Abstract
Polymerization of the intact capsid protein (CA) of HIV-1 into mature capsidlike particles at physiological ionic strength in vitro requires macromolecularly crowded conditions that approach those inside the virion, where the mature capsid is assembled in vivo. The capsid is organized as a hexameric lattice. CA subunits in each hexamer are connected through interfaces that involve the CA N-terminal domain (NTD); pairs of CA subunits belonging to different hexamers are connected through a different interface that involves the C-terminal domain (CTD). At physiological ionic strength in noncrowded conditions, CA subunits homodimerize through this CTD-CTD interface, but do not hexamerize through the other interfaces (those involving the NTD). Here we have investigated whether macromolecular crowding conditions are able to promote hexamerization of the isolated NTD and/or full-length CA (with an inactive CTD-CTD interface to prevent polymerization). The oligomerization state of the proteins was determined using analytical ultracentrifugation in the absence or presence of high concentrations of an inert macromolecular crowding agent. Under the same conditions that promoted efficient assembly of intact CA dimers, neither NTD nor CA with an inactive CTD-CTD interface showed any tendency to form hexamers or any other oligomer. This inability to hexamerize was observed even in macromolecularly crowded conditions. The results indicate that a functional CTD-CTD interface is strictly required for hexamerization of HIV-1 CA through the other interfaces. Together with previous results, these observations suggest that establishment of NTD-CTD interactions involved in CA hexamerization during mature HIV-1 capsid assembly requires a homodimerization-dependent conformational switching of CTD.
Introduction
The mature capsid of the human immunodeficiency virus (HIV-1) is assembled within the newly formed virion from capsid protein (CA) subunits released by proteolytic processing of the Gag precursor polyprotein (1–4). CA contains two independent domains: the N-terminal domain (NTD) and the C-terminal domain (CTD). The basic structural organization of both authentic mature capsids of HIV-1 and mature capsidlike particles assembled in vitro is the same, and consist of a lattice of CA hexamers (5,6). The NTDs connect neighbor subunits in each hexamer through two discrete interfaces (the NTD-NTD and NTD-CTD interfaces). The CTDs connect pairs of subunits belonging to different hexamers through a third, different interface (the CTD-CTD homodimerization interface) (1,2,7) (see Fig. 1). A fourth, very small interface has been identified between CTDs around each threefold symmetry axis in the lattice (8).
Figure 1.

Schematic models of CA oligomers obtained by x-ray crystallography and formation of the capsid hexameric lattice. (Cylinders) Helices. (Green) NTD; (red) CTD. (a) CA hexamer. Each NTD-NTD interface is defined by helices 1 and 3 of one monomer and helix 2 of the neighboring monomer (21). (b) Side-view of two neighboring subunits in the CA hexamer. The NTD-CTD interface is defined by parts of helices 4 and 7 of NTD belonging to one monomer and helices 8 and 11 of CTD belonging to the other monomer. (c) CTD dimer. The CTD-CTD interface is defined mainly by helix 9 of each monomer (14). The models were produced using the program PyMol (51). (d) A scheme of the formation of the capsid hexameric lattice from stable CA dimers. Each CA monomer is represented by two contiguous circles (green, NTD; and red, CTD). Interactions (interfaces) between CA domains, either from the same monomer or from different monomers are represented by (blue bars). (Top left) Soluble CA dimers with monomers joined through the CTD-CTD interface constitute the stable building blocks from which the capsid is assembled. (Bottom right) Establishment of NTD-NTD and NTD-CTD interactions between different CA dimers leads to formation of the hexameric rings and, thus, of the entire capsid lattice. Different pathways for the formation of each hexamer are possible, including: (i) a sixth-order reaction in which all six subunits in each ring simultaneously click into place; or (ii) a series of second-order reactions in which subunits are successively joined, until each hexameric ring is closed. The conclusions of this study are independent of which hexamerization pathway actually occurs. (e) A scheme of the predicted formation of independent hexamers from stable CAW184A/W185A monomers. (Top left) Soluble CAW184A/W185A monomers cannot dimerize because they carry mutations in the CTD-CTD interface. (Bottom right) If structuration of the NTD-NTD and NTD-CTD interacting surfaces does not depend on CA dimerization through the separate CTD-CTD interface, independent hexamers could still be formed. The results of this study show that no such hexamers are formed from CA monomers, even under macromolecular crowding conditions that increase CA activity and efficiently promote the formation of the hexameric lattice from CA dimers as in panel d.
Research on the molecular structure, conformational dynamics, and energetics of CA and CA-CA interfaces is contributing to a detailed structural understanding of HIV-1 capsid structure, assembly, and disassembly, and to the design of antiviral agents able to inhibit either process (recently reviewed in Prevelige (9), Waheed and Freed (10), and Bocanegra et al. (11)). Soluble dimers of intact CA from HIV-1 are obtained under nonpolymerizing conditions and constitute the stable building blocks from which the capsid is assembled (1–4). The CTD-CTD interface is structurally and energetically preserved in both soluble CA and the isolated CTD dimer, which allowed its description in atomic detail (8,12–15), and its thermodynamic dissection at the single-residue level (16). From that knowledge, peptides able to inhibit HIV-1 capsid assembly in vitro, and virus infectivity ex vivo, have been rationally designed (11,17,18).
In contrast to CA dimerization, the thermodynamics of CA hexamerization has not been investigated, as soluble hexamers of the HIV-1 NTD or full-length CA (with a nonfunctional CTD-CTD interface to prevent polymerization) could not be obtained (19,20). Recently, CA of HIV-1 was engineered to form soluble hexamers by crosslinking the monomers through nonnatural disulfide bridges introduced at the NTD-NTD interfaces. Dimerization of hexamers and capsid assembly were prevented by inactivating the CTD-CTD interface through a double mutation (W184A/M185A). The atomic structure of this modified CA hexamer and the NTD-NTD and NTD-CTD interfaces in it have been described in atomic detail (21–23). However, the presence of the covalent disulfide bonds in the modified hexamer may prevent a thermodynamic dissection of these interfaces. Like the disulfide-modified CA of HIV-1, NTD of CA from the related murine leukemia virus was able to hexamerize, albeit only in molecular crystals formed from highly concentrated solutions under nonphysiological conditions (24).
These observations reveal that in these far-from-physiological conditions, the presence of an active CTD-CTD interface or of the CTD itself are not strictly required for CA hexamerization. If the same were true when no disulfides are introduced, the absence of hexamerization of both NTD and CA (with an inactive dimerization interface) of HIV-1 could be explained by a simple, but untested possibility: the NTD-NTD and NTD-CTD interactions could be of such a low affinity that hexamerization would require a very high CA effective concentration, such as that found in the HIV-1 virion. This high effective concentration may have been reached in laboratory conditions by condensation of NTD molecules during crystal growth or through the formation of the disulfides in CA, but not in other in vitro experiments using closer to physiological conditions.
Two reasons account for the very high CA effective concentration in the HIV-1 virion:
-
1.
The CA nominal concentration in a typical virion may be ∼4 mM (6,25).
-
2.
The HIV-1 virion contains many copies of different viral and cellular proteins and two viral RNA molecules, and the total macromolecular concentration is ∼300 mg/mL (see Table S1 in the Supporting Material).
Thus, the interior of the HIV-1 virion is a macromolecularly crowded environment.
Macromolecular crowding is widespread in cells and biological fluids. The total concentration of macromolecules in them is extremely high (∼50–400 mg/mL), and a large fraction of the total volume (in a cell, ∼20–40%) is occupied by the macromolecular solutes, leading to significant volume exclusion effects. This condition can produce large quantitative effects in the kinetics and thermodynamics of biochemical reactions involving molecular interactions within or between macromolecules, such as protein folding or oligomerization (26–28). Crowding increases up to several orders of magnitude the chemical activity (effective concentration) of macromolecules, favoring the association equilibria between macromolecules that have any intrinsic tendency to interact with each other. This effect arises because the reduction in the volume occupied by the macromolecules in its associated state, relative to its dissociated state, reduces the total excluded volume, which decreases the free energy of the system (26,27). The calculations mentioned above indicate that the HIV-1 virion is as crowded as a cell, with perhaps ∼30% of its internal volume occupied by macromolecules. Thus, similar crowding-related effects may occur in cells and in the HIV-1 virion.
The effects of macromolecular crowding in a variety of biochemical reactions and processes involving cellular proteins have been previously investigated (28). However, the influence of the physiologically crowded environment in molecular recognition events involved in viral infection are essentially unexplored. The scarce experimental evidence so far indicates, in agreement with theoretical predictions, that macromolecular crowding may be a critical driving force for virus morphogenesis (29), and reduces the inhibitory activity of small antiviral agents targeting protein-protein interactions (30).
In previous HIV-1 capsid assembly experiments in vitro, CA concentrations used were well below 1 mM (typically, a few tens of μM). Under such conditions, CA was completely unable to polymerize into mature capsidlike particles, unless special conditions were used:
-
1.
A fusion CA-NC protein in the presence of nucleic acid (31–33); or
-
2.
A very high ionic strength (>1–2 M NaCl) (19).
The added nucleic acid may act as a CA-NC condensing agent by binding to NC; a high ionic strength may screen the electrostatic repulsions and steric effects that occur between CA subunits (16,34,35). In either case, CA assembly does occur, but under nonphysiological conditions. However, we showed that highly efficient assembly of the mature HIV-1 capsid can be achieved in vitro in the absence of NC and nucleic acid and at physiological ionic strength, by simply adding a high concentration (100–200 mg/mL) of an inert macromolecular crowding agent, to bring the chemical activity of CA closer to that found inside the virion (29). Furthermore, if the reaction is carried out at very high ionic strength, addition of the crowding agent allows the rapid and efficient assembly of CA at much lower protein concentrations than those required if no crowder is added.
In this study, we have analyzed whether hexamerization of NTD and/or the full-length CA (with an inactive CTD-CTD interface) could be likewise promoted by raising, through addition of a molecular crowding agent, the CA chemical activity. Even under exactly the same conditions that promoted efficient assembly of intact CA into capsidlike particles, and using much higher protein concentrations in crowded conditions, neither NTD nor CA with an inactive CTD-CTD interface showed any significant tendency to form any kind of oligomer. The results indicate that hexamerization of native CA of HIV-1 requires a functional CTD dimerization interface, and suggest that establishment of NTD-CTD interactions during mature HIV-1 capsid assembly requires a dimerization-dependent conformational rearrangement in CTD.
Materials and Methods
Protein expression and purification
The isolated NTD and full-length CA, either intact or with an inactivated CTD-CTD interface through mutations W184A/M185A (CAW184A/M185A), were expressed from plasmids pET21b(+)-NTD, pWISBH10 (34), and pWISBH10-W184A/M185A (a mutagenized plasmid derived from pWISBH10). The DNA segment coding for NTD (CA residues 1–145) from HIV-1 strain BH10 was amplified from plasmid pDAB72 by the PCR method. pDAB72 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, from Dr. S. Erickson-Viitanen (Erickson-Viitanen et al. (36)). The oligonucleotides used were designed to engineer NdeI and BamHI sites as well as initiation and termination codons immediately flanking the CA sequence. The amplified DNA was cloned in a recombinant plasmid pET21b(+). The isolated NTD, CA, and CAW184A/M185A were purified essentially as previously described for intact CA (16). Protein stocks were run in sodium-dodecyl-sulfate polyacrylamide gel electrophoresis gels and found to be essentially pure. Protein concentration was determined as described in Gill and von Hippel (37).
Kinetic analysis of CA polymerization
Assembly reactions were carried out essentially as described previously in del Alamo et al. (29) and Rincón et al. (30). Briefly, the polymerization mixtures contained 50 mM sodium phosphate buffer pH 7.4, 2.25 M NaCl, and 46 μM CA, or 20 μM and 100 mg/mL Ficoll-70 (GE HealthCare, Little Chalfont, UK) as a macromolecular crowding agent. The experiments were initiated by introducing an appropriate volume of a CA solution in 50 mM phosphate buffer into a spectrophotometer cuvette, and the assembly reaction was triggered by adding the appropriate volume of a solution containing 4 M NaCl and, when indicated, 179 mg/mL Ficoll-70 in 50 mM sodium-phosphate buffer pH 7.4 to get the final concentrations desired for each component in a final volume of 500 μL, followed by rapid mixing by repeated inversion of the cell. The pH of the final reaction mixture was checked in a test sample. The time-dependent increase in the optical density at 350 nm as a measure of the light scattered by the assembled particles was monitored at 25°C using a UV-1603 spectrophotometer (Shimadzu, Kyoto, Japan), with data points collected every 6 s. Traces of the variation in the turbidity were analyzed as described previously in del Alamo et al. (29) and Rincón et al. (30) using an empirical Hill function,
| (1) |
where OD is the optical density at incubation time t; ODf is the optical density at infinite time; t50 is the time at which the OD is equal to one-half the ODf; and n is a Hill coefficient/cooperativity parameter.
Analytical ultracentrifugation
Sedimentation analysis of NTD and CAW184A/M185A was performed at several protein concentrations (from 33 μM to 1 mM monomer). All samples were in 50 mM sodium-phosphate buffer pH 7.4, 2.25 M NaCl, in the absence or presence of Ficoll-70 (at 100 mg/mL unless stated otherwise). Sedimentation velocity experiments were carried out at 48,000 rpm and 20°C in a XL-A analytical ultracentrifuge (Beckman Coulter, Brea, CA) with a UV-Vis optics detection system, using an An50Ti rotor and 12-mm double-sector centerpieces except where indicated otherwise. Sedimentation profiles were registered every 3 min at 250 nm (NTD) or 300 nm (CAW184A/M185A). The sedimentation coefficient distributions were calculated by least-squares boundary modeling of sedimentation velocity data using the c(s) method (38,39) as implemented in the software SEDFIT (National Institutes of Health, Bethesda, MD). These s values were corrected to standard conditions (water, 20°C, and infinite dilution) using the software SEDNTERP (Biomolecular Interaction Technologies Center, University of New Hampshire, http://bitcwiki.sr.unh.edu/index.php/Main_Page), yielding the corresponding standard s values (s20,w).
Low-speed (18,000 and 20,000 rpm) sedimentation equilibrium experiments were performed to determine the size and self-association properties of NTD as a function of protein concentration, using short columns (80 μL) and the same experimental protocol described above. After the equilibrium scans, a high-speed centrifugation run (48,000 rpm) was conducted to estimate the corresponding baseline offsets.
For each solution composition, the whole-cell apparent weight-average buoyant molecular weight (M∗i,app) of either NTD or CAW184A/M185A was determined by fitting Eq. 2 to the experimentally measured equilibrium gradient,
| (2) |
where Si (r) is the magnitude of a signal proportional to the weight/volume concentration of NTD or CAW184A/M185A at radial position r (i.e., absorbance at 280 nm); r0 is an arbitrarily selected reference position; R is the molar gas constant; and T the temperature.
Under conditions of thermodynamic ideality (high dilution of all macromolecular species), the apparent buoyant mass M∗i,app is equal to the actual buoyant mass Mi. The average molecular weights (M∗i ) of the different proteins were obtained from the corresponding buoyant values with M∗i = Midi, where di denotes the specific density increments of Ficoll, NTD, or CAW184A/M185A. The analysis was done using the EQASSOC (40) and HETEROANALYSIS (41) programs, which yielded the same results within 5% experimental error.
The SE approach applied in this study, using short solution columns and low rotor speeds to yield shallow gradients, simplifies the analysis and interpretation of the results obtained. Under these conditions, M∗i,app becomes independent of radial distance, and is well described by the solution-average molecular weight value, which may be expressed as a function of the loading protein concentration and analyzed assuming given self-association schemes as described elsewhere in the literature (42–44).
Tracer sedimentation equilibrium
Experiments to characterize the effect of Ficoll-70 concentration on the state of association of NTD and CAW184A/M185A were performed essentially as described in Darawshe et al. (45). Sample solution was prepared in 50 mM sodium phosphate buffer pH 7.4, 2.25 M NaCl using a broad range of Ficoll-70 concentrations (up to 100 g/L) in the absence and in the presence of a fixed concentration of NTD or CAW184A/M185A. Solution samples (70 μL total volume) were loaded over 90 μL of fluorocarbon and then overlaid by an equal amount of mineral oil. Samples were centrifuged to sedimentation equilibrium at 20,000 rpm and 20°C in an Optima MAX XL preparative ultracentrifuge (Beckman-Coulter) using small polycarbonate tubes and a TLD-55 rotor, as described previously in Darawshe et al. (45). After sedimentation equilibrium was attained (elapsed time > 48 h), gradients were obtained via fractionation of tube contents with a FR-115 centrifuge tube microfractionator (Brandel, Gaithersburg, MD) (45). The concentration of protein in each fraction was determined by measuring their absorbance at 280 nm, and the concentration of Ficoll was measured by refractometry. The apparent signal-average buoyant molecular weight of Ficoll or NTD/CAW184A/M185A in each solution was determined by fitting Eq. 2 to the experimental gradients as described above.
Nonideal tracer sedimentation equilibrium analysis
The general expression that describes the condition of sedimentation equilibrium in a solution containing an arbitrary number of solute species at arbitrary concentrations states that the apparent buoyant mass of a given molecular species depends upon its interactions with all the species of the solution mixture (43)
| (3) |
where wj denotes the w/v concentration of species j, and γi is the activity coefficient of species i. The quantity (d ln γi/dwj) defines the magnitude of the thermodynamic interactions between species i and j (referred to as thermodynamic interaction factor (44)). Let us consider that the subscript 1 refers to Ficoll species, while the subscripts 2 and 3 denote NTD or CAW184A/M185A protein species, respectively. To model the dependence of M∗1,app, M∗2,app, and M∗3,app with the concentration of Ficoll, estimating the value of the thermodynamic interaction factors involved are required. This was done by using an empirical analysis in which ln γi was expanded in powers of the concentration of all solute species, as described in Fodeke and Minton (46),
| (4) |
where Bij denotes a coefficient of two-body interactions between molecules of species i and j, and Bijk is a coefficient evaluating the three-body interactions between molecules of i, j, and k species, etc.
According to this analytical procedure, the dependence of the activity coefficient of Ficoll with its own concentration is described by
| (5) |
and the dependence of the apparent buoyant mass of Ficoll with the concentration of Ficoll is described by
| (6) |
Similarly, to analyze the dependence of the apparent buoyant mass of dilute NTD protein upon Ficoll concentration, Eq. 4 reduces to
| (7) |
and Eq. 3 reduces to
| (8) |
In the case of CAW184A/M185A protein, the formulation described in Eqs. 7 and 8 applies as well if subscript 2 is replaced by subscript 3.
Equations 6 and 8 were used to globally model the dependence of M∗1,app, M∗2,app, and M∗3,app upon Ficoll concentration.
Results and Discussion
The isolated NTD of HIV-1 is unable to oligomerize even in the presence of crowding agents
As a starting point, we evaluated the ability of soluble isolated NTD monomers to form hexamers (or any other kind of oligomer) in the absence of macromolecular crowding, under conditions in which soluble full-length CA dimers are able to polymerize into capsidlike particles.
In control experiments, CA polymerization kinetics was followed by turbidimetry (Fig. 2 a) in conditions in which capsidlike particles with the organization of authentic HIV-1 capsids are assembled, as shown by electron microscopy analysis (29). The results confirmed that full-length CA does efficiently assemble into capsidlike particles in 50 mM sodium-phosphate buffer pH = 7.4, 2.25 M NaCl, even at relatively low (30–50 μM and lower) protein (monomer) concentrations (Fig. 2 a). Thus, these conditions do promote the formation of hexamers of CA dimers (which is strictly required for the formation of the capsid hexameric lattice during assembly; see Fig. 1 d). In contrast to full-length CA, the isolated NTD under those same conditions (in the absence of crowding) did not show any significant tendency to form hexamers, or any other oligomer, as determined by analytical ultracentrifugation in both sedimentation velocity and sedimentation equilibrium experiments.
Figure 2.

Analysis of CA polymerization and of NTD oligomerization in the absence of Ficoll. (a) Wild-type CA polymerization kinetics followed by turbidimetry. CA concentrations of 30 μM (squares), 40 μM (circles), and 50 μM (triangles) in 50 mM phosphate buffer, pH 7.4, 2.25 M NaCl were used. (b) Analysis of NTD oligomerization by sedimentation velocity using 80-μM protein in 50-mM phosphate buffer, pH 7.4, 1 M NaCl. The plot represents the protein concentration, c(s), as a function of the sedimentation coefficient, s20,w, as obtained from integration of sedimentation velocity curves. A monodisperse peak with a sedimentation coefficient corresponding to that of monomeric NTD is observed. (c and d) Analysis of NTD oligomerization by sedimentation equilibrium. The plots represents the absorbance at 295 nm with a 12-mm centerpiece (c) or 298 nm with a 3-mm centerpiece because the signal was too high to use the regular centerpiece (d) as a function of the centrifugation radius, using NTD concentrations of 100 μM (c) or 1 mM (d) and a centrifugation velocity of 20,000 rpm in the same buffer used in panel a. Analysis of the concentration gradients was performed with the program HETEROANALYSIS (41), and the resulting apparent molecular masses (Mapp) obtained for NTD are shown in Table 1.
In sedimentation velocity experiments using 80-μM NTD, integration of the experimental data yielded a monodisperse peak of protein (Fig. 2 b). The molecular mass Mw calculated from the sedimentation coefficient s20,w of this peak corresponded, within experimental error, to the molecular mass of the NTD monomer (16,100 Da). These results were indistinguishable from those obtained in sedimentation velocity experiments under the same conditions but at a low ionic strength (0–150 mM NaCl), conditions in which CA is unable to polymerize into capsidlike particles and hexamerization is not expected.
In sedimentation equilibrium experiments in 50 mM sodium-phosphate buffer pH = 7.4, 2.25 M NaCl using NTD concentrations from 30 μM to 300 μM, analysis of the experimental data yielded values for the apparent molecular mass Mw,app between ∼17,000 and 18,000 Da. Again, these values corresponded, within experimental error, to that of the NTD monomer (Fig. 2 c and Table 1). To favor as much as possible the self-association of the NTD monomer into oligomers, its concentration was increased up to 1 mM (16 mg/mL). Even at this very high concentration, NTD did not show any significant tendency to oligomerize (Fig. 2 d and Table 1).
Table 1.
Apparent molar mass Mw,app of CA-NTD obtained by analytical ultracentrifugation in sedimentation equilibrium experiments in the absence of crowding agent
| [NTD] (mM) | Mw,app at 18,000 rpm (kDa)a | Mw,app at 20,000 rpm (kDa)a |
|---|---|---|
| 0.033 | 17.2 ± 0.4 | 17.0 ± 0.4 |
| 0.10 | 17.4 ± 0.4 | 17.2 ± 0.5 |
| 0.30 | 18.0 ± 0.6 | 17.4 ± 0.5 |
| 1.00 | 18.6 ± 0.7 | 18.3 ± 0.6 |
The SDs are indicated.
The very high (1-mM) NTD concentration reached in the above experiments is below the CA concentration inside the typical maturing HIV-1 virion, and much lower than the CA effective concentration (chemical activity) in the macromolecularly crowded virion (see the Introduction). If NTD (or any other protein) has any intrinsic tendency to self-associate but requires a very high concentration to do so, macromolecular crowding would promote its oligomerization to a certain extent. If NTD (or any other protein) has no intrinsic tendency to self-associate, macromolecular crowding would be totally ineffective in promoting oligomerization. To test these alternatives, we added Ficoll-70 to the in vitro reaction mixtures to increase the NTD effective concentration and bring it closer to that inside the virion. Ficoll-70 (molecular mass 70,000 Da) is widely used by many researchers as a chemically defined, inert agent that leads to macromolecular crowding in simplified in vitro models. However, the use of 200–300 g/L Ficoll to nearly match in the experiments the total macromolecular concentration in the virion (∼300 mg/mL; see Table S1) would have led to strong deviations from ideality in the analytical ultracentrifugation experiments and prevented a clear interpretation of data.
Thus, we limited the Ficoll concentration to 100 mg/mL. This concentration is similar to the total macromolecular concentration in many physiological environments. Importantly, 100 mg/mL Ficoll has been shown to strongly promote macromolecular association in many other systems, and is enough to increase by several fold the CA effective concentration. In vitro assembly experiments showed that 100 mg/mL Ficoll strongly promoted polymerization of full-length CA into capsidlike particles, which under the conditions of the assay occurred efficiently and rapidly even at very low protein concentrations (10 μM and lower); this was determined following the polymerization kinetics by turbidimetry (Fig. 3 a), and confirmed by electron microscopy analysis.
Figure 3.

Analysis of CA polymerization and of NTD oligomerization in the presence of Ficoll. (a) CA polymerization kinetics followed by turbidimetry. A CA concentration of 10 μM in 50-mM phosphate buffer, pH 7.4, 2.25 M NaCl, and 100 g/L Ficoll-70 was used. (b) Analysis of NTD oligomerization in sedimentation equilibrium experiments using 1 mM protein in 50-mM phosphate buffer, pH 7.4, 2.25 M NaCl, and different concentrations of Ficoll-70. The plot represents the dependence of the apparent buoyant molar mass (M∗app) of NTD upon the concentration of Ficoll. (Symbols) Experimental data measured by tracer sedimentation equilibrium as described in Materials and Methods. (Solid line) Best-fit of the empirical analysis to estimate the dependence of the activity coefficient of NTD upon solute concentration, as detailed in Materials and Methods, with the best-fit parameter values indicated in the main text.
The ability of NTD to oligomerize in the presence of increasing concentrations of Ficoll-70 as a crowding agent, at a very high NTD concentration (1 mM, corresponding to an effective concentration of up to several mM under these crowded conditions), was then determined in nonideal sedimentation equilibrium experiments. The dependences of the apparent buoyant masses of Ficoll and NTD as functions of Ficoll concentration are, respectively, shown in Fig. S1 in the Supporting Material and in Fig. 3 b. In both cases, the buoyant mass values decrease with increasing concentration of Ficoll, indicating that interactions are predominantly repulsive. The combined data were quantitatively described by an empirical expansion of the logarithm of the activity coefficient of each species in powers of the concentration of the crowder species (see Materials and Methods), truncated after the quadratic terms.
This treatment requires specifying the buoyant masses of the species present and the corresponding two- and three-body interaction coefficients; it assumes as well that interactions between species are essentially due to excluded volume, and that single species of tracer protein and crowder are present in the solution mixture. This does not have to be necessarily the case, but it turned out that this simplifying assumption accounted quantitatively for the data. Application of Eqs. 6 and 8 to the data rendered the following best-fit parameter values (subscripts 1 and 2 refer to Ficoll and NTD, respectively): M∗1 = 23,200 ± 1500; M∗2 = 4400 ± 600; B11 = 30 ± 8 mL/g; B111 = 240 ± 40 (mL/g)2; B21 = 7 ± 3 mL/g; and B211 = 55 ± 25 (mL/g)2. The buoyant molar masses are in agreement with the values expected for the monomeric species (25,000 and 4600, respectively).
The dependence of the activity coefficient of each species with the concentration of Ficoll, calculated from the best-fit M∗i values and the corresponding Bij and Bijk coefficients using Eqs. 5 and 7, revealed that the Ficoll-Ficoll interactions are more repulsive (in very good agreement with previous data; see Fodeke and Minton (46)), than the interactions of capsid proteins and Ficoll; this finding was expected from excluded volume effects, because Ficoll has a larger volume of exclusion per mass than the capsid proteins. Therefore, the sedimentation equilibrium data can be described without the need to propose additional associations, indicating a lack of significant self-associations or hetero-associations of either component. These results revealed that the isolated NTD has no intrinsic tendency to form any kind of oligomer, even under macromolecular crowding conditions.
The full-length CA of HIV-1 with an inactive dimerization interface is unable to oligomerize even in the presence of crowding agents
One possibility to interpret the intrinsic inability of the isolated NTD from HIV-1 to hexamerize is that the association energy contributed by the NTD-CTD interfaces is strictly required for CA hexamerization (contrary to the case of CA from murine leukemia virus in which the NTD alone could hexamerize, albeit only under crystallization conditions). In such a case, it should be still possible to obtain soluble hexamers of full-length CA from HIV-1, provided that the CA effective concentration is sufficiently high and CA polymerization is prevented by inactivating the CTD-CTD dimerization interface. We tested this possibility by using a mutant full-length CAW184A/W185A containing the W184A and M185A mutations, which had been previously shown to drastically impair CTD-CTD dimerization and HIV-1 capsid assembly (14,16,47,48).
We evaluated first whether the presence of these mutations completely prevented the ability of CA to dimerize and/or polymerize under the various experimental conditions used in our study. Gel filtration analysis confirmed that purified CAW184A/W185A at physiologic ionic strength and in the absence of molecular crowding is in monomeric form, even at very high protein concentrations (>1 mM) (data not shown). In addition, polymerization kinetics analysis followed by turbidimetry showed that CAW184A/W185A was completely unable to polymerize at all, even under conditions (very high ionic strength, 2.25 M NaCl; and presence of a macromolecular crowding agent, 100 mg/mL Ficoll-70) that allowed a very rapid and efficient polymerization of nonmutated CA (Fig. 4 a).
Figure 4.

Analysis of CA polymerization and of full-length CAW184A/M185A oligomerization in the absence or presence of Ficoll. (a) Wild-type CA (solid squares) and mutant CAW184A/M185A (open squares) polymerization kinetics followed by turbidimetry. A CA concentration of 20 μM in 50-mM phosphate buffer, pH 7.4, 2.25 M NaCl, and 100 g/L Ficoll-70 was used. (b) Analysis of CAW184A/M185A oligomerization by sedimentation velocity using 100-μM protein in 50-mM phosphate buffer, pH 7.4, 2.25 M NaCl. The plot represents the protein concentration, c(s), as a function of the sedimentation coefficient, s20,w, as obtained from integration of sedimentation velocity curves. A monodisperse peak with a sedimentation coefficient corresponding to that of monomeric CAW184A/M185A is observed. (c) Analysis of CAW184A/M185A oligomerization by sedimentation equilibrium using 300-μM protein in 50-mM phosphate buffer, pH 7.4, 2.25 M NaCl, and different concentrations of Ficoll-70. The plot represents the dependence of the apparent buoyant molar mass (M∗app) of CAW184A/M185A upon the concentration of Ficoll. (Symbols) Experimental data measured by tracer sedimentation equilibrium (see Materials and Methods). (Solid line) Best-fit of the empirical analysis to estimate the dependence of the activity coefficient of CAW184A/M185A upon solute concentration (see Materials and Methods), with the best-fit parameter values indicated in the main text.
Then, we evaluated the ability of CAW184A/W185A to self-associate into hexamers (or any other kind of oligomer) in the absence of macromolecular crowding, under the same conditions in which nonmutated CA is readily able to polymerize into capsidlike particles (50 mM sodium-phosphate buffer pH = 7.4, 2.25 M NaCl). Nonmutated CA did efficiently polymerize at 30–50 μM and lower protein monomer concentrations (Fig. 2 a); in contrast, CAW184A/W185A did not show any significant tendency to oligomerize, as determined by analytical ultracentrifugation in sedimentation velocity experiments, even at higher (100 μM) protein monomer concentrations. Integration of the experimental data yielded a monodisperse peak of protein (Fig. 4 b). The molecular mass Mw calculated from the sedimentation coefficient s20,w of this peak corresponded, within experimental error, to the molecular mass of the CA monomer.
As in the equivalent experiments using NTD, the CA concentration in the above experiments is well below its concentration in the HIV-1 virion, and even much lower when compared to the CA effective concentration in the crowded virion. Again, we used Ficoll-70 as a crowding agent to get closer to the CA effective concentration inside the virion, and determine whether full-length CA with an inactive dimerization interface has any intrinsic tendency to self-associate. We added up to 100 mg/mL Ficoll-70 to the reaction mixture. Under these conditions, nonmutated CA did efficiently polymerize, even at 10 μM and lower protein monomer concentrations (Fig. 4 a).
The ability of CAW184A/W185A to oligomerize in the presence of increasing concentrations of Ficoll-70 as a crowding agent, at a very high CAW184A/W185A concentration (0.3 mM, corresponding to a severalfold higher effective concentration under crowded conditions), was then determined by analytical ultracentrifugation in sedimentation equilibrium experiments (Fig. 4 c). The data in Fig. 4 c were fitted using the same theoretical model applied for the NTD case described above. Application of Eqs. 6 and 8 to the data rendered the following best-fit parameter values (subscripts 1 and 3 refer to Ficoll and CAW184A/W185A, respectively): M∗1 = 23,200 ± 1500; M∗3 = 6200 ± 600; B11 = 30 ± 8 mL/g; B111 = 240 ± 40 (mL/g)2; B31 = 2 ± 4 mL/g; and B311 = 90 ± 30 (mL/g)2. The buoyant molar masses are in agreement with the values expected for the monomeric species (25,000 and 6500, respectively). Again, the sedimentation equilibrium data can be described without the need to propose additional associations, indicating a lack of significant self-associations or hetero-associations of either component. These results revealed that, like the NTD, CAW184A/W185A has no intrinsic tendency to form any kind of oligomer, even under macromolecular crowding conditions.
To summarize, the experiments described show that both the isolated NTD, and a full-length CA of HIV-1 unable to homodimerize though the CTD-CTD interface, are unable to form hexamers (or any other kind of oligomer) through the NTD-NTD and/or NTD/CTD interfaces, even under conditions that promote rapid and efficient polymerization of nonmutated CA into capsidlike particles. This inability of CA or its NTD of HIV-1 to form hexamers from monomers is still observed under macromolecular crowding conditions that approach those inside the HIV-1 virion. These observations indicate that a functional CTD-CTD dimerization interface and dimerization of CA through its CTD are strictly required for hexamerization of native CA of HIV-1 through the other interfaces (NTD-NTD and/or NTD/CTD).
The above results support an allosteric model for assembly of the mature HIV-1 capsid. This model contemplates that CA dimerization through the CTD acts as a conformational switch for capsid assembly by inducing a conformational rearrangement in CTD, and creating the CTD epitope involved in interactions with the NTD of another CA subunit. This allows the formation of the NTD-CTD interfaces required for CA hexamerization and the subsequent assembly of the mature capsid (see Fig. 1 d). This model is supported, in addition to the results of this study, by a number of observations:
-
1.
Mutations W184A and M185A at the CTD-CTD dimerization interface are located far away from the NTD-NTD and NTD-CTD interfaces, and cannot directly participate in them.
-
2.
Mutations W184A and M185A reduce the association energy of CTD-CTD dimerization by >10.6 kcal/mol and 7.6 kcal/mol, respectively (16). Although both single mutants W184A and M185A are monomeric except at very high concentrations, the thermodynamic data show that the W184A mutation has a dominant role in the inactivation of the CTD-CTD interface in CAW184A/W185. Biophysical analyses revealed that the stable monomeric CTDW184A mutant, and the conformationally and thermodynamically similar transient monomeric intermediate of dimerization of nonmutated CTD, lack a part of the native tertiary structure present in each subunit in the CTD dimer (49).
-
3.
In agreement with the later observation, the structure of monomeric CTD (W184A mutant) at neutral pH obtained by NMR spectroscopy revealed that the CTD monomer adopts a somewhat different conformation than each CTD subunit in the dimer. In particular, the dimerization helix 9 appears to be only transiently structured, and helices 10 and 11 have a nearly perpendicular orientation with respect to their orientation in the CTD subunit in the dimer ((50); and see Fig. 5 a). Helices 10 and 11 constitute a major part of the CTD epitope involved in the NTD-CTD dimerization interface in the mature capsid lattice ((22,23), Fig. 5 b); thus, the CTD epitope involved in the CTD-NTD interface may not be formed in the CA monomer. In the disulfide-bridged CA hexamer, the proximity between the NTD of one subunit and the CTD of the neighboring subunit strongly favors NTD-NTD association and greatly increase the collisions between NTD and CTD, which could induce-fit the CTD transition, obviating the presence of an active CTD interface for CA hexamerization.
Figure 5.
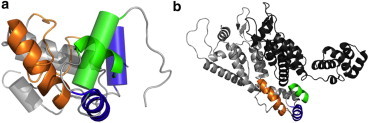
Structural differences of CTD folded as a free monomer or as a homodimer subunit, and CTD regions involved in the CTD-CTD and CTD-NTD interfaces in the mature HIV-1 capsid. (a) Structural models of the CTD monomer (mutant CAW184A) obtained by NMR (50) (with helices shown as cylinders) and of a subunit in the CTD dimer (14) (with helices shown as ribbons) are overlapped. Helix 9 (involved in the CTD-CTD dimerization interface) is colored (orange) in the subunit of the CTD dimer; this helix is not permanently structured in the CTD monomer, and the corresponding peptide segment appears in the figure as an (orange irregular ribbon). Helices 10 and 11 (involved in the CTD-NTD dimerization interface) are colored (blue or green, respectively). (b) Ribbon model of two contiguous monomers in the crystal structure of the CA hexamer (21), which are shown in (light and dark gray, respectively). In the (light gray colored monomer), helices 9-11 are colored as in panel a.
Conclusions
The results of this study indicate that dimerization of the HIV-1 capsid protein CA through a functional CTD-CTD interface is strictly required for the hexamerization of the dimers through the NTD-NTD and NTD-CTD interfaces to form the hexameric lattice of the mature virus capsid. Together with previous observations, this study suggests that establishment of NTD-CTD interactions involved in hexamerization of CA during mature HIV-1 capsid assembly requires a CTD homodimerization-dependent conformational switching event.
Acknowledgments
M.G.M. is an associate member of the Institute for Biocomputation and Physics of Complex Systems, Zaragoza, Spain.
Work in M.G.M.’s laboratory is funded by grants from the Spanish Government (grant No. BIO2009-10092 and BIO2012-37649) and Comunidad de Madrid (grant No. S-2009/MAT/1467), and by an institutional grant from Fundación Ramón Areces. Work in G.R.’s laboratory is funded by the Spanish Government (grant No. BIO2011-28941-C03).
Footnotes
Rebeca Bocanegra’s present address is Centro Nacional de Biotecnología (Consejo Superior de Investigaciones Científicas), Campus de Cantoblanco, 28049 Madrid, Spain.
Supporting Material
References
- 1.Ganser-Pornillos B.K., Yeager M., Sundquist W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganser-Pornillos B.K., Yeager M., Pornillos O. Assembly and architecture of HIV. Adv. Exp. Med. Biol. 2012;726:441–465. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs J.A., Kräusslich H.G. The molecular architecture of HIV. J. Mol. Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Sundquist W.I., Kräusslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S., Hill C.P., Finch J.T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 6.Briggs J.A., Wilk T., Fuller S.D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateu M.G. The capsid protein of human immunodeficiency virus: intersubunit interactions during virus assembly. FEBS J. 2009;276:6098–6109. doi: 10.1111/j.1742-4658.2009.07313.x. [DOI] [PubMed] [Google Scholar]
- 8.Byeon I.J., Meng X., Gronenborn A.M. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prevelige P.E., Jr. New approaches for antiviral targeting of HIV assembly. J. Mol. Biol. 2011;410:634–640. doi: 10.1016/j.jmb.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waheed A.A., Freed E.O. HIV type 1 Gag as a target for antiviral therapy. AIDS Res. Hum. Retroviruses. 2012;28:54–75. doi: 10.1089/aid.2011.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bocanegra R., Rodríguez-Huete A., Mateu M.G. Molecular recognition in the human immunodeficiency virus capsid and antiviral design. Virus Res. 2012;169:388–410. doi: 10.1016/j.virusres.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Momany C., Kovari L.C., Rossmann M.G. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 13.Gitti R.K., Lee B.M., Sundquist W.I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 14.Gamble T.R., Yoo S., Hill C.P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 15.Ganser-Pornillos B.K., Cheng A., Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 16.del Alamo M., Neira J.L., Mateu M.G. Thermodynamic dissection of a low affinity protein-protein interface involved in human immunodeficiency virus assembly. J. Biol. Chem. 2003;278:27923–27929. doi: 10.1074/jbc.M304466200. [DOI] [PubMed] [Google Scholar]
- 17.Bocanegra R., Nevot M., Mateu M.G. Rationally designed interfacial peptides are efficient in vitro inhibitors of HIV-1 capsid assembly with antiviral activity. PLoS ONE. 2011;6:e23877. doi: 10.1371/journal.pone.0023877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H., Curreli F., Debnath A.K. Antiviral activity of α-helical stapled peptides designed from the HIV-1 capsid dimerization domain. Retrovirology. 2011;8:1–18. doi: 10.1186/1742-4690-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanman J., Sexton J., Prevelige P.E., Jr. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 2002;76:6900–6908. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanman J., Lam T.T., Prevelige P.E., Jr. Identification of novel interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Mol. Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 21.Pornillos O., Ganser-Pornillos B.K., Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pornillos O., Ganser-Pornillos B.K., Yeager M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 2010;401:985–995. doi: 10.1016/j.jmb.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pornillos O., Ganser-Pornillos B.K., Yeager M. Atomic-level modeling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortuza G.B., Haire L.F., Taylor I.A. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 25.Carlson L.A., Briggs J.A., Kräusslich H.G. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minton A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 27.Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H.-X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Alamo M., Rivas G., Mateu M.G. Effect of macromolecular crowding agents on human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 2005;79:14271–14281. doi: 10.1128/JVI.79.22.14271-14281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rincón V., Bocanegra R., Mateu M.G. Effects of macromolecular crowding on the inhibition of virus assembly and virus-cell receptor recognition. Biophys. J. 2011;100:738–746. doi: 10.1016/j.bpj.2010.12.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell S., Vogt V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross I., Hohenberg H., Kräusslich H.-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 33.Ganser B.K., Li S., Sundquist W.I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 34.del Alamo M., Mateu M.G. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J. Mol. Biol. 2005;345:893–906. doi: 10.1016/j.jmb.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 35.Douglas C.C., Thomas D., Prevelige P.E., Jr. Investigation of N-terminal domain charged residues on the assembly and stability of HIV-1 CA. Biochemistry. 2004;43:10435–10441. doi: 10.1021/bi049359g. [DOI] [PubMed] [Google Scholar]
- 36.Erickson-Viitanen S., Manfredi J., Swanstrom R. Cleavage of HIV-1 Gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retroviruses. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 37.Gill S.C., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 38.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck P., Perugini M.A., Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minton A.P. Modern Analytical Ultracentrifugation. In: Schuster T.H., Lave T.H., editors. Birkhauser; Boston, MA: 1994. pp. 81–93. [Google Scholar]
- 41.Cole J.L. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivas G., Fernandez J.A., Minton A.P. Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: theory, experiment, and biological significance. Biochemistry. 1999;38:9379–9388. doi: 10.1021/bi990355z. [DOI] [PubMed] [Google Scholar]
- 43.Zorrilla S., Jiménez M., Minton A.P. Sedimentation equilibrium in a solution containing an arbitrary number of solute species at arbitrary concentrations: theory and application to concentrated solutions of ribonuclease. Biophys. Chem. 2004;108:89–100. doi: 10.1016/j.bpc.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Rivas G., Minton A.P. Beyond the second virial coefficient: sedimentation equilibrium in highly non-ideal solutions. Methods. 2011;54:167–174. doi: 10.1016/j.ymeth.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darawshe S., Rivas G., Minton A.P. Rapid and accurate microfractionation of the contents of small centrifuge tubes: application in the measurement of molecular weight of proteins via sedimentation equilibrium. Anal. Biochem. 1993;209:130–135. doi: 10.1006/abio.1993.1092. [DOI] [PubMed] [Google Scholar]
- 46.Fodeke A.A., Minton A.P. Quantitative characterization of polymer-polymer, protein-protein, and polymer-protein interaction via tracer sedimentation equilibrium. J. Phys. Chem. B. 2010;114:10876–10880. doi: 10.1021/jp104342f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Schwedler U.K., Stray K.M., Sundquist W.I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu X., Wang Q., Zheng J. Mutational analysis and allosteric effects in the HIV-1 capsid protein carboxyl-terminal dimerization domain. Biomacromolecules. 2009;10:390–399. doi: 10.1021/bm801151r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mateu M.G. Conformational stability of dimeric and monomeric forms of the C-terminal domain of human immunodeficiency virus-1 capsid protein. J. Mol. Biol. 2002;318:519–531. doi: 10.1016/S0022-2836(02)00091-8. [DOI] [PubMed] [Google Scholar]
- 50.Alcaraz L.A., del Alamo M., Neira J.L. Flexibility in HIV-1 assembly subunits: solution structure of the monomeric C-terminal domain of the capsid protein. Biophys. J. 2007;93:1264–1276. doi: 10.1529/biophysj.106.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Lano, W. L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA. http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


