Little is known about membrane remodelling during bacterial growth and sporulation. Rudner and colleagues now identify and characterize a novel protein, FisB, the first bacterial protein known to mediate this process. FisB, which localizes to reaction sites and mediates membrane fission during spore formation, is the founding member of a new class of fission proteins.
Keywords: sporulation, engulfment, fission, morphogenesis, fusion, dynamin, ESCRT-III
Abstract
How bacteria catalyze membrane fission during growth and differentiation is an outstanding question in prokaryotic cell biology. Here, we describe a protein (FisB, for fission protein B) that mediates membrane fission during the morphological process of spore formation in Bacillus subtilis. Sporulating cells divide asymmetrically, generating a large mother cell and smaller forespore. After division, the mother cell membranes migrate around the forespore in a phagocytic-like process called engulfment. Membrane fission releases the forespore into the mother cell cytoplasm. Cells lacking FisB are severely and specifically impaired in the fission reaction. Moreover, GFP-FisB forms dynamic foci that become immobilized at the site of fission. Purified FisB catalyzes lipid mixing in vitro and is only required in one of the fusing membranes, suggesting that FisB–lipid interactions drive membrane remodeling. Consistent with this idea, the extracytoplasmic domain of FisB binds with remarkable specificity to cardiolipin, a lipid enriched in the engulfing membranes and regions of negative curvature. We propose that membrane topology at the final stage of engulfment and FisB–cardiolipin interactions ensure that the mother cell membranes are severed at the right time and place. The unique properties of FisB set it apart from the known fission machineries in eukaryotes, suggesting that it represents a new class of fission proteins.
The remodeling of lipid bilayers is essential to all organisms. These remodeling events include membrane fusion, wherein two membranes merge to become one, and membrane fission, where a single lipid bilayer divides into two. Our understanding of the molecular mechanisms underlying these processes comes from extensive studies in eukaryotes and the enveloped viruses that infect them. In eukaryotes, fusion and fission play central roles in a broad array of cell biological processes, including exocytosis, endocytosis, cytokinesis, and intracellular membrane trafficking (Chernomordik and Kozlov 2003; Südhof and Rothman 2009; Kozlov et al. 2010). They are also critical for the entry and exit of enveloped viruses into (and out of) their host (Kielian and Rey 2006; Harrison 2008). In contrast, virtually nothing is known about how bacteria remodel their membranes during growth and differentiation despite the fact that they undergo membrane fission in every cell division cycle. Here, we describe the identification and characterization of a newly identified protein (FisB, for fission protein B) that functions in membrane fission during the developmental process of spore formation in the bacterium Bacillus subtilis.
From a topological perspective, fusion and fission can be seen as opposed processes. However, at the molecular level, both involve local disruption, bending, and bridging of lipid bilayers. Achieving lipid mixing during either fusion or fission requires forcing bilayers to within a critical distance of a few nanometers (Rand and Parsegian 1986; Wong et al. 1999; Kozlovsky and Kozlov 2003; Bashkirov et al. 2008; Kozlov et al. 2010). For both processes, hydration forces are thought to provide the major obstacle against lipid merger, which is alleviated by bending coupled to lipid tilting that allows hydrophobic attraction between the hydrocarbon chains of the lipids (Chernomordik and Kozlov 2003; Kozlov et al. 2010). Studies in eukaryotes and the viruses that infect them have revealed that specialized proteins generate the work required for these lipid rearrangements. In the case of eukaryotic membrane fusion, SNARE proteins anchored in apposed membranes form trans-complexes that overcome repulsive forces to bring bilayers into intimate contact. In enveloped viruses, membrane fusion proteins that are anchored in the viral membrane insert into host cell membranes and, through a series of conformational changes, bring the lipid bilayers into close proximity, leading to fusion. As for fission, the only factors described to date that participate in it are dynamin (Ferguson and De Camilli 2012) and the endosomal sorting complex required for transport (ESCRT-III complex) (Hurley 2010; Wollert and Hurley 2010; Henne et al. 2011). Dynamin and dynamin-like proteins are involved in endocytosis, multivesicular membrane budding, cytokinesis in chloroplasts, and mitochondrial genesis. These proteins are thought to function as “pinching machines”: They polymerize around the outside of the neck that connects two parts of a contiguous membrane, constricting the neck to a critical radius. GTP hydrolysis then produces conformational changes that result in fission (Ferguson and De Camilli 2012). In contrast, the ESCRT-III complex appears to assemble on the inside or at one of the openings of the neck to induce membrane constriction and, ultimately, fission (Hurley 2010; Henne et al. 2011). The ESCRT-III complex has been implicated in the biogenesis of multivesicular bodies (Gruenberg and Stenmark 2004; Hurley 2008; Wollert et al. 2009; Wollert and Hurley 2010), membrane abscission during cytokinesis (Elia et al. 2011), and budding of some enveloped viruses from the host membrane (Strack et al. 2003; Morita and Sundquist 2004; Bieniasz 2006). Interestingly, a homologous set of proteins appears to function in cytokinesis in the Crenarchaea Sulfolobus (Lindås et al. 2008; Samson et al. 2008).
The molecular mechanism underlying membrane fission during cytokinesis in bacteria remains enigmatic. It has been proposed that protein components of the cell division machinery could catalyze fission (Sharp and Pogliano 1999; Liu et al. 2006; de Boer 2010; Fleming et al. 2010). Alternatively, it has been suggested that cell wall synthesis on the outside of the cell could force the constricting membranes into close proximity, ultimately leading to fission (Weiss 2004; Judd et al. 2005; Meyer et al. 2010). To date, no protein has been directly implicated in this fission reaction.
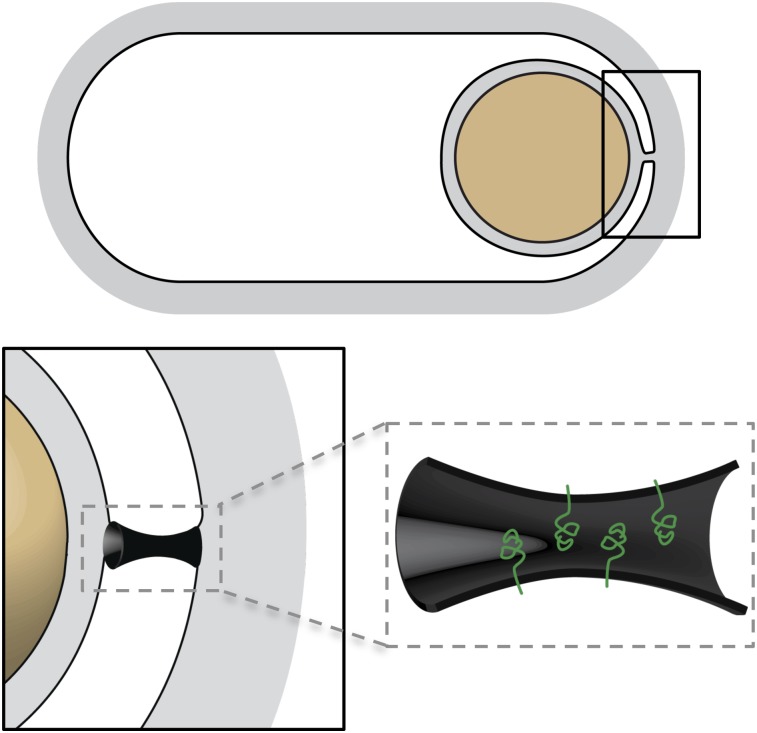
Here, we investigated a specialized membrane fission event that occurs during sporulation in B. subtilis (Fig. 1). In response to starvation, B. subtilis differentiates into a dormant spore (Stragier and Losick 1996; Errington 2003). Upon initiation of this developmental process, the cell divides asymmetrically, generating two cells of unequal size and dissimilar fate. The smaller cell is the prospective spore and is referred to as the forespore. The larger cell is called the mother cell. Initially, these two cells lie side by side, separated by a double-membrane septum. However, shortly after polar division, the mother cell membranes migrate around the forespore in a phagocytic-like process called engulfment. In the last stage of this process, the leading edges of the migrating membranes meet at the cell pole and, upon membrane fission, the forespore is released into the mother cell cytoplasm (Fig. 1). The forespore is thus surrounded by two membranes: an inner membrane that contains the forespore cytoplasm and an outer membrane derived from the mother cell membrane. The molecular mechanism underlying this membrane remodeling event is the focus of our work.
Figure 1.
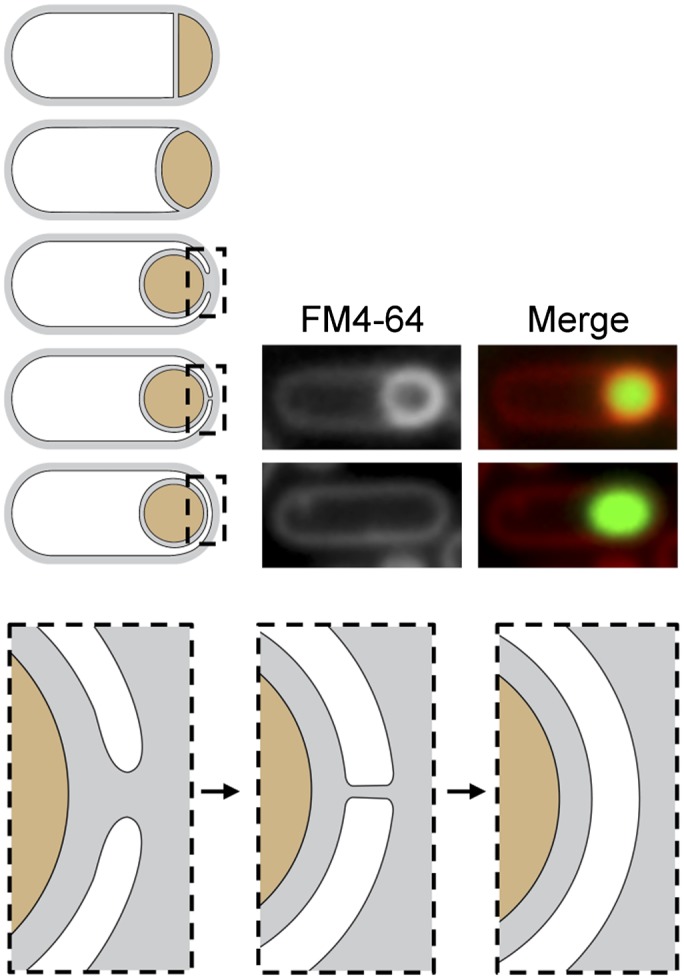
Membrane fission during spore development. Schematic representation of the morphological stages of the engulfment process. After polar division, the mother cell membranes migrate around the forespore. When they meet at (or near) the cell pole, membrane fission releases the forespore into the mother cell cytoplasm. Enlarged schematics highlight the engulfing membranes pre- and post-fission. (Middle panel) At the final stage, the membranes form a tube that is severed in the fission reaction. Fluorescent images show sporulating cells (strain BKM015) before and after membrane fission. Prior to fission, the lipophylic dye FM4-64 labels the peripheral membranes of the mother cell and the double membrane surrounding the forespore. After fission, the dye cannot access the membranes surrounding the forespore. The presence of a forespore within the mother cell is assessed by a fluorescent forespore reporter (PspoIIQ-cfp).
We describe the identification and characterization of a membrane protein, FisB, that is required for membrane fission at the last stage of engulfment. FisB is a bitopic membrane protein that is produced in the mother cell after polar division. In its absence, the migration of the mother cell membranes around the forespore proceeds normally, but engulfment stalls when the membranes reach the cell pole. In support of the idea that FisB functions in membrane remodeling, a functional GFP-FisB fusion localizes as a focus at the cell pole at the time of membrane fission. Consistent with a direct role in membrane remodeling, we show that recombinant FisB catalyzes lipid mixing in vitro. Interestingly, lipid-mixing activity required FisB in only one of the two interacting membranes, suggesting that FisB–lipid interactions drive membrane remodeling. In support of this idea, we show that the extracellular domain of FisB directly interacts with liposomes. Moreover, FisB–liposome interaction required the presence of the anionic phospholipid cardiolipin (CL). Altogether, our data support the idea that FisB directly catalyzes the membrane fission event that marks the end of engulfment. Given the topology of FisB and that of the membrane tube to be severed, we suggest that FisB promotes fission from within or near one of the openings of the tube by interacting with CL that is enriched in the engulfment membranes and in regions of high negative curvature.
Results
FisB is required for membrane fission at the last stage of engulfment
Previous studies on membrane fission during sporulation implicated the polytopic membrane protein SpoIIIE (Sharp and Pogliano 1999, 2003). SpoIIIE is a DNA transporter that is required to translocate the forespore chromosome into the forespore compartment (Wu and Errington 1994). In the absence of SpoIIIE, sporulating cells are unable to pump the forespore chromosome and fail to make viable spores. Using an innovative membrane fission assay (described below), Sharp and Pogliano (1999) discovered that cells lacking SpoIIIE are also defective in membrane fission at a late stage of engulfment. In support of the idea that SpoIIIE functions directly in catalyzing fission, a SpoIIIE-GFP fusion protein localized to the forespore pole around the time of membrane fission (Sharp and Pogliano 1999). In the course of our studies on chromosome organization and segregation during sporulation (Burton et al. 2007; Marquis et al. 2008; Sullivan et al. 2009), we observed that sporulating cells lacking SpoIIIE exhibited defects in engulfment at stages prior to membrane fission. Under our assay conditions, 80%–90% of the SpoIIIE mutant cells had bulged septal membranes and/or invaginations that appeared to prevent complete membrane migration around the forespore (Supplemental Fig. S1). These observations led us to consider the possibility that SpoIIIE might only be indirectly involved in membrane fission and that other proteins catalyze this reaction.
We began our studies by investigating two candidate factors for a role in catalyzing fission: the B. subtilis dynamin homolog DynA (Bürmann et al. 2011) and the peptidoglycan synthetic machinery (Meyer et al. 2010). B. subtilis DynA has been shown to catalyze membrane fusion in vitro; however, cells lacking DynA have virtually no phenotype during vegetative growth (Bürmann et al. 2011). To investigate whether the DynA mutant was impaired in membrane fission during sporulation, we took advantage of the fission assay developed by Sharp and Pogliano (1999). In this assay, cells are induced to sporulate synchronously by nutrient downshift. At time points after polar division, the cells are stained with the membrane dye FM4-64 and visualized by fluorescence microscopy. Since this dye is unable to cross the lipid bilayer, the membranes surrounding the forespore are only labeled in cells that have not yet undergone fission (Fig. 1). Once fission occurs and the forespore is released into the mother cell cytosol, the membrane-impermeant dye only has access to the peripheral mother cell membranes. To assess whether these cells have an engulfed forespore inside them, we used a forespore fluorescent reporter in which the gene encoding the cyan fluorescent protein (CFP) was placed under the control of the forespore-specific transcription factor σF (PspoIIQ-cfp), as described previously (Fig. 1; Meyer et al. 2010). Using this assay, we found that engulfment and membrane fission were indistinguishable in wild-type cells and cells lacking DynA (Fig. 2A). Thus, dynamin does not appear to function in membrane fission during sporulation.
Figure 2.
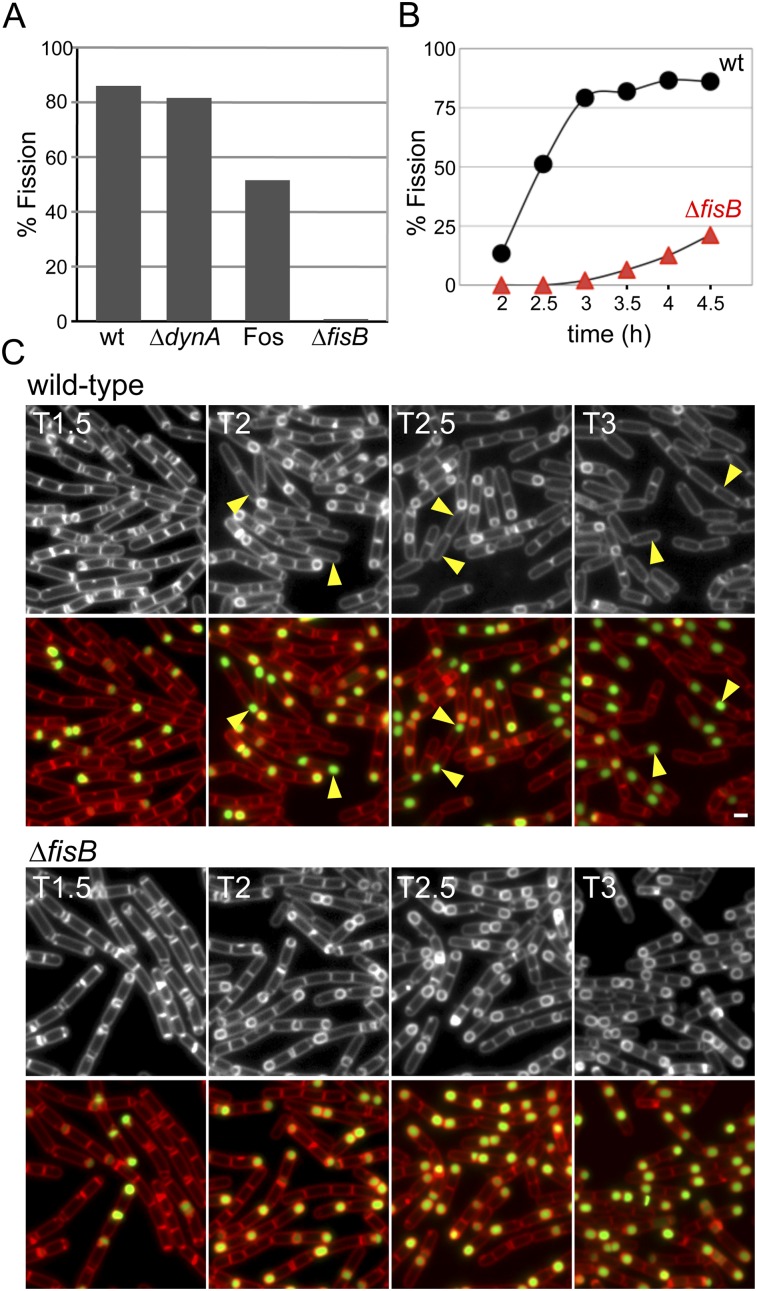
Cells lacking FisB are impaired in membrane fission. (A) Histogram shows fission efficiency (the percentage of sporulating cells that have undergone fission compared with all sporulating cells; n > 1000), 3 h after the onset of sporulation from a representative experiment. Wild type (BKM015), a DynA mutant (BAM027), wild type after the addition of 5 mM fosfomycin (Fos) that inhibits peptidoglycan synthesis at hour 1.5, and a FisB mutant (BTD3200) are shown. (B) Fission efficiency as a function of time from a representative experiment in a wild-type strain (wt, BKM15) and a FisB mutant (BTD3200). (C) Membrane fission was assessed by fluorescent microscopy during a sporulation time course in wild type (BKM015) and a FisB mutant (BTD3200). Both strains contained a fluorescent forespore reporter (PspoIIQ-cfp) to label all forespores. The membranes (false-colored red) were visualized with the fluorescent dye FM4-64 and merged with forespore CFP (false-colored green). Examples of sporulating cells in which fission has occurred are highlighted (yellow arrowheads). Time (in hours) after the initiation of sporulation is indicated. Bar, 1 μm.
Next, we investigated whether cell wall synthesis between the forespore and mother cell membranes is involved in membrane fission. Dworkin and colleagues (Meyer et al. 2010) recently proposed that the action of cell wall synthetic complexes anchored in the mother cell membrane could promote membrane migration during engulfment and membrane fission. Using a membrane fission assay similar to the one described above, these researchers reported that the addition of antibiotics that inhibit cell wall synthesis during the engulfment process resulted in a fourfold reduction in fission. To investigate whether the force generated from cell wall synthesis was the principal driver of membrane fission, we performed similar antibiotic addition experiments (see the Materials and Methods). Under our assay conditions, we observed a defect in membrane fission, although the effect was more modest than observed previously. Membrane fission was reduced from 86% to 51% when the antibiotic fosfomycin was added at hour 1.5 of sporulation (Fig. 2A). The reduction in fission was even less pronounced when the drug was added at later time points (data not shown). These results suggest that peptidoglycan synthesis and the force generated by it could help bring the membranes into close proximity but indicate that the protein (or proteins) that catalyze membrane fission remain to be discovered.
DynA was not required for membrane fission, and we were unable to identify homologs of the ESCRT-III fission machinery in the B. subtilis genome. Accordingly, we turned to the hypothesis that fission is mediated by a fusion-like mechanism. We reasoned that if such a fission protein existed, it would likely be synthesized in the mother cell at an early stage of sporulation prior to the fission reaction and would have at least one transmembrane segment, and cells lacking this protein would have reduced sporulation efficiency. The first mother cell-specific transcription factor that is induced during sporulation is the alternative σ factor σE (Stragier and Losick 1996; Piggot and Losick 2002). The σE regulon has been defined by transcriptional profiling (Eichenberger et al. 2003; Feucht et al. 2003; Steil et al. 2005), and strains harboring deletions in most of the genes in the regulon have been generated (Eichenberger et al. 2003). Using the criteria described above, we examined a subset of this ordered library of mutants using the fluorescent membrane fission assay of Sharp and Pogliano (1999). Our analysis identified one mutant (in the yunB gene) that was severely impaired in membrane fission (Fig. 2A). Based on its mutant phenotype and the characterization of the yunB gene product described below, we renamed this gene fisB for fission protein B.
To quantitatively assess the requirement for FisB in membrane fission, we compared wild type and the FisB mutant in a sporulation time course (Fig. 2B; Supplemental Fig. S2). Two hours after the initiation of sporulation, 13% of wild-type cells (n > 1000) had completed membrane fission, releasing the forespore into the mother cell cytoplasm. At this time point, there was no detectable fission in the mutant. Thirty minutes later, at hour 2.5, 51% of wild-type sporulating cells (n > 1000) had undergone membrane fission compared with <1% in the FisB mutant (n > 1000). By hour 3, 79% of wild-type sporulating cells had completed membrane fission, but only 2% had completed membrane fission in the mutant. Importantly, in cells lacking FisB, the migration of the mother cell membranes around the forespore appeared indistinguishable from wild type (Fig. 2B; Supplemental Fig. S2). Moreover, similar to wild type, membrane migration appeared complete in most cells lacking FisB by hour 2.5. These results indicate that the absence of FisB does not impact the early steps of engulfment and thus suggest that FisB plays a specific role in the membrane fission process.
Interestingly, membrane fission was not completely abolished in cells lacking FisB. By hour 4.5, ∼21% of the FisB mutant sporulating cells appeared to have undergone fission. This percentage is likely to be an overestimate because sporulating cells lacking FisB begin to lyse at this time point, reducing the total number of sporulating cells in the culture (Supplemental Fig. S2). FisB mutants have a sporulation efficiency of 12%–15%, suggesting that most of the cells that successfully undergo membrane fission are able to produce mature spores.
Altogether, these results suggest that FisB plays a specific role in membrane fission. In its absence, membrane migration around the forespore proceeds normally, but most sporulating cells fail to undergo membrane remodeling and release of the forespore into the mother cell cytoplasm. Cells that stall at the fission step ultimately lyse. Those that do complete fission do so after an ∼2-h delay.
FisB forms dynamic foci that localize to the pole at the time of membrane fission
FisB is predicted to be a bitopic membrane protein with a small N-terminal domain that resides in the mother cell cytoplasm and a larger (23-kDa) extracellular domain. To determine the subcellular localization of FisB, we generated an N-terminal monomeric GFP fusion to FisB expressed under its native promoter- and ribosome-binding site. The fusion protein complemented the FisB-null mutant for membrane fission and sporulation (data not shown). We visualized the GFP fusion by epifluorescence microscopy in a sporulation time course. For this experiment, we visualized engulfment and membrane fission using the membrane dye TMA-DPH. TMA-DPH inefficiently crosses the lipid bilayer and thus weakly labels the mother cell membrane that surrounds the forespore after fission is complete. Accordingly, using TMA-DPH, it is possible to distinguish pre- and post-fission stages without a forespore reporter. Supplemental Figure S3 shows a direct comparison of the two membrane dyes FM4-64 and TMA-DPH.
During engulfment at hour 2.5, GFP-FisB localized as discrete foci in both the peripheral membranes as well as the mother cell membranes that were engulfing the forespore (Fig. 3A). We were unable to quantify the number of FisB proteins in the foci, but the fluorescence intensity suggests that each focus is not a single protein and likely represents an oligomeric complex. Consistent with this idea, we found that both full-length FisB and the extracellular domain of FisB when purified from Escherichia coli can form oligomeric complexes in vitro (Supplemental Fig. S4). Time-lapse imaging revealed that the GFP-FisB foci were highly mobile in the membrane (Supplemental Movie M1). Importantly, at the time of membrane fission or just after it, a GFP-FisB focus was present near the cell pole adjacent to the forespore in a significant proportion of the cells. Strikingly, time-lapse imaging indicates that these polar foci remained immobile (Supplemental Movie M2). Figure 3B shows representative stills from a typical movie and an image in which each pixel is averaged over the entire time lapse. Dynamic foci are lost upon averaging, while immobile ones at the cell pole are retained.
Figure 3.
GFP-FisB forms dynamic foci that become immobilized at or near the cell pole at the time of membrane fission. (A) Images show GFP-FisB (strain BAM003) at hour 3 of sporulation. The membranes were visualized with the fluorescent dye TMA-DPH. Examples of sporulating cells with a discrete GFP-FisB focus at the cell pole are highlighted (yellow arrowheads). (B) Representative images from a time-lapse movie showing the dynamic behavior of GFP-FisB foci in the mother cell membranes at hour 2.5 of sporulation. Images were acquired every 2 min. The bottom panel (AVG) shows an image in which each pixel is averaged over the entire time lapse. Yellow arrowheads highlight immobile GFP-FisB foci at the cell pole. (C) Low levels of YFP-FisB (strain BTD3124) reveal specific localization at the cell pole at the time of fission. Fluorescent images are from hour 2 (T2) and hour 3 (T3) of sporulation. Punctate YFP-FisB localization prior to fission is highlighted with white arrowheads. Yellow arrowheads show discrete YFP-FisB foci at the cell pole at the time of membrane fission. A schematic representation of FisB localization is shown below the micrographs. Bar, 1 μm.
In some cases, lowering the level of a fluorescent fusion will reveal or highlight specific subcellular localization, perhaps by reducing nonspecific or low-affinity binding (Rudner et al. 2002; Gregory et al. 2008). Accordingly, we constructed a YFP-FisB fusion expressed at levels approximately fivefold lower than the native protein (see the Materials and Methods; data not shown). The fusion protein complemented the fisB-null, restoring sporulation efficiency to wild-type levels. Moreover, YFP-FisB supported efficient membrane fission, albeit with a 20- to 30-min delay compared with wild type (Fig. 3C; data not shown).
During early engulfment stages, YFP-FisB localized in foci in the mother cell membranes, as observed with the GFP-FisB fusion expressed at wild-type levels. These foci were more often found in the mother cell membranes that surrounded the forespore (Fig. 3C; Supplemental Fig. S5). Importantly, at hour 3, when sporulating cells were about to undergo fission or had just completed it, ∼65% of the sporulating cells (n > 1000) exhibited a YFP-FisB focus at or near the cell pole adjacent to the forespore (Fig. 3C, yellow arrowheads; Supplemental Fig. S5, yellow arrowheads). In most cases, the polar focus was brighter than the foci in the peripheral and spore membranes, consistent with the idea that FisB assembles into a larger oligomeric complex at the pole. Because of the lower expression levels, we were unable to assess FisB dynamics due to rapid photobleaching of the fusion protein.
These results suggest that FisB forms mobile oligomeric complexes in the membranes of the mother cell. When the engulfing membranes reach the cell pole, a subset of these complexes localize to this site, where they participate in the fission reaction.
FisB mediates membrane remodeling in vitro
Altogether, our results show that FisB localizes to the site of fission and is required for membrane remodeling in vivo. To investigate whether FisB plays a direct role in the fission reaction and gain insight into the mechanism, we turned to in vitro experiments. Based on tomographic cryoelectron microscopy of Caulobacter crescentus cells undergoing cytokinesis (Judd et al. 2005), the connection between daughter cells prior to fission can be viewed as a tube 20–30 nm in length. We imagine a similarly sized membrane tube exists between the peripheral membrane of the mother cell and that surrounding the forespore. Since most of the FisB protein (amino acids 39–254) is present on the extracytoplasmic side of the membrane, if FisB indeed mediates membrane fission, it is unlikely to do so via a dynamin-like constriction from outside of the tube. Instead, FisB must do work on the membrane tube from within or at one of its openings, interacting with proteins and/or lipids across the tube. This mechanism is reminiscent of those employed by eukaryotic SNAREs and viral fusion proteins to force apposing bilayers into close proximity. We therefore reasoned that if FisB catalyzes fission in vivo, purified full-length FisB inserted into artificial liposomes should be able to induce lipid mixing and membrane remodeling in vitro, as has been shown for SNAREs (Weber et al. 1998). We used a standard fluorescence dequenching assay for lipid mixing (Struck et al. 1981) in which a population of liposomes bearing the fluorescent lipid 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-PE) and its lipid-linked quencher, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (LR-PE), are mixed with unlabeled liposomes. Fusion of labeled and unlabeled liposomes increases the mean distance between the NBD-PE fluorophores and their quenchers, resulting in an increase in fluorescence (Fig. 4).
Figure 4.
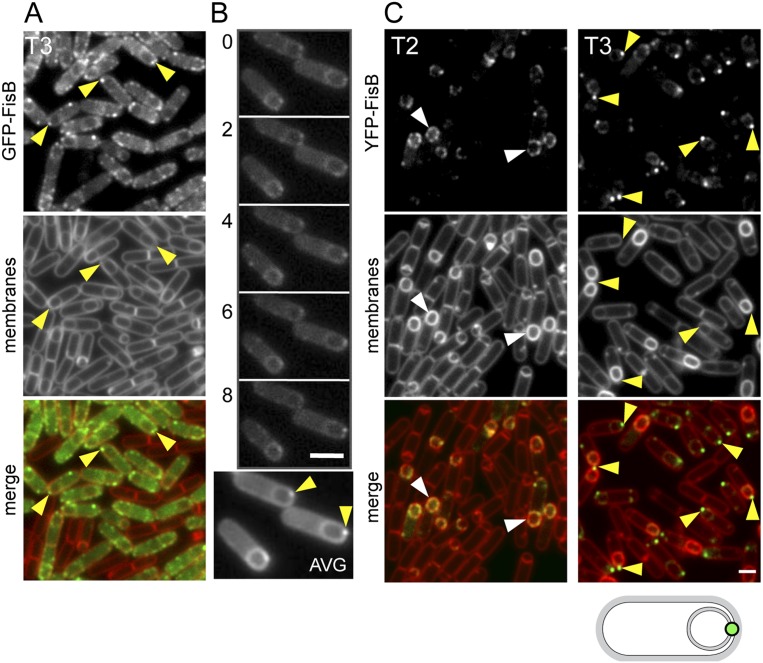
FisB catalyzes membrane remodeling in vitro. The lipid-mixing assay based on fluorescence dequenching is shown schematically above the graph. Fusion of labeled and unlabeled liposomes increases the mean distance between the NBD-PE fluorophores and their quenchers, resulting in an increase in fluorescence. See the Materials and Methods for details. Lipid mixing was monitored as an increase in NBD fluorescence as a function of time after incubation of labeled and unlabelled liposomes. Liposomes contained FisB or mock-purified proteins (mock) or were protein-free (pf). Fluorescence is expressed as the fraction of the maximum dequenching obtained after addition of detergent at the end of the experiment. The graph shows the average of four independent fusion experiments from three different reconstitutions (error bars correspond to the standard error).
Full-length his-tagged FisB was expressed in E. coli and purified from detergent-solubilized membranes using the lipid-like detergent Fos-choline-12 (see the Materials and Methods). The purified protein was then reconstituted into labeled or unlabeled liposomes composed of a mixture of phospholipids (phosphatidylcholine [PC], phosphatidylglycerol [PG], phosphatidylethanolamine [PE], and CL) found in sporulating B. subtilis cells (Griffiths and Setlow 2009) or a mixture of the minimal lipids (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine [POPC] and dioleyl phosphatidyl serine [DOPS]) commonly used for SNARE-mediated fusion (Weber et al. 1998; Karatekin et al. 2010). To control for contaminating membrane proteins in the His-FisB-purified fraction, we performed a mock purification from E. coli cells harboring the empty expression plasmid (Supplemental Fig. S6). Labeled liposomes reconstituted with FisB or with mock-purified proteins (mock) or that were protein-free were mixed with unlabeled protein-free, FisB, or mock liposomes, and lipid mixing was monitored as an increase in NBD fluorescence as a function of time.
Using liposomes composed of lipids found in sporulating B. subtilis, efficient lipid mixing was observed when FisB liposomes were mixed with either FisB or protein-free liposomes (Fig. 4). Labeled FisB liposomes and unlabeled FisB liposomes resulted in slightly less efficient fusion than between FisB and protein-free liposomes, perhaps due to surface coverage by FisB reducing the available area of target membranes to interact with FisB in trans. Importantly, all combinations involving protein-free liposomes and liposomes containing mock-purified proteins resulted in negligible lipid mixing (Fig. 4; data not shown). Interestingly, using liposomes composed of POPC/DOPS (the minimal SNARE lipid mixture), no fusion was observed between FisB–FisB or FisB–protein-free liposomes (data not shown). In contrast, neuronal SNAREs incorporated into liposomes containing POPC/DOPS resulted in efficient lipid mixing comparable with previous reports (Supplemental Fig. S7). These results show that FisB possesses an intrinsic capacity to remodel lipid membranes in vitro and strongly suggest it can also do so in vivo. The data further imply that the lipid remodeling activity of FisB critically relies on specific protein–lipid interactions.
FisB binds to CL
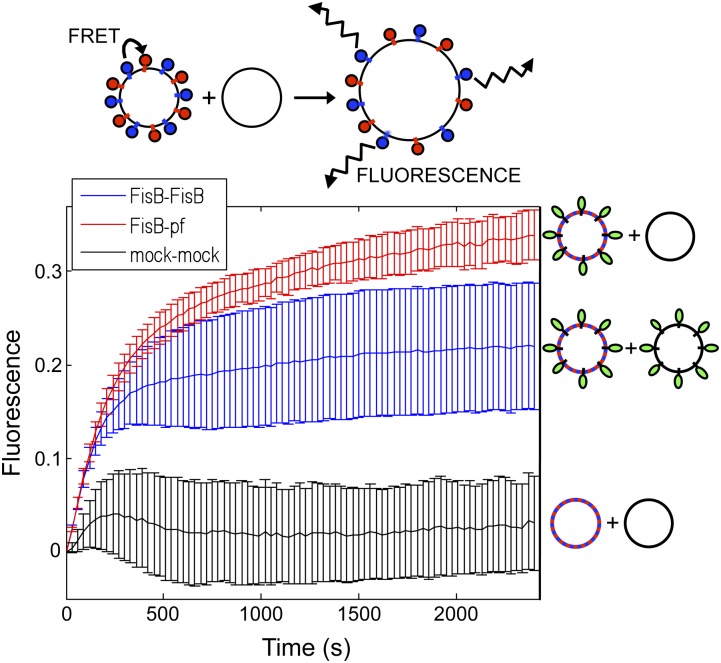
The results of the membrane fusion assay with the two different lipid mixtures suggest that an interaction between a particular lipid in the B. subtilis lipid composition was essential for FisB’s membrane remodeling activity. The simplest model is that this interaction occurs in trans and involves the extracellular domain of FisB (ECD). To test this idea and identify the lipids to which FisB binds, we used a coflotation assay (Fig. 5A; Bigay et al. 2005). The extracellular domain of FisB lacking its transmembrane segment was fused to a His tag, and the fusion protein was expressed in E. coli and purified from the soluble fraction (see the Materials and Methods). The soluble protein was then mixed with protein-free liposomes consisting of either the B. subtilis lipid mix (PG/PE/CL/PC) or the standard minimal SNARE composition (POPC/DOPS). The mixture was loaded at the bottom of a density step gradient, and the free liposomes and liposomes with bound protein were separated from unbound protein by ultracentrifugation (see the Materials and Methods). Consistent with the membrane fusion results, the His-FisB extracellular domain bound to and cofloated with the liposomes composed of the B. subtilis lipid mixture but not of the SNARE lipid mix (Fig. 5B).
Figure 5.
The extracellular domain of FisB binds CL in vitro. (A) Schematic representation of the coflotation assay. Protein-free liposomes were incubated with the soluble extracellular domain of FisB (ovals) for 2 h at room temperature. Liposomes and bound protein were separated from unbound protein using a four-step Nycodenz gradient. (B) The interface between the 0% and 30% step was collected and analyzed by SDS-PAGE stained with Coomassie blue. FisB bound to liposomes composed of a mixture of B. subtilis lipids (PG/PE/CL/PC) but not to liposomes used in SNARE fusion assays (POPC/DOPS). Input protein (20% of what was used in the binding assay) was loaded for comparison. Fractions from a step gradient lacking FisB are shown (ct). Molecular weight standards (50, 37, 25, and 20 kDa) (M) are indicated. (C,D) Coflotation assay in which the lipid composition of the liposomes was varied: PC/PG (70 mol/30 mol), PC/PE (70 mol/30 mol), PC/CL30 (70 mol/30 mol), PC/CL45 (55 mol/45 mol), and POPC/DOPS (85 mol/15 mol). (C) All liposomes contained a small percentage of NBD-PE as a fluorescent marker. Visualization of liposomes by fluorescence before (prefloat) and after (post-float) separation using a three-step idoxanol gradient (see the Materials and Methods). (D) Four fractions were collected as indicated in the post-float box and analyzed by SDS-PAGE stained with SYPRO Orange. FisB bound efficiently to liposomes containing CL but not to liposomes without CL.
To identify the lipids that FisB binds, we repeated the coflotation assay with liposomes in which we varied the lipid compositions (Fig. 5C,D). These experiments revealed that the extracellular domain of FisB binds CL but not to any appreciable degree to PC, PS, PE, or PG. The binding of FisB to CL was confirmed using both CL extracted from E. coli and synthetic tetra-oleoyl CL (data not shown). These results rule out the possibility that FisB binding was due to an impurity in the CL from E. coli and further suggest that FisB interacts with the head group of CL.
Discussion
The release of the engulfed forespore into the mother cell cytoplasm through membrane fission is a key step in the developmental process of sporulation. Here, we showed that the mother cell membrane protein FisB plays a central role in this membrane fission reaction. Collectively, our data support a model in which FisB is inserted into the mother cell membrane during the early stages of engulfment, where it forms mobile complexes. As engulfment nears completion, FisB oligomers assemble at the site of membrane closure that connects the peripheral and spore membranes. FisB then severs this membrane tube, releasing the spore into the mother cell cytoplasm (Fig. 6). Virtually all endospore-forming bacteria in the low G+C Gram-positive branch possess a FisB homolog, while the protein appears to be absent in all other bacteria. Thus, FisB is likely to function exclusively in the fission reaction that occurs at the final stage of engulfment during sporulation, taking advantage of the unusual topology of the membranes at the site of fission.
Figure 6.
Model for FisB-mediated fission. Schematic representation of the membrane tube formed at the cell pole by the engulfing mother cell membranes. Due to its high negative curvature, CL (not shown) is thought to become enriched within the tube. FisB (green) becomes immobilized at this site through its interaction with CL. Oligomerization of FisB or FisB–lipid interactions are proposed to sever the tube, releasing the spore into the mother cell cytoplasm. See the Discussion for details.
Given enough time, a subset of sporulating cells lacking FisB successfully complete engulfment and membrane fission. This implies that an additional mechanism promotes fission during sporulation. Possible candidates include peptidoglycan synthesis as proposed by Dworkin and colleagues (Meyer et al. 2010) or peptidoglycan hydrolysis carried out by the membrane-anchored peptidoglycan hydrolase complex composed of SpoIID, SpoIIP, and SpoIIM (Abanes-De Mello et al. 2002; Aung et al. 2007; Morlot et al. 2010). Either of these machines could bring the engulfing membranes into close proximity and promote fission (Judd et al. 2005), although at a slower rate than FisB. Alternatively, SpoIIIE or an as yet unidentified factor could function in an independent fission pathway.
SpoIIIE and membrane fission
Our analysis of engulfment in cells lacking the SpoIIIE DNA translocase suggests that its activity is critical for membrane migration around the forespore prior to fission. These results do not exclude the possibility that it could also function in the fission reaction. However, our analysis of the FisB protein in SpoIIIE mutants provides a simpler explanation for the original observations of Sharp and Pogliano (1999). Immunoblot analysis of sporulating cells revealed that the levels of FisB were significantly reduced in the SpoIIIE-null mutant compared with wild type (Supplemental Fig. S8A). Moreover, in a SpoIIIE point mutant (SpoIIIE36) that binds DNA but is impaired in DNA transport (Wu and Errington 1994; Wu et al. 1995), FisB protein levels were unaffected, but YFP-FisB localization was disrupted (Supplemental Fig. S8B). In accordance with this finding, Sharp and Pogliano (1999) reported that the SpoIIIE36 mutant was impaired in membrane fission but had a less severe defect than the null. Based on these observations, we favor the idea that SpoIIIE is only indirectly involved in the fission reaction, impacting membrane migration and FisB abundance and/or localization. Interestingly, it has also been proposed that SpoIIIE participates in fission during asymmetric division in sporulating cells (Liu et al. 2006; Fleming et al. 2010). This membrane remodeling event, wherein cytosol fills the gap inside the closing membrane annulus, has the opposite topology to the fission at the end of engulfment, where the extracellular medium fills the gap to be closed by the engulfing membrane (Supplemental Fig. S9). Intriguingly, SpoIIIE’s architecture (large cytoplasmic domain anchored to the membrane) is opposite to that of FisB (membrane-anchored extracytoplasmic domain). Establishing a direct role for SpoIIIE in membrane remodeling awaits in vitro reconstitution.
Interactions between the extracellular domain of FisB and CL
We found that the extracellular domain of FisB interacts with CL, and our data suggest that this interaction is important for membrane remodeling in vitro. The specificity of this interaction is remarkable for a soluble domain. Although CL has high affinity for proteins with transmembrane segments involved in oxidative phosphorylation, such as the F0F1 ATPase (Haines 2009), many soluble proteins that bind CL, such as cytochrome c (Kagan et al. 2009) and tBid (Epand et al. 2002) in eukaryotes and MinD (Mileykovskaya et al. 2003) in bacteria, also interact with other, negatively charged lipids such as PS and PG to varying extents. In contrast, we could not detect significant interaction between the extracellular domain of FisB and the acidic lipids PG and PS or with the zwitterionic lipids PE and PC (or with digalactosyldiacylglycerol) (data not shown). Thus, we expect the dynamics of FisB and CL to be intimately coupled (see below). In this regard, it is noteworthy that during sporulation, the levels of CL increase, and CL is enriched in the engulfing membranes (Kawai et al. 2004). B. subtilis possesses three genes that encode proteins homologous to CL synthases: clsA (ywnE), which encodes the major CL synthase; clsB (ywjE); and ywiE (Kunst et al. 1997; Kawai et al. 2004). The expression of both clsA and clsB increases during sporulation, consistent with the idea that CL plays an important role during this process. Cells lacking all three genes are viable and have undetectable levels of CL during vegetative growth (Kawai et al. 2004). However, CL is readily detectable in the triple mutant during the early stages of sporulation, indicating that an additional sporulation-specific CL synthase exists or that CL is produced as a side reaction by a lipid synthesis/modification enzyme (Kawai et al. 2004, 2006; Tan et al. 2012). In accordance with the presence of CL during engulfment in cells lacking ClsA, ClsB, and YwiE, we found that the triple mutant was able to undergo fission at a frequency comparable with wild type (data not shown). This result suggests that if FisB requires CL to catalyze fission in vivo, then the reduced amount of CL made in the absence of the known synthases must be sufficient. Alternatively, FisB could use another phospholipid in addition to CL in vivo.
Late forespore gene expression does not require the completion of engulfment
Previous work has shown that the developmental programs of gene expression in the mother cell and forespore are not free-running clocks but are linked to each other by cell–cell signaling pathways (Stragier and Losick 1996; Rudner and Losick 2001; Rudner and Doan 2008). Furthermore, it has been hypothesized that these cell type-specific transcriptional programs are coupled to morphogenesis. Specifically, it was proposed that the activation of the forespore transcription factor σG is coupled to the completion of engulfment and membrane fission (Stragier and Losick 1996; Rudner and Losick 2001; Rudner and Doan 2008; Regan et al. 2012). One compelling model was that the isolation of the forespore from the external environment as a result of fission served as the trigger for σG. Recently, we reported evidence that argued against the coupling of σG activation to membrane fission (Doan et al. 2009). Specifically, we found that a subset (13%) of sporulating cells that was blocked at early stages of engulfment successfully activated σG in the forespore. However, the severe defect in morphogenesis in these mutants and the small number of cells that turned on σG raised concern that the activation might not be biologically relevant. Furthermore, recent genetic experiments using large insertions into the B. subtilis chromosome that delay DNA transport into the forespore suggest that σG activation could indeed be coupled to completion of engulfment (Regan et al. 2012). The FisB mutant allowed us to revisit the link between σG activity and morphogenesis in a strain in which engulfment proceeds normally and only fission is impaired.
Analysis of σG activity in the FisB mutant revealed that 47% (n > 1000) of the sporulating cells that were stalled at the membrane fission step successfully activated σG in the forespore compartment (Supplemental Fig. S10). This observation lends further support to the idea that σG activation and the fission reaction are not coupled events. Intriguingly, we also found that sporulating FisB mutants lyse at an elevated frequency, and this lysis was suppressed in cells lacking σG (Supplemental Fig. S11). This observation suggests that activation of σG prior to the completion of membrane fission is deleterious to the sporulating cell and leads to death. Thus, although a mechanism that holds σG inactive until fission is complete would appear to be beneficial, our data suggest that B. subtilis does not employ one. We suspect that under normal circumstances (in wild-type cells), σG activation and fission are temporally distinct enough that a coupling device is not required.
How might FisB mediate membrane scission?
Although membrane fission is a fundamental and ubiquitous biological process used by species ranging from the simplest bacteria and enveloped viruses to the most sophisticated neuronal cells, only two general fission machineries have been described. The first and best understood involves constriction of a membrane neck or tube from the outside by dynamin and its homologs (Ferguson and De Camilli 2012). This constriction process relies on oligomerization of dynamin around the membrane neck and GTP hydrolysis to sever it. The second fission machinery is the ESCRT-III system (Hurley 2010; Henne et al. 2011), in which all components are soluble proteins that are recruited to the membrane via protein–protein and protein–lipid interactions. The only ESCRT-III component that seems to be absolutely required for fission is Snf7, whose oligomerization initiates the fission process, at least in vitro (Wollert and Hurley 2010). Other subunits likely terminate Snf7 filament assembly and recruit Vps4, an ATPase that disassembles the ESCRT-III complex and perhaps contributes to the fission reaction. The mechanism by which ESCRT-III mediates fission is still intensely debated (Hurley 2010; Henne et al. 2011); however, it is clear that the machinery works at the opening of and/or inside the membrane neck to achieve fission, rather than using constriction from outside the tube. Based on the topology of FisB with its large extracytoplasmic domain and very short (15-amino-acid) cytoplasmic domain, we suspect that FisB also catalyzes fission by doing work at one (or both) of the openings of the tube or from within it.
The activity of eukaryotic fusion and fission machines such as SNAREs and dynamin are exquisitely regulated in time and space by other proteins and intracellular signals. Potential regulators of FisB other than CL have not been identified. However, given the topology of the membranes during engulfment of the forespore, it is possible, in principle, that the dynamics of CL during sporulation, its propensity to associate with negatively curved membranes, FisB binding to CL, and homo-oligomerization of FisB could provide sufficient control over when and where fission occurs.
At the final stages of engulfment, the tube connecting the membrane engulfing the forespore to the rest of the mother cell membrane must shrink in diameter (Fig. 6). This will increase the negative curvature of the tube’s inner leaflet, making it increasingly favorable for CL localization. We hypothesize that FisB becomes immobilized at the fission site as a result of its interaction with CL within the membrane tube. We found that FisB forms oligomers in vitro, and one possibility is that oligomerization leads to a filling of the tube with FisB and overflow to the tube’s openings. The positive curvature of these openings would be unfavorable for CL localization. If FisB recruits CL to these positively curved regions, this could lead to the destabilization and membrane scission. In a variation of this model, FisB would localize to the highly positively curved regions at the openings of the tube by virtue of its own curvature preference and, by recruiting CL to such regions, would accumulate stresses, which could be relieved by severing the tube.
Other alternatives exist that are not mutually exclusive. For example, FisB oligomerization with concomitant conformational changes could be coupled to the destabilization of the tube, perhaps via twisting or compression. Alternatively, FisB could induce lateral segregation of CL (de Kruijff and Cullis 1980), with the resulting line tension and/or the discontinuity in elastic properties between the CL-rich and CL-poor phases leading spontaneously to fission. Such fission occurs at domain boundaries between cholesterol-rich and cholesterol-poor tubules under certain conditions (Allain et al. 2004; Roux et al. 2005). Finally, FisB might act through a mechanism reminiscent of viral fusion proteins. Although fusion and fission are topologically opposite processes, microscopically, both require cutting, bending, and rejoining of membranes. From this point of view, it is perhaps not surprising that some homologs of dynamin-1 have been described to mediate fusion, not fission (Kozlov et al. 2010; Ferguson and De Camilli 2012). In this model, FisB anchored inside the membrane tube would interact with CL across the tube via its extracytoplasmic domain. CL binding could trigger a conformational change in FisB, as in the case of cytochrome c (Kagan et al. 2009), ultimately leading to membrane fission.
Further biochemical analysis of FisB in vitro, along with the identification of other proteins that function with or regulate FisB and the missing CL synthase, will help distinguish among these models for FisB function. However, our characterization of FisB raises the intriguing possibility that it is part of a novel fission mechanism.
Materials and methods
General methods
All B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al. 1983). Sporulation was induced by resuspension at 37°C according to the method of Sterlini-Mandelstam (Harwood and Cutting 1990) or by exhaustion in supplemented DS medium (Schaeffer et al. 1965). Sporulation efficiency was determined in 36-h cultures as the total number of heat-resistant (20 min at 80°C) colony-forming units (CFUs) compared with wild-type heat-resistant CFUs. Analysis of membrane fission after inhibition of cell wall synthesis was performed as described previously (Meyer et al. 2010) except cells were sporulated at 37°C and fosfomycin (5 mM final) was added at hour 1.5. Membrane fission was assessed at hours 2, 2.5, and 3. Insertion–deletion mutations were generated by isothermal assembly (Gibson et al. 2009) of PCR products followed by direct transformation into B. subtilis. Tables of strains, plasmids, and oligonucleotide primers and a description of plasmid construction can be found online in the Supplemental Material (Supplemental Tables S1–S3; Supplemental Material).
Protein purification and antibody production
His6-FisBFL and His6-FisBECD fusion proteins were expressed in E. coli BL21 DE3 pLysS at 30°C and 16°C, respectively, and purified by affinity chromatography on Ni2+-NTA agarose (Qiagen). MBP-FisBECD fusion protein was expressed in E. coli NB42 and purified by affinity chromatography on amylose resin (New England Biolabs). A complete description of the purifications can be found in the Supplemental Material. MBP-FisBECD peak fractions were pooled and used to generate rabbit polyclonal antibodies (Covance). Crude serum was affinity-purified as described (Campo and Rudner 2006) using His6-FisBECD.
Immunoblot analysis
Whole-cell lysates from sporulating cells induced by resuspension were prepared as described (Doan and Rudner 2007). Samples were heated for 10 min at 50°C prior to loading. Equivalent loading was based on OD600 at the time of harvest. Proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels, electroblotted onto Immobilon-P membranes (Millipore), and blocked in 5% nonfat milk in phosphate-buffered saline (PBS) and 0.5% Tween-20. The blocked membranes were probed with affinity-purified anti-His (1:1000; Santa Cruz Biotechnology), anti-FisB (1:10,000), and anti-σF (15,000) (Carniol et al. 2004) diluted into 3% BSA in 1× PBS-0.05% Tween-20. Primary antibodies were detected using horseradish peroxidase-conjugated goat anti-rabbit G (Bio-Rad) and the Western Lightning reagent kit as described by the manufacturer (PerkinElmer).
In vitro lipid mixing assay
The protocol described in Weber et al. (1998) was followed with minor modifications. All lipids were purchased from Avanti Polar Lipids. For fluorescent donor liposomes, 100 μL of 3 mM lipids (L-α-PG [E. coli; sodium salt], CL [E. coli; sodium salt], L-α-phosphatidylethanolamine [E. coli, PE], L-α-PC [egg, chicken], NBD-PE [ammonium salt; 18:1], LR-PE [ammonium salt; 18:1]) in a molar ratio of 30:45:13:10:1:1 dissolved in chloroform were dried under nitrogen gas, placed under a vacuum dessicator for 1 h, and then dissolved in reconstitution buffer (RB; 25 mM Hepes at pH 7.4, 100 mM KCl, 10% glycerol, 1 mM DTT) supplemented with 1% (w/v) octyl-β-D-glucopyranoside (OG) and either FisB protein, the mock purification, or nothing to obtain protein-free liposomes. Unlabeled acceptor liposomes were prepared using 100 μL of 15 mM lipids as above in a molar ratio of 30:45:15:10. FisB protein was reconstituted into the lipids at a 1:1000 (protein:lipid) molar ratio, and then the OG was removed by overnight dialysis (6000- to 8000-Da molecular weight cutoff). Unincorporated protein was removed by flotation of the liposomes through a Nycodenz (Accurate Chemicals) step gradient. Labeled liposomes were recovered in a 150-μL volume (2 mM lipid). Unlabeled liposomes were collected in 400 μL (3.75 mM lipid).
The lipid-mixing assay was performed in white 96-well Maxisorp plates (Nunc). Liposomes (10 μL of labeled and 45 μL of unlabeled) were mixed on ice and then placed in a room temperature SpectraMax M5 Microplate Reader (Molecular Devices), and NBD fluorescence was recorded with filters set at 460 nm (excitation) and 538 nm (emission) at 30-sec intervals for 45 min. To determine the maximum NBD fluorescence, 10 μL of 2.5% (w/v) n-dodecyl-β-D-maltoside (DM) (Thermo Scientific) was added at the end of the experiment, and fluorescences were monitered for 15 min.
Flotation assay
Natural (PG, PE, and CL, all from E. coli, or egg PC) lipids or synthetic lipids (POPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt) , 1′,3′-bis(1,2-dioleoyl-sn-glycero-3-phospho)-sn-glycerol (sodium salt; 18:1 CL), and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) ) from Avanti Polar Lipids were used. All liposomes included 0.2 mole percent NBD-PE (ammonium salt; 18:1) as a fluorescent marker. The lipids were dissolved in a 2:1 (v/v) mixture of chloroform and methanol. Lipids (1 μmol total) were mixed at desired ratios, and the mixtures were dried under nitrogen flow. Residual organic solvent was removed under vacuum for at least 2 h in a vacuum dessicator. Lipids were hydrated in 1 mL of RB-EDTA buffer [25 mM Hepes at pH 7.4, 140 mM KCl, 1 mM EDTA, 0.2 mM tris(2-carboxyethyl)phosphine] by shaking for >30 min. The lipid suspension was then immersed in liquid nitrogen and then allowed to thaw in a water bath at 37°C. This freeze–thaw cycle was repeated five additional times to form large unilamellar vesicles (LUVs) (Pick 1981). The LUVs were either flash-frozen in liquid nitrogen and stored at −80°C or used fresh for extrusion. The LUV stock was extruded 19 times through 50-nm pore size polycarbonate filters (Avanti) at 40°C using a miniextruder.
The extruded liposomes (40 nmol total lipid) were incubated with 40 pmol of FisB extracytoplasmic domain in a total volume of 100 μL for 1 h at room temperature. Two-hundred milliliters of 60% iodoxanol density gradient medium (Optiprep, Sigma-Aldrich) was added to make a 40% iodoxanol solution (density, 1.215 g/mL) that was layered at the bottom of a 5-mm × 41-mm Beckman Ultra-Clear ultracentrifuge tube (Beckman Coulter, Inc.). This was overlaid with 200 μL of 20% iodoxanol solution (diluted in RB-EDTA; density, 1.110 g/mL), followed by a top layer of 200 μL of RB-EDTA. To assess liposome redistribution after floatation, NBD-PE fluorescence was measured before and after centrifugation as follows: Step gradients were prepared in parallel, and the tubes were placed in a rack in a gel-imaging instrument (ImageQuant LAS 4000, GE Healtcare). A mirror was placed at a 45° angle next to the rack, reflecting the image of the tubes to the camera placed above (Fig. 5C, pre-float; Bigay and Antonny 2005; Bigay et al. 2005). Blue epi-illumination was used, and fluorescence was collected through a green long-pass filter (Y515 Di). After imaging, the tubes were spun at 48,000 rpm (269,000g) for 1.5 h at 20°C. After floatation, the tubes were imaged again to assess the position of the liposomes in the gradient, and then fractions were collected from the bottom of each tube as indicated in Figure 5C, post-float. Because FisB could have varying affinities for the different lipids, we quantified both lipid and protein in all fractions. The amount of lipid in each fraction was quantified using NBD-PE fluorescence, and the amount of protein was quantified by PAGE (Novex mini Nu-PAGE Bis-tris, 12%, 1 mm thick; Invitrogen), stained with SYPRO Orange (Invitrogen). The sum of the protein from each fraction was usually less than the total initially mixed with liposomes (Fig. 5D). This is probably because some of the protein sticks to the walls of the ultracentrifuge tube. However, this was not an issue when comparing relative amounts of protein in different fractions (Bigay and Antonny 2005; Bigay et al. 2005).
Fluorescence microscopy
Fluorescence microscopy was performed on an Olympus BX61 microscope as previously described (Doan et al. 2005). Fluorescent signals were visualized with a phase-contrast objective UplanF1 100× and captured with a monochrome CoolSnapHQ digital camera (Photometrics) using Metamorph software (Molecular Device). Exposure times were typically 500 msec for CFP and GFP and 2000 msec for YFP-FisB. Membranes were stained with either TMA-DPH or FM4-64 (Molecular Probes) at a final concentration of 0.01 mM and 3 μg/mL, respectively. Exposure times were typically 200 msec. Images were analyzed, adjusted, and cropped using Metamorph software. Cells were concentrated by centrifugation (8000 rpm for 30 sec) prior to visualization. This step had no impact on the localization of the fusion proteins.
Acknowledgments
We thank members of the Rudner laboratory (past and present) for advice and encouragement, Tom Rapoport and Jochen Zimmer for advice on membrane protein purification, Tom Bernhardt and Tsuyoshi Uehara for help with gel filtration, Mike Strauss and Chris Rodrigues for stimulating discussions, Annick Turbé-Doan for help with figures, Bruno Antonny for discussions and advice on lipid–protein interactions, and Rich Losick for generously providing the library of σE regulon mutants. E.K. thanks Jim Rothman for a home to do the fusion and flotation experiments. T.D. thanks Anne Galinier for her patience and generosity. Support for this work comes from the National Institutes of Health Grant GM086466 (to D.Z.R.) and CNRS and Marie-Curie International Reintegration Grant PIRG08-GA-2010-276750 (to T.D.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.209049.112.
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev 16: 3253–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain JM, Storm C, Roux A, Ben Amar M, Joanny JF 2004. Fission of a multiphase membrane tube. Phys Rev Lett 93: 158104. [DOI] [PubMed] [Google Scholar]
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K 2007. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol 65: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA 2008. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 135: 1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD 2006. Late budding domains and host proteins in enveloped virus release. Virology 344: 55–63 [DOI] [PubMed] [Google Scholar]
- Bigay J, Antonny B 2005. Real-time assays for the assembly-disassembly cycle of COP coats on liposomes of defined size. Methods Enzymol 404: 95–107 [DOI] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, Mesmin B, Antonny B 2005. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J 24: 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ 2007. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell 131: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürmann F, Ebert N, van Baarle S, Bramkamp M 2011. A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol Microbiol 79: 1294–1304 [DOI] [PubMed] [Google Scholar]
- Campo N, Rudner DZ 2006. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell 23: 25–35 [DOI] [PubMed] [Google Scholar]
- Carniol K, Eichenberger P, Losick R 2004. A threshold mechanism governing activation of the developmental regulatory protein σF in Bacillus subtilis. J Biol Chem 279: 14860–14870 [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM 2003. Protein–lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem 72: 175–207 [DOI] [PubMed] [Google Scholar]
- de Boer PA 2010. Advances in understanding E. coli cell fission. Curr Opin Microbiol 13: 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kruijff B, Cullis PR 1980. Cytochrome c specifically induces non-bilayer structures in cardiolipin-containing model membranes. Biochim Biophys Acta 602: 477–490 [DOI] [PubMed] [Google Scholar]
- Doan T, Rudner DZ 2007. Perturbations to engulfment trigger a degradative response that prevents cell-cell signalling during sporulation in Bacillus subtilis. Mol Microbiol 64: 500–511 [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol 55: 1767–1781 [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Rudner DZ 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5: e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, González-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, et al. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327: 945–972 [DOI] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci 108: 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RF, Martinou JC, Fornallaz-Mulhauser M, Hughes DW, Epand RM 2002. The apoptotic protein tBid promotes leakage by altering membrane curvature. J Biol Chem 277: 32632–32639 [DOI] [PubMed] [Google Scholar]
- Errington J 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1: 117–126 [DOI] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P 2012. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol 13: 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht A, Evans L, Errington J 2003. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology 149: 3023–3034 [DOI] [PubMed] [Google Scholar]
- Fleming TC, Shin JY, Lee SH, Becker E, Huang KC, Bustamante C, Pogliano K 2010. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev 24: 1160–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Pogliano K 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev 22: 3475–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths KK, Setlow P 2009. Effects of modification of membrane lipid composition on Bacillus subtilis sporulation and spore properties. J Appl Microbiol 106: 2064–2078 [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H 2004. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 5: 317–323 [DOI] [PubMed] [Google Scholar]
- Haines TH 2009. A new look at cardiolipin. Biochim Biophys Acta 1788: 1997–2002 [DOI] [PubMed] [Google Scholar]
- Harrison SC 2008. Viral membrane fusion. Nat Struct Mol Biol 15: 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C, Cutting S 1990. Molecular biological methods for Bacillus. Wiley, New York [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD 2011. The ESCRT pathway. Dev Cell 21: 77–91 [DOI] [PubMed] [Google Scholar]
- Hurley JH 2008. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 20: 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH 2010. The ESCRT complexes. Crit Rev Biochem Mol Biol 45: 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH 2005. Distinct constrictive processes, separated in time and space, divide caulobacter inner and outer membranes. J Bacteriol 187: 6874–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang J, Stoyanovsky DA, Wipf P, Kochanek PM, et al. 2009. Cytochrome c/cardiolipin relations in mitochondria: A kiss of death. Free Radic Biol Med 46: 1439–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatekin E, Di Giovanni J, Iborra C, Coleman J, O’Shaughnessy B, Seagar M, Rothman JE 2010. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc Natl Acad Sci 107: 3517–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K 2004. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol 186: 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F, Hara H, Takamatsu H, Watabe K, Matsumoto K 2006. Cardiolipin enrichment in spore membranes and its involvement in germination of Bacillus subtilis Marburg. Genes Genet Syst 81: 69–76 [DOI] [PubMed] [Google Scholar]
- Kielian M, Rey FA 2006. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat Rev Microbiol 4: 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, McMahon HT, Chernomordik LV 2010. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci 35: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky Y, Kozlov MM 2003. Membrane fission: Model for intermediate structures. Biophys J 85: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390: 249–256 [DOI] [PubMed] [Google Scholar]
- Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R 2008. A unique cell division machinery in the Archaea. Proc Natl Acad Sci 105: 18942–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K 2006. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol 59: 1097–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis KA, Burton BM, Nollmann M, Ptacin JL, Bustamante C, Ben-Yehuda S, Rudner DZ 2008. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes Dev 22: 1786–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K, Dworkin J 2010. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol 76: 956–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W 2003. Effects of phospholipid composition on MinD–membrane interactions in vitro and in vivo. J Biol Chem 278: 22193–22198 [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI 2004. Retrovirus budding. Annu Rev Cell Dev Biol 20: 395–425 [DOI] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ 2010. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev 24: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick U 1981. Liposomes with a large trapping capacity prepared by freezing and thawing of sonicated phospholipid mixtures. Arch Biochem Biophys 212: 186–194 [DOI] [PubMed] [Google Scholar]
- Piggot P, Losick R 2002. Sporulation genes and intercompartmental regulation. In Bacillus subtilis and its closest relatives: From genes to cells (ed. A Sonenshein et al.), pp. 483–517. ASM Press, Washington, D.C [Google Scholar]
- Rand RP, Parsegian VA 1986. Mimicry and mechanism in phospholipid models of membrane fusion. Annu Rev Physiol 48: 201–212 [DOI] [PubMed] [Google Scholar]
- Regan G, Itaya M, Piggot PJ 2012. Coupling of σG activation to completion of engulfment during sporulation of Bacillus subtilis survives large perturbations to DNA translocation and replication. J Bacteriol 194: 6264–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B 2005. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J 24: 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D, Doan T 2008. Intercompartmental signal transduction during sporulation in Bacillus subtilis. In Chemical communication among bacteria (ed. S Winans, B Bassler), pp. 3–12. ASM Press, Washington, D.C [Google Scholar]
- Rudner DZ, Losick R 2001. Morphological coupling in development: Lessons from prokaryotes. Dev Cell 1: 733–742 [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci 99: 8701–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD 2008. A role for the ESCRT system in cell division in archaea. Science 322: 1710–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP 1965. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci 54: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci 96: 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K 2003. The membrane domain of SpoIIIE is required for membrane fusion during Bacillus subtilis sporulation. J Bacteriol 185: 2005–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steil L, Serrano M, Henriques AO, Völker U 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151: 399–420 [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Göttlinger HG 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114: 689–699 [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30: 297–341 [DOI] [PubMed] [Google Scholar]
- Struck DK, Hoekstra D, Pagano RE 1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry 20: 4093–4099 [DOI] [PubMed] [Google Scholar]
- Südhof TC, Rothman JE 2009. Membrane fusion: Grappling with SNARE and SM proteins. Science 323: 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ 2009. Recruitment of SMC by ParB–parS organizes the origin region and promotes efficient chromosome segregation. Cell 137: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z 2012. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci 109: 16504–16509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, Rothman JE 1998. SNAREpins: Minimal machinery for membrane fusion. Cell 92: 759–772 [DOI] [PubMed] [Google Scholar]
- Weiss DS 2004. Bacterial cell division and the septal ring. Mol Microbiol 54: 588–597 [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464: 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH 2009. Membrane scission by the ESCRT-III complex. Nature 458: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JY, Park CK, Seitz M, Israelachvili J 1999. Polymer-cushioned bilayers. II. An investigation of interaction forces and fusion using the surface forces apparatus. Biophys J 77: 1458–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264: 572–575 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J 1995. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev 9: 1316–1326 [DOI] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci 80: 2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]