmids encoding either the D. melanogaster or R. baltica PTPMT1 were grown in YPD at 30 °C to A600 of 2. The purified mitochondria were isolated as previously described (25). Briefly, cells were resuspended in sorbitol buffer, converted to spheroblasts by zymolase treatment, and homogenized. Crude mitochondria were isolated via a low-speed centrifugation (1,500 × g) followed by a high-speed spin (12,000 × g). Further purification was achieved by histodenz gradient centrifugation.
Immunofluorescence Analysis
The plasmid for the immunofluorescence assay was constructed by ligating the PCR product of R. baltica PTPMT1 into the pEGFP-N1 vector. The plasmid was transfected into COS1 cells using Fugene 6 (Roche). MitoTracker Red was added to cells in DMEM to a concentration of 100 nm and incubated at 37 °C for 30 min before fixation with 4% paraformaldehyde and permeabilization with ice-cold acetone.
RESULTS
Identification of a Putative PTPMT1 Ortholog in R. baltica
PTPMT1 is a member of the DSP family and its putative orthologs have been identified in many species of mammals, plants, protists, and even bacteria (26). Among the bacterial sequences identified by searching the NCBI database, the R. baltica and Blastopirellula marina proteins display the closest homology to the Homo sapiens PTPMT1, with 48 and 46% sequence similarity, respectively (Fig. 1B). They also contain other PTPMT1-like features such as a predicted transmembrane domain, an active site CX5R motif with basic residues, and a WPD loop shown to be important for the formation of the thiophosphoryl intermediate and its subsequent hydrolysis (Fig. 1B) (18). Notably, Rhodopirellula and Blastopirellula belong to the bacterial phylum Planctomycetes and contain many unique eukaryote-like features, such as intracellular compartmentalization (27, 28). Some studies have suggested that planctomycetes might be an evolutionary intermediate in the transition from prokaryotes to eukaryotes (29). Because of the conserved requirement for CL in both bacteria and eukaryotes, it is of interest to determine whether PTPMT1 orthologs in planctomyocytes are functional PGP phosphatases.
To obtain insight into the sequence homology of PTPMT1-like phosphatases, we built a phylogenetic tree of human DSPs (Fig. 1C). Homo sapiens PTPMT1 clustered distantly from phosphatases that are known to dephosphorylate proteins, such as the MAP kinase phosphatases (MKP) (30). Interestingly, Rhodopirellula and Blastopirellula PTPMT1 orthologs grouped relatively close to human PTPMT1. However, because the phylogenetic tree does not guarantee orthology or functional homology, we sought to characterize the enzymatic activity of the PTPMT1-like phosphatase in bacteria.
The Rhodopirellula PTPMT1 Dephosphorylates PGP in Vitro
To test the functional conservation of PTPMT1 from diverse species, we compared the phosphatase activities of the R. baltica, D. melanogaster, and M. musculus orthologs. Each gene was cloned into bacterial expression vectors, expressed, and affinity purified. The pH/rate profiles of these recombinant proteins against the artificial phosphotyrosine analog pNPP were obtained. The catalytic activities of M. musculus, D. melanogaster, and R. baltica PTPMT1 variants were most efficient at pH 5.5, 7.0, and 8.5, respectively (data not shown). Subsequently, the specific activities of all orthologs were measured at their respective optimum pH using pNPP as the substrate (Fig. 2A). Although the M. musculus and D. melanogaster PTPMT1s were more active, the bacterial variant still possessed measurable phosphatase activity against pNPP. As expected, active site cysteine to serine mutants (CS) were catalytically inactive. The specific activity of PTPMT1 orthologs against pNPP was relatively low compared with other PTPs that dephosphorylate proteins, such as dPTP61F and VHR (19). However, the inability to efficiently dephosphorylate pNPP is shared by other lipid phosphatases, like PTEN and myotubularins (19).
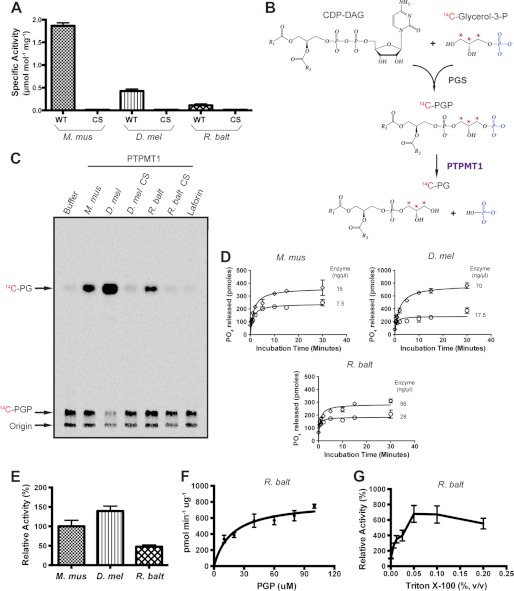
FIGURE 2.
Analysis of bacterial PTPMT1 activity in vitro. A, specific activities of recombinant murine (M. mus), fly (D. mel), and bacterial (R. balt) PTPMT1 are measured using pNPP as substrate. Data were calculated from the change in absorbance at 410 nm and represented as the mean ± S.D. of triplicate measurements. CS, catalytically inactive mutant. B, synthesis of 14C-PGP. Highlighted in blue is the phosphate group cleaved by PTPMT1 and marked with red asterisks are the radiolabled carbons. C, activities of wild-type and catalytically inactive PTPMT1 orthologs against PGP. Lipid products from the reactions were separated by TLC and analyzed by autoradiography. A trace amount of PGP phosphatase activity is seen in the background, because a contaminant E. coli PGP phosphatase co-purified with the recombinant PGS proteins (18). D, phosphatase assay conditions are determined for each indicated enzyme by measuring the amount of free phosphate released using the malachite green reagent. The reaction was carried out at 30 °C and in the presence of 100 μm PGP. Enzyme concentrations are indicated in the insets. E, comparison of enzymatic activities. Phosphatase activity was calculated under initial linear rate conditions (30 °C, 100 μm PGP, and 30 s of incubation time), compared with that of M. mus PTPMT1, and represented as the mean ± S.D. from three independent experiments. F, saturation curve of the Rhodopirellula PTPMT1 ortholog. The phosphatase assay was carried out under initial linear rate conditions and in the presence of various PGP concentrations. A nonlinear regression analysis was performed to indicate the saturation. G, effect of Triton X-100 on Rhodopirellula PTPMT1 PGP phosphatase activity.
We next tested whether these proteins could dephosphorylate PGP, the physiological substrate of mammalian PTPMT1. We enzymatically synthesized radiolabeled PGP by incubating sn-[U-14C]glycerol-3-phosphate with CDP-DAG in the presence of purified recombinant PGP synthase from E. coli (Fig. 2B) (18, 20). The resulting [14C]PGP was then extracted and incubated with each PTPMT1 ortholog. The reaction components were separated by thin-layer chromatography (TLC) and visualized by autoradiography (Fig. 2C). Non-radiolabeled PG standards were observed to migrate toward the middle of the TLC plate, whereas PGP stayed close to the origin (data not shown). Like the M. musculus enzyme, Drosophila, and Rhodopirellula PTPMT1 successfully converted PGP to PG (Fig. 2C). Their active site CS mutants along with the glucan phosphatase Laforin, displayed no activity toward PGP.
To further support this finding, we compared the PGP phosphatase activities of M. musculus, D. melanogaster, and R. baltica PTPMT1 orthologs. Determination of enzyme kinetic parameters was hindered by the lack of unlabeled water-soluble substrates. Nonetheless, we examined the activity of each enzyme by measuring the amount of phosphate released in the reaction through a malachite green assay (21). The initial linear rate was first determined by varying enzyme concentration and incubation times (Fig. 2D). After establishing the initial rate condition where less than 10% of PGP was dephosphorylated during the course of the reaction, we calculated the phosphatase activities of all three enzymes. Compared with M. musculus PTPMT1, the Drosophila protein was slightly more active, 139% of the mouse enzyme value (Fig. 2E). Whereas the R. baltica phosphatase contained considerable activity toward PGP, it was less efficient (∼50%) than mouse PTPMT1. These results are consistent with the data shown in Fig. 2C, supporting the notion that PTPMT1 orthologs in Drosophila and Rhodopirellula dephosphorylate PGP in vitro.
To further investigate the phosphatase activity of the Rhodopirellula PTPMT1 ortholog, we determined saturating substrate levels by incubating the enzyme with various concentrations of PGP. As shown in Fig. 2F, the affinity of R. baltica phosphatase for PGP was well below 100 μm, indicating that the PGP concentration utilized in the initial rate determination was not limiting.
Previous studies of E. coli and yeast PGP phosphatases have demonstrated that detergents, in particular Triton X-100, significantly stimulate their in vitro enzymatic activities (31, 32). To examine the effect of Triton X-100 on the Rhodopirellula PTPMT1 ortholog, we performed phosphatase assays in the presence of increasing amounts of the detergent. The addition of Triton X-100 substantially increased the PGP phosphatase activity of Rhodopirellula PTPMT1 (Fig. 2G).
The Rhodopirellula PTPMT1 Functions as a PGP Phosphatase in Vivo
To further validate that the Rhodopirellula and Drosophila PTPMT1 function as PGP phosphatases, we utilized an in vivo yeast complementation system. A PTPMT1 ortholog is not found in fungi (26). However, Osman et al. reported that the mitochondrial matrix protein GEP4 possesses PGP phosphatase activity in Saccharomyces cerevisiae (24). Deletion of the GEP4 gene results in an increase of PGP along with a reduction of CL levels. Therefore, we complemented GEP4-knock-out yeast with a control plasmid or plasmids encoding either wild type or catalytically inactive forms of PTPMT1 orthologs. The expression of those PTPMT1 orthologs was confirmed by Western blot analysis using anti-FLAG M2 antibody (Fig. 3A).
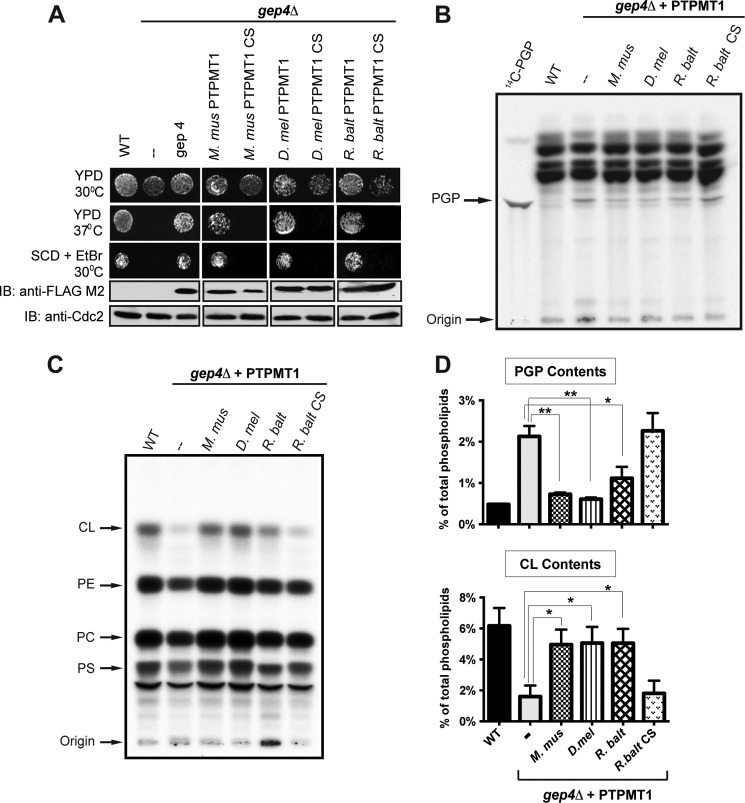
FIGURE 3.
Bacterial PTPMT1 functionally compensates for loss of yeast gep4 in vivo. A, wild-type murine, fly, and bacterial PTPMT1 complement the growth deficiency of GEP4-null cells. The expression levels of FLAG-tagged PTPMT1s and GEP4 are indicated by Western blot analysis. YPD, yeast extract-peptone-glucose medium; SCD, synthetic completed medium with dextrose; EtBr, ethidium bromide. CDC2 levels are shown as the loading control. Detection of steady state PGP (B) and CL (C) levels in gep4Δ yeast cells complemented with control plasmid or plasmids encoding PTPMT1 orthologs. Cells were labeled with [32P]orthophosphate at 10μCi/ml for 12 h. Lipids were extracted, separated via TLC, and viewed by autoradiography. D, PGP and CL contents are determined by the ratio of 32P incorporated into PGP versus total phospholipids, and represented as the mean ± S.D. from three independent experiments. **, p ≤ 0.01; *, p ≤ 0.05.
Yeast cells that lack cardiolipin synthase (Crd1p) and PGP synthase (Pgs1p) have respectively been shown to display significant growth deficiency at elevated temperatures or when cultured on media containing ethidium bromide, an agent that induces loss of mitochondrial DNA and defects in the cell wall (33–35). Notably, gep4Δ cells also exhibit growth deficiency under these stress conditions (Fig. 3A) (24). If PTPMT1 orthologs function as PGP phosphatases, they should rescue this GEP4-knock-out phenotype. Indeed, expression of WT PTPMT1 orthologs effectively restored normal growth, whereas expression of their catalytically inactive CS mutants failed to complement the loss of GEP4 (Fig. 3A). Our results indicate that PTPMT1 orthologs from both Drosophila and bacteria serve as functional equivalents of GEP4 in yeast.
To examine the steady-state levels of PGP and CL in yeast, total lipids were extracted from [32P]orthophosphate labeled cells and separated by TLC. As previously reported, a detectable amount of PGP has been observed in gep4Δ cells, while it was barely measurable in wild type cells (Fig. 3B) (24). To further quantify changes in CL levels, we measured the percentage of radioactivity incorporated into phospholipids and subsequently determined the ratio of 32P in PGP versus total phospholipids. Loss of GEP4 led to a significant accumulation of PGP, from 0.48% in wild type cells to 2% in gep4Δ cells (Fig. 3D, top panel). More importantly, the individual complementation of mouse, fly, or bacterial PTPMT1 in gep4Δ cells alleviated the accumulation of PGP (Fig. 3B and top panel of Fig. 3D). In contrast, the CS mutant of R. baltica PTPMT1 was unable to lower PGP levels (Fig. 3, B and D), consistent with their in vitro PGP phosphatase activities. Furthermore, the expression of PTPMT1 orthologs restored the CL deficiency in gep4Δ cells (Fig. 3C and lower panel of Fig. 3D). Conversely, the CS mutant of R. baltica PTPMT1 failed to rescue the defect. Taken together, these results indicate that the bacterial and fly PTPMT1 possess PGP phosphatase activity in vivo and are crucially involved in maintaining CL level.
The Rhodopirellula PTPMT1 Localizes to the Mitochondrion of Eukaryotes
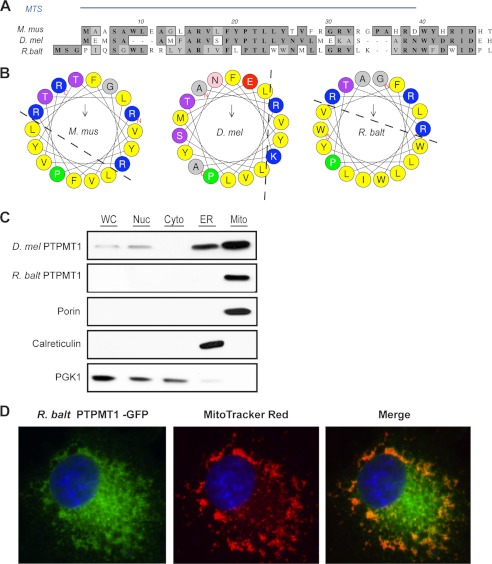
To compensate for the loss of GEP4 in yeast, bacterial PTPMT1 had to be correctly localized to the mitochondria to access its substrate, PGP. The proper targeting of newly synthesized proteins to the mitochondria is often mediated by a mitochondrial targeting sequence (MTS) comprised of 20–80 amino acids located at the N terminus of the protein (36). Although these peptide signals do not have a conserved primary sequence, they are generally enriched with the positively charged amino acid arginine, have a high percentage of leucine and serine, and contain few if any acidic residues (37, 38). The spacing between the residues often allows the formation of an amphiphilic α-helix, with positively charged and hydrophobic residues on the opposite faces. It has been shown that the first 37 amino acids of M. musculus PTPMT1 encompass a MTS (8). When fused to a Green Fluorescent Protein (GFP), these 37 amino acids direct the fluorescent signal exclusively to the mitochondrion. Therefore, we compared the N-terminal regions of D. melanogaster, and R. baltica PTPMT1 orthologs to that of the mouse protein (Fig. 4A). Notably, many basic and hydrophobic residues are conserved among PTPMT1 orthologs. We next plotted the amino acids comprising the regions potentially forming amphiphilic helices onto a helical wheel (Fig. 4B). In both M. musculus and R. baltica PTPMT1, the positive charges grouped on one side, and the hydrophobic residues clustered on the other, thus fitting the MTS model described above.
FIGURE 4.
Mitochondrial localization of the Rhodopirellula PTPMT1. A, N-terminal sequences of PTPMT1 orthologs are aligned using PROMALS3D. Amino acid similarities are shaded in light gray and identities in dark gray. The MTS of mouse PTPMT1 has been previously characterized (8) and is indicated by the blue bar. B, N-terminal region of bacterial PTPMT1 forms an amphiphilic helix. 18 amino-terminal residues were plotted on the helical wheel using HeliQuest (51). Basic residues are colored blue, and hydrophobic residues are colored yellow. C, subcellular fractionation of yeast gep4Δ cells expressing fly or bacterial PTPMT1. 30 μg of protein from each fraction were analyzed by SDS-PAGE and immunoblotting. Porin, Calreticulin, and PGK1 serve as marker proteins for mitochondria, ER, and cytosol, respectively. D, bacterial PTPMT1 localizes to the mitochondria of mammalian COS-1 cells. 24 h post-transfection, cells were stained with MitoTracker Red, fixed, and analyzed by immunofluoresence imaging.
To determine whether the Rhodopirellula PTPMT1 resides in the mitochondria in an in vivo setting, we cultured gep4Δ yeast cells that were complemented with PTPMT1 orthologs and performed subcellular fractionation. As shown in Fig. 4C, both the R. baltica and Drosophila PTPMT1s are highly enriched in the histodenz gradient-purified mitochondria. The purity of each fraction was further validated by the presence of the mitochondrial marker Porin, the endoplasmic reticulum (ER) marker Calreticulin, as well as the cytoplasma marker Phosphoglycerate Kinase 1/Pgk1. The localization of a bacterial protein to the yeast mitochondria likely indicates the conservation of the MTS and is consistent with the bacterial origin of mitochondria. We then postulated that this MTS would behave similarly in mammalian cells. To test our hypothesis, a C-terminal GFP-tagged bacterial PTPMT1 was introduced into primate COS-1 cells. Mitochondria were subsequently labeled with MitoTracker Red and the cells were fixed for immunofluorescence analysis. A substantial amount of the R. baltica PTPMT1 targeted to the mitochondria as indicated by the co-localization of the two fluorescent signals (Fig. 4D). Together, our results suggest that the R. baltica PTPMT1 contains a conserved MTS-like sequence, even though R. baltica does not have mitochondria. It is not known whether R. baltica has any CL-enriched lipid domain, particularly in its internal membrane structures.
DISCUSSION
Raetz et al. previously identified three distinct bacterial PGP phosphatases, pgpA, pgpB, and pgpC in E. coli, which do not share any significant domain structure and sequence homology with PTPMT1 (Fig. 1D) (39–41). A pgpA, pgpB, or pgpC-like phosphatase was not found in R. baltica (Fig. 1E). The yeast PGP phosphatase GEP4 belongs to the larger haloacid dehalogenase (HAD)-like family and contains a conserved DXDX(T/V) motif in the active site (Fig. 1D) (24, 42). Orthologs of GEP4 exist in fungi and plants, as well as several protists, but not in bacteria (9, 24). Our analysis revealed that the prokaryotic R. baltica utilizes a PTPMT1-like PGP phosphatase, as opposed to using those existent in yeast (GEP4) or E. coli (pgpA, pgpB, and pgpC) to dephosphorylate PGP, suggesting an important role of this phosphatase in Rhodopirellula.
R. baltica is a marine bacterium whose genome is first sequenced in 2003 (43). Cultivable stains have not been isolated until recently (44). As such, the phospholipid composition and CL abundance in this organism are presently unknown. Interestingly, our bioinformatics search of Rhodopirellula genome identified orthologs of PGP synthase (PGS) and CL synthase (CLS) with confidence (Fig. 1E). PGS is the first enzyme of CL de novo synthesis which facilitates the nucleophilic attack of a glycerol-3-phosphate (G3P) on CDP-diacylglycerol (CDP-DAG) to generate PGP (Fig. 1A) (10). Following the action of PGP phosphatase, CLS converts PG into CL. The presence of all three enzymes of CL de novo synthesis in Rhodopirellula genome likely indicates active CL biosynthesis in this organism. To date, genetic manipulation of Rhodopirellula genome has not been achieved. Using a yeast mutant strain lacking the PGP phosphatase GEP4, we demonstrated that R. baltica PTPMT1 complemented the loss of GEP4, and the rescue is dependent on its phosphatase activity (Fig. 3A), suggesting that Rhodopirellula PTPMT1 is a functional PGP phosphatase in vivo.
R. baltica belongs to the bacterial phylum Planctomycetes. Like other plantomycetes, it possesses distinctive features that are uncommon among bacteria, such as peptidoglycan-less cell wall, complex internal structures, and partial compartmentalization. It also encodes endocytosis-like protein uptake and membrane coat-like proteins (27, 28). The unique eukaryote-like features of planctomycetes challenge the current hypothesis regarding the origin of eukaryotic organelles (27). Because of their complex endomembrane system, Planctomycyetes together with Verrucomicrobia and Chlamydiae form the PVC superphylum in eubacteria and are thought to represent the intermediates of prokaryote and eukaryote evolution (29). Notably, PTPMT1-like sequences were also found in two species of Chlamydiae, Simkania negevensis and Candidatus Protochlamydia amoebophila. Our results suggested that Rhodopirellula contains a unique mammalian-type PGP phosphatase that could potentially play an important role in the organism CL metabolism.
Ectopically expressed R. baltica PTPMT1 was localized to the mitochondria of yeast and mammalian cells (Fig. 4, C and D), suggesting the existence of a conserved region that could function as a MTS in eukaryotes. In mammals, the mitochondrial proteome contains over 1000 proteins, of which the majority are encoded in the nucleus and subsequently transported into the mitochondria (45). Many imported proteins have a characteristic MTS composed of amphiphilic α-helix forming amino acids and recognized by the Translocase of the Outer Membrane (TOM) and Translocase of the Inner Membrane (TIM) complexes of mitochondria (46). Our sequence alignment and helical wheel analysis revealed that the N terminus of R. baltica PTPMT1 contains several conserved positively charged and hydrophobic amino acids, which would form an amphiphilic helix (Fig. 4A). Interestingly, a number of bacterial proteins have been shown to contain functional MTS when expressed in mammalian cells, including the enteropathogenic E. coli effectors EspF and Map (47). The existence of MTS in bacterial proteins may reflect an evolutionarily conserved mechanism for selective protein transport and domain organization.
In summary, we have identified a mammalian-type PGP phosphatase in the bacterium R. baltica. We performed biochemical and gene complementation assays to subsequently validate our initial bioinformatic search. The ability of the bacterial phosphatase to target to eukaryotic mitochondria suggests the existence of a conserved mitochondrial targeting sequence despite the absence of mitochondrion in bacteria. The R. baltica PTPMT1 may function at the organization center for enzymes involved in the biosynthesis of CL in this organism.
Acknowledgments
We thank Drs. Gregory Taylor, David Pagliarini, Matthew Gentry, Sandra Wiley, Claudia Kent, and members of the Dixon laboratory for reagents, technical assistance, and discussion.
This work was supported by Grants DK18024 and DK18849 (to J. E. D.), HG004164 (to G. M.) from the National Institutes of Health, the Barth Syndrome Foundation research grant and the American Heart Association Scientist Development Grant #12SDG12070058 (to J. Z.).
Cardiolipin analogues are found in Archaea but contain unique diphytanylglycerol diether lipid, in contrast to the largely diacylglycerol-derived membrane lipids in eukaryotes and bacteria (48).
- PTP
- protein tyrosine phosphatase
- DSP
- dual specificity phosphatase
- PTPMT1
- PTP localized to mitochondrion 1
- PGP
- phosphatidylglycerophosphate
- PG
- phosphatidylglycerol
- CL
- cardiolipin.
REFERENCES
- 1. Hunter T. (1987) A thousand and one protein kinases. Cell 50, 823–829 [DOI] [PubMed] [Google Scholar]
- 2. Alonso A., Sasin J., Bottini N., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 [DOI] [PubMed] [Google Scholar]
- 3. Tonks N. K., Neel B. G. (2001) Combinatorial control of the specificity of protein tyrosine phosphatases. Curr. Opin. Cell Biol. 13, 182–195 [DOI] [PubMed] [Google Scholar]
- 4. Fauman E. B., Saper M. A. (1996) Structure and function of the protein tyrosine phosphatases. Trends Biochem. Sci 21, 413–417 [DOI] [PubMed] [Google Scholar]
- 5. Tagliabracci V. S., Turnbull J., Wang W., Girard J. M., Zhao X., Skurat A. V., Delgado-Escueta A. V., Minassian B. A., Depaoli-Roach A. A., Roach P. J. (2007) Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 19262–19266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Worby C. A., Gentry M. S., Dixon J. E. (2006) Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 281, 30412–30418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maehama T., Dixon J. E. (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 8. Pagliarini D. J., Wiley S. E., Kimple M. E., Dixon J. R., Kelly P., Worby C. A., Casey P. J., Dixon J. E. (2005) Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta cells. Mol Cell 19, 197–207 [DOI] [PubMed] [Google Scholar]
- 9. Zhang J., Guan Z., Murphy A. N., Wiley S. E., Perkins G. A., Worby C. A., Engel J. L., Heacock P., Nguyen O. K., Wang J. H., Raetz C. R., Dowhan W., Dixon J. E. (2011) Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab 13, 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlame M. (2008) Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 49, 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cronan J. E. (2003) Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57, 203–224 [DOI] [PubMed] [Google Scholar]
- 12. Tan B. K., Bogdanov M., Zhao J., Dowhan W., Raetz C. R., Guan Z. (2012) Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 16504–16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pei J., Tang M., Grishin N. V. (2008) PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 36, W30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchler-Bauer A., Anderson J. B., Cherukuri P. F., DeWeese-Scott C., Geer L. Y., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Marchler G. H., Mullokandov M., Shoemaker B. A., Simonyan V., Song J. S., Thiessen P. A., Yamashita R. A., Yin J. J., Zhang D., Bryant S. H. (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33, D192–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., Gasteiger E., Bairoch A., Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao J., Engel J. L., Zhang J., Chen M. J., Manning G., Dixon J. E. (2011) Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 11860–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor G. S., Maehama T., Dixon J. E. (2000) Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. U.S.A. 97, 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dowhan W. (1992) Phosphatidylglycerophosphate synthase from Escherichia coli. Methods Enzymol. 209, 313–321 [DOI] [PubMed] [Google Scholar]
- 21. Maehama T., Taylor G. S., Slama J. T., Dixon J. E. (2000) A sensitive assay for phosphoinositide phosphatases. Anal. Biochem. 279, 248–250 [DOI] [PubMed] [Google Scholar]
- 22. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 23. Chang S. C., Heacock P. N., Clancey C. J., Dowhan W. (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9829–9836 [DOI] [PubMed] [Google Scholar]
- 24. Osman C., Haag M., Wieland F. T., Brugger B., Langer T. (2010) A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glick B. S., Pon L. A. (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223 [DOI] [PubMed] [Google Scholar]
- 26. Pagliarini D. J., Worby C. A., Dixon J. E. (2004) A PTEN-like phosphatase with a novel substrate specificity. J. Biol. Chem. 279, 38590–38596 [DOI] [PubMed] [Google Scholar]
- 27. Fuerst J. A., Sagulenko E. (2011) Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413 [DOI] [PubMed] [Google Scholar]
- 28. Lindsay M. R., Webb R. I., Strous M., Jetten M. S., Butler M. K., Forde R. J., Fuerst J. A. (2001) Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175, 413–429 [DOI] [PubMed] [Google Scholar]
- 29. Devos D. P., Reynaud E. G. Evolution (2010) Intermediate steps. Science 330, 1187–1188 [DOI] [PubMed] [Google Scholar]
- 30. Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 31. Chang Y. Y., Kennedy E. P. (1967) Phosphatidyl glycerophosphate phosphatase. J. Lipid Res. 8, 456–462 [PubMed] [Google Scholar]
- 32. Kelly B. L., Greenberg M. L. (1990) Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1046, 144–150 [DOI] [PubMed] [Google Scholar]
- 33. Janitor M., Subik J. (1993) Molecular cloning of the PEL1 gene of Saccharomyces cerevisiae that is essential for the viability of petite mutants. Curr Genet 24, 307–312 [DOI] [PubMed] [Google Scholar]
- 34. Jiang F., Ryan M. T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M. L. (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 [DOI] [PubMed] [Google Scholar]
- 35. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 36. Omura T. (1998) Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 123, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 37. Roise D., Horvath S. J., Tomich J. M., Richards J. H., Schatz G. (1986) A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 5, 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. von Heijne G. (1986) Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5, 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Icho T., Raetz C. R. (1983) Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J. Bacteriol. 153, 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Funk C. R., Zimniak L., Dowhan W. (1992) The pgpA and pgpB genes of Escherichia coli are not essential: evidence for a third phosphatidylglycerophosphate phosphatase. J. Bacteriol. 174, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu Y. H., Guan Z., Zhao J., Raetz C. R. (2011) Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 286, 5506–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Collet J. F., Stroobant V., Pirard M., Delpierre G., Van Schaftingen E. (1998) A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J. Biol. Chem. 273, 14107–14112 [DOI] [PubMed] [Google Scholar]
- 43. Glöckner F. O., Kube M., Bauer M., Teeling H., Lombardot T., Ludwig W., Gade D., Beck A., Borzym K., Heitmann K., Rabus R., Schlesner H., Amann R., Reinhardt R. (2003) Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. U.S.A. 100, 8298–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winkelmann N., Harder J. (2009) An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J. Microbiol. Methods 77, 276–284 [DOI] [PubMed] [Google Scholar]
- 45. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Habib S. J., Neupert W., Rapaport D. (2007) Analysis and prediction of mitochondrial targeting signals. Methods Cell Biol. 80, 761–781 [DOI] [PubMed] [Google Scholar]
- 47. Papatheodorou P., Domańska G., Oxle M., Mathieu J., Selchow O., Kenny B., Rassow J. (2006) The enteropathogenic Escherichia coli (EPEC) Map effector is imported into the mitochondrial matrix by the TOM/Hsp70 system and alters organelle morphology. Cell Microbiol. 8, 677–689 [DOI] [PubMed] [Google Scholar]
- 48. Corcelli A. (2009) The cardiolipin analogues of Archaea. Biochim. Biophys. Acta 1788, 2101–2106 [DOI] [PubMed] [Google Scholar]
- 49. Pei J., Kim B. H., Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 51. Gautier R., Douguet D., Antonny B., Drin G. (2008) HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102 [DOI] [PubMed] [Google Scholar]