Abstract
A number of drugs target the DNA repair pathways and induce cell kill by creating DNA damage. Thus, processes to directly measure DNA damage have been extensively evaluated. Traditional methods are time consuming, expensive, resource intensive and require replicating cells. In contrast, the comet assay, a single cell gel electrophoresis assay, is a faster, non-invasive, inexpensive, direct and sensitive measure of DNA damage and repair. All forms of DNA damage as well as DNA repair can be visualized at the single cell level using this powerful technique.
The principle underlying the comet assay is that intact DNA is highly ordered whereas DNA damage disrupts this organization. The damaged DNA seeps into the agarose matrix and when subjected to an electric field, the negatively charged DNA migrates towards the cathode which is positively charged. The large undamaged DNA strands are not able to migrate far from the nucleus. DNA damage creates smaller DNA fragments which travel farther than the intact DNA. Comet Assay, an image analysis software, measures and compares the overall fluorescent intensity of the DNA in the nucleus with DNA that has migrated out of the nucleus. Fluorescent signal from the migrated DNA is proportional to DNA damage. Longer brighter DNA tail signifies increased DNA damage. Some of the parameters that are measured are tail moment which is a measure of both the amount of DNA and distribution of DNA in the tail, tail length and percentage of DNA in the tail. This assay allows to measure DNA repair as well since resolution of DNA damage signifies repair has taken place. The limit of sensitivity is approximately 50 strand breaks per diploid mammalian cell 1,2. Cells treated with any DNA damaging agents, such as etoposide, may be used as a positive control. Thus the comet assay is a quick and effective procedure to measure DNA damage.
Keywords: Neuroscience, Issue 70, Genetics, Cellular Biology, Molecular Biology, Medicine, Cancer Biology, Anatomy, Physiology, DNA, DNA damage, double strand break, single strand break, repair, neurons, comet assay, cell culture
Protocol
1. Cell Culture
Culture neuronal cells and treat them as needed.
Harvest cells into 15 ml tubes: aspirate media, rinse with phosphate buffered saline (PBS, calcium and magnesium free), add trypsin, collect in 15 ml tubes and neutralize trypsin with appropriate serum containing media.
Spin at 1,000 x g for 5 min.
Aspirate media.
Resuspend cells in PBS.
Spin at 1,000 x g for 5 min.
Aspirate media and resuspend cells in fresh PBS.
Count cells using a hemacytometer or your preferred cell counter.
Cell samples should be prepared immediately before starting the assay.
All samples should be handled in the dark or yellow light to prevent DNA damage from ultraviolet light.
2. Comet Assay
1. Slide Preparation
Melt 1% agarose (1 g / 100 ml in 1X Tris Base, Boric acid, EDTA, TBE) in a microwave for 3 min until all granules disappear.
Dip slides into the molten agarose and wipe a side clean with a kimwipe. Allow the agarose to air-dry to a transparent film. This can be done in advance and slides can be stored.
2. Neutral Comet Assay
Chill lysis solution (2.5 M NaCl, 100 mM EDTA pH 10, 10 mM Tris Base, 1% sodium lauryl sarcosinate, and 1% Triton X-100, pH 10) at 4 °C for at least 1 hr before use.
2.2.2. Melt 1% low melting point agarose (1 g / 100ml in 1X Tris Base, Boric acid, EDTA) in a microwave for 3 min until all granules disappear. The agarose needs to be cooled to 37 °C in a water bath to avoid artificial induction of comet tail. Ideally, the agarose needs to be cooled for half an hour before use.
2.2.3. Dilute the cell suspension so that there are 100,000 cells per ml. Combine the cell suspension with the low melting point agarose (at 37 °C) at a ratio of 1:10 (v/v), vortex briefly, and immediately pipette 50 μl onto the Comet Slide. Use the side of the pipette tip to spread the cell suspension evenly over the sample area. Each treatment group should be at least in triplicate.
Place slides flat in refrigerator for 30 min until a circle appears in the periphery of the slide.
Prechill the lysis solution and submerge slides in this solution for 30 min in the dark at 4 °C (or in a refrigerator).
Pour off or aspirate the lysis solution and add 1X neutral electrophoresis buffer (Tris base, Boric acid, EDTA, 1X TBE). Leave slides in this buffer for half an hour in the refrigerator.
Add prechilled 1X Neutral Electrophoresis Buffer (TBE) in electrophoresis chamber, place slides in electrophoresis slide tray. Align slides so that they are equidistant from electrodes.
Pour 1X neutral electrophoresis buffer up to 0.2 inches above slides. Excess buffer will interfere with electrophoresis.
Set power supply voltage to 1 V per cm (measured electrode to electrode) and run for 30 min at 4 °C (or the cold room) in the dark.
3. Alkaline Comet Assay
Chill lysis solution (2.5 M NaCl, 100 mM EDTA pH 10, 10 mM Tris Base, 1% sodium lauryl sarcosinate, and 1% Triton X-100, pH 10) at 4 °C for at least 1 hr before use.
Melt 1% low melting point agarose (1 g / 100 ml in 1X Tris Base, Boric acid, EDTA) in a microwave for 3 min until all granules disappear. Then cool in a 37 °C water bath for at least 30 min.
Combine cells at 1 x 105 / ml with molten low melting point agarose (at 37 °C) at a ratio of 1:10 (v/v), vortex briefly, and immediately pipette 50 μl onto the Comet Slide. Use the side of the pipette tip to spread the cell suspension evenly over the sample area. Each treatment group should be at least in triplicate.
Place slides flat in refrigerator for 30 min until a circle appears in the periphery of the slide.
Immerse slides in prechilled lysis solution and leave at 4 °C for 1 hr to overnight in the dark.
Remove the slides from the lysis solution, drain the slides and rinse once with cold neutralization buffer for 5 min to remove residual detergent and salts prior to the alkali-unwinding step.
Place slides in a gel electrophoresis chamber filled with prechilled freshly made electrophoresis Buffer (300 mM NaOH, 1 mM EDTA, pH>13) not to exceed 0.5 cm above slides. Align slides so that they are equidistant from electrodes.
Let slides sit in the alkaline buffer for 30 min in the dark to allow for unwinding of the DNA and the expression of alkali-liable damage.
Set power supply voltage to 1 V per cm (measured electrode to electrode) and run for 30 min at 4 °C (or the cold room).
4. Fixing and Staining Cells
Drain excess Electrophoresis Buffer
Immerse slides in pre-chilled distilled water for 5 min at RT.
Immerse slides in pre-chilled 70% ethanol for 5 min at RT.
Dry samples overnight. Do not expose slides to bright light. Samples may be stored for months at room temperature prior to scoring at this stage.
Stain slides by immersing in Sybr green (1X diluted in PBS) for 20 min in the refrigerator.
Remove slides and allow them to dry completely in the dark. The agarose will become transparent when completely dry.
4. Image Acquisition and Analysis
Acquire images using fluorescent microscope set to the green filter (Zeiss AxioVision) and analyze using Comet Assay software (Perceptive Instruments).
Click on the "comet head" (the nucleus) and the software calculates the parameters including the mean tail moment and amount of DNA in the nucleus.
Analyze at least 200 cells per treatment.
Export data to Microsoft Excel.
Calculate mean tail moment for each treatment group and plot data as appropriate.
5. Representative Results
An example of comet assay analysis on neuronal cells is shown in Figure 2 and Figure 3. In this case, irradiation of the neuronal cells induces DNA damage. As the cells are subjected to the electrical field, the DNA migrates at different rates due to differences in size which is subsequently analyzed using the Comet Assay software. The more the DNA damage, the farther the DNA migrates out of the nucleus. This decreases the fluorescent intensity in the nucleus which is subsequently picked up by the software and results in higher tail moment. Table 1 depicts a representative table from comet assay analysis and Figure 4 shows a representative graph comparing DNA damage in neuronal cells following radiation as measured by the comet assay.
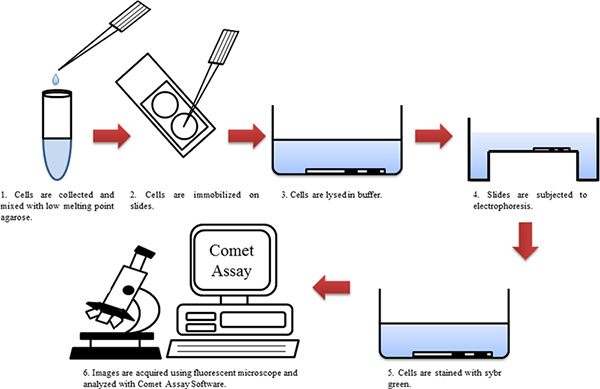 Figure 1. Flow chart of the comet assay. Click here to view larger figure.
Figure 1. Flow chart of the comet assay. Click here to view larger figure.
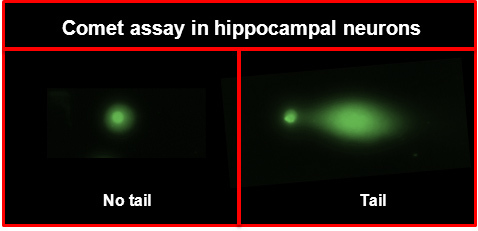 Figure 2. Representative images of neuronal cells (A) without and (B) with comet tail.
Figure 2. Representative images of neuronal cells (A) without and (B) with comet tail.
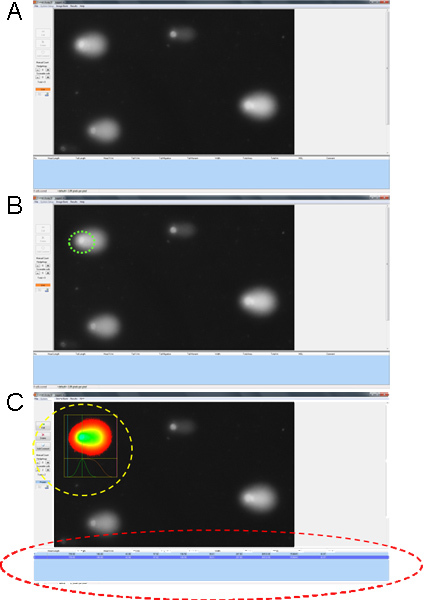 Figure 3. Comet assay analysis using Comet Assay software. (A) Representative screen shots of image acquired using Carl Zeiss fluorescent microscope and analyzed using Comet Assay software. (B) Clicking on the nucleus or the "comet head" (circled in green) generates a fluorescent map and graph (circled in yellow) and a (C) data table (circled in red). Click here to view larger figure.
Figure 3. Comet assay analysis using Comet Assay software. (A) Representative screen shots of image acquired using Carl Zeiss fluorescent microscope and analyzed using Comet Assay software. (B) Clicking on the nucleus or the "comet head" (circled in green) generates a fluorescent map and graph (circled in yellow) and a (C) data table (circled in red). Click here to view larger figure.
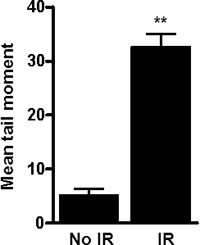 Figure 4. Representative graph obtained by plotting the mean tail moment obtained by analyzing DNA damage in irradiated and non-irradiated neuronal cells using neutral comet assay. As expected, 3Gy radiation (x-ray) induces DNA damage as depicted by the higher mean tail moment in the irradiated neuronal cells. Shown is the mean tail moment (+/- Standard error), **P<0.01.
Figure 4. Representative graph obtained by plotting the mean tail moment obtained by analyzing DNA damage in irradiated and non-irradiated neuronal cells using neutral comet assay. As expected, 3Gy radiation (x-ray) induces DNA damage as depicted by the higher mean tail moment in the irradiated neuronal cells. Shown is the mean tail moment (+/- Standard error), **P<0.01.
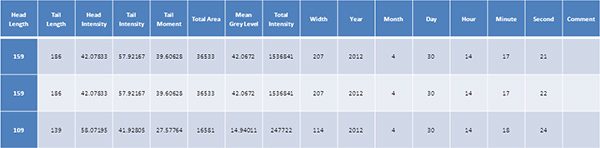 Table 1. Representative table obtained by comet assay analysis on irradiated and non-irradiated neuronal cells. Click here to view larger figure.
Table 1. Representative table obtained by comet assay analysis on irradiated and non-irradiated neuronal cells. Click here to view larger figure.
Discussion
The comet assay has the unique capacity of analyzing individual cells. This is advantageous in identification of subpopulations of cells that demonstrate differential response to cytotoxic agents. A few practical limitations have to be taken into account. The number of cells that can be evaluated individually may vary depending on the individual. The sample size needs to be increased if there is variance in DNA damage within a population. Viable single-cell suspension is critical for this assay since predominant presence of necrotic or apoptotic cells will add to error. The lysis and electrophoresis steps gets rid of apoptotic cells, small DNA fragments (smaller than 50kb), and mitochondrial DNA. Thus, they are not detected by the comet assay 1,3-11.
As mentioned above, the comet assay is an efficient way of detecting single- and double-strand breaks, including alkali-labile sites and DNA-DNA/DNA-protein cross-links on the DNA. Here we have outlined two distinct protocols: the alkaline assay allows the detection of single strand breaks as well as double strand breaks, whereas the neutral comet assay allows only the detection of double strand breaks. Enzyme-modified comet assay can been applied to detect oxidative DNA adducts. For example, treatment with Endo III (endonuclease) and hOGG1 (glycosylase) recognize oxidized pyrimidines including thymine glycol and uracil glycol 12,13 and 8-oxo-7,8-dihydroguanine (8-oxoGua) 14,15 respectively. Formamidopyrimidine DNA-glycosylase (FPG) can be used similarly. For this modified assay, treatment with the enzymes precedes the unwinding step. The addition of lesion-specific endonucleases increases the sensitivity of the comet assay as well 13.
Here we use tail moment as the descriptor of DNA damage. Tail moment is calculated using the following formula: percentage of DNA in the tail multiplied by the length of the comet tail 1,3-7. The length of the comet tail (tail length) is the distance from the center of the nucleus to the farthest point of migration of the DNA. Head length is the diameter of the nucleus. Head intensity and tail intensity are the mean pixel intensities in the head and tail of the comet, respectively. Total area is the overall surface area of the comet 1,3-7.
Though there are other methods of measuring DNA damage such as γ-H2AX foci staining and pulsed-field gel electrophoresis, these assays have their drawbacks as well 4,6,11,16,17. For instance, replication stress is often a confounder in the measurement of γ-H2AX levels 18. Since formation of γ-H2AX foci is dependent on recruitment of DNA repair proteins, failure to recruit DNA repair proteins and a condensed chromatin structure may lead to falsely low γ-H2AX foci levels as well. Similarly, pulsed-field gel electrophoresis is very time intensive and is only beneficial for separation of large DNA molecules 19,20. Thus, the comet assay is a relatively easy, efficient, and inexpensive, direct and sensitive measure of DNA damage and repair 10,11,21.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by the IMPACT Award from the Department of Radiation Oncology, University of Alabama-Birmingham Comprehensive Cancer Center, the Fighting Children's Cancer Foundation, and the Gabrielle's Angel Foundation (to ESY.).
References
- Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan GG. Fluorescence in situ hybridization in combination with the comet assay and micronucleus test in genetic toxicology. Mol. Cytogenet. 2010;3 doi: 10.1186/1755-8166-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL. The comet assay. An overview of techniques. Methods Mol. Biol. 2002;203:179–194. doi: 10.1385/1-59259-179-5:179. [DOI] [PubMed] [Google Scholar]
- Olive PL. Detection of DNA damage in individual cells by analysis of histone H2AX phosphorylation. Methods Cell Biol. 2004;75:355–373. doi: 10.1016/s0091-679x(04)75014-1. [DOI] [PubMed] [Google Scholar]
- Olive PL. Impact of the comet assay in radiobiology. Mutat. Res. 2009;681:13–23. doi: 10.1016/j.mrrev.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP. Kinetics of H2AX phosphorylation after exposure to cisplatin. Cytometry B Clin. Cytom. 2009;76:79–90. doi: 10.1002/cyto.b.20450. [DOI] [PubMed] [Google Scholar]
- Olive PL, Durand RE. Heterogeneity in DNA damage using the comet assay. Cytometry A. 2005;66:1–8. doi: 10.1002/cyto.a.20154. [DOI] [PubMed] [Google Scholar]
- Collins AR. The comet assay for DNA damage and repair. Molecular biotechnology. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- Fairbairn DW, Olive PL, O'Neill KL. The comet assay: a comprehensive review. Mutation Research/Reviews in Genetic Toxicology. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- Yang ES, Nowsheen S, Wang T, Thotala DK, Xia F. Glycogen synthase kinase 3B inhibition enhances repair of DNA double-strand breaks in irradiated hippocampal neurons. Neuro-Oncology. 2011;13:459–470. doi: 10.1093/neuonc/nor016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ES, et al. Lithium-mediated protection of hippocampal cells involves enhancement of DNA-PK dependent repair in mice. The Journal of clinical investigation. 2009. pp. 119–1124. [DOI] [PMC free article] [PubMed]
- Collins AR, Harrington V, Drew J, Melvin R. Nutritional modulation of DNA repair in a human intervention study. Carcinogenesis. 2003;24:511–515. doi: 10.1093/carcin/24.3.511. [DOI] [PubMed] [Google Scholar]
- Smith CC, O'Donovan MR, Martin EA. hOGG1 recognizes oxidative damage using the comet assay with greater specificity than FPG or ENDOIII. Mutagenesis. 2006;21:185–190. doi: 10.1093/mutage/gel019. [DOI] [PubMed] [Google Scholar]
- Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proceedings of the National Academy of Sciences. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banath JP, Klokov D, MacPhail SH, Banuelos CA, Olive PL. Residual gammaH2AX foci as an indication of lethal DNA lesions. BMC Cancer. 2010;10:4. doi: 10.1186/1471-2407-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- Bonner WM, et al. [gamma]H2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M. Pulsed Field Gel Electrophoresis. Current protocols in molecular biology. 2000. [DOI] [PubMed]
- Electrophoresis FI, et al. Pulsed-field gel electrophoresis. 2000.
- Nowsheen S, Bonner JA, Yang ES. The poly (ADP-Ribose) polymerase inhibitor ABT-888 reduces radiation-induced nuclear EGFR and augments head and neck tumor response to radiotherapy. Radiotherapy and Oncology. 2011. [DOI] [PMC free article] [PubMed]


