Abstract
Superfamily I helicases are nonhexameric helicases responsible for the unwinding of nucleic acids. However, whether they unwind DNA in the form of monomers or oligomers remains a controversy. In this study, we addressed this question using direct single-molecule fluorescence visualization of Escherichia coli UvrD, a superfamily I DNA helicase. We performed a photobleaching-step analysis of dye-labeled helicases and determined that the helicase is bound to 18-basepair (bp) double-stranded DNA (dsDNA) with a 3′ single-stranded DNA (ssDNA) tail (12, 20, or 40 nt) in a dimeric or trimeric form in the absence of ATP. We also discovered through simultaneous visualization of association/dissociation of the helicase with/from DNA and the DNA unwinding dynamics of the helicase in the presence of ATP that these dimeric and trimeric forms are responsible for the unwinding of DNA. We can therefore propose a new kinetic scheme for the helicase-DNA interaction in which not only a dimeric helicase but also a trimeric helicase can unwind DNA. This is, to our knowledge, the first direct single-molecule nonhexameric helicase quantification study, and it strongly supports a model in which an oligomer is the active form of the helicase, which carries important implications for the DNA unwinding mechanism of all superfamily I helicases.
Introduction
Helicases are highly conserved enzymes that are involved in DNA replication, repair, and recombination, as well as in the genome stability of prokaryotes, eukaryotes, bacteriophages, and viruses. They are classified into six superfamilies (SF1–6) on the basis of their primary structures, called helicase motifs (1). Helicases are also classified into two main classes according to their functional forms. One class is known to function as hexameric ring structures that can encircle DNA (2,3), whereas the other class, which includes the SF1 and SF2 helicases, functions in a nonhexameric form. Among the SF1 helicases, the tertiary structures of the Escherichia coli UvrD and Rep and the Bacillus stearothermophilus PcrA have been resolved by x-ray crystallography (4–7). These structures make it clear that these helicases share high structural homology (40%). Due to their high homology, the SF1 DNA helicases are believed to be responsible for the unwinding of DNA through a similar mechanism. A number of different methodologies, including single-molecule analytical methods (5,8–16), have elucidated various key aspects of these SF1 helicases, such as their unwinding, translocation, processivity, and conformational changes. However, two conflicting models have been proposed for the unwinding of DNA by nonhexameric helicases. One is the monomeric helicase model that has been proposed for the PcrA (7), E. coli UvrD (17), and SF2 hepatitis C viral NS3 RNA helicases (18). The other model is the dimeric helicase model, proposed for the Rep (6,19,20), PcrA (21), UvrD (22), and NS3 helicases (23).
E. coli UvrD is an SF1 DNA helicase that plays a crucial role in both nucleotide excision repair and methyl-directed mismatch repair (24). Using ATP hydrolysis energy, this enzyme unwinds a duplex DNA starting from its 3′ end ssDNA tail, a gap, or a nick. Previous biochemical studies have suggested that this enzyme has optimal activity in its oligomeric form (22), and this hypothesis is supported by a single-molecule DNA manipulation study using magnetic tweezers (13). However, crystal structures of UvrD-DNA complex have been resolved only for monomeric UvrD (4).
In this study, we initially employed a photobleaching-step analysis (25–28) to quantify the number of helicases that bind to DNA in the absence of ATP and found that the helicase in its oligomeric form binds to 18-bp dsDNA with a 12-, 20-, or 40-nt 3′-ssDNA tail. Then, to determine whether the helicase unwinds DNA in the form of a monomer or oligomer in the presence of ATP, we performed simultaneous single-molecule visualization studies of DNA unwinding events that are driven by the helicase and of association/dissociation events between the helicase and DNA. The results of these experiments, conducted using DNA with a 20-nt 3′-ssDNA tail, suggest that the helicase completely unwinds the DNA in a few seconds after two or more of the helicases are bound to it, which strongly supports the model in which the active form of the helicase is an oligomer. In addition, the determined dissociation and association rates increase as the number of helicases bound to DNA increases. We can therefore propose a new kinetic scheme for the helicase-DNA interaction, in which not only a dimeric helicase but also a trimeric helicase can unwind DNA. Although a dozen DNA-binding proteins, including helicases, have been directly visualized at the single-molecule level to date (29–36), quantification of the number of helicases has, to our knowledge, not yet been performed using direct-visualization analyses. Thus, this is the first study that we know of to utilize direct single-molecule visualization to quantify the number of nonhexameric helicases, and it demonstrates that the nonhexameric helicase unwinds DNA in the form of an oligomer.
Materials and Methods
Detailed information on preparation of DNA substrates and UvrD proteins, PEGylation of glass substrates, single-molecule imaging assays, and microscopy is available in the Supporting Material.
DNA substrates
The duplex DNA substrates were prepared through hybridization reactions and stored in the form of aliquots at −20°C until use.
UvrD proteins
The UvrD proteins with Cys-to-Ala mutations were expressed and purified as described previously (35) and labeled with Cy5 maleimide (PA25001, GE Healthcare, Little Chalfont, UK) at a UvrD/dye molar ratio of 1:3 in 500 mM NaCl, 10% (v/v) glycerol, and 20 mM HEPES (pH 7.0) for 20 h at 4°C. After the unlabeled dyes were removed, the labeled proteins were aliquoted, quickly frozen in liquid nitrogen, and stored at −80°C until use.
Single-molecule imaging assays
All single-molecule experiments reported in this article were performed at 25°C using either of two types of flow cells with different total internal reflection fluorescence methods: a nail-polish-sealed flow cell with prism-type total internal reflection microscopy (Fig. 1 A) or a double-sided tape-sealed flow cell with objective-type total internal reflection microscopy (see Fig. 3 A).
Figure 1.
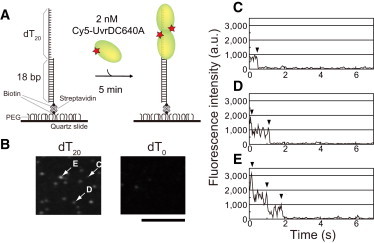
Single-molecule visualization of Cy5-UvrDC640A bound to a duplex DNA substrate with a 3′-ssDNA tail in the absence of nucleotide. (A) Schematic drawing of the experiment. First, 18-bp dsDNA with a 20-nt 3′-ssDNA tail and a biotin at one end was attached via streptavidin-biotin interactions to streptavidin that was immobilized on a PEGylated quartz slide. Then, 50 μl of 2.0 nM Cy5-UvrDC640A in buffer U (6 mM NaCl, 2.5 mM MgCl2, 10% (v/v) glycerol, and 25 mM Tris-HCl (pH 7.5)) with an oxygen scavenger system was infused. After 5 min, the single-molecule visualization of Cy5-UvrDC640A was performed by prism-type total-internal fluorescence microscopy in which Cy5 was excited at 637 nm. (B) Fluorescence images (average of 0.5 s) of Cy5-UvrDC640A bound to DNA with a 20-nt 3′-ssDNA tail (left) and blunt-end dsDNA (right). Scale bar, 10 μm. (C–E) Time courses of the fluorescence intensity (average of three moving frames (0.1 s)) of the Cy5-UvrDC640A fluorescent spots indicated in B by arrows labeled C–E. The photobleaching processes indicated by the arrowheads occurred in one (C), two (D), and three (E) steps.
Figure 3.
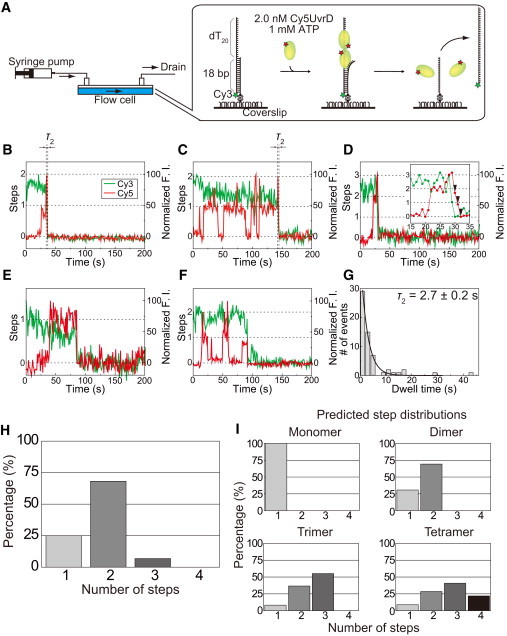
Simultaneous single-molecule visualization of the unwinding of DNA driven by the helicase and the association/dissociation events between the helicase and DNA in the presence of 1 mM ATP. (A) Schematic drawing of the experiment. Lasers to excite Cy3 at 532 nm and Cy5 at 632.8 nm were incident on the sample plane with objective-type total internal fluorescence microscopy. Single-molecule fluorescence signals from Cy3-DNA and Cy5-UvrDC640A were simultaneously imaged using a dual-view apparatus. For a more detailed explanation, see the text. (B and C) Typical time traces of the Cy3 and Cy5 fluorescence intensities (F. I.) in which the Cy5 fluorescence intensity increases in a two-step manner just before the unwinding of the DNA and the Cy3 fluorescence intensity decreases to its background level. (D) Time traces of the Cy3 and Cy5 fluorescence intensities in which the Cy5 fluorescence intensity just before the DNA unwinding process changes in a three-step manner; specifically, the Cy5 fluorescence increases in two steps but decreases in three steps. The three-step fluorescence decrease is indicated by arrowheads. (E and F) Typical time traces of the Cy3 and Cy5 fluorescence intensities in which the Cy5 fluorescence intensity increases in a single step just before the DNA unwinding process. (G) Dwell time distribution of the second step (τ2) just before the DNA unwinding process for traces that exhibit two steps. The mean dwell time, which was obtained using a single-exponential fit, is 2.7 ± 0.2 s. (H) Experimentally obtained distribution of the number of step changes that the Cy5 fluorescence undergoes just before or after DNA unwinding. (I) Theoretically predicted distributions of the number of steps that the Cy5 fluorescence undergoes prior to DNA unwinding.
Microscope
The prism-type and objective-type of total internal reflection microscopy was conducted using two identical inverted microscopes (IX71, Olympus, Tokyo, Japan) and lasers to excite Cy3 at 532 nm and Cy5 at 637 nm or 632.8 nm. The fluorescence signals from the samples were passed through dichroic mirrors to separate fluorescences from Cy3 and Cy5, and through barrier filters (580DF30 for Cy3 and 670DF40 for Cy5, Omega Optical, Brattleboro, VT) to eliminate the background light. The filtered fluorescence signals (565–595 nm for Cy3 and 650–690 nm for Cy5) were imaged using a dual-view apparatus and recorded with a high-sensitivity CCD camera. The recorded images were analyzed using Image-Pro PLUS (Media Cybernetics, Rockville, MD).
The time resolution of the experiments with prism-type total internal reflection microscopy was 33 ms, whereas the time resolution of the experiments conducted using objective-type total internal reflection microscopy was 1 s. The lasers exciting Cy3 and Cy5 were simultaneously incident on the sample plane for 100 ms/s using mechanical shutters (LS3, Uniblitz, Rochester, NY) to minimize photobleaching of the dyes.
Results
UvrD protein binds to the ss/ds DNA junction
To quantify the number of UvrD proteins that bind to DNA, we used a Cy5-labeled Cys-Ala mutant (Cy5-UvrDC640A) in which Cys52 was labeled with high specificity (Figs. S1, S2, Table S3, and Supplementary Results 1 and 2 in the Supporting Material). Fig. 1 B shows single-molecule fluorescence images of Cy5-UvrDC640A at a concentration of 2.0 nM in the presence of 18-bp dsDNA with a 20-nt 3′-ssDNA tail (22,35,37–39) and blunt-end 18-bp dsDNA immobilized on the surface via streptavidin-biotin interactions (Fig. 1 A). We used buffer U (6 mM NaCl, 2.5 mM MgCl2, 10% (v/v) glycerol, and 25 mM Tris-HCl, pH 7.5) in all of the experiments in this study (35). In agreement with a previous report (22), as well as with our previous study (35), Cy5-UvrDC640A had a high affinity to the ss/ds DNA junction; therefore, many fluorescent spots were observed with the dsDNA with a 20-nt 3′-ssDNA tail but not with the blunt-end dsDNA. Fig. 1, C–E, shows the time courses of the fluorescence intensity of three different fluorescent spots in which a different number of photobleaching events occurred (one, two, or three steps); these spots are indicated by arrowheads in Fig. 1 B.
Distribution of photobleaching steps of Cy5 labeled to UvrDC640A
A study that used single-turnover DNA unwinding experiments reported that longer DNA tail lengths result in higher fractions of DNA being unwound (22). To investigate the relationship between DNA tail length and the number of UvrD proteins that are bound to DNA, we first analyzed Cy5-UvrDC640A bound to 18-bp DNA substrates with 12-nt, 20-nt, or 40-nt 3′-ssDNA tails in the presence of 0.5, 1.0, or 2.0 nM Cy5-UvrDC640A but in the absence of nucleotide. The shortest tail length that has been reported for the unwinding of duplex DNA is 12 nt (13,22); therefore, we used DNA substrates with 3′-ssDNA tail lengths of 12 nt or greater. In addition, using an assay similar to the one illustrated in Fig. 1 A, we confirmed that Cy5-UvrDC640A (≥1.0 nM) can unwind the DNA substrates efficiently in the presence of ATP using a Cy3-labeled oligo (Fig. S3). Fig. 2 A shows the distributions of the number of photobleaching steps observed in the presence of 0.5, 1.0, or 2.0 nM Cy5-UvrDC640A in solution. The longer 3′-ssDNA tail lengths decreased the ratio of single photobleaching steps to multiple photobleaching steps and increased the ratios of the multiple photobleaching steps, which demonstrates that more UvrD proteins are bound to DNA substrates that have longer 3′-ssDNA tails.
Figure 2.

Experimentally obtained and theoretically predicted distributions of the number of photobleaching steps for the Cy5-UvrDC640A that was bound to 18-bp DNA with a 12-, 20-, or 40-nt 3′-ssDNA tail. (A and B) Experimentally obtained distributions derived from solutions containing Cy5-UvrDC640A alone (A) or Cy5-UvrDC640A and 1 mM ATPγS (B). The total number of analyzed fluorescent spots for each condition is indicated. (C) Theoretically predicted distributions of the number of photobleaching steps for the monomer, dimer, trimer, and tetramer models.
We then performed the same experiments in the presence of adenosine 5′-(γ-thio)triphosphate (ATPγS), a nonhydrolyzable ATP analog (Fig. 2 B). The photobleaching-step analysis revealed that the longer 3′-ssDNA tail lengths again decreased the ratio of single photobleaching steps to multiple photobleaching steps and increased the ratios of multiple photobleaching steps, as was observed in the distributions obtained with the UvrD protein alone. However, the photobleaching-step distributions shifted to a larger value compared to those obtained with the UvrD protein alone, which suggests that the presence of ATPγS increases the number of UvrD proteins that binds to DNA (22,38).
Multiple UvrD proteins are bound to DNA in the absence of ATP
To derive from the obtained distributions information on the number of UvrD proteins that are bound to DNA, we calculated the predicted distributions for monomer, dimer, trimer, and tetramer models of the UvrD protein bound to DNA (Fig. 2 C, Supplementary Result 5). These calculations were based on the estimated number of Cy5 dyes per UvrDC640A protein (Table S4), which was based on the labeling ratios of Cy5-UvrDC640A (75%) and Cy5-UvrDC52A/C640A (10%). To determine which model best fits the experimentally obtained distributions, we performed goodness-of-fit tests in which the χ2 values of the models were calculated for each distribution. All of the distributions were fitted least with the monomer model (Fig. S4)—the χ2 values for the monomer model had the highest values under all analyzed conditions. This result suggests that multiple UvrD proteins are bound to the DNA. Table 1 lists the models that minimized the χ2 values under the different conditions. As shown, longer DNA tail lengths, higher UvrD concentrations, and the presence of ATPγS increased the number of bound UvrD proteins. These results demonstrate that at least two and at most three UvrD proteins bind to the same DNA substrate and that the presence of ATPγS promotes the binding of more UvrD proteins to the DNA.
Table 1.
Models that best fit the histograms
| Cy5-UvrDC640A |
Cy5-UvrDC640A + 1 mM ATPγS |
|||||
|---|---|---|---|---|---|---|
| 0.5 nM | 1.0 nM | 2.0 nM | 0.5 nM | 1.0 nM | 2.0 nM | |
| dT12 | Dimer | Dimer | Trimer | Dimer | Trimer | Trimer |
| dT20 | Dimer | Trimer | Trimer | Trimer | Trimer | Trimer |
| dT40 | Dimer | Trimer | Trimer | Dimer | Trimer | Trimer |
The best-fitting models minimize the χ2 values, as determined through goodness-of-fit tests (Fig. S4).
Visualization of an increase in the Cy5 fluorescence just before DNA unwinding
Although the determined amount of Cy5-UvrDC640A that is bound to DNA implies that the helicase unwinds the DNA in the form of an oligomer, the data did not provide any pertinent information on the number of helicases that are bound to the DNA as it is unwound in the presence of ATP. Therefore, using a flow cell, we performed simultaneous single-molecule visualization studies of the unwinding of DNA, which is driven by the helicase, and of the association/dissociation events between the helicase and DNA in the presence of ATP (Fig. 3 A). We performed this experiment using 2.0 nM Cy5-UvrDC640A with a high labeling ratio of 90% and the DNA substrate with a 20-nt 3′-ssDNA tail (22,35,37–39). The unwinding of the DNA was monitored through the disappearance of the fluorescence of Cy3, which was attached to one of the two oligonucleotides that form the DNA.
Although the UvrD and DNA concentrations were lower than those used in the biochemical studies by Maluf et al. (22,39), the high rate constants and high equilibrium constants of the UvrD-DNA interaction enabled us to visualize the interaction at a single-molecule resolution (Supplementary Discussion 1). We performed the experiment using a time-lapse imaging method featuring two electronic shutters that control the laser excitations (Cy3 at 532 nm and Cy5 at 632.8 nm), thus minimizing the photobleaching of the dyes; specifically, the excitation laser beams were periodically incident on the sample plane for 100 ms at 1-s intervals (see details in Materials and Methods). We observed many traces in which the Cy3 and Cy5 fluorescences decreased almost simultaneously to their respective background levels (Fig. 3, B–F), which shows that the UvrD protein unwound the DNA and then immediately dissociated from it. We could thus exclude from our data analysis the traces in which Cy3 photobleached before the DNA was completely unwound.
We found that the Cy5 fluorescence intensity, which decreased almost simultaneously with the Cy3 fluorescence intensity, increased either in multiple steps (Fig. 3, B–D) or in a single step (Fig. 3, E and F) just before the completion of the DNA unwinding process. The traces can be further divided into two categories: some exhibit no significant Cy5 fluorescence intensity change before these step(s) (Fig. 3, B and E), and others exhibit at least one Cy5 fluorescence intensity increase-decrease cycle before these step(s) (Fig. 3 C, D, and F). Note that some traces exhibit a three-step fluorescence increase or decrease (Fig. 3 D). Of the 88 analyzed traces, the traces that exhibit multiple and single steps account for 66 and 22 traces, respectively. The 66 traces, including six traces that show three steps, consist of 17 traces that do not exhibit any prior Cy5 fluorescence intensity change and 49 traces that do show prior Cy5 fluorescence intensity change. The 22 traces that show a single-step increase include 17 that do and five that do not exhibit prior Cy5 fluorescence intensity change.
DNA unwinding is complete immediately after additional UvrD protein(s) bind to the DNA
Intriguingly, most of the traces with multiple steps exhibit rapid DNA unwinding immediately after additional UvrD protein(s) bind to the DNA. Fig. 3 G shows the distribution of the dwell time of the second step (τ2) in traces that show the two-step Cy5 fluorescence increase just before completion of the DNA unwinding process. The mean dwell time, which was obtained using single-exponential fit, was 2.7 ± 0.2 s. In traces that show a three-step Cy5 fluorescence increase or decrease, the last step of the increase in Cy5 fluorescence before the unwinding of the DNA had a similar mean dwell time of 2.3 ± 1.5 s. The dwell time has larger uncertainty, because it was calculated through simple averaging of the limited amount of data for these traces (n = 6).
These results suggest that a single UvrD protein binds to the DNA, waits for an additional UvrD protein(s) to bind, forms a protein complex with the additional UvrD protein(s) on the DNA, and completes the DNA unwinding process in a few seconds. The traces that show a three-step fluorescence increase or decrease suggest that three UvrD proteins are bound to the DNA in the presence of ATP and are responsible for the unwinding of the DNA, as indicated by the earlier experiments in the absence of ATP (Table 1).
UvrD unwinds the DNA in the form of an oligomer
The traces with multiple steps clearly demonstrate that the UvrD protein unwinds DNA in the form of an oligomer. We can explain the traces with the single step using the oligomer model even though they initially seem to support the monomer model, because the ratios of the step numbers of the Cy5 fluorescence changes (Fig. 3 H) are better reproduced by the dimer model than by the monomer model (Fig. 3 I). The ratios of the models were calculated based on the estimated number of Cy5 dyes per UvrDC640A protein (Supplementary Result 7). The percentages of the one- and two-step fluorescence changes in the experimentally obtained traces (22/(66 + 22) = 25% and 60/(60 + 22) = 68%) are closest to those predicted by the dimer model (31% and 69%, respectively). We can therefore conclude that the DNA unwinding is performed not by a UvrD monomer but rather mostly by a UvrD dimer, and in some cases by a UvrD trimer, because the dimer model best fits the experimental step distribution and we obtained some traces that indicated DNA unwinding by a UvrD trimer.
We did not take into account in the above estimate the effect of Cy5 photobleaching, because the dimer model explains the ratios satisfactorily and we assumed that Cy5 rarely photobleached before completion of the DNA unwinding process. Thus, Cy5 photobleaching would have little, if any, effect on the data analysis. In the highly unlikely situation in which the Cy5 photobleaching effect is significant, the oligomer model would not be overturned but rather would be supported, because the analysis of step number in the presence of photobleached Cy5-UvrDC640A proteins never leads to an overestimate of the number of UvrD proteins.
Association/dissociation rates of the UvrD-DNA interaction
Direct visualization of the Cy5-labeled UvrD protein can also be used to determine almost all of the kinetic rate constants of the UvrD-DNA interaction. To determine approximate values of the rate constants, we interpreted the Cy5 fluorescence increase/decrease steps as UvrD association/dissociation events. This approximate determination is possible because >80% of the UvrD proteins are labeled with Cy5 (Table S4). We obtained traces with various compositions of one-, two-, and three-step Cy5 fluorescence increases or decreases, which indicate sequential UvrD monomer association/dissociation. We also obtained traces that display simultaneous binding to or dissociation from DNA by UvrD dimers or trimers. We used the two- or three-step increase or decrease of the Cy5 fluorescence intensity before or after the association/dissociation as a criterion for confirmation of the steps that represent the association/dissociation of the UvrD dimer or trimer (Fig. 3 D). We can therefore propose a new kinetic scheme of UvrD-DNA interactions in which not only two but also three UvrD proteins are involved (Fig. 4).
Figure 4.
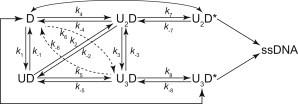
Kinetic scheme of the UvrD-DNA interaction. This scheme was conceived based on the assumption that the number of UvrD proteins involved in the interaction is at most three. The UvrD protein initially forms a complex with DNA in the form of UD, U2D, or U3D, undergoes a number of UvrD association/dissociation events, and then forms the UvrD oligomer. The oligomer isomerizes along the DNA to become an active complex (U2D∗ or U3D∗) that unwinds the DNA.
Fig. 5 A shows the dwell-time distributions of the indicated states in the association and dissociation reactions between UvrD proteins and DNA in which up to two UvrD proteins were involved. The distributions do not include a dwell time for the dissociation that occurs just before completion of the DNA unwinding process. Each distribution is fitted satisfactorily with a single exponential, which yields the corresponding mean dwell time. Intriguingly, the dwell times of both the dissociation and association processes decrease as the number of UvrD proteins that are involved increases (Fig. 5 B).
Figure 5.
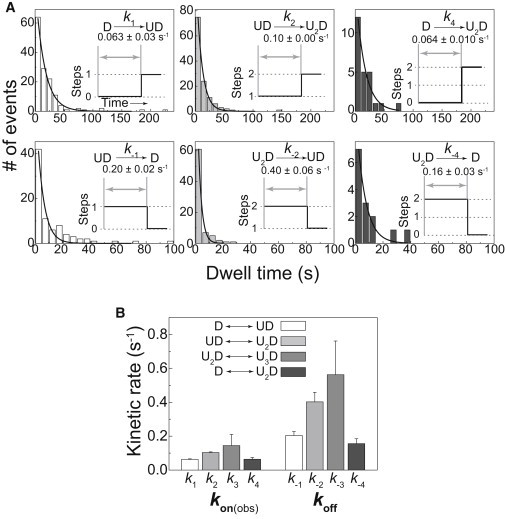
Association and dissociation rate constants. (A) Dwell-time distributions of the indicated states. Each distribution was fitted satisfactorily with a single exponential, which yielded the corresponding association and dissociation rate constants. The distributions do not include a dwell time for dissociation process during the unwinding of the DNA. (B) Comparison of the obtained rate constants. k3 and k−3, which were obtained using only a small amount of the data, were calculated through simple averaging and not by single-exponential fit. The error bars represent the standard errors of k1, k2, k4, k−1, k−2, and k−4 and the standard deviations of k3 and k−3.
Comparison of the rate constants of the sequential association of two UvrD monomers (D→UD and UD→U2D) and the association of a UvrD dimer (D→U2D) and comparison of the rate constants of the sequential dissociation of two UvrD monomers (U2D→UD and UD→D) and the dissociation of a UvrD dimer (U2D→D) indicate that a preassembled UvrD dimer association/dissociation is being visualized in our experiments (Supplementary Result 8). We thus obtained approximate association/dissociation rate constants of the UvrD-DNA interaction in the presence of ATP, in which not only two but also three UvrD proteins are involved (Table S5).
Discussion
Number of UvrD proteins bound to DNA in the absence of ATP
We used a photobleaching-step analysis of single-molecule fluorescence images to determine the number of UvrD proteins that are bound to DNA substrates with 12-, 20-, or 40-nt 3′-ssDNA tails in the absence of nucleotide. This number did not decrease for concentrations of Cy5-UvrDC640A below 1.0 nM (Table 1), which did not result in an efficient unwinding in the presence of ATP of the dsDNA with a 20-nt 3′-ssDNA tail that was immobilized on a glass surface (Fig. S3 B). These results are in agreement with those from a biochemical study in which UvrD dimers were stabilized when bound to DNA (40). A goodness-of-fit analysis revealed that the number of bound UvrD proteins was at most three (Fig. S4) and showed that this number is not proportional to the concentration of Cy5-UvrDC640A or the 3′-ssDNA tail length. This result is consistent with the number of UvrD proteins bound to a DNA substrate with a 20-nt 3′-ssDNA tail, which was determined through analytical ultracentrifugation under saturating UvrD concentration conditions (38) when the DNA and UvrD concentrations used were 0.1–0.5 μM and a >3 molar excess of the DNA, respectively. UvrD proteins must directly interact with one another on ssDNA because the UvrD protein has a low affinity for blunt duplex DNA (Fig. 1 B) and the lower limit of the estimated UvrD site size on poly(dT) of 10 ± 2 nt precludes the possibility of multiple UvrD monomers binding in tandem (40).
The presence of 1 mM ATPγS slightly increased the number of UvrD proteins that bound to DNA (three at most), which also agrees with the results obtained from an analytical ultracentrifugation study, in which ADP and ATPγS did not significantly affect the dimerization equilibrium constant (38). The effects of nucleotide binding on the UvrD protein conformation were examined using trypsin and chymotrypsin as probes (41). It has been reported that the presence of nucleotides (ATP or ATPγS) or ssDNA (M13mp11ssDNA) stabilizes the 72-kDa tryptic polypeptides with respect to further cleavage, which indicates that either the UvrD conformation is altered or that the cleavage site is protected by occlusion. These results demonstrate that the UvrD protein in the presence of DNA and ATPγS assumes a similar conformation to that obtained in the presence of DNA and ATP.
Kinetic mechanism of the UvrD-DNA interaction
Simultaneous single-molecule visualization of the DNA unwinding process and the association/dissociation dynamics of the UvrD protein in the presence of ATP suggests that the helicase unwinds DNA in the form of an oligomer. In addition, we were able to use these studies to determine almost all of the approximate association/dissociation rates and equilibrium constants of the UvrD-DNA interaction that had not been determined in previous studies (Table S5). The oligomeric model was originally proposed by Maluf et al., who used chemical quenched-flow methods to measure the DNA unwinding activity of the helicase under various UvrD concentrations and 3′-ssDNA overhang lengths (22). They were able to observe the dynamic interaction and found that the overall extent of DNA unwinding has a nonlinear sigmoidal dependence on [UvrD]/[DNA] (22). These researchers also examined the kinetic mechanisms for the formation of the active, dimeric helicase-DNA complex (39). A study of the DNA unwinding activity, which was measured using magnetic tweezers, also supports the oligomer model. Sun et al. measured the on time as the time span of an unwinding burst and the off time as the time between two adjacent bursts (13). These researchers found the on-time distributions to be exponential. The off-time distributions, however, were fitted with the convolution of two exponentials, which indicated that the two UvrD proteins subsequently bound to the DNA. The association/dissociation rate constants obtained in this study (Fig. 5 A, Table S5) are comparable to those obtained in the single-molecule DNA manipulation studies reported by Sun et al. (13) (k1 = 0.10 s−1, k2 = 0.15 s−1, and k−1 = 0.12 s−1 for [UvrD] = 2 nM), but they are lower than those obtained with the chemical quenched-flow methods (k1 = 0.3 s−1, k2 > 0.3 s−1, k−1 = 0.025 s−1, and k−2 = ∼3 s−1for [UvrD] = 2 nM) (39).
Although the rates obtained in this study are lower than the lower limits that were estimated by Maluf et al. for k1, k2, and k−2 and larger than their upper-limit estimate for k−1 (Supplementary Discussion 2), the rate constants in the present study have similar internal relationships to those determined by Maluf et al. from their double-mixing quenched-flow experiments (39); specifically, k2 is larger than k1 and k−2 is larger than k−1. Note that no correlation was found between k1, k2, k−1, and k−2. This result demonstrates that there is neither positive nor negative cooperativity between the binding and dissociation events of the UvrD monomers, which is in agreement with the results previously reported based on the chemical quenched-flow method (22).
Three kinetic steps are supposed to occur during the mean dwell time of multiple UvrD bound states (2.7 ± 0.2 or 2.3 ± 1.5 s) before the completion of the DNA unwinding process (Fig. 3 G). These include the translocation of the late-coming UvrD protein(s) that was (were) bound to the ssDNA to form a UvrD oligomer with those that had been bound previously, the isomerization of the nonproductive oligomer to become productive, and the unwinding of the DNA. The isomerization process should comprise most of the dwell time, because the other two processes should be completed in <1 s (Supplementary Discussion 3) and the dwell time is quite close to the inverse of the isomerization rate estimated by Maluf et al. (39) (1/0.337 s−1 = 3.0 s) (Table S5).
We obtained traces that indicate the sequential binding of UvrD monomers, as well as traces that indicate that a preassembled UvrD dimer unwinds the DNA. Maluf et al. claimed that the dimer is also a functional helicase that can unwind DNA in <50 ms without undergoing an isomerization process on the DNA (39). Due to the lack of time resolution and the small amount of data (Supplementary Discussion 4), our single-molecule imaging assays cannot be used to determine whether the preassembled dimer can unwind DNA without the isomerization process. We also obtained several traces that suggest that a UvrD trimer unwinds the DNA. The trimeric form, which was proposed based on the maximum UvrD-DNA-binding stoichiometry of 3 that was measured by sedimentation equilibrium (38), was also observed through the quantification of the number of UvrD proteins that bound to DNA in the absence of ATP (Table 1). Maluf et al. reported that a DNA unwinding model that incorporates both a UvrD dimer and a UvrD trimer better fits the nonlinear sigmoidal dependence of the total extent of DNA unwinding as a function of [UvrD]/[DNA] (22). Although the length of the ss-DNA tail (20 nt) may be insufficient for the trimer to bind, it is reasonable to incorporate a UvrD trimer in the kinetic scheme of the UvrD-DNA interaction (Fig. 4).
The single-molecule imaging assays that were performed in this study support the oligomeric model with regard to the unwinding of DNA by the UvrD protein. It is hypothesized that the UvrD protein functions through an active mechanism (22,42,43) in which a helicase actively unwinds duplex DNA using energy from ATP hydrolysis (Supplementary Discussion 5). Maluf et al. showed that the data from their DNA unwinding time courses cannot be explained by the independent monomer model (22), which is in agreement with the results from this study. Therefore, it is strongly suggested that the UvrD protein binds to the ss/ds DNA junction with high affinity and unwinds the DNA in the form of an oligomer through the active mechanism (Fig. 6). This explanation is in contrast to the monomeric model previously proposed based on genetic complementation assays (44) and crystal structures (4). Both of those studies used a UvrD truncation mutant, UvrDΔ40C (40 amino acid residues were deleted from the C terminus), which preferentially functions as a monomer in vitro (44). However, the dimer model is more plausible, because UvrDΔ40C may form a dimer in a low-salt buffer (38), and the ssDNA tail length of the DNA substrates used for the crystallization of monomeric UvrD-DNA complexes was 7 or 8 nt, which is shorter than the length required for efficient DNA unwinding (≥12 nt) (22) (Supplementary Discussion 6). Therefore, taken with solution studies, it indicates that the monomer bound to this DNA complex represents a dead-end complex.
Figure 6.
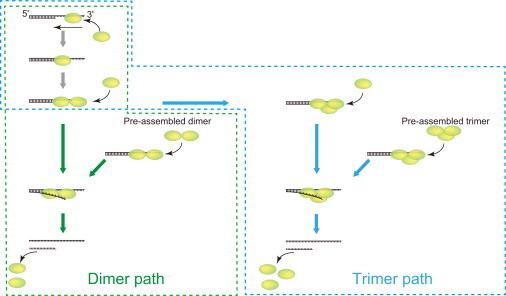
Model of the unwinding of DNA by the UvrD protein.
Although our results strongly indicate that UvrD unwinds DNA in the form of an oligomer, we cannot completely rule out the possibility that UvrD monomers have helicase activity. Traces with a single step just before the DNA unwinding process (Fig. 3, E and F) may be explained by the monomer model. Therefore, we can conclude that most DNA unwinding is performed by two or more UvrD proteins in the form of an oligomer. Maluf et al. mentioned that 3% of the DNA unwinding activity, which is below their experimental uncertainty, may arise from monomeric UvrD (22).
Conclusion
This study will promote our understanding of the general characteristics of SF1 helicases (Supplementary Discussion 7) that are responsible for the unwinding of duplex DNA. In addition, this study will also enhance the capabilities of single-molecule fluorescence studies on helicase-DNA interactions, as well as on other protein molecules that form multimolecule complexes to fulfill their functions in a concerted manner.
Acknowledgments
This work was supported by grants from JSPS KAKENHI (Grant numbers 18770145 and 20770131), PRESTO at the Japan Science and Technology Agency, Astellas Foundation for Research on Medical Resources, Kowa Life Science Foundation, and The Uehara Memorial Foundation, which were awarded to HY.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References
- 1.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Singleton M.R., Sawaya M.R., Wigley D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 3.Enemark E.J., Joshua-Tor L. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J.Y., Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia H., Korolev S., Lohman T.M. Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J. Mol. Biol. 2011;411:633–648. doi: 10.1016/j.jmb.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korolev S., Hsieh J., Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 7.Velankar S.S., Soultanas P., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 8.Ha T., Rasnik I., Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 9.Rasnik I., Myong S., Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J. Mol. Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Dessinges M.N., Lionnet T., Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl. Acad. Sci. USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumont S., Cheng W., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myong S., Bruno M.M., Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun B., Wei K.J., Xi X.G. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 2008;27:3279–3287. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fili N., Mashanov G.I., Molloy J.E. Visualizing helicases unwinding DNA at the single molecule level. Nucleic Acids Res. 2010;38:4448–4457. doi: 10.1093/nar/gkq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balci H., Arslan S., Ha T. Single-molecule nanopositioning: structural transitions of a helicase-DNA complex during ATP hydrolysis. Biophys. J. 2011;101:976–984. doi: 10.1016/j.bpj.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd A.K., Matlock D.L., Raney K.D. Dda helicase tightly couples translocation on single-stranded DNA to unwinding of duplex DNA: Dda is an optimally active helicase. J. Mol. Biol. 2012;420:141–154. doi: 10.1016/j.jmb.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mechanic L.E., Latta M.E., Matson S.W. A region near the C-terminal end of Escherichia coli DNA helicase II is required for single-stranded DNA binding. J. Bacteriol. 1999;181:2519–2526. doi: 10.1128/jb.181.8.2519-2526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.L., Morgenstern K.A., Caron P.R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 19.Wong I., Lohman T.M. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 20.Cheng W., Hsieh J., Lohman T.M. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y., Dou S.X., Xi X.G. Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res. 2008;36:1976–1989. doi: 10.1093/nar/gkm1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maluf N.K., Fischer C.J., Lohman T.M. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 23.Sikora B., Chen Y., Raney K.D. Hepatitis C virus NS3 helicase forms oligomeric structures that exhibit optimal DNA unwinding activity in vitro. J. Biol. Chem. 2008;283:11516–11525. doi: 10.1074/jbc.M708125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 25.Shu D., Zhang H., Guo P. Counting of six pRNAs of ϕ29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao F., Zhang H., Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulbrich M.H., Isacoff E.Y. Subunit counting in membrane-bound proteins. Nat. Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain A., Liu R., Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harada Y., Funatsu T., Yanagida T. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 1999;76:709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myong S., Rasnik I., Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 31.Blainey P.C., van Oijen A.M., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granéli A., Yeykal C.C., Greene E.C. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc. Natl. Acad. Sci. USA. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorman J., Chowdhury A., Greene E.C. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J.H., Larson R.G. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokota H., Han Y.W., Harada Y. Single-molecule visualization of binding modes of helicase to DNA on PEGylated surfaces. Chem. Lett. 2009;38:308–309. [Google Scholar]
- 36.van Mameren J., Modesti M., Wuite G.J. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali J.A., Maluf N.K., Lohman T.M. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J. Mol. Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- 38.Maluf N.K., Lohman T.M. Self-association equilibria of Escherichia coli UvrD helicase studied by analytical ultracentrifugation. J. Mol. Biol. 2003;325:889–912. doi: 10.1016/s0022-2836(02)01276-7. [DOI] [PubMed] [Google Scholar]
- 39.Maluf N.K., Ali J.A., Lohman T.M. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J. Biol. Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 40.Runyon G.T., Wong I., Lohman T.M. Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II) Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- 41.Chao K., Lohman T.M. DNA and nucleotide-induced conformational changes in the Escherichia coli Rep and helicase II (UvrD) proteins. J. Biol. Chem. 1990;265:1067–1076. [PubMed] [Google Scholar]
- 42.Ali J.A., Lohman T.M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 43.Manosas M., Xi X.G., Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mechanic L.E., Hall M.C., Matson S.W. Escherichia coli DNA helicase II is active as a monomer. J. Biol. Chem. 1999;274:12488–12498. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- 45.Hermanson G.T. 2nd ed. Academic Press; New York: 2008. Bioconjugate Techniques. [Google Scholar]
- 46.Drapeau G.R. Substrate specificity of a proteolytic enzyme isolated from a mutant of Pseudomonas fragi. J. Biol. Chem. 1980;255:839–840. [PubMed] [Google Scholar]
- 47.Harada Y., Sakurada K., Yanagida T. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J. Mol. Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. [DOI] [PubMed] [Google Scholar]
- 48.Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohman T.M., Chao K., Runyon G.T. Large-scale purification and characterization of the Escherichia coli rep gene product. J. Biol. Chem. 1989;264:10139–10147. [PubMed] [Google Scholar]
- 50.Fischer C.J., Maluf N.K., Lohman T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Tomko E.J., Fischer C.J., Lohman T.M. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomko E.J., Fischer C.J., Lohman T.M. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J. Mol. Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohman T.M., Tomko E.J., Wu C.G. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 54.Nanduri B., Byrd A.K., Raney K.D. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc. Natl. Acad. Sci. USA. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjornson K.P., Amaratunga M., Lohman T.M. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry. 1994;33:14306–14316. doi: 10.1021/bi00251a044. [DOI] [PubMed] [Google Scholar]
- 56.Dillingham M.S., Wigley D.B., Webb M.R. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 57.Dillingham M.S., Wigley D.B., Webb M.R. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 58.Niedziela-Majka A., Chesnik M.A., Lohman T.M. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


