To determine the regulatory processes in the progression of normal cells to pancreatic intraepithelial neoplasia, Reichert et al. investigated the role of Prrx1 in regulating duct cell properties during development, injury, and neoplastic transformation. They provide new mechanistic insight by showing that the isoform Prrx1b fosters ductal cell proliferation and self-renewal and regulates Sox9 gene expression, a factor implicated in metaplastic changes in the pancreas. Their results provide a possible explanation for why pancreatic cancer skews toward a ductal fate.
Keywords: Prrx1, Sox9, acinar-ductal metaplasia, pancreatic ductal epithelial cell, pancreatitis
Abstract
Pancreatic exocrine cell plasticity can be observed during development, pancreatitis with subsequent regeneration, and also transformation. For example, acinar–ductal metaplasia (ADM) occurs during acute pancreatitis and might be viewed as a prelude to pancreatic intraepithelial neoplasia (PanIN) and pancreatic ductal adenocarcinoma (PDAC) development. To elucidate regulatory processes that overlap ductal development, ADM, and the progression of normal cells to PanIN lesions, we undertook a systematic approach to identify the Prrx1 paired homeodomain Prrx1 transcriptional factor as a highly regulated gene in these processes. Prrx1 annotates a subset of pancreatic ductal epithelial cells in Prrx1creERT2-IRES-GFP mice. Furthermore, sorted Prrx1+ cells have the capacity to self-renew and expand during chronic pancreatitis. The two isoforms, Prrx1a and Prrx1b, regulate migration and invasion, respectively, in pancreatic cancer cells. In addition, Prrx1b is enriched in circulating pancreatic cells (Pdx1cre;LSL-KrasG12D/+;p53fl/+;R26YFP). Intriguingly, the Prrx1b isoform, which is also induced in ADM, binds the Sox9 promoter and positively regulates Sox9 expression. This suggests a new hierarchical scheme whereby a Prrx1–Sox9 axis may influence the emergence of acinar–ductal metaplasia and regeneration. Furthermore, our data provide a possible explanation of why pancreatic cancer is skewed toward a ductal fate.
The pancreas has two compartments; namely, endocrine and exocrine. The exocrine pancreas comprises acinar, ductal, and centroacinar cells. An undifferentiated pancreatic trunk epithelium (also called pancreatic cords) is evident in the mouse embryo up to day 13 (embryonic day 13 [E13]). Then, there is a phase designated as “secondary transition” that commences at E13.5–E14.5 with accompanying differentiation of cells. These cells proliferate and expand into the endocrine and exocrine lineages. Ducts mature by E17–E18 (Gittes 2009; Wescott et al. 2009). Ductal cells are relatively quiescent in the adult pancreas and form an intricate network of ducts from terminal ducts to intralobular ducts to interlobular ducts to the main duct (Bardeesy and Depinho 2002). This ductal tree is the conduit for the flow of digestive enzymes secreted by acinar cells. Acinar cells constitute the preponderant cell type in the pancreas, and it appears that acinar cells have intrinsic plasticity. Acinar cells have the capacity to undergo metaplasia to ductal cells or duct-like cells (termed acinar–ductal metaplasia [ADM]) in the setting of acute or chronic pancreatitis, representing an important link to pancreatic ductal adenocarcinoma (PDAC) (Reichert and Rustgi 2011). It has been suggested that ADM occurs in a subset of metaplastic lesions based on genetic lineage tracing tools in mice. ADM might constitute a prelude to pancreatic intraepithelial neoplasia (PanIN) and PDAC development (Strobel et al. 2007). For example, chronic cerulein administration in adult K-Ras+/LSLG12Vgeo;Elas-tTA/tetO-Cre mice results in the formation of ADM, PanIN, and PDAC lesions, suggesting that pancreatitis is important in mediating tumorigenesis (Guerra et al. 2007). The proinflammatory microenvironment has been demonstrated to be critical in PDAC development (Bayne et al. 2012; Pylayeva-Gupta et al. 2012; Rhim et al. 2012).
We reasoned there might be a transcriptional program governing embryonic ductal development, ADM, and PanIN formation, since all three processes share a ductal phenotype. Using techniques to isolate ductal cells and comprehensive RNA microarray approaches, we identified the nuclear Prrx1 homeodomain transcriptional factor as a novel gene that might unify these three processes. Alternative splicing of the Prrx1 results in two isoforms—Prrx1a and Prrx1b—that differ in abundance and expression patterns. Prrx1a, the canonical Prrx1 transcript, is translated into a 245-amino-acid protein, whereas Prrx1b encodes for a 217-amino-acid product that is identical from the N terminus to amino acid 199. This overlap includes the homeobox domain (94–153 amino acids). The DNA-binding homeodomain shows similarities to that of other paired families of transcription factors, most notably prd (paired) and gsb (gooseberry); however, Prrx1 lacks the paired domain. At their C termini, Prrx1a and Prrx1b differ from each other. Prrx1a harbors a so-called OAR (otp, aristaless, and rax) domain, named after three proteins that share this 15-amino-acid region. The OAR domain appears to be involved in modifying transactivation ability (Simeone et al. 1994; Norris and Kern 2001). The C-terminal end of Prrx1b lacks the OAR domain and does not contain any other known protein domains and therefore is designated as having an alternative C terminus. Interestingly, the amino acid sequence is 100% conserved in their human orthologs, underscoring their potential biological importance. Prrx1 has an important role during embryonic development, as Prrx1−/− mice die soon after birth. The phenotype of Prrx1−/− mice consists of craniofacial defects, limb shortening, and incomplete penetrant spina bifida (Martin and Olson 2000). Prrx1 may be a regulator of sonic hedgehog and controls cell proliferation during mandibular arch morphogenesis (ten Berge et al. 2001). Functionally, it is known that Prrx1 expression induced by FAK promotes tenascin C-dependent fibroblast migration (McKean et al. 2003).
We found that Prrx1 has novel functions in pancreatic ductal cells, including the finding that Prrx1a and Prrx1b promote ductal cell migration and invasion, respectively. Prrx1b fosters ductal cell proliferation and self-renewal. Importantly, Prrx1b appears to be critical for regeneration after cerulein-induced pancreatitis and annotates a subpopulation of ductal cells with self-renewal capacity. Furthermore, Prrx1b positively regulates Sox9 gene expression and binds the Sox9 promoter. The Sox9 transcriptional factor is associated with ADM and regulates a transcriptional program in pancreatic progenitor cells during development (Lynn et al. 2007; Furuyama et al. 2011; Kopp et al. 2012; Prévot et al. 2012). These findings help to unravel new mechanistic insights into the regulation of the ductal cell fate in pancreatic regeneration and carcinogenesis.
Results
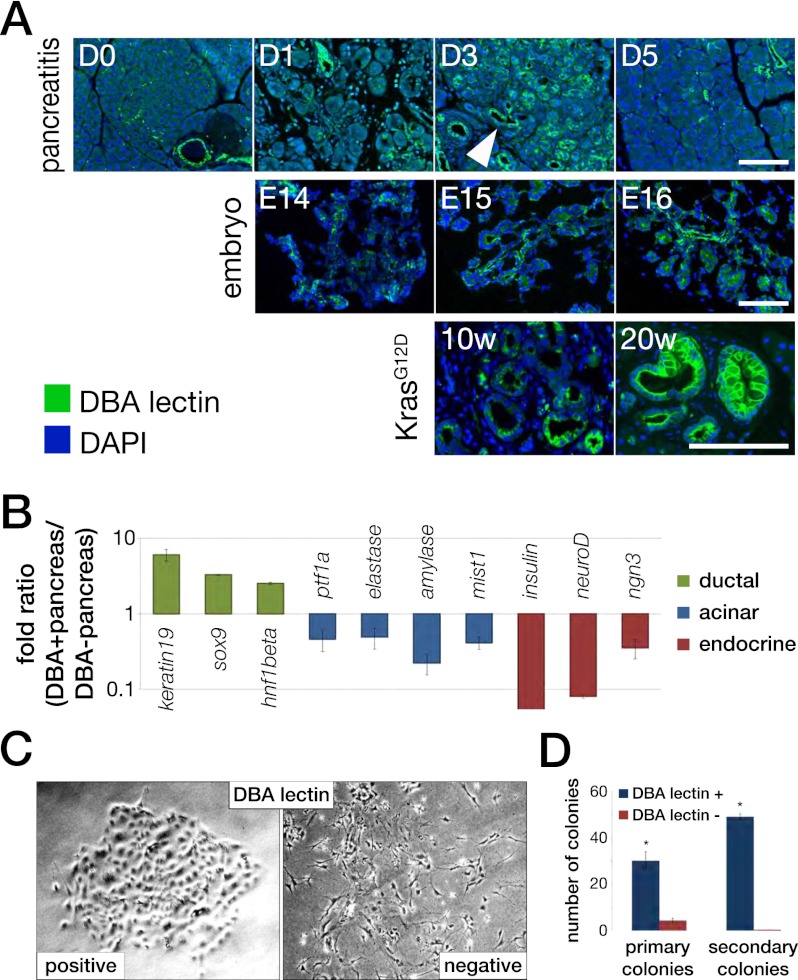
To determine the molecular basis of gene regulation in pancreatic ductal epithelial cells, we developed methods for the isolation of this cell population during mouse development and normal adult homeostasis as well as in conditions with ductal features (ADM, PanIN, and PDAC). Our technique uses the specificity of Dolichos biflorus agglutinin (DBA) lectin marking the entire normal ductal tree, including terminal intercalated ducts (putative sites of stem or progenitor cells) and ductal structures in ADM and PanIN (Fig. 1A). We used ferromagnetic-labeled DBA lectin to isolate ductal structures. DBA lectin+ cells are enriched for ductal gene markers (Sox9, HNF1β, and Keratin 19) but not acinar (Ptf1a, Mist1, elastase, and amylase) or neuroendocrine (insulin, neuroD, and neurogenin 3 [ngn3]) cell-specific genes (Fig. 1B). DBA lectin+ cells can be cultured and passaged and form epithelial colonies (Fig. 1C,D). We next reasoned there may be overlapping mechanisms in the molecular regulation of gene expression in pancreatic ductal cells during development, ADM, and PanIN. Ductal cells were isolated under the following conditions: (1) embryonic development in wild-type mice (E14, E15, E16, and postnatal day 1 [P1]); (2) injury and regeneration (pancreatitis) 0, 1, 3, and 5 d following cerulein-induced acute pancreatitis (cerulein is a cholecystokinin analog that produces a self-limited pancreatitis with injury and subsequent regeneration and repair, completed 5 d after insult) (Fendrich et al. 2008); and (3) Pdx1-Cre;LSL-KrasG12D/+ mice aged 10 and 20 wk (termed KrasG12D) that harbor PanIN lesions and a subset develop PDAC (Aguirre et al. 2003; Hingorani et al. 2003, 2005). Ductal/PanIN cells were isolated from these mice with appropriate control mice (Pdx1-Cre;Kras+/+).
Figure 1.
Duct and duct-like cells in pancreatic development, pancreatitis, and preneoplasia. (A) Immunofluorescence using a FITC-labeled DBA lectin (green) and DAPI (blue) during cerulein-induced acute pancreatitis, embryonic development, and KrasG12D-driven carcinogenesis. The top panel displays staining of sections of acute pancreatitis controls (D0) and at day 1 (D1), day 3 (D3), and day 5 (D5) after onset of injury. Arrowhead at day 3 indicates DBA lectin-labeled tubular structures. The middle panel shows staining with DBA lectin at E14, E15, and E16. The bottom panel depicts DBA lectin staining of precancerous lesions using the Pdx1cre;KrasG12D/+ mouse model at 10 wk (10w) and 20 wk (20w) of age. Bars, 50 μm. (B) Enrichment of ductal cells using DBA lectin labeling followed by magnetic beads sorting from normal adult pancreas. RT-qPCR of transcripts specifically expressed within the ductal (keratin19, sox9, and hnf1β), acinar (ptf1a, elastase, amylase, and mist1), or endocrine (insulin, neuroD, and ngn3) lineage. (C) Phase-contrast photograph of cell cultured after DBA lectin sorting. DBA lectin-positive cells form epithelial colonies, whereas the DBA-negative fraction fails to establish colonies. (D) Quantification of colony-forming ability of DBA lectin-positive and DBA lectin-negative fractions. (*) P < 0.05 (Student’s t-test).
All DBA+ cells from the preceding conditions were subjected to RNA isolation and purification, cDNA synthesis, and Affymetrix microarrays. Transcriptional signatures were then compared across pancreatic development, pancreatitis, and PanIN lesions. During development (E14, E15, and E16), DBA lectin+ cells have a gene signature that is distinct from those genes from P1 DBA lectin+ cells (Supplemental Fig. 1A), as indicated by higher ngn3 and ptf1a levels in the former, whereas prox1, hnf1β, and hhex are enriched in the latter, suggesting a more ductal lineage-committed state (Supplemental Fig. 1B). During acute pancreatitis, the transcriptional profiles of DBA+ cells 1 and 3 d after cerulein injection, when the pancreas is undergoing regeneration, were similar (Supplemental Fig. 1C). Interestingly, the signature of DBA+ cells from control pancreas prior to cerulein injection mirrored the signature from cells taken 5 d after pancreatitis, when regeneration had completed. Further confirming the robustness of our approach, the gene signatures from mouse PanIN lesions (Supplemental Fig. 1D) overlap with those described in laser-captured microdissected human PanIN 1B/2 lesions (Supplemental Fig. 1E; Prasad et al. 2005).
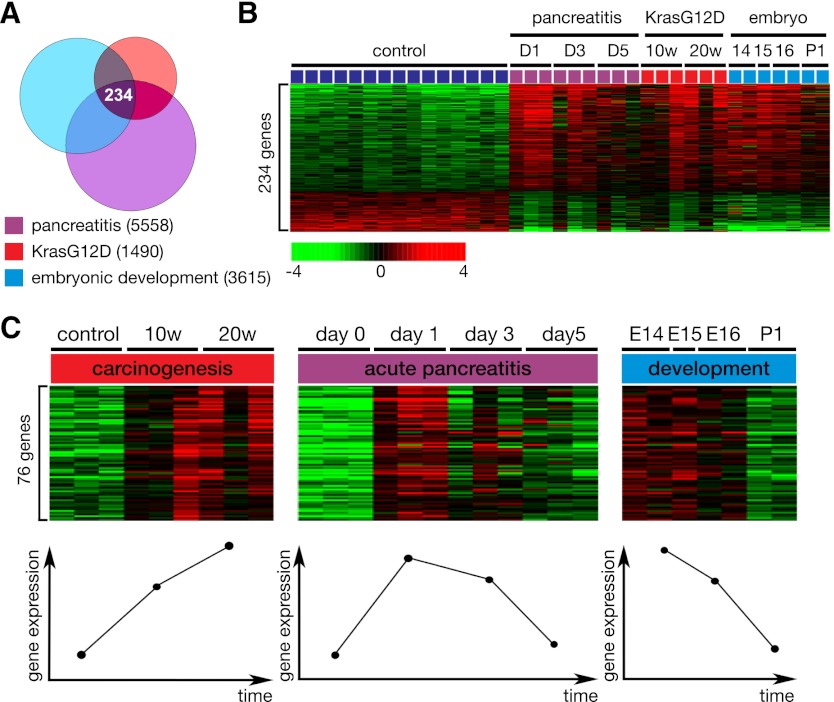
A Venn diagram shows that there are 234 genes that are differentially regulated (cutoff of P < 0.001) in all three cohorts of pancreatic development, pancreatitis, and PanIN (Fig. 2A; Supplemental Table 1). Gene expression patterns are depicted according to relevant time points (Fig. 2B). Genes were first selected to have a positive correlation (r > 0.5) with time points in the KrasG12D data set. Then, genes were further selected to have a negative correlation (r < −0.5) with the different time points in embryonic pancreatic development. This led to a restricted pattern of 76 genes that is correlated significantly with each given data set (Fig. 2C; Supplemental Table 2). Further correlation with pancreatitis showed the highest peak at day 1 for the selected 76 genes. This approach was performed in order to identify a unique and novel gene signature present in a population of cells that might serve as potential progenitors in each of these three processes.
Figure 2.
Gene expression arrays of acute cerulein-induced pancreatitis (control and days 1, 3, and 5), embryonic development (E14, E15, E16, and P1), and early KrasG12D-driven carcinogenesis (Pdx1creKrasG12D/+) at 10 and 20 wk of age. (A) Venn diagram showing 234 differentially expressed genes shared in development, carcinogenesis (Kras), and pancreatitis. Univariate test (two-sample t-test) with multivariate permutation test (10,000 random permutations) was applied; P < 0.001. Numbers of differentially expressed genes are indicated. (B) Heat map depicting set of 234 genes. The bar represents a log2-transformed scale. (C) To identify genes that follow a particular pattern in which they are up-regulated in carcinogenesis, transiently up-regulated during acute pancreatitis, and down-regulated as terminal differentiation occurs, genes were first selected to have positive correlation (r > 0.5) with time points in the KrasG12D data set. Then, genes were further selected to have negative correlation (r < −0.5) with the different time points in embryonic pancreatic development. This led to a restricted pattern of 76 genes that is significantly correlated with each given data set. Further correlation with pancreatitis showed the highest peak at day 1 for the selected 76 genes.
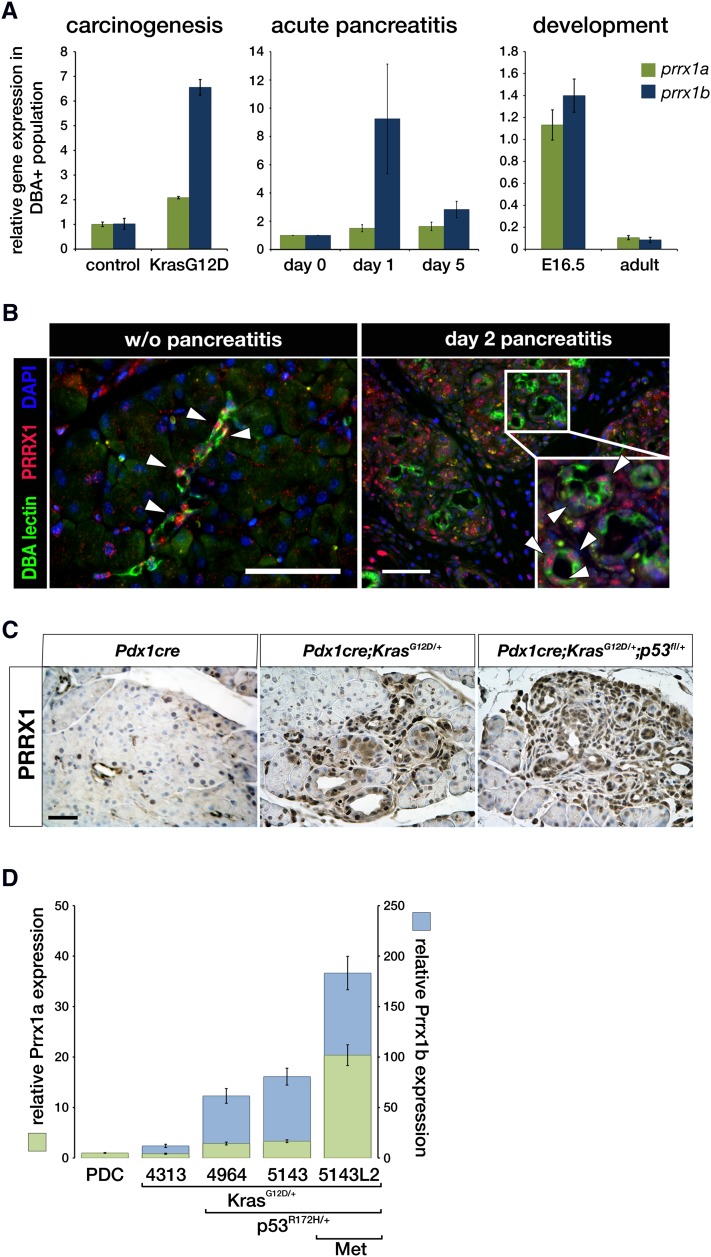
We elected to focus on transcriptional factors as potential regulators of these processes. To that end, the Prrx1 transcription factor emerged as the most regulated transcription factor in all three processes (Supplemental Table 2). We next evaluated the potential differences and common features between the Prrx1 isoforms (Prrx1a and Prrx1b) in our model systems. There is enrichment of the Prrx1b isoform in RNA isolated from ADM lesions (especially day 1), whereas both isoforms are enriched at E16 during pancreatic ductal development and in KRasG12D pancreatic lesions (Fig. 3A). Analysis of ADM lesions from wild-type mice with acute pancreatitis reveals that the number of Prrx1+ cells is increased based on immunofluorescence staining (Fig. 3B). Prrx1+ cells also expand during the progression from normal pancreas to PanIN and PDAC lesions in mice (Fig. 3C). A similar expression pattern for Prrx1 can be observed in human tissues and for the Prrx1 isoforms in human pancreatic cancer cell lines (Supplemental Fig. 2A–D). Additionally, both isoforms are increased in the progression from primary pancreatic ductal cell (PDC) lines derived from the wild-type mouse pancreas, from the Pdx1-Cre;LSL-KrasG12D/+ pancreas (called the 4313 cell line), and from the Pdx1-Cre;LSL-KrasG12D/+;p53R172H/+ pancreas (called the 4964 and 5143 primary tumor cell lines, respectively) (Fig. 3D). The metastatic cell line (5143 liver metastasis) derived from the same animal (5143 primary tumor) shows even greater Prrx1a and Prrx1b mRNA levels (Fig. 3D).
Figure 3.
PRRX1 expression patterns. (A) Relative gene expression of Prrx1a and Prrx1b in DBA lectin+ population in KrasG12D-driven precarcinogenesis (Pdx1cre;LSL-KrasG12D/+, 10 wk of age) during cerulein-induced acute pancreatitis and embryonic development (E16.5) compared with controls. (B) Immunofluorescence for PRRX1 (red), DBA lectin (green), and DAPI (blue) in normal and pancreatitis at day 2. Bar, 50 μm. (C) PRRX1 immunohistochemistry in mouse sections with the indicated genotypes. Bar, 50 μm. (D). Primary pancreatic ductal cell lines derived from a wild-type mouse (PDC), Pdx1cre;KrasG12D/+ mouse (4313), and Pdx1cre;KrasG12D/+;p53R172H/+ mouse (4964 and 5143) and from a liver metastasis (Met) of mouse 5143 (5143L2).
Prrx1a and Prrx1b mediate different functional properties
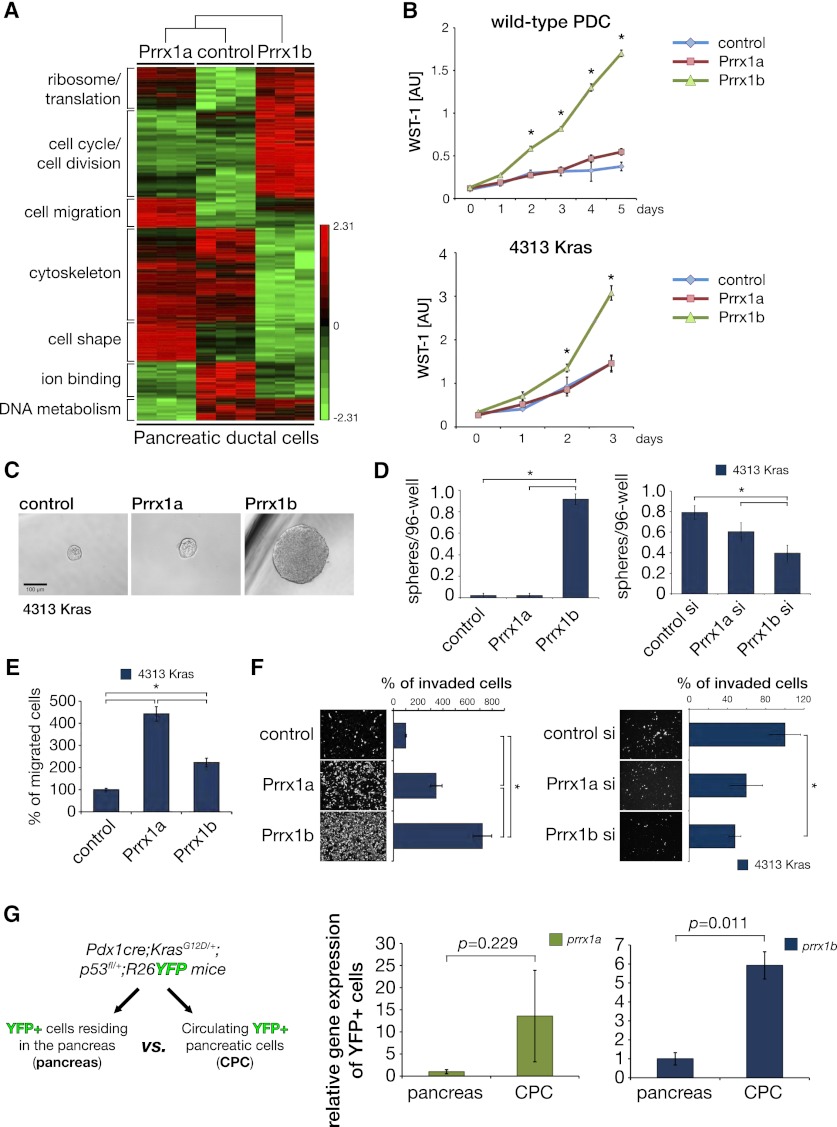
We generated low-passage wild-type PDC lines with stable Prrx1a or Prrx1b overexpression and subjected them to RNA microarray analysis and functional assays. Prrx1a and Prrx1b induce different gene signatures based on our gene ontology (GO) evaluation (Fig. 4A; Huang et al. 2009a,b). For example, Prrx1a is associated with induction of genes involved in cell migration/motility, whereas Prrx1b is associated with genes involved in cell cycle/cell division (Fig. 4A). Indeed, Prrx1b overexpression in PDCs (wild-type) and 4313 cells (PanIN) increases proliferation significantly, whereas no effect was observed with Prrx1a overexpression (Fig. 4B; Supplemental Fig. 3A,B). Interestingly, costaining of Prrx1 and Ki67 in sections derived from a Pdx1cre;KrasG12D/+ mouse shows substantial overlap, especially in reactive ducts and ADMs, respectively (Supplemental Fig. 3C, insert). Given the induction of Prrx1b expression during pancreatitis and ADM, we evaluated Prrx1b in the pancreatosphere assay, a measure of cell self-renewal capacity (Rovira et al. 2010; Rhim et al. 2012). Prrx1b overexpression in 4313 cells increases sphere formation in a pancreatosphere assay (Fig. 4C,D); however, Prrx1a overexpression does not (Fig. 4C,D). Conversely, transient knockdown of Prrx1b using RNAi strategies decreases sphere size and the number of spheres significantly (control siRNA scrambled sequence does not) (Fig. 4D; Supplemental Fig. 3D). Importantly, Prrx1a does not show significant differences in the pancreatosphere assay compared with control cells upon transient knockdown of Prrx1a (Fig. 4D). We further investigated Prrx1a’s and Prrx1b’s effects on migration in 4313 cells. Both Prrx1a and Prrx1b increase migration; however, Prrx1a appears to have a more pronounced effect (Fig. 4E), consistent with the GO analysis. We found that Prrx1b induces more cell invasion than Prrx1a does (Fig. 4F). Knockdown of Prrx1b diminishes the invasive capacity of 4313 (Fig. 4F). As a means to evaluate the effects of the Prrx1 isoforms upon cell polarity, we employed three-dimensional (3D) cyst assays (Wescott et al. 2009). PDCs form well-organized cysts (Supplemental Fig. 4A). However, the organization of these cysts is disturbed by Prrx1b overexpression and, to a lesser extent, Prrx1a overexpression (Supplemental Fig. 4A,B). Recently, we developed a technique to detect circulating pancreatic cells (CPCs) in mice (Rhim et al. 2012). Analysis of CPCs from Pdx1-Cre;LSL-KrasG12D/+;p53fl/+;R26YFP mice reveals significantly increased Prrx1b mRNA levels (P = 0.011) compared with cells resident in the pancreas (Fig. 4G). Prrx1a shows a similar trend but is not statistically significant (P = 0.229).
Figure 4.
Prrx1a and Prrx1b display distinct functional properties. (A) Heat map of RNA microarrays of early-passage Prrx1a- and Prrx1b-overexpressing PDCs and control PDCs. GO was performed with DAVID (Huang et al. 2009a,b) and revealed differences in classes of genes related to protein translational machinery, cell cycle/division, cell migration, cytoskeleton, cell shape, ion binding, and DNA metabolism. (B) WST-1 proliferation assay reveals that PDC-Prrx1b cells and 4313-Prrx1b cells have increased proliferation, but not PDC-Prrx1a cells or 4313-Prrx1a cells or control cells. (C,D) Spheroid assay (sphere is defined as diameter >10 cells). 4313-Prrx1b cells form a greater number of spheres than do either 4313-Prrx1a cells or control cells. siRNA Prrx1b knockdown results in a smaller number of spheres. (*) P < 0.05 is statistically significant (Mann-Whitney-Wilcoxon test). (E) Migration assay using 4313 cells overexpressing Prrx1a or Prrx1b, respectively. (F) Boyden chamber invasion assay using 4313 (PanIN). Prrx1a or Prrx1b overexpression in 4313 cells leads to increased invasion, although a greater effect is observed with Prrx1b overexpression. Knockdown of Prrx1b in 4313 cells decreases invasion. (*) P < 0.05 (Mann-Whitney-Wilcoxon test). (G) Prrx1a and Prrx1b mRNA expression (qPCR) in YFP+ CPCs versus YFP+ resident pancreas cells (pancreas) from Pdx1cre;KrasG12D/+;p53fl/+-;Rosa26YFP at a PanIN stage. P-values (paired Student’s t-test) are indicated.
Prrx1+ cells are enriched for self-renewal properties
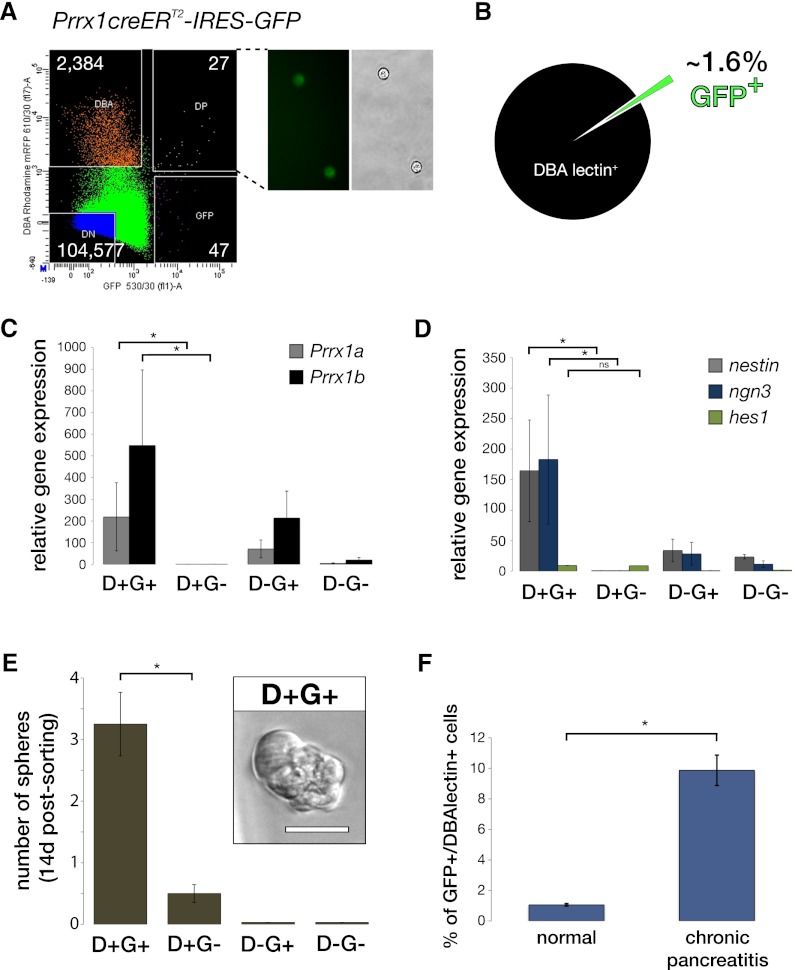
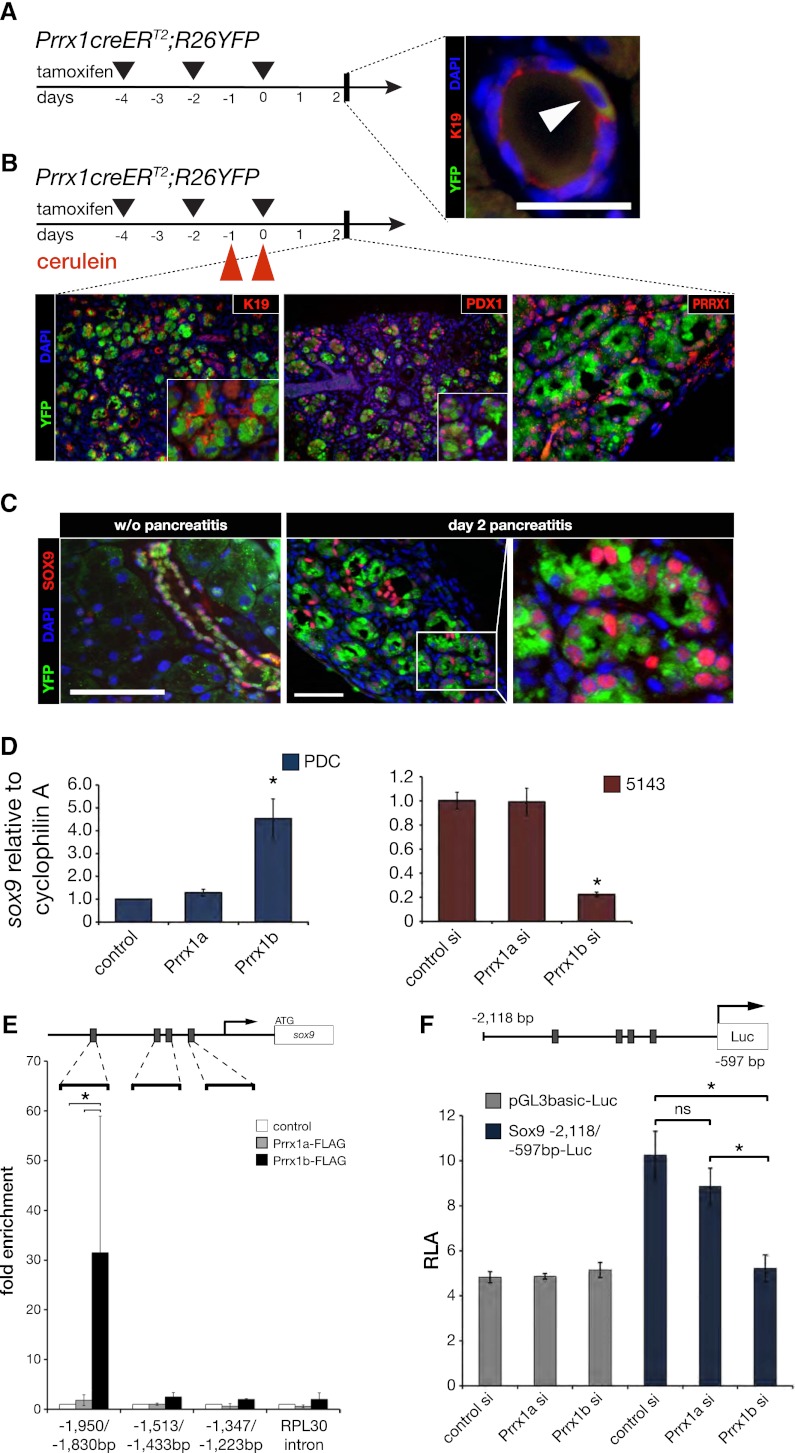
To understand Prrx1’s role in vivo, we characterized pancreatic cell populations from adult (2-mo-old) Prrx1creERT2-IRES-GFP mice (Kawanami et al. 2009). We were able to identify a distinct DBA lectin+(ductal)/GFP+ population (Fig. 5A). DBA lectin labeling of pancreatic cells reveals that ∼1.6% of ductal cells are GFP+ in the normal pancreas (Fig. 5B). Moreover, sorted cells that are GFP+-positive express high Prrx1 levels (both Prrx1a and Prrx1b) compared with GFP− cells by quantitative PCR (qPCR) (Fig. 5C). Interestingly, DBA lectin+/GFP+ cells show exceptionally high Ngn3 and Nestin mRNA levels compared with DBA lectin+/GFP− cells, suggesting that the Prrx1-expressing subpopulation of duct cells may be less differentiated (Fig. 5D). It has been demonstrated that Ngn3 gene expression is induced within Sox9+ duct cells during pancreatitis (Kopp et al. 2011). Of note, Hes1 is not increased in DBA lectin+/GFP+ cells (Fig. 5D). Among the fluorescent-absorbed cell sorting (FACS)-sorted cell fractions, DBA lectin+/GFP+ cells formed spheres to a much greater extent than the DBA lectin+/GFP− fraction (Fig. 5E). The DBA lectin−/GFP+ and DBA lectin−/GFP− cells failed to establish spheres after 2 wk of culture (Fig. 5E). In addition, mice that were challenged with low-dose chronic cerulein administration show an ∼10-fold increase of GFP+ cells within the DBA lectin+ population (Fig. 5F). We next evaluated Prrx1creERT2;Rosa26YFP mice in the absence and presence of pancreatitis to determine the status of YFP+ cells (demonstrating recombination) (Fig. 6A). We found that a small percentage of ductal cells are YFP-labeled 48 h after tamoxifen administration. When tamoxifen administration is combined with cerulein-induced acute pancreatitis, we observed YFP+ cells within ADM lesions 2 d after cerulein administration (Fig. 6B). Importantly, the YFP+ cells express K19, PDX1, and PRRX1 (Fig. 6B). We next crossed the Prrx1creERT2;Rosa26YFP mice with the LSL-KrasG12D/+ mice, treated them with chronic cerulein for 50 d, and evaluated the percentage of YFP+ cells. At day 50, there is a robust expansion of YFP+ cells, suggesting that Prrx1+ cells expand during chronic pancreatitis and are susceptible to the biological effects of KrasG12D/+ mutation (Supplemental Fig. 5). We next found that SOX9 and YFP costain in a small subset of normal ducts and the majority of ADM lesions (Fig. 6C). Additionally, Prrx1b expression correlates with Sox9 expression in our murine pancreatic cell lines (R2 = 0.9445). Given the colocalization of YFP and SOX9 in ADM, we sought to determine whether there is any potential mechanistic interplay between Prrx1b and Sox9 given their expression in ADM lesions in the Prrx1creERT2;Rosa26YFP mice with pancreatitis and the importance of Sox9 in ADM (Kopp et al. 2012; Prévot et al. 2012). Also, Sox9 is increased in PDC-Prrx1b cells (Fig. 4A, RNA microarray). Therefore, we evaluated Sox9 expression upon Prrx1a or Prrx1b overexpression in wild-type PDCs by RT-qPCR. Indeed, Sox9 expression is up-regulated significantly in PDC-Prrx1b cells but not PDC-Prrx1a cells (Fig. 6D). When we used a transient knockdown of either Prrx1a or Prrx1b in 5143 cells that harbor high endogenous Prrx1 levels, Sox9 expression is decreased upon Prrx1b knockdown and remains unchanged in the Prrx1a knockdown (Fig. 6D). To test whether Prrx1b regulates Sox9 directly, we performed a chromatin immunoprecipitation (ChIP) assay. This revealed that Prrx1b, but not Prrx1a, binds a specific region of the Sox9 promoter (between −1950 and −1830 base pairs [bp]) that has a highly conserved putative Prrx1 DNA-binding motif (Fig. 6E). We next generated a Sox9 luciferase reporter gene construct that contains this portion of the Sox9 promoter (Fig. 6F). Prrx1b knockdown diminishes significantly Sox9 luciferase activity (Fig. 6F). Of note, Sox9 knockdown reduces sphere formation in Prrx1b-4313 cells, suggesting an avenue of potential functional interplay between Prrx1 and Sox9 in self-renewal (Supplemental Fig. 6A,B). This is reinforced further by the finding that Sox9 expression is reduced ∼50% in DBA+ pancreatic cells isolated from E16.5 Prrx1+/− pancreatic tissues compared with E16.5 Prrx1+/+ pancreatic tissues (Supplemental Fig. 7; Lu et al. 1999). These data suggest a novel and exciting potential regulation of Sox9 by Prrx1b and have implications on a hierarchy of transcriptional governance in ADM.
Figure 5.
Characterization of Prrx1-expressing cells in the pancreas from Prrx1creERT2-IRES-GFP mice. (A) FACS analysis of cells isolated from Prrx1creERT2-IRES-GFP mice based on GFP and DBA lectin rhodamine labeling. Representative FACS plot (n = 6) demonstrated four populations of cells: DBA lectin+GFP− (designated as DBA; 2384 cells), DBA lectin+GFP+ or double-positive (designated as DP; 27 cells), DBA lectin−GFP+ (designated as GFP; 47 cells), and DBA lectin−GFP− (designated as double negative or DN; 104,577 cells). (B) Pie diagram illustrates that 1.6% of the DBA lectin+ ductal population is GFP+. (C) Relative Prrx1a and Prrx1b gene expression (by qPCR) in the four populations of cells ([D] DBA lectin; [G] GFP) with expected enrichment of Prrx1 isoforms in GFP+ cells. (D) Nestin and ngn3 are enriched in the DBA lectin+GFP+ (D+G+) cells, but not hes1. (E) The DBA lectin+GFP+ (D+G+) cells demonstrate increased capacity to form spheres compared with the other three populations of cells. The insert reveals a representative photomicrogaph of a sphere from the DBA lectin+GFP+ (D+G+) cells. Bar, 50 μm. (F) The cells were sorted with magnetic beads from Prrx1creERT2-IRES-GFP mice with and without pancreatitis and subjected to FACS analysis for GFP expression. (*) P < 0.05 is statistically significant (Mann-Whitney-Wilcoxon test).
Figure 6.
Prrx1b binds the Sox9 promoter. (A) Prrx1creERT2;R26YFP mice. Coimmunofluorescence staining of YFP (green), K19 (red), and DAPI (blue) in representative normal pancreatic intralobular duct (white arrowhead) 48 h after induction with tamoxifen with indicated administration schedule by oral gavage. Bar, 50 μm. (B) Prrx1creERT2;R26YFP mice induced with tamoxifen and treated with cerulein (see the Materials and Methods). Coimmunofluorescence staining of YFP (green); K19, PDX1, or PRRX1 (red); and DAPI (blue) in representative ADM lesions 48 h after cessation of cerulein administration. Each panel shows 200× magnification, and inserts are 400× magnification. (C) Coimmunofluorescence staining in Prrx1creERT2;R26YFP mice of YFP (green), SOX9 (red), and DAPI (blue) in a intralobular duct without cerulein administration or in ADM lesions 48 h after cessation of cerulein administration. Bar, 50 μm. (D) Sox9 expression (qPCR) in PDC-Prrx1a and PDC-Prrx1b cells and Sox9 expression in siRNA to Prrx1a or Prrx1b in 5143 cells (high endogenous Prrx1 level). (E) Schematic diagram of Sox9 upstream 5′ untranslated region. Gray boxes indicate predicted Prrx1-binding sites. Primers were designed for ChIP qPCR flanking the indicated regions. The RPL30 gene intron served as a negative control. There is >30-fold enrichment of Prrx1b binding (Flag immunoprecipitation of PDC-Prrx1b-Flag cells compared with control non-Flag-PDC cells) to the −1950 to −1830 region of the Sox9 gene. There is no enrichment of Prrx1b in regions −1513 to −1433 and −1347 to −1223. Of note, there is no enrichment of Prrx1a to any of the three regions. (F) Prrx1b knockdown (siRNA) reduces Sox9 luciferase reporter activity, whereas Prrx1a knockdown (siRNA) does not. (*) P < 0.05 is statistically significant (Mann-Whitney-Wilcoxon test).
Discussion
Regulation of pancreatic ductal epithelial cell differentiation is critical in development, regeneration, and neoplasia. Having developed innovative techniques for the isolation of pancreatic ductal epithelial cells and duct-like cells, we used RNA microarray and bioinformatic approaches to identify genes that follow a particular pattern of expression (overlap of increased expression of genes in primitive embryonic duct-like epithelial cells at E14, increased gene expression at day 1 of cerulein-induced acute pancreatitis with corresponding enrichment of ADM lesions, and increased expression of genes during progression from normal ducts to early PanIN lesions). We focused on the paired-related homeobox transcription factor Prrx1 as the highest-ranked transcription factor (Supplemental Table 2). The importance of Prrx1 is underscored by its conserved sequence across species (100% homology at amino acid level between mice and humans) and the fact that Prrx1−/− mice die in P1 with a number of developmental abnormalities (Martin and Olson 2000).
Pancreatitis is a common disease in humans. Acute pancreatitis can be modeled in the rodent through several means, one of which is through the use of cerulein, which is highly reproducible. Following cerulein administration, there is rapid induction (day 1) of acinar cell injury, emergence of intermediate ADM structures (days 1 and 3), and inflammatory cellular infiltration and edema in the stroma, all consistent with acute pancreatitis. This is followed by a recovery phase with regeneration of epithelial cells (by day 5) and resolution of the inflammation. We found that Prrx1b RNA, but not Prrx1a RNA, is specifically induced in ADM lesions in experimental acute pancreatitis but is restored to the baseline level upon recovery and regeneration.
We used Prrx1-creERT2-IRES-GFP mice in combination with isolation of DBA lectin+ ductal cells. This reveals that a small subset of DBA lectin+ cells that expresses Prrx1 has enriched self-renewal capacity based on the pancreatosphere assay, reinforced by the finding that this subpopulation of cells has high ngn3 and nestin expression. It is tempting to speculate that Prrx1+ duct cells that express progenitor markers (e.g., Ngn3 and nestin) reside in a certain niche within the ductal compartment, which is maintained by Hgf, since we found that Hgf expression is increased in the DBA+GFP+ cells (data not shown). Notably, the pancreas is a relatively quiescent organ (Bardeesy and Depinho 2002). We found that the DBA+GFP+ cells form a greater number of spheres. The number of DBA+GFP+ cells increase during mild chronic pancreatitis, perhaps suggesting that such a small subpopulation of cells expands during inflammatory stress and is susceptible to the biological effects of KrasG12D/+ mutation.
Interestingly, during experimental pancreatitis induced in Sox9-IRES-creERT2;R26LacZ mice, there is expansion of Sox9+ cells that express ngn3. Sox9 is a transcription factor whose expression is largely restricted to ductal cells in the postnatal and adult pancreas (Kopp et al. 2011). Sox9 is expressed also in ADM lesions (Kopp et al. 2012; Prévot et al. 2012). We found that Prrx1b overexpression results in the induction of Sox9 expression; conversely, in cells with high endogenous Prrx1a and Prrx1b, knockdown of Prrx1b specifically decreases Sox9 expression. To explore a potential functional mechanistic relationship between Prrx1 and its isoforms with Sox9, we performed ChIP assays. This reveals that Prrx1b, not Prrx1a, binds specifically to the Sox9 promoter, specifically in a region from −1950 to −1830 bp. This region contains a highly conserved sequence (AACAAATTACA) that is predicted to be bound by Prrx1 (Hooghe et al. 2008). Interestingly, while both Prrx1a and Prrx1b share an identical homeobox domain, Prrx1a does not bind Sox9, thereby suggesting that the OAR domain in Prrx1a (not found in Prrx1b) is interfering potentially with DNA binding to Sox9. Indeed, a mechanism by which the OAR domain is modulating DNA-binding ability has been described (Norris and Kern 2001). Furthermore, Prrx1 binding of the Sox9 promoter is restricted to −1950 to −1830 but is not found in −1512 to −1433 and −1247 to −1223, although these latter two regions harbor sequences with conserved CAAT motifs. Functionally, the Sox9 promoter luciferase reporter gene is regulated by Prrx1b, and that relationship is underscored by their coordinated expression in murine pancreatic cell lines and in Prrx1+/− E16 pancreatic ductal cells in vivo.
Our results suggest a model in which pancreatitis and ensuing regeneration involve Prrx1b-mediated induction of Sox9, a novel finding. The interplay between Prrx1b and Sox9 might be critical for a subpopulation of cells (either ductal cells or ADM cells) that are able to self-renew and contribute to regeneration. Malignant transformation may be viewed as another example of cellular stress. Prrx1b overexpression is associated with increased proliferation, self-renewal of pancreatic ductal epithelial cells, and misshapen cysts in 3D culture. Whereas Prrx1b preferentially results in increased invasion, Prrx1a appears to have a greater capacity to induce migration of pancreatic cancer cells. Interestingly, both isoforms are up-regulated in metastatic pancreatic cancer cells. Notably, Prrx1b is increased significantly in CPCs. CPCs have been noted to have increased self-renewal capacity (Rhim et al. 2012). Perhaps Prrx1b is enriched in cellular clones that have the ability to survive after escaping the primary pancreatic cancer microenvironment, enter the circulation, and eventually metastasize. Whether the same population of cells or a different population of cells participates in self-renewal in pancreatitis versus PDAC is not clear, but nevertheless, the ductal cell appears to be a default phenotype regardless of the cells of origin.
Materials and methods
Pancreatic ductal cell isolation
Single-cell suspensions from the mouse pancreas were done as described previously (Schreiber et al. 2004). Cells were resuspended in a buffer containing PBS (pH 7.2), 0.5% bovine serum albumin, and 2 mM EDTA. The buffer solution was divided into 106 cell fractions. One fraction was excluded from the following separation process and kept as a presorting sample. Fluorescein-labeled DBA lectin or biotin-labeled DBA (Vector Laboratories), respectively, was applied at a 1:400 dilution for 10 min on a rotor at 4°C. Cells were washed in the same buffer and spun down at 1000 rpm for 10 min. After discarding the supernatant, 106 cells were resuspended in 90 μL of buffer. Ten microliters of anti-FITC or streptavidin Microbeads (Miltenyi Biotec), respectively, was added and incubated on a rotor for 15 min at 4°C. Separation was performed using MS columns (Miltenyi Biotec), according to the manufacturer’s protocol. RNA from each of the three fractions (DBA-positive, DBA-negative, and presorting) was isolated using the RNAqueous microkit (Ambion).
The colony formation assay after DBA lectin separation was performed with the cell culture conditions described below. Five-hundred viable cells were plated onto a collagen-coated 24-well plate. Colonies were quantified after 7 d. At this time, cells on collagen were dissociated and replated at the same density and culture condition. Secondary colony formation was assessed 7 d later. Error bars represent the standard error of the mean (SEM). (*) P < 0.05 is statistically significant (Student’s t-test).
Cell culture
Primary pancreatic cells were cultured and maintained as described previously (Schreiber et al. 2004). Dr. Sunil Hingorani kindly provided murine PanIN, PDAC, and metastatic cell lines. The 4313 cell line was isolated from a mouse harboring PanIN lesions (KrasG12D/+;Cre). The 4964 and 5143 cell lines (KrasG12D/+;p53R172H/+;Cre) were established from a mouse with PDAC (Hingorani et al. 2005). The 5143L2 cell line (KrasG12D/+;p53R172H/+;Cre) was isolated from a liver metastasis in mouse 5143. 3D organotypic culture was performed as described previously (Wescott et al. 2009). Cysts were cultured for a period of 10 d before they were quantified. Assays have been performed independently at least three times. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
Migration and invasion assays
Cell migration and invasion assays were performed using Boyden chambers with the BD Falcon FluoroBlok 24-Multiwell Insert System (BD Biosciences) or BD BioCoat Matrigel Invasion Chamber (8-μm pore size; BD Biosciences). We suspended 4 × 105 cells per milliliter in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma Chemical) supplemented with 0.5% fetal bovine serum (FBS). DMEM supplemented with 10% FBS served as a chemo-attractant in the lower chamber. Cells were incubated in 5% CO2 for 22 h at 37°C for the migration assay and for 48 h at 37°C for the invasion assay. Cells were stained by Calcein AM fluorescent dye (BD Biosciences), according to the manufacturer’s instructions and counted manually in a blinded fashion of four fields or read at wavelength 485/528 nm with the FLX800 multidetection microplate reader (BioTek Instruments). Each experiment was performed independently in quadruplicate for a total of three experiments. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
Cell proliferation assay
A total of 2.0 × 103 mouse primary pancreatic cell lines were cultured in 96-well plates and incubated in 5% CO2 at 37°C. Cells were quantified by a colorimetric cell proliferation assay using the WST-1 reagent (Roche) according to the manufacturer’s instructions. Absorbance was measured on a microplate reader (Tecan Sunrise, Tecan Group) at a wavelength of 450 nm. Each experiment was performed independently in quadruplicate for a total of three experiments. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
Pancreatosphere assay
Cells were grown in suspension as described before (Rovira et al. 2010). Sphere assays with 4313 cells were plated at a concentration of 10 cells per well by serial dilution. Accuracy was determined immediately after plating. Overexpression studies were assayed 5 d after plating. RNAi studies were assayed 8 d after plating. Primary cells from the Prrx1creERT2-IRES-GFP mice were sorted directly onto ultralow-attachment 96-well plates with a concentration of 50 cells per well. Spheres were cultivated for 14 d. Sphere medium was added every other day. Spheres were quantified with the Leica DMIRB inverted microscope. Assays were performed independently at least three times. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
Immunofluorescence/immunohistochemistry
Immunostaining was described previously (von Burstin et al. 2010). In brief, 4% PFA-fixed paraffin-embedded tissues were sectioned (6 μm). Antigen retrieval was performed using citric buffer (pH 6.0) and pressure cooking. Sections were washed with PBS and blocked using PBS + 0.3% Triton-X + 5% normal goat serum (Jackson ImmunoResearch) for 1 h at room temperature. Primary antibodies were applied and incubated overnight at 4°C. After two washes with PBS, secondary antibodies were applied and incubated for 30 min at 37°C. DAPI counterstaining was performed (Sigma-Aldrich).
The following primary antibodies were used: anti-K19 (dilution 1:100; TROMA-III, Developmental Studies Hybridoma Bank, University of Iowa), anti-PDX1 (dilution 1:1000; A-17, Santa Cruz Biotechnology), anti-PRRX1 (dilution 1:250; NBP1-06067, Novus Biologicals), anti-PRRX1 (dilution 1:20; HPA051084, Sigma), anti-GFP (dilution 1:250; ab13970, Abcam), and anti-SOX9 (dilution 1:1000; AB5535, EMD Millipore).
qPCR
One microgram of RNA was primed using random hexamers and transcribed into cDNA (TaqMan reverse transcription reagents, Applied Biosystems). SYBR Green was detected, and quantitative analysis was performed on the StepOnePlus system (Applied Biosystems). Primer sequences are listed in Supplemental Table S4. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
RNAi transfection, lentiviral transduction, and vector constructs
Cells were transfected using Lipofectamine RNAiMAX (Life Technologies) and a final concentration of 2 nM siRNA in Opti-MEM I reduced serum medium (GIBCO, Life Technologies). siRNAs were custom-designed and purchased from Invitrogen (Supplemental Table 4). As control siRNA, siGENOME Non-Targeting siRNA Pool 1 (Dharmacon) was used. Mouse Sox9 siRNA was purchased from Dharmacon (Sox9 On-Target Plus Smart Pool, Dharmacon).
Coding sequences of Prrx1a and Prrx1b were amplified from pancreatic ductal cell cDNA (Supplemental Table 4). Following PCR amplification, coding sequences were digested using XhoI and EcoRI or AgeI and MluI, respectively, and subcloned into pIRES2-EGFP (Clonetech) and pTRIPZ (RHS4743; Open Biosystems). Both Prrx1a and Prrx1b were Flag-tagged at the N terminus (Supplemental Table 4). Human pTRIPZ-shPrrx1 (RHS4696-99354863, RHS4696-99682631, and RHS4696-99703533, Open Biosystems) and mouse pGIPZ (V3LMM_514313, Open Biosystems) sh-Prrx1 constructs were purchased.
ChIP
ChIP in primary pancreatic ductal epithelial cells was performed according to the protocol established at the University of Pennsylvania Functional Genomics Core (FGC; http://fgc.genomics.upenn.edu) with the following modifications: Ten micrograms of chromatin was diluted with ChIP dilution buffer to final volume of 1 mL. Two percent of the sample was used as input chromatin. Remaining chromatin was incubated with 2 μg of Flag-M2 antibody (F1804, Sigma-Aldrich) overnight at 4°C. ChIP-grade magnetic beads (9006, Cell Signaling) were used for the pull-down. Reverse-cross-linked chromatin was purified using the QIAquick PCR purification kit (28106, Qiagen). ChIP primer sequences are listed in Supplemental Table 4. Immunoprecipitated chromatin was normalized to input chromatin. Enrichment was calculated relative to non-Flag-tagged control. ChIP was done in at least three independent immunoprecipitations. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
Luciferase assay
The luciferase reporter assay was performed as described previously using the Dual-Luciferase assay kit (Promega) (von Burstin et al. 2010). In brief, 4313 cells were plated onto 24-well plates in a density of 5 × 104 cells per well. Two nanograms of renilla luciferase (pRL-SV40 vector; Promega) and 200 ng of pGL3-basic (Promega) or pGL3-Sox9-2118/-597bp (primer sequences are listed in Supplemental Table 4), respectively, were cotransfected with control, Prrx1a siRNA, and Prrx1b siRNA. The assay was performed 48 h after transfection and read with the GloMax multidetection system (Promega). Assays were performed independently at least three times in quadruplicates. P < 0.05 was statistically significant (Mann-Whitney-Wilcoxon test). Error bars represent the SEM.
FACS
Cells from pancreata isolated from Prrx1creERT2-IRES-GFP mice were brought into single-cell suspension in buffer containing PBS (pH 7.2), 0.5% bovine serum albumin, and 2 mM EDTA as described earlier. Ductal cells were labeled using rhodamine-DBA lectin (dilution 1:400; RLK-2200, Vector Laboratories). DAPI (1 μg/mL) was added to assess cell viability. The FACSDiVa sorter (BD Biosciences) was used within the University of Pennsylvania Flow Cytometry and Cell Sorting Facility. Cells were directly sorted into 300 μL of lysis provided with the RNAqueous microkit (Ambion) for RNA isolation. Cells were sorted directly into ultralow-attachment 96-well plates for sphere assays in suspension. Epithelial cells from Prrx1creERT2;Rosa26YFP;LSL-KrasG12D/+ mice were labeled using anti-mouse CD326 (EpCAM) APC (eBiosciences) and analyzed using the FACSCalibur (BD Biosciences).
Animals and experimental procedures
The Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania approved all procedures (protocols #802868, #803699 and #804209). C57Bl/6 and Rosa26YFP mice were purchased from the Jackson Laboratory (JAX). Prrx1creERT2-IRES-GFP animals were kindly provided by Dr. Shunichi Murakami and described before (Kawanami et al. 2009). The Pdx1cre (Gu et al. 2002), LSL-KrasG12D/+ (Jackson et al. 2001), and Pdx1-Cre;KrasG12D/+;p53fl/+(Rhim et al. 2012) mice were used for studies. Acute pancreatitis was induced as described previously (Algül et al. 2007; Siveke et al. 2008). In brief, after a fasting period of 18 h, 8- to 10-wk old C57Bl/6 mice were injected intraperitoneally with cerulein (C9026, Sigma-Aldrich). Mice were injected over two consecutive days with eight hourly injections per day. Two-hundred microliters of 10 μg/mL cerulein (∼50 μg/kg bodyweight) was administered per injection. Control animals received 0.9% saline. As for the chronic pancreatitis, mice received 100 μL of 50 μg/mL cerulein for 5 d per week for the indicated time periods (Guerra et al. 2007). Tamoxifen (Sigma-Aldrich) was dissolved in peanut oil (30 mg/mL). Two-hundred microliters was administered via oral gavage.
Analysis of CPCs
YFP+ cells were isolated from 10-wk-old Pdx1cre;LSL-KrasG12D/+;p53fl/+;Rosa26YFP mice as described previously (Rhim et al. 2012). The paired Student’s t-test was used (P < 0.05 was statistically significant).
Statistical analysis of microarray data
Partek Genomics suite (Partek Inc.) or Biometric Research Branch (BRB)-ArrayTools were used for statistical analysis of the gene expression data (Simon et al. 2007), and all other statistical analyses were performed in the R language environment (http://www.r-project.org). All gene expression data were generated using the Affymetrix platform (GeneChip Mouse Gene 1.0 ST array) and normalized using a robust multiarray averaging method in BRB-ArrayTools (Irizarry et al. 2003). We identified genes that were differentially expressed between the two classes using a random variance t-test. Differences in gene expression between the two classes were considered statistically significant if P < 0.001. Heat maps of gene expression patterns were generated as described previously (Eisen et al. 1998).
Selection of genes from RNA microarray analysis
To uncover genes whose expression pattern is significantly different across three experimental groups (acute pancreatitis, embryonic ductal development, and Pdx1cre;LSL-KrasG12D/+ [referred to as KrasG12D]) compared with control, we first applied a stringent cutoff (P < 0.001) in each comparison (i.e., acute pancreatitis vs. control) to avoid inclusion of potential false-positive genes. When a Venn diagram approach was applied to gene lists from the three comparisons, 234 genes showed a common expression pattern across all three experimental groups.
In order to select genes whose expression patterns are correlated with KrasG12D sample sets, gene expression data from KrasG12D samples were first ranked according to the specific KrasG12D group (wild type < KRAS10 < KRAS20, where 10 = 10-wk-old mice, and 20 = 20-wk-old mice). Correlation of expressed gene levels with ranked KrasG12D was then computed to identify potential candidate genes. Likewise, embryonic samples were ranked according to developmental stages (E14 > E15 > E16 > P1). Levels of genes were correlated with the specific developmental stages. When the two gene lists were compared with each other, expression of only 76 genes was correlated positively with KRAS activity (r > 0.5) as well as negatively correlated with developmental stages (r < −0.5).
Acknowledgments
We express our gratitude to the Microarray Core Facility (Don Baldwin and John Tobias), Functional Genomics Core Facility (Jonathan Schug and Klaus Kaestner), and Flow Cytometry and Cell Sorting Facility. We are grateful to Dr. S. Murakami for the Prrx1creERT2-IRES-GFP mice, and Dr. Sunil Hingorani for the 4313, 4964, 5143, 5143L2 cell lines. Finally, we are grateful for discussions with members of the Rustgi laboratory (Blair Madison and Basil Bakir) and the Penn Pancreas Group. This work was supported by the NIH DK060694 (to M.R., S.T., S.H., and A.K.R.), National Pancreas Foundation (M.R. and A.D.R.), Honjo International Scholarship Foundation (S.T.), Deutsche Krebshilfe (S.H.), NIH/NIDDK P30-DK050306 Center for Molecular Studies in Digestive and Liver Diseases (and Molecular Pathology and Imaging, Molecular Biology/Gene Expression, Cell Culture, and Transgenic and Chimeric Mouse Cores), and American Society Grant RP-10-033-01-CCE. This work was also supported by other NIH grants (DK088945 and DK007066 to A.D.R.; and CA117969, DK083355, and DK083111 to B.Z.S.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.204453.112.
References
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, Depinho RA 2003. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 17: 3112–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algül H, Treiber M, Lesina M, Nakhai H, Saur D, Geisler F, Pfeifer A, Paxian S, Schmid RM 2007. Pancreas-specific RelA/p65 truncation increases susceptibility of acini to inflammation-associated cell death following cerulein pancreatitis. J Clin Invest 117: 1490–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Depinho RA 2002. Pancreatic cancer biology and genetics. Nat Rev Cancer 2: 897–909 [DOI] [PubMed] [Google Scholar]
- Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, Vonderheide RH 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 21: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrich V, Esni F, Garay MVR, Feldmann G, Habbe N, Jensen JN, Dor Y, Stoffers D, Jensen J, Leach SD, et al. 2008. Hedgehog signaling is required for effective regeneration of exocrine pancreas. Gastroenterology 135: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. 2011. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41 [DOI] [PubMed] [Google Scholar]
- Gittes G 2009. Developmental biology of the pancreas: A comprehensive review. Dev Biol 326: 4–35 [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA 2002. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447–2457 [DOI] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, Dubus P, Sandgren EP, Barbacid M 2007. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11: 291–302 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4: 437–450 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483 [DOI] [PubMed] [Google Scholar]
- Hooghe B, Hulpiau P, van Roy F, De Bleser P 2008. ConTra: A promoter alignment analysis tool for identification of transcription factor binding sites across species. Nucleic Acids Res 36: W128–W132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009a. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15: 3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanami A, Matsushita T, Chan YY, Murakami S 2009. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochem Biophys Res Commun 386: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M 2011. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development 138: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, von Figura G, Mayes E, Liu F-F, Dubois CL, Morris JP, Pan FC, Akiyama H, Wright CVE, Jensen K, et al. 2012. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22: 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MF, Cheng HT, Kern MJ, Potter SS, Tran B, Diekwisch TG, Martin JF 1999. prx-1 functions cooperatively with another paired-related homeobox gene, prx-2, to maintain cell fates within the craniofacial mesenchyme. Development 126: 495–504 [DOI] [PubMed] [Google Scholar]
- Lynn FC, Smith SB, Wilson ME, Yang KY, Nekrep N, German MS 2007. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci 104: 10500–10505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JF, Olson EN 2000. Identification of a prx1 limb enhancer. Genesis 26: 225–229 [PubMed] [Google Scholar]
- McKean DM, Sisbarro L, Ilic D, Kaplan-Alburquerque N, Nemenoff R, Weiser-Evans M, Kern MJ, Jones PL 2003. FAK induces expression of Prx1 to promote tenascin-C-dependent fibroblast migration. J Cell Biol 161: 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris RA, Kern MJ 2001. The identification of Prx1 transcription regulatory domains provides a mechanism for unequal compensation by the Prx1 and Prx2 loci. J Biol Chem 276: 26829–26837 [DOI] [PubMed] [Google Scholar]
- Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, Hruban RH, Goggins M, Leach SD 2005. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res 65: 1619–1626 [DOI] [PubMed] [Google Scholar]
- Prévot P-P, Simion A, Grimont A, Colletti M, Khalaileh A, Van den Steen G, Sempoux C, Xu X, Roelants V, Hald J, et al. 2012. Role of the ductal transcription factors HNF6 and Sox9 in pancreatic acinar-to-ductal metaplasia. Gut 61: 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D 2012. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21: 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Rustgi AK 2011. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest 121: 4572–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. 2012. EMT and dissemination precede pancreatic tumor formation. Cell 148: 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira M, Scott S-G, Liss AS, Jensen J, Thayer SP, Leach SD 2010. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci 107: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber FS, Deramaudt TB, Brunner TB, Boretti MI, Gooch KJ, Stoffers DA, Bernhard EJ, Rustgi AK 2004. Successful growth and characterization of mouse pancreatic ductal cells: Functional properties of the Ki-RAS(G12V) oncogene. Gastroenterology 127: 250–260 [DOI] [PubMed] [Google Scholar]
- Simeone A, D’Apice MR, Nigro V, Casanova J, Graziani F, Acampora D, Avantaggiato V 1994. Orthopedia, a novel homeobox-containing gene expressed in the developing CNS of both mouse and Drosophila. Neuron 13: 83–101 [DOI] [PubMed] [Google Scholar]
- Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y 2007. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform 3: 11–17 [PMC free article] [PubMed] [Google Scholar]
- Siveke JT, Lubeseder-Martellato C, Lee M, Mazur PK, Nakhai H, Radtke F, Schmid RM 2008. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology 134: 544–555 [DOI] [PubMed] [Google Scholar]
- Strobel O, Dor Y, Alsina J, Stirman A, Lauwers G, Trainor A, Castillo CFD, Warshaw AL, Thayer SP 2007. In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology 133: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Reijnen MJ, van Raaij EJ, Verbeek F, Gaffield W, Meijlink F 2001. Prx1 and Prx2 are upstream regulators of sonic hedgehog and control cell proliferation during mandibular arch morphogenesis. Development 128: 2929–2938 [DOI] [PubMed] [Google Scholar]
- von Burstin J, Reichert M, Wescott MP, Rustgi AK 2010. The pancreatic and duodenal homeobox protein PDX-1 regulates the ductal specific keratin 19 through the degradation of MEIS1 and DNA binding. PLoS ONE 5: e12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wescott MP, Rovira M, Reichert M, von Burstin J, Means A, Leach SD, Rustgi AK 2009. Pancreatic ductal morphogenesis and the Pdx1 homeodomain transcription factor. Mol Biol Cell 20: 4838–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]