Abstract
Background
To assess the association between MTHFR polymorphism and cervical cancer risk, a meta-analysis was performed.
Methods
Based on comprehensive searches of the PubMed, Embase, and Web of Science databases, we identified outcome data from all articles estimating the association between MTHFR polymorphism and cervical cancer risk. The pooled odds ratio (OR) with 95% confidence intervals (CIs) were calculated.
Results
A total of 12 studies with 2,924 cases (331 cervical intraepithelial neoplasia (CIN) I, 742 CIN II/III, 1851 invasive cervical cancer) and 2,581 controls were identified. There was no significant association between MTHFR C677T polymorphism and CIN I risk (T vs. C, OR = 1.10, 95% CI = 0.92–1.31; TT vs. CC, OR = 1.14, 95% CI = 0.78–1.68; TT+CT vs. CC, OR = 1.22, 95% CI = 0.94–1.58; TT vs. CT+CC, OR = 0.99, 95% CI = 0.70–1.40). For the CIN II/III, lack of an association was also found (T vs. C, OR = 1.08, 95% CI = 0.95–1.23; TT vs. CC, OR = 1.15, 95% CI = 0.87–1.52; TT+CT vs. CC, OR = 1.13, 95% CI = 0.94–1.35; TT vs. CT+CC, OR = 1.07, 95% CI = 0.83–1.38). The T allele had significant association to susceptibility of invasive cervical cancer in recessive model (TT vs. CT+CC, OR = 1.23, 95% CI = 1.02–1.49). On subgroup analysis by ethnicity, similarly significant differences in T vs. C, TT vs. CC, and recessive model were found in Asians.
Conclusion
The present meta-analysis suggested that MTHFR C677T polymorphism were to substantially contribute to invasive cervical cancer in recessive model.
Introduction
Cervical cancer continues a serious threat to women throughout the world [1]. As the third most common cancer in women, it is estimated that there are nearly 530,232 new cases and 275,008 deaths die of cervical cancer in 2008 [2]. Epidemiological observations have established an aetiological association between human papillomavirus (HPV) infection and cervical cancer [3]–[4]. However, only a small percentage of infected women will ever develop cervical cancer. Therefore, infection with HPV alone is not sufficient for the development of cervical cancer and host genetic susceptibility, combined with lifestyle factors, may play a crucial role in exploring the progression of disease [5].
Methylenetetrahydrofolate reductase (MTHFR), a homodimeric enzyme, catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahyd- rofolate [6]. The enzyme plays a critical role in regulating the metabolism of folate and methionine, both being involved in DNA methylation and DNA synthesis required for normal development and growth [7]. The most common polymorphism, C-to-T transition at nucleotide 677 (C677T), is located on chromosome 1p36. This transition had been found to affect the catalytic domain of the MTHFR, thus reduce folate levels and elevate homocysteine levels [8]–[9]. Low folate levels may cause several cancers by influence DNA methylation and DNA synthesis [10]–[11]. Therefore, the MTHFR gene might be one of the candidate genes for susceptibility of cervical cancer.
A relatively large number of studies evaluated the association between MTHFR C677T polymorphism and cervical cancer risk. However, the MTHFR C677T polymorphism’s association with cervical cancer, or the lack thereof, remain inconclusive. To derive a more comprehensive and precise estimation of the relationship, we carried out a meta-analysis on all eligible case-control studies to estimate the effect of MTHFR polymorphism on the risk of cervical cancer.
Results
Study Characteristics
Twelve publications, including 2,924 cases (331 cervical intraepithelial neoplasia (CIN) I patients, 742 CIN II/III patients, 1851 invasive cervical cancer patients) and 2,581 controls, met the inclusion criteria [11]–[22]. A flowchart detailing the process for study identification and selection is shown in Fig. S1. The sample sizes ranged from 95 to 1546 patients (median 260.5, Interquartile range 161.5–777.5). Five of the 12 included studies evaluated the association between MTHFR C677T polymorphism and susceptibility of CIN I [12], [18], [20]–[22]. Six studies evaluated the association between MTHFR C677T polymorphism and susceptibility of CIN II/III [12], [14], [18], [20]–[22]. Eleven studies evaluated the association between MTHFR C677T polymorphism and susceptibility of cervical cancer [11]–[21]. The Newcastle-Ottawa Scale (NOS) scores ranged from 6 to 9, which indicated that the methodological quality was generally good. The genotype distribution in the controls of all studies was in agreement with Hardy-Weinberg equilibrium (HWE). The main characteristics of the studies were shown in Table 1.
Table 1. Association between individual study characteristics and MTHFR C677T polymorphism.
| Study | Country | Ethnicity | Genetic type | Mean age, yearcases/controls | CIN I | CIN II/III | Invasive cancer | Control | Scores | ||||||||
| CC | CT | TT | CC | CT | TT | CC | CT | TT | CC | CT | TT | ||||||
| Mostowska et al. | Poland | Caucasian | C677T | 54.6/53.3 | 56 | 59 | 9 | 69 | 81 | 18 | 9 | ||||||
| Prasad et al. | India | Mixed | C677T | NA/NA | 57 | 5 | 0 | 116 | 8 | 1 | 6 | ||||||
| Tong et al. | Kerea | Asian | C677T | 50.8/45.7 | 52 | 82 | 25 | 54 | 74 | 32 | 53 | 65 | 28 | 152 | 198 | 77 | 8 |
| Kohaar et al. | India | Caucasian | C677T | 49.4/48.2 | 28 | 11 | 0 | 113 | 47 | 4 | 161 | 65 | 5 | 7 | |||
| Nandan et al. | India | Mixed | C677T | NA/NA | 36 | 0 | 26 | 53 | 0 | 24 | 8 | ||||||
| Shekari et al. | India | Caucasian | C677T | 48.6/48.8 | 125 | 68 | 7 | 170 | 28 | 2 | 7 | ||||||
| Ma et al. | China | Asian | C677T | 52.5/50.6 | 20 | 53 | 38 | 33 | 60 | 18 | 7 | ||||||
| Kang et al. | Kerea | Asian | C677T | NA/NA | 27 | 32 | 20 | 30 | 32 | 12 | 7 | ||||||
| Zoodsma et al. | Netherlands | Caucasian | C677T | NA/NA | 27 | 21 | 6 | 121 | 120 | 23 | 357 | 230 | 49 | 273 | 262 | 57 | 8 |
| Sull et al. | Kerea | Asian | C677T | 50.3/46.2 | 10 | 22 | 8 | 50 | 90 | 36 | 73 | 115 | 58 | 153 | 221 | 80 | 7 |
| Lambropoulos et al. | Greece | Caucasian | C677T | 33.2/33.2 | 20 | 28 | 5 | 27 | 29 | 8 | 11 | 8 | 2 | 42 | 37 | 12 | 6 |
| Piyathilake et al. | USA | Mixed | C677T | 30.4/23.9 | 6 | 13 | 6 | 11 | 23 | 5 | 16 | 12 | 3 | 7 | |||
Abbreviations and definitions: CIN, cervical intraepithelial neoplasia; MTHFR, methylenetetrahydrofolate reductase; NA, not available.
The MTHFR C677T Polymorphism and CIN I Susceptibility
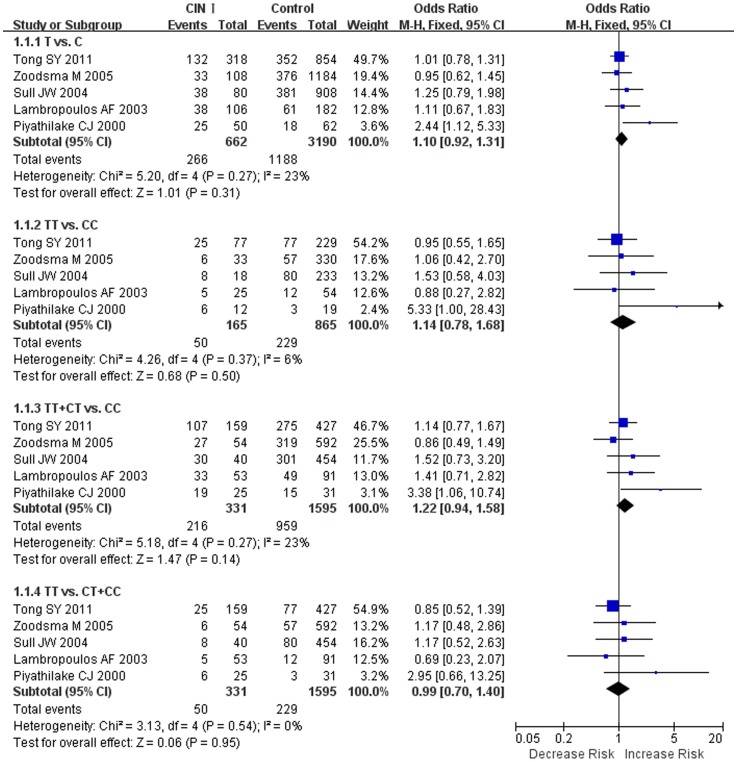
Fixed effects models were used to calculate the pooled OR in all genetic models. Overall, the combined results showed that no significant association was found in all genetic models (OR = 1.10, 95% CI = 0.92–1.31 for T vs. C, OR = 1.14, 95% CI = 0.78–1.68 for TT vs. CC, OR = 1.22, 95% CI = 0.94–1.58 for TT+CT vs. CC, and OR = 0.99, 95% CI = 0.70–1.40 for TT vs. CT+CC). Forest plots on the basis of all studies were shown in Fig. 1.
Figure 1. Forest plot of the overall risk of CIN I associated with the MTHFR C677T polymorphism.
No significant association was found between the MTHFR C677T polymorphism and CIN I risk in all genetic models. A, T vs. C; B, TT vs. CC; C, dominant genetic model; D, recessive genetic model. Error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR.
The MTHFR C677T Polymorphism and CIN II/III Susceptibility
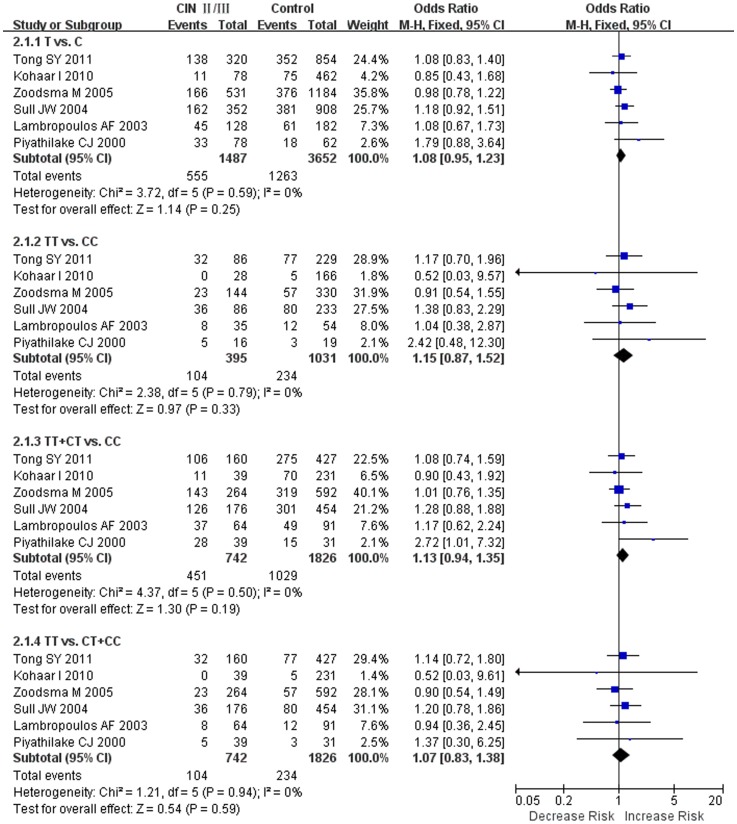
The results on the MTHFR C677T polymorphism indicated that the T allele had no significant association to CIN II/III susceptibility as compared to the C allele under the fixed effects models (Fig. 2). The results were as followed: T vs. C (OR = 1.08, 95% CI = 0.95–1.23), TT vs. CC (OR = 1.15, 95% CI = 0.87–1.52), TT+CT vs. CC (OR = 1.13, 95% CI = 0.94–1.35), TT vs. CT+CC (OR = 1.07, 95% CI = 0.83–1.38).
Figure 2. Forest plot of the overall risk of CIN II/III associated with the MTHFR C677T polymorphism.
No significant association was found between the MTHFR C677T polymorphism and of CIN II/III risk in all genetic models. A, T vs. C; B, TT vs. CC; C, dominant genetic model; D, recessive genetic model. Error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR.
The MTHFR C677T Polymorphism and Invasive Cervical Cancer Susceptibility
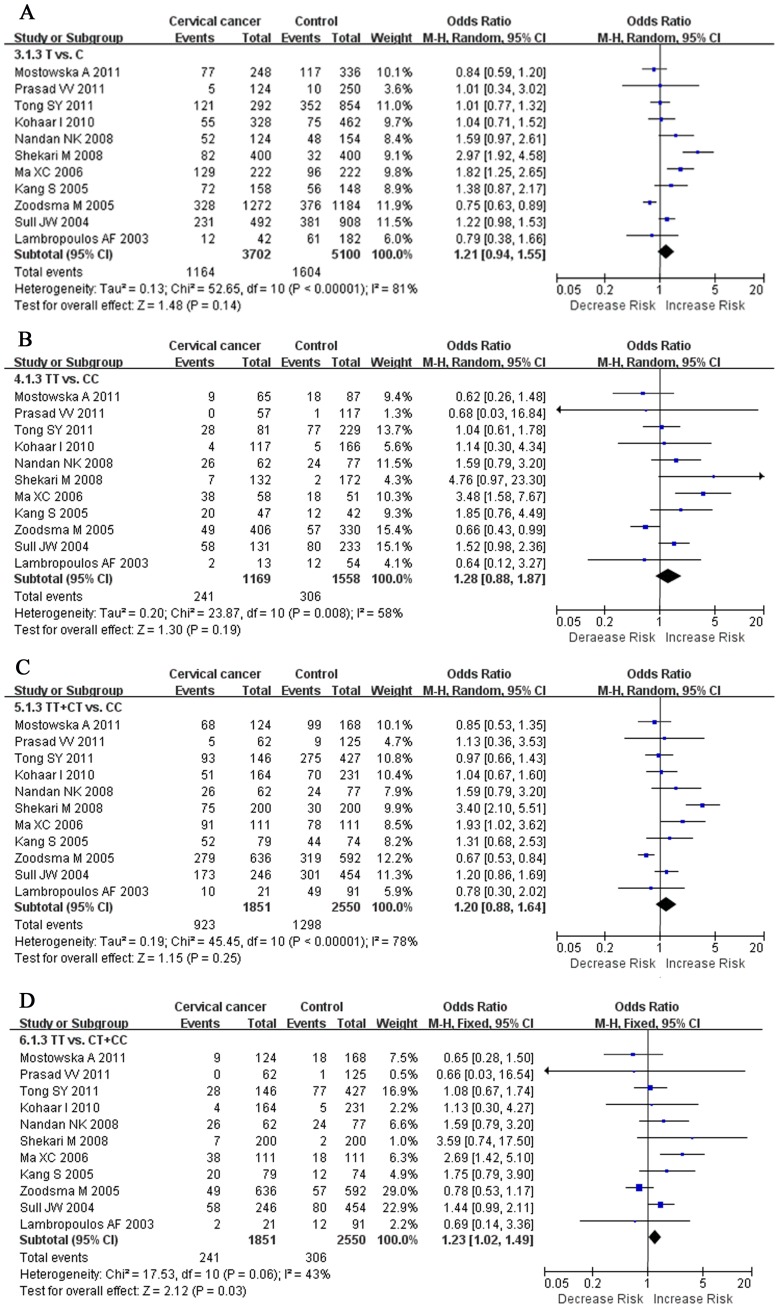
Fig. 3 showed that MTHFR C677T polymorphism was no significantly associated with invasive cervical cancer in T vs. C (OR = 1.21, 95%CI = 0.94–1.55), TT vs. CC, (OR = 1.28, 95% CI = 0.88–1.87), and TT+CT vs. CC (OR = 1.20, 95% CI = 0.88–1.64). The combined results showed significant differences in recessive model (TT vs. CT+CC, OR = 1.23, 95% CI = 1.02–1.49). When stratified by ethnicity, we observed a wide variation of T allele frequencies between the controls across different ethnicities. The result of One-way ANOVA indicated that the T allele frequencies were significant difference in Caucasians, Asians, and Mixed populations (P = 0.015). When meta-analysis was performed to assess association between MTHFR C677T polymorphism and different ethnicities, the T allele of MTHFR C677T polymorphism had significant association with invasive cervical cancer susceptibility in Asians. The results were showed in Table 2.
Figure 3. Forest plot of the overall risk of cervical cancer associated with the MTHFR C677T polymorphism.
Significant association was found between the MTHFR C677T polymorphism and cervical cancer risk in recessive genetic model. A, T vs. C; B, TT vs. CC; C, dominant genetic model; D, recessive genetic model. Error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR.
Table 2. Meta-analyses of MTHFR C677T polymorphism and risk of cervical cancer in each subgroup.
| Category | T vs. C | TT vs. CC | Dominant model | Recessive model | ||||
| OR(95%CI) | I 2(%) | OR(95%CI) | I 2 (%) | OR(95%CI) | I 2(%) | OR(95%CI) | I 2(%) | |
| Ethnicity | ||||||||
| Caucasian | 1.09(0.68–1.74) | 88 | 0.76(0.54–1.06) | 36 | 1.10(0.61–1.99) | 89 | 0.84(0.61–1.17) | 0 |
| Asian | 1.28(1.02–1.62) | 53 | 1.66(1.05–2.62) | 53 | 1.20(0.96–1.50) | 11 | 1.51(1.17–1.94) | 42 |
| Mixed | 1.48(0.94–2.31) | 0 | 1.53(0.78–3.01) | 0 | 1.45(0.80–2.62) | 0 | 1.53(0.77–3.01) | 0 |
| SA | 1.18(0.90–1.54) | 85 | 1.19(0.79–1.81) | 68 | 1.18(0.84–1.66) | 82 | 1.16(0.96–1.41) | 59 |
Abbreviations and definitions: CI, 95% confidence intervals; OR, odds ratio; SA: sensitivity analysis.
Heterogeneity Analysis
For the association between MTHFR C677T polymorphism and invasive cervical cancer susceptibility, there were statistically significant heterogeneity in T vs. C (I 2 = 81%, P Q<0.00001), TT vs. CC (I 2 = 61%, P Q = 0.005), dominant genetic model (I 2 = 78%, P Q<0.00001), and recessive genetic model (I 2 = 50%, P Q = 0.03).
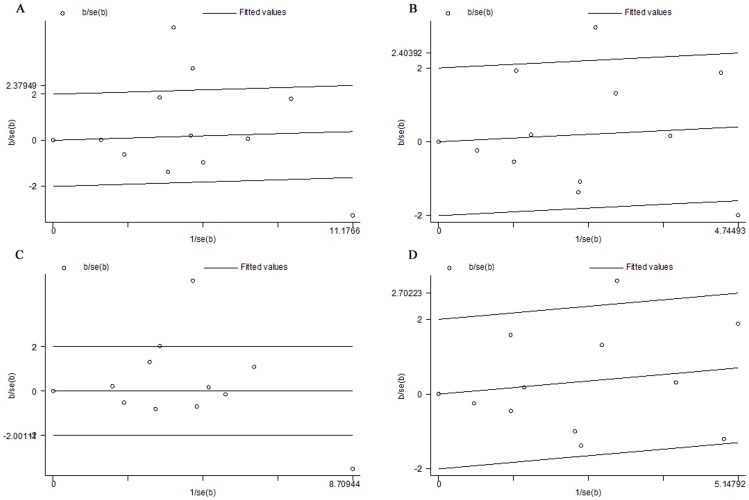
To explain the heterogeneity, Galbraith plots were performed in all genetic models. Galbraith plots [23] provide a graphical display to obtain a visual impression of the amount of heterogeneity from a meta-analysis. The position of each trial on the horizontal axis gives an indication of the weight allocated to it in a meta-analysis. The position on the vertical axis gives the contribution of each trial to the Q statistic for heterogeneity. In the absence of heterogeneity, we could expect all the points to lie within the confidence bounds (positioned 2 units over and below the regression line). In this meta-analysis, the three studies of Shekari M et al., Ma XC et al., and Zoodsma M et al. were outliers in the T vs. C and dominant genetic model (Fig. 4A, C). The two studies of Ma XC et al. and Zoodsma M et al. were outliers in the TT vs. CC (Fig. 4B). The study of Ma XC et al. was outliers in the recessive genetic model (Fig. 4D). When the studies of Shekari M et al., Ma XC et al., and Zoodsma M et al. were excluded respectively, all I 2 values were less than 50% and P Q were greater than 0.1 (Table 3). The significant of pooled OR showed significant differences in TT vs. CC (OR = 1.31, 95% CI = 1.01–1.69).
Figure 4. Galbraith plot of MTHFR C677T polymorphism and cervical cancer risk.
. A, The three studies of Shekari M et al., Ma XC et al., and Zoodsma M et al. were outliers in the T vs. C; B, The two studies of Ma XC et al. and Zoodsma M et al. were outliers in the TT vs. CC; C, The three studies of Shekari M et al., Ma XC et al., and Zoodsma M et al. were outliers in dominant genetic model; D, The study of Ma XC et al. was outliers in the recessive genetic model.
Table 3. Meta-analyses of MTHFR C677T polymorphism and cervical cancer susceptibility after omitting the studies.
| Polymorphism | OR (95% CI) | Z | P OR | I 2 (%) | P Q | Effect model |
| T vs. Ca | 1.11 (0.97, 1.26) | 1.55 | 0.12 | 6 | 0.38 | F |
| TT vs. CCb | 1.31 (1.01, 1.69) | 2.05 | 0.04 | 5 | 0.40 | F |
| TT+CT vs. CCa | 1.12 (0.95, 1.33) | 1.34 | 0.18 | 0 | 0.56 | F |
| TT vs. CT+CCc | 1.13 (0.92, 1.38) | 1.20 | 0.23 | 19 | 0.27 | F |
Abbreviations and definitions: CI, 95% confidence intervals; OR, odds ratio; P Q, P value of Q test for heterogeneity; F, fixed-effect models.
MTHFR C677T polymorphism and cervical cancer susceptibility after excluding the three studies of Shekari M et al., Ma XC et al., and Zoodsma M et al.
MTHFR C677T polymorphism and cervical cancer susceptibility after excluding the two studies of Ma XC et al. and Zoodsma M et al.
MTHFR C677T polymorphism and cervical cancer susceptibility after excluding the study of Ma XC et al.
Sensitivity Analysis
Robustness of our results with regard to different assumptions was examined by performing a sensitivity analysis. Sensitivity analysis was performed based on the high NOS score (≥7). Two studies with relatively low NOS score (<7) were excluded from the sensitivity analysis. The sensitivity analysis indicated the results of our meta-analysis were relatively consistent even when some studies were excluded. The results were shown in Table 2.
Publication Bias
Publication bias was estimated by the funnel plots. As shown in Fig. S2, the shape of the funnel plots revealed asymmetry in some degree due to the limited number of literatures. Then, Egger’s linear regression test was used to provide statistical evidence of funnel plots asymmetry. The result still did not suggest any evidence of publication bias.
Discussion
Worldwide study has indicated that folate levels show a protective role in a variety of cancers. Owing to the importance of MTHFR in maintaining folate homeostasis, the MTHFR C677T polymorphism has been investigated in certain types of cancer, which included Colorectal, Thyroid, Breast, Ovarian, and cervical cancers [24]. The association between MTHFR C677T polymorphism and cervical cancer risk was first reported in a mixed populations by Piyathilake et al [22]; however, as discussed above, conflicting data regarding the role of MTHFR in cervical cancer susceptibility and presentation have been reported by series of case-control studies [11]–[14], [16], [18]–[21]. Against this backdrop, we performed a meta-analysis to clarify the relationship between MTHFR C677T polymorphism and cervical cancer risk.
In this meta-analysis, 12 studies (5 subgroups for CIN I, 6 subgroups for CIN II/III, and 11 subgroups for invasive cervical cancer) on MTHFR C677T polymorphism were performed to provide the most comprehensive assessment of the relationship between polymorphism and cervical cancer risk. The T allele of MTHFR C677T polymorphism had no association with the CIN I susceptibility for the T vs. C, TT vs. CC, dominant genetic model, and recessive genetic model in overall populations. Lack of an association was also found in CIN II/III and cervical cancer. In view of the complex effect of genetic polymorphisms on disease progression, the lack of an association between MTHFR C677T polymorphism and invasive cervical cancer susceptibility may attribute to other polymorphisms in MTHFR gene promoter which could affect the activity of MTHFR. Ulvik A et al. [25] demonstrated that MTHFR A1298T polymorphism was associated with reduced MTHFR activity. Meanwhile, the MTHFR C677T and A1298T polymorphisms appeared to interact with folate in determining cancer risk. Strong correlation between MTHFR C677T and A1298T polymorphisms was observed in cervical dysplasia as compared to normal cervical cytology [26]. In current study, we also performed meta-analysis to identify the association between MTHFR A1298T polymorphism and cervical cancer risk. There was no association between MTHFR A1298T polymorphism and cervical cancer risk (Table S1 and S2). Thus, the interaction between gene and gene might influence the association of MTHFR gene polymorphism with cervical cancer risk.
To explore a more precise relationship between MTHFR C677T polymorphism and invasive cervical cancer susceptibility, subgroup analysis by ethnicity was performed. First, we detected whether there was T allele frequency of variation in different ethnicities. The T allele frequency has significant differences in different populations. Next, the association between MTHFR C677T polymorphism and invasive cervical cancer risk in different ethnicities was explored. Lack of an association was also found in all genetic models.
In our meta-analysis, obvious heterogeneity was observed for the association between MTHFR C677T polymorphism and invasive cervical cancer risk. Then, we used the Galbraith plots to explore the sources of heterogeneity. We found all of the I 2 values were less than 50% and P Q were greater than 0.1 after excluding the studies of Shekari M et al., Ma XC et al., and Zoodsma M et al. respectively. The results indicated that the three studies might be the major source of the heterogeneity for the association between MTHFR C677T polymorphism and cervical cancer risk. The results of subgroup analysis revealed that the ethnicity might contribute to the potential heterogeneity.
There are some limitations to this meta-analysis. Firstly, the retrieved literature is potentially not comprehensive enough. Studies included in our meta-analysis were limited to published articles. We did not track the unpublished articles to obtain data for analysis. Secondly, as many other factors such as age, parity, smoking, and alcohol consumption may participate in the progression of disease, we did not carry out subgroup analysis based on these factors due to limited data. Thirdly, the small sample sizes in some subgroup analyses limited the ability to draw more solid conclusions.
Conclusively, MTHFR C677T polymorphism may associate with genetic susceptibility of invasive cervical cancer in recessive model based on the current published studies. Similarly significant differences in T vs. C, TT vs. CC, and recessive model were found in Asians. Moreover, further studies with large sample size of different ethnic populations will be necessary to combine genetic factors together with age, parity, smoking, and alcohol consumption.
Materials and Methods
Data Sources and Search Strategy
This meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) criteria [27]. Two investigators (L.Y. and K.C.) independently performed a systematic electronic search of the PubMed, Embase, Web of Science databases for original articles published until 1 April, 2012 to identify potentially relevant articles and abstracts. Search terms used were “Methylenetetrahydrofolate reductase or MTHFR” and “cervical cancer or cervical carcinoma or uterine cervix cancer or cervical neoplasia or cervical dysplasia” and “polymorphism or mutation or variant”. There were no language restrictions. We reviewed the bibliographies of all selection articles to identify additional relevant studies.
Selection of Publications
Two reviewers independently screened titles and abstracts of all studies for relevancy. Disagreements were resolved by a third opinion. Full-text publications were retrieved for relevant articles. The strength of the individual studies was weighed for relevance, based on the following items: (1) evaluation of the MTHFR C677T polymorphism and cervical cancer or its precursor lesion, CIN, (2) case-control studied, (2) sufficient data for estimating an odds ratio (OR) with 95% confidence intervals (CIs), (3) genotype distribution of control population in HWE, and (4) studies written in English or Chinese. For the studies with the same or overlapping data by the same authors, the most recent or largest population was selected.
Data Extraction
Data were extracted independently from each study by two reviewers according to the inclusion criteria listed above. Agreement was reached after discussion for conflicting data. The following data were collected from each study: first author’s name, publication year, original country, ethnicity, control source, sample size, genotyping method, and genotype number in cases and controls.
Quality Assessment
The quality of included studies was assessed independently by the same two investigators using the NOS [28]. The NOS uses a ‘star’ rating system to judge quality based on 3 aspects of the study: selection of study groups, comparability of study groups and ascertainment of the exposure of interest. Studies with a score of 7 stars or greater were considered to be of high quality.
Statistical Analysis
The strength of association between MTHFR polymorphism and susceptibility of cervical cancer or CIN was estimated by OR and corresponding 95% CIs. The pooled OR was calculated respectively for T vs. C, TT vs. CC, dominant genetic model (TT+CT vs. CC), and recessive genetic model (TT vs. CT+CC). Between-study heterogeneity was assessed by the Q-test and I 2 test, P Q <0.10 and I 2>50% indicated evidence of heterogeneity. Then, the random-effects model (the DerSimonian and Laird method)[29]–[30] was used to calculate the pooled OR. Otherwise, the fixed-effects model (Mantel-Haenszel) was adopted [31]. The forest plots were inspected to indicate the overall results, which show information from the individual studies that were included in the meta-analysis, and an estimate of the overall results. It also allows a visual assessment of the amount of variation between the results of the studies (heterogeneity).
Subgroup analyses were performed by ethnicity of study population. Sensitivity analysis was performed based on the high quality studies (according to the NOS score). Asymmetry funnel plots were inspected to assess potential publication bias. The Egger’s linear regression test was also used to assess publication bias statistically [32].
Data were analyzed by using STATA 11.0 (Stata Corporation, College Station, TX, USA) and Revman 5.0 (The Cochrane Collaboration).
Supporting Information
Flow diagram of the selection of eligible studies.
(TIF)
Funnel plots of all genetic models in overall studies. A. T vs. C; B. TT vs. CC; C. dominant model (TT+CT vs. CC); D. recessive model (TT vs. CT+CC). Funnel plots of dominant model seemed asymmetry. Each point represents a separate study for the indicated association.
(TIF)
Association between individual study characteristics and MTHFR A1298T polymorphism.
(DOC)
Meta-analyses of MTHFR A1298T polymorphism and risk of cervical cancer.
(DOC)
Checklist Association between methylenetetrahydrofolate reductase C677T polymorphism and susceptibility of cervical cancer.
(DOC)
Funding Statement
This study was supported by the Chinese National Natural Science Foundation (No. 81071428, 81070505). The authors have declared that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Echelman D, Feldman S (2012) Management of cervical precancers: a global perspective. Hematol Oncol Clin North Am 26: 31–44. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J SH, Bray F (2011) GLOBOCAN 2008 v1.2, cancer incidence and mortality worldwide: IARC CancerBase No.10 [Internet]. Lyon (France): International Agency for Research on Cancer; 2010. Available at: http://globocan.iarc.fr. Accessed November 6, 2011.
- 3. Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, et al. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189: 12–19. [DOI] [PubMed] [Google Scholar]
- 4. Sankaranarayanan R, Thara S, Esmy PO, Basu P (2008) Cervical cancer: screening and therapeutic perspectives. Med Princ Pract 17: 351–364. [DOI] [PubMed] [Google Scholar]
- 5. Josefsson AM, Magnusson PK, Ylitalo N, Sorensen P, Qwarforth-Tubbin P, et al. (2000) Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 355: 2189–2193. [DOI] [PubMed] [Google Scholar]
- 6. Stankova J, Lawrance AK, Rozen R (2008) Methylenetetrahydrofolate reductase (MTHFR): a novel target for cancer therapy. Curr Pharm Des 14: 1143–1150. [DOI] [PubMed] [Google Scholar]
- 7. Misra UK, Kalita J, Srivastava AK, Agarwal S (2010) MTHFR gene polymorphism and its relationship with plasma homocysteine and folate in a North Indian population. Biochem Genet 48: 229–235. [DOI] [PubMed] [Google Scholar]
- 8. Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, et al. (1994) Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet 7: 195–200. [DOI] [PubMed] [Google Scholar]
- 9. Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, et al. (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10: 111–113. [DOI] [PubMed] [Google Scholar]
- 10. Kim YI (2009) Role of the MTHFR polymorphisms in cancer risk modification and treatment. Future Oncol 5: 523–542. [DOI] [PubMed] [Google Scholar]
- 11. Prasad VVTS, Wilkhoo H (2011) Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie 34: 422–426. [DOI] [PubMed] [Google Scholar]
- 12. Tong SY, Kim MK, Lee JK, Choi SW, Friso S, et al. (2011) Common polymorphisms in methylenetetrahydrofolate reductase gene are associated with risks of cervical intraepithelial neoplasia and cervical cancer in women with low serum folate and vitamin B12. Cancer Causes & Control 22: 63–72. [DOI] [PubMed] [Google Scholar]
- 13. Mostowska A, Myka M, Lianeri M, Roszak A, Jagodzinski PP, et al. (2011) Folate and choline metabolism gene variants and development of uterine cervical carcinoma. Clinical Biochemistry 44: 596–600. [DOI] [PubMed] [Google Scholar]
- 14. Kohaar I, Kumar J, Thakur N, Hussain S, Niyaz MK, et al. (2010) Homocysteine levels are associated with cervical cancer independent of methylene tetrahydrofolate reductase gene (MTHFR) polymorphisms in Indian population. Biomarkers 15: 61–68. [DOI] [PubMed] [Google Scholar]
- 15. Shekari M, Sobti RC, Kordi Tamandani DM, Suri V (2008) Impact of methylenetetrahydrofolate reductase (MTHFR) codon (677) and methionine synthase (MS) codon (2756) on risk of cervical carcinogenesis in North Indian population. Arch Gynecol Obstet 278: 517–524. [DOI] [PubMed] [Google Scholar]
- 16. Nandan NK, Wajid S, Biswas S, Juneja SS, Rizvi M, et al. (2008) Allelic variations in 5, 10-methylenetetrahydrofolate reductase gene and susceptibility to cervical cancer in Indian women. Drug Metab Lett 2: 18–22. [DOI] [PubMed] [Google Scholar]
- 17. Ma XC WJ, Zhou Q (2006) Relationship between Methylenetetrahydrofolate reductase polymorphismand cervical cancer susceptibility. Chin J Public Health Dec 22: 1427–1428. [Google Scholar]
- 18. Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der Steege G, et al. (2005) Methylenetetrahydrofolate reductase (MTHFR) and susceptibility for (pre)neoplastic cervical disease. Hum Genet 116: 247–254. [DOI] [PubMed] [Google Scholar]
- 19. Kang S, Kim JW, Kang GH, Park NH, Song YS, et al. (2005) Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecologic Oncology 96: 173–180. [DOI] [PubMed] [Google Scholar]
- 20. Sull JW, Jee SH, Yi S, Lee JE, Park JS, et al. (2004) The effect of methylenetetrahydrofolate reductase polymorphism C677T on cervical cancer in Korean women. Gynecologic Oncology 95: 557–563. [DOI] [PubMed] [Google Scholar]
- 21. Lambropoulos AF, Agorastos T, Foka ZJ, Chrisafi S, Constantinidis TC, et al. (2003) Methylenetetrahydrofolate reductase polymorphism C677T is not associated to the risk of cervical dysplasia. Cancer Letters 191: 187–191. [DOI] [PubMed] [Google Scholar]
- 22. Piyathilake CJ, Macaluso M, Johanning GL, Whiteside M, Heimburger DC, et al. (2000) Methylenetetrahydrofolate reductase (MTHFR) polymorphism increases the risk of cervical intraepithelial neoplasia. Anticancer Res 20: 1751–1757. [PubMed] [Google Scholar]
- 23. Galbraith RF (1988) A note on graphical presentation of estimated odds ratios from several clinical trials. Stat Med 7: 889–894. [DOI] [PubMed] [Google Scholar]
- 24. Prasad VV, Wilkhoo H (2011) Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie 34: 422–426. [DOI] [PubMed] [Google Scholar]
- 25. Ulvik A, Ueland PM, Fredriksen A, Meyer K, Vollset SE, et al. (2007) Functional inference of the methylenetetrahydrofolate reductase 677C>T and 1298A>C polymorphisms from a large-scale epidemiological study. Hum Genet 121: 57–64. [DOI] [PubMed] [Google Scholar]
- 26. Goodman MT, McDuffie K, Hernandez B, Wilkens LR, Selhub J (2000) Case-control study of plasma folate, homocysteine, vitamin B(12), and cysteine as markers of cervical dysplasia. Cancer 89: 376–382. [DOI] [PubMed] [Google Scholar]
- 27. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 62: 1006–1012. [DOI] [PubMed] [Google Scholar]
- 28. Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 29. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 30. DerSimonian R, Kacker R (2007) Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 28: 105–114. [DOI] [PubMed] [Google Scholar]
- 31. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 32. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow diagram of the selection of eligible studies.
(TIF)
Funnel plots of all genetic models in overall studies. A. T vs. C; B. TT vs. CC; C. dominant model (TT+CT vs. CC); D. recessive model (TT vs. CT+CC). Funnel plots of dominant model seemed asymmetry. Each point represents a separate study for the indicated association.
(TIF)
Association between individual study characteristics and MTHFR A1298T polymorphism.
(DOC)
Meta-analyses of MTHFR A1298T polymorphism and risk of cervical cancer.
(DOC)
Checklist Association between methylenetetrahydrofolate reductase C677T polymorphism and susceptibility of cervical cancer.
(DOC)