Summary
Hematopoietic stem cells (HSC) and an earlier wave of definitive erythroid/myeloid progenitors (EMPs) differentiate from hemogenic endothelial cells in the conceptus. EMPs can be generated in vitro from embryonic or induced pluripotent stem cells, but efforts to produce HSCs have largely failed. The formation of both EMPs and HSCs requires the transcription factor Runx1 and its non-DNA binding partner core binding factor β(CBFβ). Here we show that the requirements for CBFβ in EMP and HSC formation in the conceptus are temporally and spatially distinct. Pan-endothelial expression of CBFβ in Tek-expressing cells was sufficient for EMP formation, but was not adequate for HSC formation. Expression of CBFβ in Ly6a-expressing cells, on the other hand, was sufficient for HSC but not EMP formation. The data indicate that EMPs and HSCs differentiate from distinct populations of hemogenic endothelial cells, with Ly6a expression specifically marking the HSC-generating hemogenic endothelium.
Introduction
Hematopoietic stem cells (HSCs), which reside in the bone marrow of adult mammals, differentiate from a small population of endothelial cells in the conceptus referred to as “hemogenic endothelium” (Bertrand et al.; Boisset et al., 2010; Chen et al., 2009; Jaffredo et al., 2005; Kissa et al., 2008; Zovein et al., 2008). Before HSCs appear, committed erythroid/myeloid progenitors (EMPs) emerge in the yolk sac, also from hemogenic endothelium. HSCs capable of repopulating adult mice are first detected at 10.5 days post coitis (dpc; 35 somite pair stage, or 35s) in the dorsal aorta, vitelline and umbilical arteries, and at 11.0–11.5 dpc in the fetal liver, yolk sac, and placenta (de Bruijn et al., 2000; Gekas et al., 2005; Medvinsky et al., 1996; Müller et al., 1994; Ottersbach and Dzierzak, 2005). Hemogenic endothelium is located in all sites of EMP and HSC emergence including the ventral aspect of the dorsal aorta, vitelline and umbilical arteries, yolk sac, and placenta. The process by which blood forms from hemogenic endothelium has been most well studied in the dorsal aorta and in ES cell cultures, and involves an endothelial to hematopoietic cell transition during which individual cells round up and detach from the endothelial layer (Bertrand et al.; Boisset et al., 2010; Kissa et al., 2008). This rounding up (budding) process can be towards the lumen of an artery (prominent in mammals and birds) (Jaffredo et al., 2005), where many newly formed hematopoietic cells collect in clusters that are presumably released into the circulation. Alternatively, the budding can occur in the opposite direction, into the subaortic mesenchyme, after which single hematopoietic cells enter the circulation through the axial vein by intravasation (zebrafish) (Bertrand et al.; Kissa et al., 2008).
Hemogenic endothelium is distinguished from all other endothelial cells by the presence of a transcription factor called Runx1 (North et al., 1999). Runx1 is expressed in hemogenic endothelial cells in early somite pair stage conceptuses prior to the formation of clusters, in the clusters themselves, and in all functional EMPs and HSCs (North et al., 2002; North et al., 1999). Fetuses lacking Runx1 have no EMPs, HSCs, or intra-arterial clusters (Cai et al., 2000; North et al., 2002; North et al., 1999; Okuda et al., 1996; Wang et al., 1996a). In zebrafish Runx1 morphants, most aortic endothelial cells did not attempt the endothelial to hematopoietic cell transition, and those that did self-destructed by what appeared to be an apoptotic-like process (Kissa and Herbomel, 2010).
We showed that deletion of a conditional Runx1 allele in endothelial cells with Cre recombinase driven from Cdh5 (encoding vascular endothelial cadherin) or Tek (encoding the angiopoietin receptor Tie2) regulatory sequences blocked EMP, HSC, and intra-arterial cluster formation, indicating that Runx1 is required in the hemogenic endothelium for the endothelial to hematopoietic cell transition (Chen et al., 2009; Li et al., 2006). A complementary study (Liakhovitskaia et al., 2009) demonstrated that restoring Runx1 function in endothelial cells in Runx1 deficient embryos rescued both EMP and HSC formation. The latter experiment involved reactivating an inactive Runx1 allele with Tek-Cre, allowing Runx1 to be appropriately expressed from its endogenous locus in Tek+ hemogenic endothelial cells, and all of their progeny throughout the remainder of hematopoietic development.
Runx1 function during hematopoiesis requires the activity of a non-DNA binding subunit, core binding factor β (CBFβ), which allosterically regulates DNA binding by Runx1 (Sasaki et al., 1996; Wang et al., 1996b; Yan et al., 2004). Fetuses lacking CBFβ had dramatically impaired hematopoiesis, although some residual EMPs remained (Wang et al., 1996b). Fetuses with only 15% of wildtype CBFβ levels had functional EMPs and HSCs (Talebian et al., 2007), thus very low levels of CBFβ are needed to support EMP and HSC formation. We previously showed that restoring CBFβ expression in Tek+ cells would rescue EMP formation in the fetus (Miller et al., 2002). Our rescue strategy was different from that employed by Liakhovitskaia et al. (Liakhovitskaia et al., 2009), in that we expressed CBFβ (fused to the green fluorescent protein, GFP/CBFβ) as a transgene from Tek regulatory sequences in CBFβ deficient fetuses (Miller et al., 2002). Although GFP/CBFβ was detected in endothelial cells, its level in hematopoietic cells, including Sca1+ or c-Kit+ fetal liver cells, was low (Miller et al., 2002). Fetal liver EMPs were rescued by the Tek-GFP/Cbfb transgene, but GFP/CBFβ was not expressed at later stages of blood cell differentiation, and consequently the terminal differentiation of all hematopoietic cells, with the exception of erythrocytes, was impaired.
Another marker of endothelial cells in hematopoietic sites is the HSC cell surface protein Sca-1, which is encoded by the Ly6a gene (de Bruijn et al., 2002; Ottersbach and Dzierzak, 2005). Surface Sca-1 is found on only 50% of AGM HSCs (de Bruijn et al., 2002; North et al., 2002). However, expression of green fetal fluorescent protein (GFP) from the Ly6a regulatory sequences marked all functional HSCs in the dorsal aorta, fetal liver, and placenta (de Bruijn et al., 2002; Ottersbach and Dzierzak, 2005). GFP+ cells were found amongst the endothelial cells of the dorsal aorta, vitelline and umbilical arteries, and placenta, but not all endothelial cells in these sites were GFP+. A recently published live imaging study that utilized the Ly6a-GFP transgene tracked the budding of GFP+ hematopoietic cells into the lumen of the dorsal aorta in slice cultures from 10.5 dpc fetuses (Boisset et al., 2010).
Here we show that a GFP/CBFβ fusion protein expressed from the Tek regulatory sequences in CBFβ deficient mice is sufficient to restore EMPs formation, but insufficient for HSC formation. Conversely, expressing GFP/CBFβ from Ly6a regulatory sequences will rescue HSC formation, but not EMP formation. These data indicate that EMP and HSC formation can be uncoupled, and that Ly6a expression specifically marks a subset of hemogenic endothelial cells that are destined to produce HSCs.
Results
Expression of GFP/CBFβ from the Tek regulatory sequences cells fails to rescue HSCs in Cbfb−/− fetuses
We previously showed that a GFP/CBFβ fusion protein, when expressed from a Tek-driven transgene, could rescue EMPs in the fetal liver of 12.5 dpc Cbfb−/− fetuses (Miller et al., 2002). The number of EMPs was lower than in wild type fetuses, but was sufficient for Cbfb−/−;Tek-GFP/Cbfb conceptuses to complete gestation (lethality was at postnatal day 1). Cbfb−/− fetuses, on the other hand, died at midgestation with almost no EMPs (Miller et al., 2002; Wang et al., 1996b). We also showed that 17.5 dpc Cbfb−/−;Tek-GFP/Cbfb fetuses contained both B and T lymphocytes, but in reduced numbers (Miller et al., 2002). We assessed the B and T lymphoid potential of 14.5 dpc Cbfb−/−;Tek-GFP/Cbfb fetuses by isolating fetal liver lymphoid progenitors (Lin− c-Kit+Flt3+) and co-culturing them with either OP9 stromal cells or Notch-ligand expressing OP9-DL4 stromal cells. Cbfb−/−;Tek-GFP/Cbfb progenitors yielded small numbers of CD19+B220+ B cells in OP9 cultures, and Thy1+CD25+ T cells in OP9-DL4 cultures (Figure 1A). Limit dilution analysis determined that Cbfb−/−;Tek-GFP/Cbfb fetal livers contained approximately 30 fold fewer B cell and 15 fold fewer T cell progenitors within the Lin− c-Kit+Flt3+ population as compared to Cbfb+/+;Tek-GFP/Cbfb (heretofore referred to as Tek-GFP/Cbfb) fetal livers (Figure 1B). Methylcellulose colony assays using unfractionated fetal liver cells similarly showed that Cbfb−/−;Tek-GFP/Cbfb fetuses contained 11 fold fewer B progenitors than Tek-GFP/Cbfb fetuses (not shown). We transplanted 14.5 dpc fetal liver cells from Cbfb−/−;Tek-GFP/Cbfb conceptuses into irradiated adult mice to assess HSCs and observed no donor contribution to the peripheral blood or bone marrow (Figure 1C). Thus fetal liver lymphoid progenitors and HSCs are poorly rescued by the Tek-GFP/Cbfb transgene.
Figure 1. Expression of GFP/CBFβ from the Tek regulatory sequences poorly rescues lymphoid progenitor and HSC formation.
A. Fetal liver progenitors (Lin−Kit+Flt3+) were sorted from 14.5 dpc embryos and plated on OP9 or OP9-DL4 stromal cells. Number of cells plated were 400 and 200 Tek-GFP/Cbfb on OP9 and OP9-DL1, respectively; 2000 Cbfb−/−; Tek-GFP/Cbfb on both OP9 and OP9-DL4. OP9 cultures contained 5 ng/ml Flt3L, 10ng/ml IL7. OP9-DL4 cultures contained 5 ng/ml Flt3L, 1 ng/ml IL7. Cells were analyzed for the expression of lymphoid markers 7 days later.
B. Limit dilution analysis of fetal liver progenitors cultured as described above.
C. Percent and number of recipients containing ≥ 5% donor-derived blood 16 weeks following transplantation with 2 × 105, 5 × 105, or 1 × 10 6 14.5 dpc fetal liver cells from wild type (+/+), Cbfb−/− (−/−), and Cbfb−/−; Tek-GFP/Cbfb (−/−;Tek) conceptuses.
D. Representative scatter plots of 13.5 dpc fetal liver HSCs (LSK cells).
E. Total number of fetal liver and LSK cells at 13.5 dpc. Data are compiled from 2–5 fetuses. Error bars represent standard error of the mean (SEM).
F. Representative histograms showing GFP/CBFβ expression in the LSK and LT-HSC populations of 13.5 dpc Tek-GFP/Cbfb fetal livers. The two green traces and percentages in the histograms represent two independent fetuses.
Examination of the fetal liver revealed that there were lower numbers of cells overall, and even fewer Lin− Sca1+ c-Kit+ (LSK) cells (Figure 1D). We previously showed by Western blot that the amount of GFP/CBFβ fusion protein produced from the Tek-GFP/Cbfb transgene in embryonic skin was equivalent to that of endogenous CBFβ, but was undetectable in CD45+ or c-Kit+ fetal liver cells (Miller et al., 2002). We extended those findings by examining GFP/CBFβ levels in phenotypic HSCs and LT-HSCs by FACS. Fewer than 10% of phenotypic HSCs (LS Mac1+) and LT-HSCs (CD48− CD150+ LS Mac1+) in the liver of Tek-GFP/Cbfb fetuses were GFP/CBFβ+ (Figure 1E). Thus, GFP/CBFβ expression from the Tek-GFP/Cbfb transgene is not sustained in the majority of newly formed HSCs, which, as discussed later, may contribute to its failure to rescue HSC formation in Cbfb−/− fetuses.
Expression of GFP/CBFβ from the Ly6a regulatory sequences fails to rescue EMPs
We next tried to rescue EMP and HSC formation in Cbfb−/− fetuses using a Ly6a-driven transgene. Three Ly6a-GFP/Cbfb founders were generated, all of which produced GFP/CBFβ at levels similar to that of endogenous CBFβ in unfractionated adult bone marrow cells (Figure 2A). Essentially 100% of adult bone marrow LSK and CD48− CD150+ LSK cells in Ly6a-GFP/Cbfb mice were GFP/CBFβ+, although the mean fluorescence intensity differed in independent founder lines (line A>B>C) (Figure 2B). Expression from the Ly6a-GFP/Cbfb transgene at 11.5 dpc mirrored that described for Ly6a-GFP (de Bruijn et al., 2002; Ottersbach and Dzierzak, 2005). GFP/CBFβ+ cells were present in the placental and umbilical vasculature, vitelline artery, and AGM region (including the mesonephric tubules and Wolffian/Mullerian ducts) (Figure 2C). Fetal liver CD48− CD150+ LSK cells were ~80% GFP/CBFβ+ in the strongest-expressing Ly6a-GFP/Cbfb line (Figure 2D).
Figure 2. Expression of GFP/CBFβ in endothelial cells and hematopoietic cells from the Ly6a-GFP/Cbfb transgene.
A. Western blot analysis of whole bone marrow from wild type, Ly6a-GFP, and three independent Ly6a-GFP/Cbfb lines using a monoclonal antibody recognizing CBFβ. Endogenous CBFβ and the GFP/CBFβ fusion protein are indicated. Whole cell lysates from 2 × 105 cells were loaded.
B. Representative histograms showing GFP/CBFβ expression in the LSK and LT-HSC populations in the bone marrow (BM) of thee independent lines of adult Ly6a-GFP/Cbfb mice.
C. (Left) Whole mount immunofluorescence of an 11.5 dpc Ly6a-GFP/Cbfb conceptus. The fetus is outlined with a dashed white line. Pl, placenta; UA, umbilical artery. (Right) An isolated AGM region with expression in the dorsal aorta (DA), vitelline artery (VA), UA, mesonephric tubules (MT), and Wolffian/Mullerian ducts (W/M), identical to that reported for the Ly6a-GFP transgene (de Bruijn et al., 2002).
D. Representative histograms showing GFP/CBFβ expression in the majority phenotypic LT-HSC in a 13.5 dpc Ly6a-GFP/Cbfb fetal liver (FL, Line A).
The three independent Ly6a-GFP/Cbfb transgenes were crossed onto the Cbfb−/− background. Surprisingly, Cbfb−/−;Ly6a-GFP/Cbfb conceptuses died in utero at the same time as Cbfb−/− conceptuses (Figure S1). Cbfb−/−;Ly6a-GFP/Cbfb fetal livers were pale, suggesting that colonization by yolk sac EMPs had not occurred. Hematopoietic assays were performed to examine EMPs in Cbfb−/−;Tek-GFP/Cbfb versus Cbfb−/−;Ly6a-GFP/Cbfb versus Cbfb−/−;Tek-GFP/Cbfb;Ly6a-GFP/Cbfb conceptuses (Figure 3). Two-way comparisons using Student’s t-test revealed significantly higher numbers of EMPs in Cbfb−/−;Tek-GFP/Cbfb versus Cbfb−/− conceptuses (fetal liver and yolk sac), and in Cbfb−/−;Tek-GFP/Cbfb versus Cbfb−/−;Ly6a-GFP/Cbfb conceptuses (fetal liver and placenta) at 11.5 dpc (Figure 3). Although the number of EMPs in Cbfb−/−;Tek-GFP/Cbfb fetal livers was initially low, it increased considerably over the next few days, and by 14.5 dpc EMPS were only 33% fewer in number than in Tek-GFP/Cbfb conceptuses (Figure 3). Cbfb−/− fetuses carrying both the Tek-GFP/Cbfb and Ly6a-GFP/Cbfb transgenes had significantly higher numbers of EMPs in all tissues at 11.5 dpc (Figure 3), indicating that the two transgenes complemented each other. In summary, the Tek-GFP/Cbfb transgene rescues EMPs and fetal viability significantly better than the Ly6a-GFP/Cbfb transgene. We conclude that the Ly6a-GFP/Cbfb transgene is not expressed at the correct time, or in the correct cells to adequately restore EMP formation and rescue the viability of Cbfb−/− fetuses.
Figure 3. EMPs in Cbfb−/− conceptuses are rescued upon expression of GFP/CBFβ from the Tek but not the Ly6a-driven transgene. See also Figure S1.
Shown are the numbers of hematopoietic progenitors (colonies) per embryo equivalent (ee) in four hematopoietic tissues: fetal liver, yolk sac, AGM region combined with umbilical and vitelline arteries (A+U+V), and placenta. Numbers are compiled from 3–14 11.5 or 14.5 dpc conceptuses per genotype. P values in boxes indicate significant differences in total progenitor numbers in Cbfb−/− conceptuses (−/−, indicated by #) versus Cbfb−/− conceptuses carrying transgenes as determined by one-way ANOVA. Error bars represent SEM. Asterisks indicate significant differences compared to −/− (P < 0.01) according to Dunnett’s Multiple Comparison test. Differences between Cbfb−/−, Cbfb−/−; Tek-GFP/Cbfb (−/−; Tek) and Cbfb−/−; Ly6a-GFP/Cbfb (−/−; Ly6a) were determined by unpaired two-tailed Student’s t test. Cbfb−/−; Tek-GFP/Cbfb; Ly6a-GFP/Cbfb is −/−; Tek+Ly6a.
Expression of GFP/CBFβ from the Ly6a regulatory sequences rescues HSCs
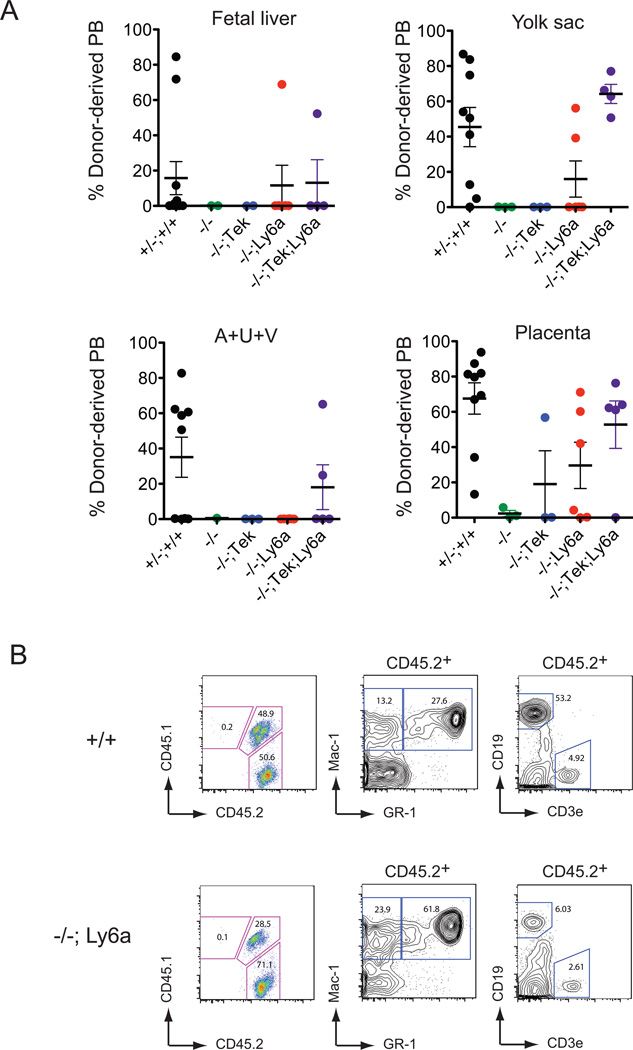
Since most Cbfb−/−;Ly6a-GFP/Cbfb fetuses died by 13.5 dpc, we assayed hematopoietic tissues at 11.5 dpc for the presence of HSCs. Quite surprisingly, the Ly6a-GFP/Cbfb transgene restored HSC formation in all hematopoietic tissues except for the AGM region, vitelline and umbilical arteries (A+U+V) (Figure 4A). High-level reconstitution by Cbfb−/−;Ly6a-GFP/Cbfb HSCs was observed in multiple recipients (6 out of 17), with contribution to both lymphoid and myeloid lineage cells (Figure 4B). Cbfb−/− fetuses expressing both the Tek-GFP/Cbfb and Ly6a-GFP/Cbfb transgenes had HSCs in all hematopoietic sites including the A+U+V (Figure 4A), thus the two transgenes complemented each other in HSC formation. We observed reconstitution of only one recipient by Cbfb−/−;Tek-GFP/Cbfb HSCs that had received 0.9 embryo equivalents of placental cells. These results confirm previous data showing that HSCs are associated with endothelium marked by Ly6a transgene expression (de Bruijn et al., 2002; Ottersbach and Dzierzak, 2005), and further show that CBFβ function in Ly6a-expressing cells is sufficient for HSC, but not for EMP formation.
Figure 4. Expression of GFP/CBFβ from the Ly6a regulatory sequences rescues HSCs in Cbfb−/− conceptuses.
A. Repopulation by 0.9 ee of 11.5 dpc cells from conceptuses of the indicated genotypes at 4 months post transplant. Percent engraftment represents the percent donor-derived blood; each dot is an individual transplant recipient. Error bars represent SEM.
B. Multi-lineage engraftment by donor derived wild type and Cbfb−/−; Ly6a-GFP/Cbfb HSCs in a recipient of placenta cells.
Comparison of Tek-GFP/Cbfb and Ly6a-GFP/Cbfb expression
Since the Tek-driven transgene selectively rescued EMP formation, and the Ly6a-driven transgene restored only HSC formation, comparison of their expression patterns during embryonic development should reveal when and in what cells CBFβ is required. We first considered why the Ly6a-GFP/Cbfb transgene failed to rescue EMP formation. EMPs are detected in the yolk sac beginning at 8.25 dpc (2–6s) (Palis et al., 1999) as clusters of CD41bright cells at the border between the yolk sac blood islands and the capillary bed leading to the embryo proper (Ferkowicz et al., 2003). No GFP/CBFβ+ cells were found anywhere in Ly6a-GFP/Cbfb conceptuses at this time or in slightly older 10–11s conceptuses (Figure 5B). In contrast, the Tek-GFP/Cbfb transgene, which rescued EMP formation, was highly expressed in endothelial cells throughout 6–7s conceptuses, in the yolk sac vascular plexus, dorsal aortae, heart (endocardial tube), allantois, and placenta (Figure 5A). Therefore the Tek-GFP/Cbfb transgene was expressed in endothelial cells at the time of EMP emergence in the yolk sac, but the Ly6a-GFP/Cbfb transgene was not.
Figure 5. Expression of the Tek-GFP/Cbfb and Ly6a-GFP/Cbfb transgenes in 8.5–9.5 dpc conceptuses analyzed by confocal microscopy. See also Figure S2 and Movie S2.
A. Whole mount immunofluorescence of a 7s Tek-GFP/Cbfb conceptus using antibodies recognizing CD31 and GFP (10X). Scale bars for all panels = 100µm. Figure represents the MAX of six consecutive 3µm z-sections for CD31 and GFP. DA, dorsal aorta; YS, yolk sac; Al, allantois; Pl, placenta; H, heart.
B. A 11s Ly6a-GFP/Cbfb conceptus, analyzed as described in A.
C. A 16s Tek-GFP/Cbfb conceptus reconstructed as the standard deviation (STD) of 13 consecutive 1.3µm z-sections for CD31, and the SUM for GFP (10X).
D. A 16s Ly6a-GFP/Cbfb conceptus, analyzed as described in C. Expression is observed in endothelial cells in the caudal aspect of the dorsal aortae, placenta, yolk sac (primarily caudal region), vitelline artery (VA), vitelline vein (VV), foregut (Fg), and in the hindgut (not shown). See also Movie S1.
The fact that the Ly6a-GFP/Cbfb transgene rescued HSC formation means that the onset of its expression could define the earliest time point at which CBFβ is required to specify HSCs. Initiation of Ly6a-GFP/Cbfb transgene expression was found in a subset of endothelial cells between 11s and 16s (~9 dpc) (Figure 5D and Movie S1). These included endothelial cells in the dorsal aorta, the vitelline vasculature, placenta, and the yolk sac vascular plexus primarily (although not exclusively) at the caudal end of the conceptus. A half day later (24s, 9.5 dpc), expression of Ly6a-GFP/Cbfb in the yolk sac is confined to the major vitelline vessels (Figure S2 and Movie S2). The appearance of Ly6a-GFP/Cbfb expressing endothelial cells precedes the presence of both yolk sac HSCs capable of repopulating busulfan-conditioned neonatal recipients (13–20s) (Yoder et al., 1997) and AGM HSCs that can repopulate adult irradiated recipients (>34s, 10.5 dpc) (Medvinsky and Dzierzak, 1996; Müller et al., 1994). Thus, CBFβ is not necessary for HSC formation until, at the very earliest, the 11–16s stage. Further, the inability of the Ly6a-GFP/Cbfb transgene to rescue EMP formation indicates that CBFβ is required before the 16s stage for EMP formation, and/or that EMPs develop from Ly6a negative endothelial cells.
Transgene expression was also examined at 10.5 dpc, when adult repopulating HSCs appear. Ly6a-GFP/Cbfb expression was intense in a subset of endothelial cells located in the regions of HSC emergence, including the dorsal aorta, umbilical and vitelline vasculature (Figure 6B and Movie S3). GFP/CBFβ+ cells in the yolk sac were mostly confined to the larger vitelline vessels, as reported previously (Figure 6D) (de Bruijn et al., 2002). In contrast to the Ly6a-driven transgene, which marks a relatively restricted population of endothelial cells that include a subset of hemogenic endothelial cells, the Tek-GFP/Cbfb transgene was expressed throughout the vasculature of the 10.5 dpc conceptus, although not uniformly in all endothelial cells (Figure 6A,E). Tek-GFP/Cbfb was also expressed the yolk sac at 10.5 dpc in the vascular plexus as well as in the larger vitelline vessels (Figure 6C). FACS analysis of A+U+V of 11.5 dpc Tek-GFP/Cbfb and Ly6a-GFP/Cbfb conceptuses indicated that 18.2% and 20.2%, respectively, of CD144+ CD45− endothelial cells were GFP/CBFβ+ (Figure S3, panel C). Fetuses expressing both transgenes had a significantly higher percentage of endothelial cells that were GFP/CBFβ+ (33.5%) suggesting that the two transgenes were expressed in partially non-overlapping endothelial cell populations. Thus, the differences in the percentage of endothelial cells expressing Tek-GFP/Cbfb versus Ly6a-GFP/Cbfb do not underlie the distinct abilities of these two transgenes to rescue EMPs and HSCs. The failure of the Tek-GFP/Cbfb transgene to rescue HSCs must either be due to suboptimal expression in endothelial cells that give rise to HSCs, or premature extinction of its expression in HSCs.
Figure 6. Expression of the Tek-GFP/Cbfb and Ly6a-GFP/Cbfb transgenes in 10.5 dpc conceptuses. See also Figures S3, S5.
A. Whole mount immunofluorescence of a 36s Tek-GFP/Cbfb specimen using antibodies recognizing CD31 and GFP (5X). Scale bars = 100µm. View represents AVG of fourteen 10µm z-sections. DA, dorsal aorta; UA, umbilical artery; VA, vitelline artery, Fl, fetal liver.
B. A 38s Ly6a-GFP/Cbfb specimen, analyzed as described in A. The Ly6a-GFP/Cbfb transgene is also highly expressed in the tail. MT, Mullerian tubules; HG, hindgut. See also Movie S3.
C. Yolk sac from the Tek-GFP/Cbfb fetus shown in A (5X). View represents SUM of 32 9.7µm z-sections. Asterisk shows area of expression in the yolk sac vascular plexus.
D. Yolk sac from the Ly6a-GFP/Cbfb fetus shown in B, analyzed as described in C. Arrows show expression in large vitelline vessels.
E. Umbilical artery from the Tek-GFP/Cbfb fetus shown in A (20X). View represents SUM of six 0.38µm z-sections.
F. Umbilical artery from the Ly6a-GFP/Cbfb fetus shown in B (20X), analyzed as described in E.
G. Intra-arterial hematopoietic cluster from the umbilical artery of a Tek-GFP/Cbfb fetus (20X).
H. Intra-arterial hematopoietic cluster from the umbilical artery of a Ly6a-GFP/Cbfb fetus, as in G.
We examined whether other HSC/endothelial markers could be used as surrogates for Ly6a-driven transgenes. Endothelial specific adhesion molecule (ESAM) is expressed on fetal liver HSCs, and in the AGM region on c-Kit+ cells and lymphoid progenitors (Yokota et al., 2009). The vast majority (>95%) of GFP/CBFβ+ hematopoietic and endothelial cells in the 10.5 dpc A+U+V were ESAM+, demonstrating that cell surface ESAM marks HSCs and HSC-producing endothelium (Figure 7A,B). However, fewer than 15% of vascular endothelial cadherin (CD144+) ESAM+ endothelial or hematopoietic cells were GFP/CBFβ+. Therefore ESAM cannot substitute for Ly6a-driven transgenes to enrich for HSCs or HSC-producing endothelium. We found the same to be true for endoglin (CD105) and Tek, i.e. most GFP/CBFβ+ hematopoietic or endothelial cells in the A+U+V expressed these markers, but fewer than 15% of cells expressing these markers were GFP/CBFβ+ (Figure 7A,C). The percentage of Flk1+ cells is relatively low compared to ESAM, CD105, and Tek, and less useful for isolation or analysis of HSCs and HSC-producing hemogenic endothelium (Figure 7A,C). We obtained similar results with Ly6a-GFP embryos (data not shown), indicating that the Ly6a regulatory sequences faithfully reproduce this expression pattern in independently derived transgenic mice. In summary, Ly6a-driven transgenes mark a subset of endothelial cells (CD31+, CD144+, ESAM+, CD105+, Tek+) and hematopoietic cells (c-Kit+ and presumably Runx1+) and thus enrich for HSCs and HSC-producing endothelium. Ly6a transgene expression does not, however, remain restricted to this small population of endothelial cells throughout development, as at later gestational ages its expression expands to other cell types including some veins, epithelial cells of the metanephric kidney, endothelial and other cells in the fetal liver, lung macrophages (Figure S4) and a variety of other adult tissues (Holmes and Stanford, 2007; Ma et al., 2002b).
Figure 7. Ly6a transgene expression marks a subset of CD144+, ESAM+, CD105+, Tek+, Flk1+, and c-Kit+ cells. See also Figure S4.
A. Live (DAP1 negative) cells from the 10.5 dpc A+U+V were gated for GFP/CBFβ in the plot furthest to the left, and GFP/CBFβ+ cells are shown as red dots in this and all other plots.
B. Live cells are gated for CD31 and c-Kit expression and divided into endothelial cells (CD31+ c-Kit−) and hematopoietic cells (CD31+ c-Kit+). Each population in turn is analyzed for ESAM and CD144 expression, and all ESAM+ cells for CD144 and GFP/CBFβ expression. Black numbers represent the percentage of all cells in a particular gate, while red numbers represent percentages of GFP/CBFβ+ cells in that gate.
C. Analysis of CD105+, Tek+, and Flk1+ populations, gated as described in A,B.
D. Diagram illustrating that Ly6a-GFP expression marks the HSC-producing population of Runx1+ hemogenic endothelium.
Ly6a expression is not regulated by CBFβ
The restricted endothelial expression of the Ly6a transgene in sites of HSC emergence suggested it might be a target of Runx1-CBFβ. However, we found no evidence for this. There were GFP/CBFβ+ cells in the umbilical and placental vasculature in both Ly6a-GFP/Cbfb and Cbfb−/−; Ly6a-GFP/Cbfb 11.5 dpc conceptuses, and a similar percentage of Sca1+ GFP/CBFβ+ cells in the vasculature (A+U+V, Figure S5 A,B). The percentage of GFP/CBFβ+ endothelial (CD45−CD144+) cells in the A+U+V was also equivalent (Figure S5 C,D), and GFP/CBFβ+ CD31+ cells were clearly visible in the endothelial layer of the dorsal aorta of 10.5 dpc Cbfb−/−; Ly6a-GFP/Cbfb fetuses (Figure S5 E). Thus Runx1 and Ly6a are regulated independently. Runx1 expression marks all hemogenic endothelium that gives rise to EMPs and HSCs. Ly6a expression, on the other hand, marks the subset of hemogenic endothelium that gives rise to HSCs (Figure 7D).
Discussion
It was previously not possible to study the genetic requirements of HSC development in the conceptus in the absence of earlier waves of EMP formation. Here we demonstrate that the formation of EMPs and HSCs can be uncoupled. The emergence of both EMPs and HSCs requires Runx1 and its non-DNA binding subunit CBFβ. EMP formation in conceptuses lacking CBFβ could be rescued by expressing a functional GFP/CBFβ fusion protein from Tek regulatory sequences (Miller et al., 2002). HSCs could be rescued by expressing GFP/CBFβ from Ly6a regulatory sequences. Thus EMPs and HSCs have different temporal and/or spatial requirements for CBFβ. Ly6a-GFP/Cbfb expressing endothelial cells in the yolk sac are not yet present at the time EMPs emerge, and therefore CBFβ is required before the onset of Ly6a expression in the yolk sac for EMP formation. On the other hand, Ly6a-GFP/Cbfb expression in the dorsal aorta, vitelline and umbilical vasculature precedes the emergence of HSCs, and Ly6a-GFP/Cbfb could rescue HSCs. That the rescue of HSCs by the Ly6a-GFP/Cbfb transgene was only partial could reflect a slightly earlier requirement for CBFβ in a subset of HSC precursors. Alternatively, suboptimal expression levels of Cbfb expression could be responsible. Previous studies showed that Runx1 haploinsufficiency differentially affected AGM HSC activity as compared to that in yolk sac and placenta, thus suboptimal expression levels could underlie the selective failure of the Ly6a-GFP/Cbfb transgene to rescue aorta, vitelline and umbilical HSCs (Cai et al., 2000; Robin et al., 2006).
Activation of an inactive, floxed Runx1 allele with Tek-Cre restored both EMP and HSC formation, thus the Tek regulatory sequences should have been expressed sufficiently early in the cells that give rise to EMPs and HSCs to rescue (Liakhovitskaia et al., 2009). The most likely reason that Tek-GFP/Cbfb only partially rescued EMPs, and failed to rescue HSC formation in Cbfb−/− conceptuses is inadequate expression in endothelial cells, or in newly formed EMPs or HSCs. Partial EMP rescue by Tek-GFP/Cbfb could also reflect a requirement for CBFβ in cells in which it is normally expressed and Tek is not, such as the primitive streak (Bollerot et al., 2005). Cbfb is expressed in the posterior aspect of the primitive steak in early and intermediate streak embryos, while Runx1 expression initiates approximately 10 hours later in the area opaca (blood islands). Since the Runx proteins are reliant on CBFβ for high affinity DNA binding, it would make sense for CBFβ to be present before Runx1 to ensure that, once present, Runx1 is active. A likely explanation for the failure of Tek-GFP/Cbfb to rescue HSCs is that, unlike Runx1, sustained CBFβ expression may be required in HSCs. In fact, preliminary data indicate that the number of HSCs in mice in which CBFβ is deleted in the fetal liver with Vav1-Cre is decreased approximately 25 fold, greater than the approximate 4-fold decrease observed upon Runx1 deletion with Vav1-Cre (not shown). The Ly6a-driven transgene, in contrast, provided sustained expression of GFP/CBFβ in fetal liver HSCs. We point out that although low levels of cell surface Tek is present on fetal liver HSCs (Hsu et al., 2000), this does not necessarily mean that expression from a Tek-driven transgene would supply adequate levels of CBFβ to rescue HSC formation or function.
Ly6a encodes a glycoprotein-I-linked cell-surface glycoprotein Sca1, which is a marker of stem/progenitor populations in multiple organs (Holmes and Stanford, 2007). There are two mouse strain-specific Ly6a alleles, only one of which is expressed in all HSCs (Spangrude and Brooks, 1993). Despite early work on Ly6a expression (DNaseI hypersensitivity) in the FDCP1 multipotent progenitor and other hematopoietic cell lines (Sinclair and Dzierzak, 1993), relatively little is known about the regulation of Ly6a expression in endothelial cells, and nothing at the transcriptional level. However, two mutant mouse models showed decreased numbers of Ly6a-GFP expressing aortic endothelial cells. Embryos haploinsufficient for Gata2 (HSC numbers were reduced) showed a 10-fold decrease in GFP+ aortic endothelial cells (Ling et al., 2004) and conceptuses deficient for Jagged1, a ligand for Notch, had a 2-fold decrease (Robert-Moreno et al., 2008). Ly6a is regulated in HSCs by interferon α signaling through STAT1 (Essers et al., 2009), thus signaling through JAK-STAT is a possible mechanism for activating Ly6a expression in endothelium. We showed that Ly6a expression is not, however, dependent on Runx1-CBFβ. A recent ChIP-Seq analysis of multiple transcription factors involved in hematopoietic development (Tal1, Lyl1, Lmo2, Gata2, Runx1, and Pu.1) found that the Ly6a gene was occupied only by Pu.1 in a hematopoietic progenitor cell line (Wilson et al., 2010). Although EMP cell lines are fairly far removed from endothelium, the indication that Ly6a is not a direct Runx1 target is consistent with our observation that Ly6a expression in endothelium is unaffected by CBFβ deficiency. Although expression of Ly6a in HSCs is strain-specific, and a human equivalent does not exist, it would nevertheless be interesting to identify the signals that activate expression of the mouse transgene in the subset of endothelial cells that give rise to HSCs, as these may provide clues as to how HSCs, as opposed to EMPs, are specified. A comparison of gene expression profiles between Ly6a-GFP+ and Ly6a-GFP− endothelium might reveal other markers that could be used to follow the development of the comparable endothelium in human embryos and ES cell cultures.
Most importantly, our data imply that there is a discrete endothelial population in the mouse conceptus that gives rise to HSCs, and the murine Ly6a regulatory sequences are active in these endothelial cells. Importantly, and quite unexpectedly, EMPs do not emerge if CBFβ expression is restricted to Ly6a-expressing cells, indicating that the Ly6a-driven transgene is not active in the hemogenic endothelium that gives rise to EMPs at the time Runx1-CBFβ is required. This interpretation is entirely consistent with known markers on EMPs and HSCs in the murine conceptus. EMPs in the yolk sac are cell surface Sca1− (Huang and Auerbach, 1993), whilst HSCs in the Ly6a-GFP AGM region and placenta are in the GFP+ population (de Bruijn et al., 2002; Huang and Auerbach, 1993; Ottersbach and Dzierzak, 2005). This has important implications for ongoing efforts to produce HSCs ex vivo, from ES/induced pluripotent stem cells and fibroblasts. Many investigators have generated EMPs and lymphoid progenitors from these cell sources, but no functional HSCs have been produced without introducing genetic modifications (Cho et al., 1999; Irion et al., 2010; Lengerke and Daley, 2010; Schmitt et al., 2004; Szabo et al., 2010). EMP formation proceeds through an endothelial intermediate in ES cell cultures (Eilken et al., 2009; Lancrin et al., 2009), and presumably HSC formation will as well. It would therefore be useful to follow the generation of Ly6a-GFP+ endothelial cells in vitro when optimizing conditions for HSC production.
Materials and Methods
Generation of Ly6a-GFP/Cbfb transgenes
We generated Ly6a-GFP/Cbfb mice by inserting a cDNA coding for a GFP/CBFβ fusion protein into the Ly6a promoter/enhancer sequences. We introduced silent mutations into the cDNA for GFP/CBFβ (Adja et al., 1998) (a gift from Paul Liu) to eliminate the SalI, XhoI, BglII, XhoI, and HindIII sites in order to facilitate cloning. An artificial intron and poly-A signal was added to the 3’ end of the coding region. The GFP/CBFβ insert was amplified by PCR to introduce ClaI sites at both ends then inserted into the ClaI site of pPOLYIII-Ly6A14 (Ma et al., 2002a). The insert was flanked by 4 kb of upstream and 10 kb of downstream genomic sequences containing Ly6a regulatory elements. The transgene was excised from the vector by BamHI and injected into fertilized oocytes from C57BL/6 × 129S1/SVImJ F1 mice by the Brigham and Women’s transgenic facility. Six independent founders were identified, and three further characterized. Tek-GFP/Cbfb and Cbfb+/− (Cbfbtm1Spe/+) mice were described previously (Miller et al., 2002; Wang et al., 1996b).
PCR genotyping protocol and primers
Animals were genotyped by PCR using tail DNA from adults, and yolk sac snips or limb buds from conceptuses. The Tek-GFP/Cbfb transgene was detected by the following primers: Tek-PF263: 5’AGC GGG AAG TCG CAA AGT TGT G, GFP-R138: 5’CTT CAG GGT CAG CTT GCC GTA G. The Ly6a-GFP/Cbfb transgene was detected by: Ly6A-PF55: 5’AGA ACT TGC CAC TGT GCC TGC and GFP-R138: 5’CTT CAG GGT CAG CTT GCC GTA G. Cbfb+/− was detected by three primers: CBFbex5R: 5’CTC TCA CCT CCA TTT CCT CCC G. CBFbex5F: 5’TAT TTC ATG TAC CCT GAC AGC AGG. PGKP-R-475: 5’ATG CTC CAG ACT GCC TTG GG. The Cbfb deleted allele yields a ~400 bp band, and the wild type Cbfb allele yields a ~110 bp band. All PCR reactions were run using the following program: 5 min denaturation at 94°C followed by 35 cycles of denaturation (30 sec at 94°C), annealing (30 sec at 60°C), and elongation (1 min 30 sec at 72°C).
Embryo and mouse generation
Conceptuses were generated from timed matings. Detection of the vaginal plug is 0.5 dpc. Ly6a-GFP/Cbfb and Cbfb−/−;Ly6a-GFP/Cbfb conceptuses were generated by crossing Cbfb+/− ;Tg males with Cbfb+/− females. The same strategy was used to generate Cbfb−/−;Tek-GFP/Cbfb conceptuses.
Cell Preparation
Embryonic cells were prepared for flow cytometry, methylcellulose and transplant assays as described previously (Chen et al., 2009; North et al., 2002).
Flow cytometry
We used PerCP-Cy5.5 anti-Sca-1 (D7), eFluor® 450 lineage specific cocktail [(anti-TER119 (TER119), anti-Gr-1 (RB6-8C5), anti-B220 (RA3-6B2), anti-CD3 (17A2), anti-Mac-1 (M1/70)], APC anti-CD48 (HM48-1), APC- eFluor® 780 anti-c-Kit (2B8) (eBioscience), and PE-Cy7 anti-CD150 (SLAM) (BioLegend) to stain adult bone marrow. Fetal liver cell staining is as above except for removal of eFluor® 450 anti-Mac-1 from the lineage cocktail, and staining with PE anti-Mac-1 (M1/70). Dead cells were excluded with DAPI (Molecular Probes). To analyze peripheral blood engraftment we used PerCP-Cy5.5 anti-Gr-1 (RB6-8C5), APC anti-CD3ε (145-2C11), PE anti-CD19 (1D3), PE-Cy7 anti-Mac-1 (M1/70) (BD Pharmingen), V450 anti-CD45.2 (104) (BD Horizon), and PE-Cy5 anti-CD45.1 (A20). Dead cells were excluded with Fixable Viability Dye eFluor® 780 (eBioscience). To examine HSC/endothelial marker expression we used PE-Cy7-anti-CD31 (390), Alexafluor® 647-anti-CD144 (eBioBV13), PE-anti-CD105 (MJ7/18), PE-anti-Tie2 (TEK4) (eBioscience); PE-anti-ESAM (1G8/ESAM) and PE-anti-Flk1 (89B3A5) (BioLegend). Dead cells were excluded by DAPI staining. We analyzed cells on an LSR II flow cytometer (BD Biosciences, San Jose CA), and data by FlowJo (Tree Star, Ashland, OR). Cells were sorted on a FACSAria (BD Biosciences).
Cell culture
OP9 and OP9-DL4 cells were essentially used as described (Schmitt and Zuniga-Pflucker, 2006). For OP9-DL4 cultures, IL-7 was added at a final concentration of 1 ng/ml and Flt3 ligand at 5 ng/ml. OP9 cultures included IL-7 at a final concentration of 10 ng/ml. Stromal cells were plated 2 days before initiation of culture at a concentration of 20,000 cells/ml in 96-well plates. Cultures were examined after 7 days of culture by flow cytometry. For in vitro limiting-dilution analysis, the progenitor frequency was calculated using the method of maximum likelihood applied to Poisson distribution with L-Calc software (StemCell Technologies).
Transplant analyses
129S1/SvImJ × B6.SJL-Ptprca Pepcb/BoyJ F1 mice (CD45.1/CD45.2) were subjected to a split dose of 900 cGy, 3 hours apart. Each recipient received 0.9 embryo equivalent of donor fetal liver, yolk sac, placenta, or AGM and arteries, and 2×105 carrier spleen cells via tail vein injection. All donor fetuses were of a mixed C57BL/6J and 129S1/SVImJ background and expressed CD45.2. Spleen cells were prepared from B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mice. We assessed donor engraftment of peripheral blood 16 weeks later.
Western-blot analyses
Bone marrow was isolated and resuspended at 1× 106 cells per ml in lysis buffer (150 mM NaCl, 50 mM Tris, pH8.0, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 0.2 mM EDTA, 2.0 mM EGTA plus 1 µg/ml pepstatin A, 1 µM Pefablock, 2 µg/ml leupeptin, 2 µg/ml aprotinin). Lysates equivalent to 2 × 105 cells were boiled in SDS loading buffer, resolved by SDS-PAGE through 4–12% Bis-Tris gels (Invitrogen), proteins transferred to nitrocellulose, the blot probed with mouse monoclonal antibodies to CBFβ (β141.4.1 + β141.2.2.5), and developed with ECL reagents (Pico kit, Pierce, Rockford, IL).
Microscopy and Imaging
Images of dissected intact conceptuses in phosphate-buffered saline were visualized using a Leica MZFIII stereomicroscope equipped with a Plan 1X 0.14(.025–.125) objective lens (Leica, Heerbrugg, Switzerland). A Color Mosaic 11.2 camera and Spot Insight 4.0 acquisition software (Diagnostic Instruments, Sterling Heights, MI) were used to capture images, which were processed with Adobe Photoshop CS4 software (Adobe Systems, San Jose, CA).
Whole mount immunohistochemistry was performed as described (Yokomizo and Dzierzak, 2010; Yokomizo et al., 2011). Biotinylated rat anti-mouse CD31 (MEC13.3, BD Biosciences) and rabbit anti-GFP polyclonal antibody (MBL, Naka-ku Nagoya) were used with Cy-3-conjugated streptavidin (Jackson Immuno Research, West Grove PA) and Alexafluor® 647 goat anti-rabbit IgG (Invitrogen). Specimens were analyzed with a Leica DM 6000 upright confocal microscope with HCX PL FLUOTAR 5.0 × 0.15, NA 0.15/ HC PL APO CS 10.0 × 0.4, NA 0.4 objectives using multi-track sequential mode. Pinhole diameter was set at 1 Airy unit and steps were 1.3–9.7µm per z-section.
Supplementary Material
Acknowledgements
The authors thank Tomomasa Yokomizo for teaching us how to perform the whole mount immunohistochemistry. We are especially grateful to Andrea Stout for her help with confocal microscopy. This work was supported by R01HL091724 (NAS) and Netherlands Institute of Regenerative Medicine (ED). Core services were supported by the Abramson Family Cancer Research Institute and the Abramson Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adja N, Stacy T, Speck NA, Liu PP. The leukemic protein CBFβ-SMMHC sequesters CBFα2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bollerot K, Romero S, Dunon D, Jaffredo T. Core binding factor in the early avian embryo: cloning of Cbfbeta and combinatorial expression patterns with Runx1. Gene Expr Patterns. 2005;6:29–39. doi: 10.1016/j.modgep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Cai Z, de Bruijn MFTR, Ma X, Dortland B, Luteijn T, Downing JR, Dzierzak E. Haploinsufficiency of AML1/CBFA2 affects the embryonic generation of mouse hematopoietic stem cells. Immunity. 2000;13:423–431. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:889–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Webber TD, Carlyle JR, Nakano T, Lewis SM, Zuniga-Pflucker JC. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc Natl Acad Sci U S A. 1999;96:9797–9802. doi: 10.1073/pnas.96.17.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn M, Ma X, Robin C, Ottersbach K, Sanchez M-J, Dzierzak E. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 2002;16:673–683. doi: 10.1016/s1074-7613(02)00313-8. [DOI] [PubMed] [Google Scholar]
- de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Ferkowicz MJ, Starr M, Xie X, Li W, Johnson SA, Shelley WC, Morrison PR, Yoder MC. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Holmes C, Stanford WL. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 2007;25:1339–1347. doi: 10.1634/stemcells.2006-0644. [DOI] [PubMed] [Google Scholar]
- Hsu HC, Ema H, Osawa M, Nakamura Y, Suda T, Nakauchi H. Hematopoietic stem cells express Tie-2 receptor in the murine fetal liver. Blood. 2000;96:3757–3762. [PubMed] [Google Scholar]
- Huang H, Auerbach R. Identification and characterization of hematopoietic stem cells from the yolk sac of the early mouse embryo. Proc Natl Acad Sci U S A. 1993;90:10110–10114. doi: 10.1073/pnas.90.21.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Clarke RL, Luche H, Kim I, Morrison SJ, Fehling HJ, Keller GM. Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells. Development. 2010;137:2829–2839. doi: 10.1242/dev.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo T, Nottingham W, Liddiard K, Bollerot K, Pouget C, de Bruijn M. From hemangioblast to hematopoietic stem cell: an endothelial connection? Exp Hematol. 2005;33:1029–1040. doi: 10.1016/j.exphem.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Kissa K, Murayama E, Zapata A, Cortes A, Perret E, Machu C, Herbomel P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C, Daley GQ. Autologous blood cell therapies from pluripotent stem cells. Blood Rev. 2010;24:27–37. doi: 10.1016/j.blre.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen MJ, Stacy T, Speck NA. Runx1 function in hematopoiesis is required in cells that express Tek. Blood. 2006;107:106–110. doi: 10.1182/blood-2005-05-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakhovitskaia A, Gribi R, Stamateris E, Villain G, Jaffredo T, Wilkie R, Gilchrist D, Yang J, Ure J, Medvinsky A. Restoration of Runx1 expression in the Tie2 cell compartment rescues definitive hematopoietic stem cells and extends life of Runx1 knockout animals until birth. Stem Cells. 2009;27:1616–1624. doi: 10.1002/stem.71. [DOI] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, de Bruijn M, Robin C, Peeters M, Kong-A-San J, Snoija C, Dzierzak E. Expression of the Ly-6A (Sca-1) lacZ transgene in mouse hematopietic stem cells and embryos. Br J Haematol. 2002a;116:401–408. doi: 10.1046/j.1365-2141.2002.03250.x. [DOI] [PubMed] [Google Scholar]
- Ma X, Robin C, Ottersbach K, Dzierzak E. The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem Cells. 2002b;20:514–521. doi: 10.1634/stemcells.20-6-514. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Medvinsky AL, Gan OI, Semenova ML, Samoylina NL. Development of day-8 colony-forming unit-spleen hematopoietic progenitors during early embryogenesis: spatial and temporal mapping. Blood. 1996;87:557–566. [PubMed] [Google Scholar]
- Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, Speck NA. The core-binding factor β subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32:645–649. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- Müller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- North TE, Gu T-L, Stacy T, Wang Q, Howard L, Binder M, Marín-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A, Guiu J, Ruiz-Herguido C, Lopez ME, Ingles-Esteve J, Riera L, Tipping A, Enver T, Dzierzak E, Gridley T, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. Embo J. 2008;27:1886–1895. doi: 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin C, Ottersbach K, Durand C, Peeters M, Vanes L, Tybulewicz V, Dzierzak E. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev Cell. 2006;11:171–180. doi: 10.1016/j.devcel.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zuniga-Pflucker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. T-cell development, doing it in a dish. Immunol Rev. 2006;209:95–102. doi: 10.1111/j.0105-2896.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- Sinclair AM, Dzierzak EA. Cloning of the complete Ly-6E.1 gene and identification of DNase I hypersensitive sites corresponding to expression in hematopoietic cells. Blood. 1993;82:3052–3062. [PubMed] [Google Scholar]
- Spangrude GJ, Brooks DM. Mouse strain variability in the expression of the hematopoietic stem cell antigen Ly-6A/E by bone marrow cells. Blood. 1993;82:3327–3332. [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010 doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- Talebian L, Li Z, Guo Y, Gaudet J, Speck ME, Sugiyama D, Kaur P, Pear WS, Maillard I, Speck NA. T lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBF{beta} dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marín-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996a;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller JD, Lewis AF, Huang X, Bories J-C, Bushweller JH, Alt FW, Binder M, Marín-Padilla M, et al. The CBFβ subunit is essential for CBFα (AML1) function in vivo. Cell. 1996b;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Yan J, Liu Y, Lukasik SM, Speck NA, Bushweller JH. CBFbeta allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat Struct Mol Biol. 2004;11:901–906. doi: 10.1038/nsmb819. [DOI] [PubMed] [Google Scholar]
- Yoder MC, Hiatt K, Mukherjee P. In vivo repopulating hematopoietic stem cells are present in the murine yolk sac at day 9.0 postcoitus. Proc Natl Acad Sci U S A. 1997;94:6776–6780. doi: 10.1073/pnas.94.13.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–3661. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, Yamada-Inagawa T, Yzaguirre AD, Chen MJ, Speck NA, Dzierzak E. Whole mount 3-dimensional imaging of internally-localized immunostained cells within mouse embryos. Nat Protocols. 2011 doi: 10.1038/nprot.2011.441. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Oritani K, Butz S, Kokame K, Kincade PW, Miyata T, Vestweber D, Kanakura Y. The endothelial antigen ESAM marks primitive hematopoietic progenitors throughout life in mice. Blood. 2009;113:2914–2923. doi: 10.1182/blood-2008-07-167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.