Abstract
Brassica rapa is an important crop species that produces vegetables, oilseed, and fodder. Although many studies reported quantitative trait loci (QTL) mapping, the genes governing most of its economically important traits are still unknown. In this study, we report QTL mapping for morphological and yield component traits in B. rapa and comparative map alignment between B. rapa, B. napus, B. juncea, and Arabidopsis thaliana to identify candidate genes and conserved QTL blocks between them. A total of 95 QTL were identified in different crucifer blocks of the B. rapa genome. Through synteny analysis with A. thaliana, B. rapa candidate genes and intronic and exonic single nucleotide polymorphisms in the parental lines were detected from whole genome resequenced data, a few of which were validated by mapping them to the QTL regions. Semi-quantitative reverse transcriptase PCR analysis showed differences in the expression levels of a few genes in parental lines. Comparative mapping identified five key major evolutionarily conserved crucifer blocks (R, J, F, E, and W) harbouring QTL for morphological and yield components traits between the A, B, and C subgenomes of B. rapa, B. juncea, and B. napus. The information of the identified candidate genes could be used for breeding B. rapa and other related Brassica species.
Keywords: Brassica rapa, quantitative trait loci (QTL), morphological traits, single nucleotide polymorphism (SNP), conserved genome blocks
1. Introduction
Brassica rapa (2n = 20, AA) is an important Brassica species that is grown widely to produce leafy vegetables in Korea, China, and Japan, for vegetable oil in India, China, and Canada, and as a fodder crop in Europe. Chinese cabbage, pak choi, sarson, and turnips are distinct morphotypes of B. rapa belonging to different subspecies that are adapted to different geographical regions and climatic conditions. During the last two decades, studies by different laboratories using diverse germplasm led the development of several molecular markers and genetic linkage maps1,2 besides mapping quantitative trait loci (QTL) for erucic acid,3 glucosinolates,4,5 disease resistance,6,7 plant morphology, flowering time, and yield component traits.8,9 Furthermore, comparative mapping between different cultivated Brassica species revealed the structural conservation between the homoeologous chromosomes of the A, B, and C subgenomes that were originally derived from three diploid Brassica species, namely, the Brassica rapa, Brassica nigra (BB, n = 8), and Brassica oleracea (CC, n = 9) genomes, and their polyploid derivatives, i.e. the important oilseed crops Brassica juncea (AABB, n = 18) and Brassica napus (AACC, n = 19), respectively.5,10,11 Furthermore, several comparative mapping studies between Brassica and Arabidopsis thaliana revealed the triplicate nature of the Brassica genome, with an average of three copies of each chromosomal segment of A. thaliana, which resulted from the triplication of the whole Brassica genome at ∼11–12 MYA,12 although segmental conservation between Brassica and A. thaliana chromosomes has been observed at the gross level.10,11,13,14
Therefore, to decode the whole genome sequences of the complicated Brassica genomes to be used in breeding programmes and to study the divergence of gene function and genome evolution associated with polyploidy and extensive duplications, the ‘Multinational Brassica Genome Sequencing Project Consortium’ was initiated in 2003. The first Brassica genome to be sequenced among the six cultivated Brassica species was the A genome of the B. rapa Chinese cabbage Chiifu-401 cultivar that has a comparatively small genome (529 Mb) among the cultivated Brassica species,15 and the draft genome sequence was published in 2011.16
With the availability of recently developed advanced next-generation sequencing (NGS) technology to sequence the whole genome of crop plants in a short time span, the emphasis is being shifted to genomics-assisted breeding from traditional crop breeding using conventional molecular markers. The combined use of QTL mapping, which detects functional loci for traits of interest, and whole genome sequence information to identify candidate genes and their variation between the parental lines of B. rapa will greatly supplement the development of gene-specific molecular markers for breeding this crop with desired plant architecture and quality. Many oleifera and sarson types were exploited for breeding high seed yield component traits in B. rapa, but the vegetable types were not exploited for this purpose. Although several QTL have been mapped for leaf morphology, yield components, and other quality traits, most of the genes underlying trait variation have not been identified.9,17
Therefore, in the present study, we used chromosome-specific bacterial artificial chromosome-derived simple sequence repeats (BAC-SSRs) and gene specific markers, e.g. intron polymorphisms (IPs) and expressed sequenced tag-derived SSRs (EST-SSRs) to map QTL governing morphological and yield component traits. Using whole genome next-generation sequence information data of the parental lines in combination with comparative alignment with the A. thaliana genome, potential candidate genes and single nucleotide variations within some of the potential candidate genes were identified. Furthermore, the chromosomal regions of B. rapa containing clusters of QTL were aligned with the QTL regions of B. juncea and B. napus to identify structural and functional conservation between the A, B, and C subgenomes, so that the candidate gene information of B. rapa could be used for breeding these crops.
2. Materials and methods
2.1. Plant materials, growth conditions, and trait measurement
The genetic map developed earlier by us2,18 using a CRF2 mapping population that was derived by crossing the diverse parental lines ‘Chiifu 401–42,’ a vegetable-type Chinese cabbage, and ‘rapid cycling B. rapa’ (hereafter referred to as ‘RCBr’), was used for QTL mapping in this study. For phenotypic investigation, 12 F3 plants derived by selfing each of the 190 F2 plants were planted per replication in 3 replications in 2008, 2009, and 2010 from March to July. The seeds were germinated in cell trays in a greenhouse for 1 month. In 2009 and 2010, two sets were grown, one set was vernalized after 20 days of germination for 1 month and the other set was grown without vernalization. All the plants were transplanted to the open field of Chungnam National University, Daejeon, Korea. Fourteen morphological and yield component traits (Table 1) were recorded in the CRF3 mapping population and parental lines. Four plants from the middle of each row from each replication were used for phenotypic data measurement for each family, and the average value of three replications was taken as trait data. Flowering time was the only trait that was recorded in the vernalized plants.
Table 1.
Details of traits measurement in CRF3 population
| Trait name | ABS | Trait description | Scale | Year |
|---|---|---|---|---|
| Flowering time | FT | Days from sowing to opening of the first flower | Days | 2008/2009/2010 |
| Bolting time | DB | Days from sowing to the emergence of bud | Days | 2009/2010 |
| Flowering time after vernalization | FT* | Days from sowing to opening of the first flower after vernalization for 1 month | Days | 2009/2010 |
| Plant height | PH | Height from ground to the stem top when first flower opens | cm | 2008/2009/2010 |
| Leaf length | LL | From base of petiole to the tip of leaf | cm | 2008/2009 |
| Leaf width | LW | Leaf width at the widest point | cm | 2008/2009 |
| Midrib length | MRL | Length from the bottom to the apex of midrib | cm | 2008/2009 |
| Midrib width | MRW | Width of the bottom of midrib | cm | 2008/2009 |
| Petiole length | PL | From base of petiole to the bottom of lamina | cm | 2009 |
| Siliqua length | SQL | Length from pedicel of siliqua to the top of seed beak | cm | 2008/2009/2010 |
| Siliqua width | SQW | Width at the lengthwise midpoint of each siliqua | cm | 2008/2009/2010 |
| Siliqua beak length | SBL | Length from the top of siliqua to the top of beak | cm | 2009/2010 |
| Seeds per siliqua | SSQ | Number of seed per siliqua | 2009/2010 | |
| Seed weight | SW | Seed weight of 100 seeds each line | mg | 2009/2010 |
* Represent the vernalization treatment.
2.2. Statistical analysis and QTL mapping
The SAS 9.0 program (SAS institute, Inc., Cary, NC, USA) was used for correlation coefficient analysis. The previously described genetic map18 was used for QTL mapping. WinQTL cartographer version 2.519 was used to perform QTL analysis using the composite interval mapping function as described previously.20 Tests for the presence of QTL were performed at 2-cM intervals using a 10-cM window. Significant QTL-defining logarithm of odds (LOD) values were calculated by 1000 permutations for phenotypic traits derived from each year.
2.3. Comparative map alignment between the A, B, and C subgenomes
To identify the functionally conserved loci between B. rapa with B. juncea and B. napus for morphological and yield component traits, comparative alignment of QTL maps was performed between these three species. B. juncea contains the A and B subgenomes while B. napus contains the A and C subgenomes. For comparison, the B. juncea QTL map of Ramchiary et al.20 and the B. napus QTL maps of Quijada et al.21 and Udall et al.22 were used. The marker sequences from the respective maps21,22 were downloaded from the National Center for Biotechnology Information and aligned with the A. thaliana genome using BLAST analysis, and crucifer building blocks14 containing important trait QTL were defined. The updated map of B. juncea based on IP markers11 was used to redraw the QTL map of Ramchiary et al.20 For QTL map alignment, the homoeologous A, B, and C subgenome groups of Brassica species defined by Panjabi et al.11 were used.
2.4. Whole genome resequencing, identification of SNPs in candidate genes, and semi-quantitative RT-PCR analysis
Whole genome resequencing of parental line RCBr was performed using an Illumina GAII next-generation sequencer. Sequence assembly and single nucleotide polymorphism (SNP) identification were performed in a stepwise manner: (i) the scaffold sequences from each linkage group (LG) of the B. rapa Chiifu 401–42 cultivar genome (http://brassicadb.org/brad/) were used as reference sequence, (ii) Augustus program23 was used to predict candidate genes in QTL regions, (iii) short-read sequences generated from RCBr were aligned to the reference B. rapa genome using Bowtie (http://bowtie.cbcb.umd.edu)24 and mapped onto each reference LG, (iv) alignments from Bowtie were transformed to MAQ25 to produce consensus short-read sequences and identify SNPs.
Because we previously demarcated 24 crucifer building blocks in the B. rapa genome by comparing it with A. thaliana,18 we selected already characterized candidate genes from A. thaliana and looked for orthologous genes in the B. rapa genome in those blocks harbouring important trait QTL using a homology search approach and synteny analysis. SNPs were identified between the start and stop codons through comparative alignment between the sequences of the parental lines. Gene-specific primer pairs were designed for SNP validation, with an amplicon size of not more than 300 bp. DNA extraction, PCR using 1 × LC Green Plus (Idaho Technologies), and mapping functions were performed as described previously.18 The PCR conditions were 4 min at 94°C, 45 cycles of 20 s at 94°C, 20 s at 55–60°C, and 20 s at 72°C, with a final extension step of 7 min at 72°C. For the detection of SNPs between the parental lines and the F2 mapping population, we used a Light Scanner System (Idaho Technologies), as described previously.26 RNA was extracted from 20-day-old leaf samples, and semi-quantitative RT-PCR analysis was performed following Lee et al.27 using gene-specific primer pairs listed in Supplementary Table S4.
3. Results
3.1. Variation of phenotypic traits in the parental lines and segregating populations
The parental line Chiifu is a heading-type Chinese cabbage, whereas RCBr is a short life cycle Brassica plant, also known as the Wisconsin fast plant. The two parental lines showed significant differences for flowering, leaf, siliqua, and seed traits (Table 2). Chiifu showed higher phenotypic values for all the traits studied. The CRF3 population showed transgressive segregation for some of the traits, e.g. plant height in 2008, midrib length and width, siliqua length and width, siliqua beak length, seeds per siliqua, and seed weight. For flowering and bolting time, the transgression phenomenon trend was towards RCBr because Chiifu did not flower until seed harvesting time. As the bolting and flowering time were influenced by temperature and photoperiod, the mean value of flowering time measured in spring (2009 and 2010) was ∼30 days shorter than in winter (2008), even when they were grown in a heated greenhouse. The mean values of the F3 lines for the leaf traits and plant height evaluated in spring 2009 and 2010 were also lower than those of the F3 lines grown in 2008. Distribution analysis of the phenotypic values of 14 traits in the mapping population showed a normal distribution for all the traits, suggesting that each trait was governed by many genes. Pearson's correlation coefficient analysis showed moderate to strong positive correlation among leaf traits (LL, LW, MRL, and MRW, correlation coefficient r = 0.53–0.92). Siliqua length, width, and beak length were significantly positively correlated with each other, but seed weight was not correlated with other siliqua traits (Supplementary Table S1). In addition, we found a positive correlation between bolting time, flowering time, and four leaf traits (Supplementary Table S1).
Table 2.
Phenotypic trait values of parental lines and the CRF3 mapping population of B. rapa observed in different years
| Traits | Mean trait value of parent Chiifu |
Mean trait value of parent RCBr |
Range in F3 population |
Mean value in F3 population |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2008 | 2009 | 2010 | 2008 | 2009 | 2010 | 2008 | 2009 | 2010 | |
| Flowering time | –a | –a | –a | 39.0 | 45.0 | 45.0 | 33.0–139.0 | 30.0–59.0 | 45.2–64.0 | 70.6 | 41.9 | 53.4 |
| Bolting time | – | 90.0 | 95.0 | – | 29.0 | 29.0 | – | 11.0–44.7 | 34.0–65.0 | – | 26.5 | 42.3 |
| Flowering time after vernalization | – | 135.0 | 145.0 | – | 30.0 | 30.0 | – | 54.2–68.8 | 66.0–83.5 | – | 57.9 | 73.0 |
| Plant height | 65.0 | 100.0 | 98.0 | 17.0 | 20.0 | 18.5 | 5.0–98.0 | 3.6–34.5 | 4.0–24.8 | 43.0 | 13.6 | 13.1 |
| Leaf length | 28.0 | 20.0 | – | 9.0 | 6.5 | – | 14.0–49.0 | 2.8–8.0 | – | 30.2 | 5.0 | – |
| Leaf width | 14.0 | 16.0 | – | 3.8 | 3.0 | – | 4.5–19.0 | 2.2–5.8 | – | 11.2 | 3.5 | – |
| Midrib length | 14.0 | 7.7 | – | 7.4 | 4.5 | – | 9.0–36.0 | 3.3–10.0 | – | 20.8 | 5.8 | – |
| Midrib width | 3.0 | 2.5 | – | 0.5 | 0.3 | – | 0.5–3.5 | 0.3–1.5 | – | 1.7 | 0.6 | – |
| Petiole length | – | 0.0 | – | – | 6.5 | – | – | 0.0–6.3 | – | – | 3.3 | – |
| Siliqua length | 3.0 | 3.8 | 3.5 | 2.8 | 3.2 | 3.0 | 1.5–6.2 | 2.0–6.8 | 2.2–6.7 | 3.7 | 4.2 | 4.1 |
| Siliqua width | 0.5 | 0.5 | 0.6 | 0.2 | 0.3 | 0.2 | 0.2–0.7 | 0.3–0.8 | 0.2–0.8 | 0.5 | 0.5 | 0.4 |
| Siliqua beak length | – | 1.0 | 1.0 | – | 0.7 | 0.5 | – | 0.3–1.6 | 0.2–1.5 | – | 0.8 | 0.7 |
| Seeds per siliqua | – | 20.0 | 25.0 | – | 11.0 | 15.0 | – | 4.1–27.1 | 2.0–24.9 | – | 14.3 | 12.6 |
| Seed weight | – | 320.0 | 345.0 | – | 105.0 | 20.0 | – | 115.0–337.0 | 30.0–257.0 | – | 211.0 | 141.4 |
a‘Chiifu’ did not flower until populations seed harvest.
‘–’, Not measured in the corresponding year.
3.2. QTL mapping and identification of crucifer building blocks in the B. rapa genome
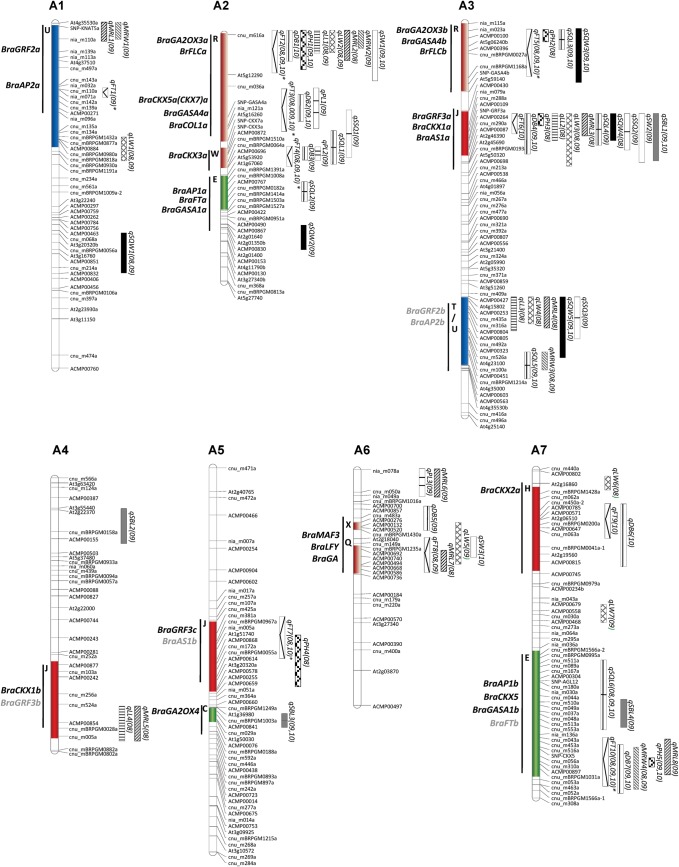
QTL mapping identified a total of 95 QTL for 14 traits in the CRF3 mapping population. It was observed that almost all the trait-enhancing alleles, except for flowering time, were contributed by the Chiifu parental line. The number of QTL detected ranged from 4 QTL for petiole length, seed number per siliqua and seed weight to 14 QTL for flowering traits, and the confidence interval ranged from 1 to 25 cM (Fig. 1 and Supplementary Table S2).
Figure 1.
Distribution of QTL for morphological and yield component traits in B. rapa genome. QTL names are indicated by abbreviations of trait names as shown in Table 1. The numbers in parenthesis indicate the year of QTL detection. The crucifer building blocks in each LGs of B. rapa, which are homologous to five chromosomes of A. thaliana At C1-At C5, are indicated by different colours. Putative candidate genes identified within the QTL blocks are shown in bold black letters on the left of each LG, and those outside QTL intervals are shown in bold grey letters.
Plant height
A total of seven QTL were detected for plant height, one in each LG, i.e. A2 and A3 (R block), A3 and A5 (J block), A7 (E block), A9 (H block), and A10 (A block). The QTL on A2 (qPH1) and A7 (qPH4) showed higher LOD values and phenotypic explanation and were detected both in 2009 and 2010 (Fig. 1 and Supplementary Table S2). The percentage of phenotypic variation explained by individual QTL ranged from 5 to 19%.
Leaf traits
Five leaf traits, i.e. leaf length and width, midrib length and width, and petiole length, were used for QTL analysis (Fig. 1 and Supplementary Table S2). A total of six QTL for leaf length were detected in four LGs. qLL1 and qLL5 in the R blocks of A3 and A10, respectively, were consistently detected in two consecutive years (2008 and 2009). The percentage of phenotypic variation explained by individual QTL ranged from 7 to 49%. For leaf width, 10 QTL were detected, of which qLW1 in the U block of A1, qLW2 in the R block of A2, and qLW3 in the J block of A3 were consistently found in 2008 and 2009 (Supplementary Table S2). A total of 10 QTL were detected for midrib length, of which QTL in the J (qMRL3) and T (qMRL4) blocks of A3 and qMRL6 in the A block of A6 showed comparatively higher LOD values, but none of them were detected consecutively in different years. For midrib width, five QTL were detected, of which QTL in A2 (qMRW2) and A10 (qMRW5) were detected in the R blocks of both LGs. qMRW3 in T/U block of A3 and qMRW4 in E block of A7 were detected both in 2008 and 2009. Four QTL, two on A2 (qPL1 in R block and qPL2 in W block) and one each on LG A6 (qPL3) and A10 (qPL4 in R block) were detected for petiole length. The QTL on qPL3 was major QTL showing the highest LOD value of 11.8. The phenotypic variation explained by individual QTL ranged from 6 to 13%.
Bolting and flowering traits
A total of 10 QTL were detected for bolting time, of which qDB2 in the R block of A2, qDB4 in the J block of A3, qDB7 in the E block of A7, qDB8 in the A block of A8, and qDB10 in the R block of A10 were detected in 2008 and 2009 (Fig. 1 and Supplementary Table S2). The QTL on A2, A7, and A10 were major loci, and the percentage of phenotypic variation explained by individual QTL ranged from 4 to 29%.
For flowering time in the non-vernalized plants, a total of 12 QTL were identified in 8 LGs (Supplementary Table S2 and Fig. 1). Of these, QTL mapped to the R (qFT2 and qFT3) and W (qFT4) blocks of A2, R block of A3 (qFT5), E block of A7 (qFT10), N block of A9 (qFT13), and R block of A10 (qFT15) were major loci and were detected consecutively for 3 yrs. A total of nine QTL, including two new QTL, i.e. qFT1 in the U block of A1 and qFT13 in the A block of A10, were detected in vernalized population in addition to the already detected QTL in the non-vernalized population (marked by asterisks, Supplementary Table S2 and Fig. 1). qFT2 and qFT3, both on A2, qFT10 on A7, qFT13 on A9, and qFT14 on A10, were major flowering time QTL detected in the vernalized and non-vernalized populations.
Siliqua traits
A total of nine QTL were detected for siliqua length, two QTL on A2 (qSQL1 in the R/W block and qSQL2 in E block), three QTL on A3 (qSQL3 in R block, qSQL4 in J block, and qSQL5 in T/U block), and one each on A7 (qSQL6 in the E block), A8 (qSQL7 in the B block), and A9 (qSQL8 in B block), and A10 (qSQL9 in the W block) (Fig. 1 and Supplementary Table S2). qSQL3, qSQL5, and qSQL9 were detected in two consecutive years while qSQL6 on A7 was detected in three consecutive years. The QTL on A3 (qSQL5), A7 (qSQL6), and A10 (qSQL9) were major loci, contributing to phenotypic variation ranging from 7 to 19% in different years.
Seven QTL, one each on LGs A1 (qSQW1 in the F block), A2 (qSQW2 in the K Block), A8 (qSQW6 in the C block), and A9 (qSQW7 in the N block) and three on A3 (qSQW3 in the R, qSQW4 in the J, and qSQW5 in the T/U blocks, respectively), were detected for siliqua width. qSQW1 on A1 and qSQW3 and qSQW5 on A3 were major loci that were detected in two consecutive years and explained 3–16% of the phenotypic variation.
For siliqua beak length, five QTL were detected, one each on A3 (qSBL1 in the J block), A4 (qSBL2 in the N Block), A5 (qSBL3 in the C block), A7 (qSBL4 in the E block), and A9 (qSBL5 in H block). The QTL on A3 (qSBL1) and A5 (qSBL3) were comparatively major loci with high LOD values and were detected in two different years (2009 and 2010).
Seed traits
The seed numbers per siliqua and seed weight are important traits for increasing yield (Fig. 1 and Supplementary Table S2). For the number of seeds per siliqua, one QTL each on A2 (qSSQ1 in the R/W block) and A10 (qSSQ4 in the W block) and two QTL on A3 (qSSQ2 in the J block and qSSQ3 in the T block) were detected. Only qSSQ4 on A10 was detected in two years, although the other three QTL also showed significantly high LOD values. For seed weight, one QTL each was detected on A2 (qSW1 in the R block), A3 (qSW2 in the J block), A6 (qSW3 in the X block), and A10 (qSW4 in the R block). Of these, only qSW1 and qSW4 were detected in more than 1 yr (2009 and 2010). The phenotypic variation explained by individual QTL ranged from 6 to 17%.
3.3. Identification of potential candidate genes in the QTL blocks
As synteny between our B. rapa map and A. thaliana was established,21 we searched for orthologs of previously characterized A. thaliana genes in the B. rapa genome (Fig. 1 and Table 3). The most important candidate genes, identified in the R blocks of A2, A3, and A10 harbouring the major QTL clusters for flowering and bolting, leaf, and siliqua traits, were orthologous to genes in the gibberellic acid (GA) oxidase 3 (AtGA20OX3),28 cytokinin oxidase/dehydrogenase 5 (AtCKX5),29,30 flowering locus C (FLC),8,9 GA-stimulated Arabidopsis (GASA4),31 and constans-like (COL) gene families. The Brassica homologs BraFLC and BraGA20OX3 were identified in the R blocks of A2, A3, and A10. However, some paralogs were differentially present on Brassica genomes such as BraCKX5 on A2 and A10, but not on A3, BraGASA4 only on A2 and A3, BraCOL1 on A2 and A10, etc. (Fig. 1). These suggest the preferential retention and fractionation of paralogous genes in duplicated segments of the B. rapa genome.
Table 3.
Putative candidate genes identified in the QTL blocks of B. rapa genome and SNPs observed between gene sequences of the parental lines, Chiifu and RCBr
| Gene family | Gene name | At gene ID | B. rapa gene ID | Block (LGs) | QTL co-mapping | SNP location (number) |
|---|---|---|---|---|---|---|
| Gibberellin biosythesis | AtGA2OX4 | AT1G47990 | Bra032238 | C (A5) | qSBL3 | Intron & exon (4) |
| AtGA20OX3 | AT5G07200 | Bra028706 | R (A2) | qFT2,qDB1,qPH1,qLL1,qLW2,qMRL2,qMRW2,qSW1 | – | |
| Bra005927 | R (A3) | qFT5,qSQW3 | Exon (1) | |||
| Bra009285 | R (A10) | qFT14,qDB10,qMRL10,qMRW4,qSW4,qLL5, qPL4 | – | |||
| Cytokinin oxidase/dehydrogenases | AtCKX5 | AT1G75450 | Bra015842 | E (A7) | qFT10,qDB7,qMRL8,qMRW3,qPH4 | Intron & exon (2) |
| AtCKX2 | AT2G19500 | Bra036719 | H (A9) | qSBL5 | Intron & exon (3) | |
| Bra040677 | H (A7) | qLW6,qDB5,qFT9 | Intron (1) | |||
| AtCKX1 | AT2G41510 | Bra000229 | J (A3) | qPH2,qLL2,qLW3,qMRL3,qDB4,qFT6,qSQL3,qSQW4,qSBL1,qSSQ2, qSW2 | – | |
| Bra016928 | J (A4) | qLL4,qMRL5 | Intron & exon (6) | |||
| AtCKX5 (CKX7) | AT5G21482 | Bra020157 | R (A2) | qFT3,qDB2,qPL1 | Intron & exon (7) | |
| Bra002371 | R (A10) | – | Intron & exon (4) | |||
| AtCKX3 | AT5G56970 | Bra035640 | W (A2) | qFT4,qDB3,qPL2,qSQL1,qSSQ1 | Intron (1) | |
| Bra002777 | W (A10) | qSQL8,qSSQ4 | Intron & exon (2) | |||
| Gast1 protein homolog | GASA4 | AT5G15230 | Bra023513 | R (A2) | qFT3,qDB2,qPL1 | Intron & exon (3) |
| Bra006291 | R (A3) | qFT5,qSQW3 | Intron (1) | |||
| GASA1 | AT1G75750 | Bra008222 | E (A2) | qSQL2 | – | |
| Bra015820 | E (A7) | qFT10,qDB7,qMRL8,qMRW3,qPH4 | Exon (1) | |||
| Bra003743 | E (A7) | qFT10,qDB7,qMRL8,qMRW3,qPH4 | Intron (2) | |||
| Growth-regulating factor | AtGRF2 | AT4G37740 | Bra011781 | U (A1) | qMRL1,qMRW1 | Exon (3) |
| Bra017851 | U (A3) | – | Exon (4) | |||
| AtGRF3 | AT2G36400 | Bra023066 | J (A3) | qPH2,qLL2,qLW3,qMRL3,qDB4,qFT6,qSQL3,qSQW4,qSBL1,qSSQ2, qSW2 | Intron (1) | |
| Bra017240 | J (A4) | – | Intron (4) | |||
| Bra005268 | J (A5) | qFT7,qPH3 | Intron & exon (3) | |||
| Apetala1 | AP1 | AT1G69120 | Bra004007 | E (A7) | qSQL5,qSBL4 | Intron (6) |
| Bra038326 | E (A2) | qSQL2 | – | |||
| Apetala2 | AP2 | AT4G36920 | Bra011741 | U (A1) | qFT1 | Intron & exon (5) |
| Bra017809 | U (A3) | – | – | |||
| Asymmetric leaves 1 | AS1 | AT2G37630 | Bra000011 | J (A3) | qPH2,qLL2,qLW3,qMRL3,qDB4,qFT6,qSQL3,qSQW4,qSBL1,qSSQ2, qSW2 | Exon (1) |
| Bra005177 | J (A5) | – | – | |||
| Constans like | COL1 | AT5G15850 | Bra023541 | R (A2) | qFT3,qDB2,qPL1 | Exon (2) |
| Bra008668 | R (A10) | – | Exon (7) | |||
| Flowering locus T | FT | AT1G65480 | Bra022475 | E (A2) | qSQL2 | – |
| Bra004117 | E (A7) | – | – | |||
| Leafy | LFY | AT5G61850 | Bra019619 | X (A6) | qDB5 | – |
| GA-insensitive dwarf 1C | GA | AT5G27320 | Bra009970 | Q (A6) | qFT8,qMRL7,qLW5,qSW3 | Intron & exon (6) |
| Mads affecting flowering 3 | MAF3 | AT5G65060 | Bra024350 | X (A6) | qDB4 | Intron & exon (4) |
| Flowering locus C | FLC | AT5G10140 | Bra028599 | R (A2) | qFT2,qDB1,qPH1,qLL1,qLW2,qMRL2,qMRW2,qSW1 | Intron (1) |
| Bra006051 | R (A3) | qFT5,qSQW3 | – | |||
| Bra009055 | R (A10) | qFT14,qDB10,qMRL10,qMRW4,qSW4,qLL5,qPL4 | Intron (1) |
We detected two W blocks, one each on A2 and A10, that harboured QTL for petiole length, seeds per siliqua, leaf traits, days to bolting and flowering, and siliqua length (Fig. 1). These two blocks harbour paralogous BraCKX3 genes (Table 3). The E block on A2 only contained QTL for siliqua traits, but the paralogous block on A7 contained major QTL clusters for leaf traits, flowering and bolting, plant height, and siliqua traits. We detected GASA1, flowering locus T (BraFT), and apetala 1 (BraAP1) on both LGs. FT and AP are components of the flowering gene network..BraCKX5 was found only on A7 (Fig. 1).
The J block of A3 contained many QTL for leaf, siliqua, and seed traits, plant height, and days to bolting and flowering, whereas the same blocks in A4 harboured QTL for leaf length and midrib length, and A5 contained QTL for flowering time and plant height (Fig. 1). A. thaliana growth-regulating factor32 (BraGRF3) was found in all three LGs. However, BraCKX1 paralogs were found only on A3 and A4, whereas asymmetric leaves 133 (BraAS1) paralogs were found only on A3 and A5 (Fig. 1).
The candidate gene BraGA20X434 was identified in the C block of A5, which contains QTL for siliqua beak length. The H block of A7 contained QTL for days to flowering, bolting, and leaf width, whereas the paralogous block of A9 contained QTL for siliqua beak length. BraCKX2 paralogs were found in the QTL regions of both LGs (Fig. 1). The U block of A1 contained QTL for flowering and leaf width, whereas the paralogous block on A3 (U/T block) contained QTL for leaf width, length, and seeds per siliqua. The candidate genes BraGRF2 and BraAP2 were detected in both LGs. The X block in A6 contained QTL for days to bolting and harboured candidate gene LEAFY and mads affecting flowering time 3. The Q block harboured QTL for seed weight, leaf, and flowering traits and contained the GA-insensitive 1 gene (BraGA) that interacts with GA and is involved in floral transition and GA signalling.
3.4. Identification, validation, and functional significance analysis of SNPs in candidate genes
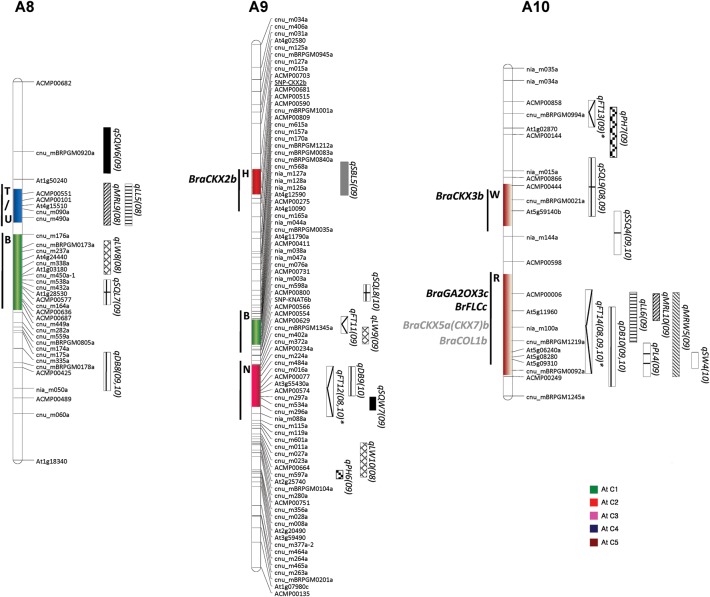
To develop gene-specific SNP markers for breeding morphological and yield-related traits, we aligned all the identified candidate gene sequences from the parental lines and identified varying number of SNPs, ranging from one to many, between them (Fig. 2 and Table 3). As expected, we found more SNPs in the intronic regions than in the exonic regions. However, we did not detect any SNPs in a few genes, e.g. BraGA20OX3 in the R blocks of A2 and A10 and BraCKX1 in the J block of A03 (Fig. 2 and Table 3).
Figure 2.
SNPs identified for different cytokinin oxidases/dehydrogenases (CKX) paralogs between Chiifu and RCBr derived from whole genome resequenced data. Exons and introns are represented by blue rectangular bars and lines, respectively. Numbers indicated by arrows show the SNP positions from the start codon for Chiifu (left nucleotide) and RCBr (right nucleotide).
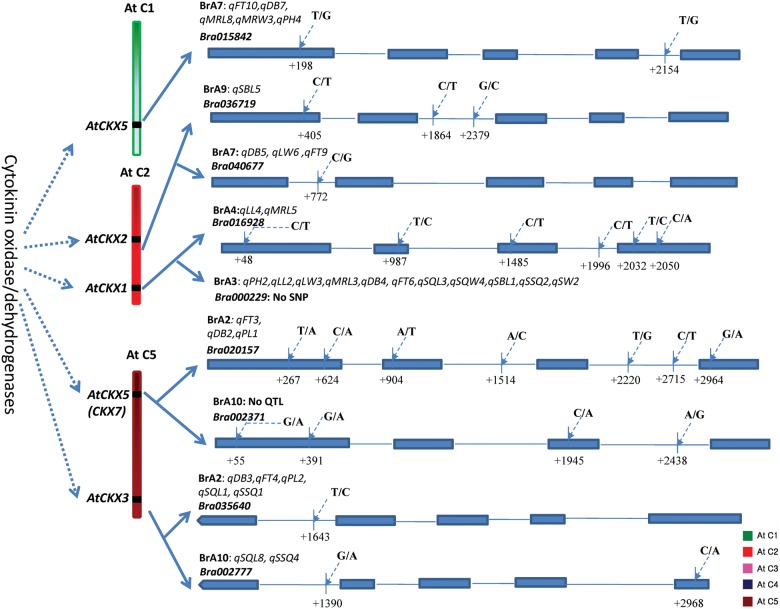
To validate these SNPs experimentally, we designed a total of 12 primer pairs, 1 each from different genes flanking the SNPs and genotyped the parental lines and mapping population. Of the 12 SNPs, 10 primer pairs revealed clear polymorphisms (Supplementary Table S3). We genotyped using these 10 primer pairs in the mapping population and mapped them precisely in the B. rapa genome, which indicated their co-localization with the QTL positions on the respective chromosomes (Figs 1 and 3). Association analysis with the different phenotypes of the F3 population showed a correlation with the phenotypes and their co-localization in the QTL regions. We sequenced some of the genes containing SNPs using the resequenced data to validate whether the NGS data of the parental lines were correct. Resequencing of 10 genes showed the exact sequences as shown by NGS. We observed that the SNPs in coding regions of some potential candidate genes affected amino acid sequence, suggesting possible functional roles in phenotypic differences (Supplementary Fig. S1). The SNP in exon 2 of BraGA20OX3b codes for isoleucine in Chiifu and leucine in RCBr, methionine/threonine in exon 2 of BraCKX1b while serine was found in Chiifu, but no counterpart amino acid was observed in RCBr in exon 5 of BraCKX3b. Two amino acid changes were observed in exon 1 of BraGA2OX4 (arginine/serine and serine/threonine), one each in exons 2 (aspartate/valine), and four (alanine/threonine) of BraCKX7a, two each in exon 1 (valine/isoleucine and glycine/serine) of BraCKX7b and exon 4 (isoleucine/serine and threonine/alanine) of BraGRF2a, one each in exon 2 (leucine/arginine) and exon 4 (glycine/serine) of BraGRF3c, and two in exon 1 (methionine /valine and threonine/serine) of BraCOL1 gene.
Figure 3.
Amplicon scanning analysis of the CRF2 mapping population for SNP markers designed from candidate gene BraGASA4b located in A3 using Light Scanner. Plot A represents the curve-shifted plot, and B represents a difference plot showing ‘Chiifu’ type in red colour, ‘RCBr’ type in grey colour, and hetero type in green colour.
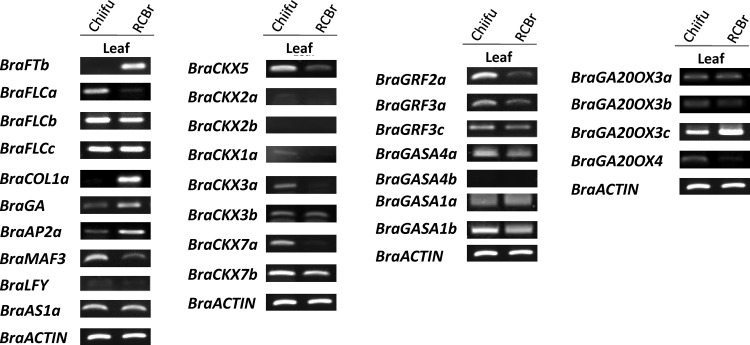
Semi-quantitative RT-PCR analysis was used to evaluate expression differences of candidate genes between the parental lines. The gibberellin biosynthesis and oxidase, cytokinin oxidases/dehydrogenases and growth-regulating factor gene families that affect leaf, plant, and flower development, showed differential gene expression between Chiifu and RCBr lines (Fig. 4). BraCKX5, BraCKX3a, BraCKX1a, and BraCKX7a showed higher expression in Chiifu, whereas very little or no expression was seen in RCBr. Genes that are involved in flowering pathways, such as FLC, FT, COL, and GA, were also analysed for differential expression between the parental lines. Of the three BraFLC paralogs, only BraFLCa on A2 did not show any expression in RCBr while the other two BraFLCs showed no difference in expression. BraMAF3 on A6 showed higher expression level in Chiifu when compared with RCBr. BraAP2a on A1, BraCOL1a on A2, BraGA on A6 and BraFTb on A7 showed strong expression in RCBr, but very little to no expression in Chiifu. BraGRF2a, BraGRF3a, and BraGA2OX4 showed comparatively higher expression in Chiifu while BraGA20OX3c showed higher expression in RCBr. This differential gene expression and the amino acid changes suggest possible roles in phenotypic differences between the parental lines and in the segregating population.
Figure 4.
Semi-quantitative RT-PCR analysis of candidate genes using RNA samples extracted from 20-day-old seedlings of Chiifu and RCBr. Actin gene amplicons were used as control for RNA quantity.
3.5. Functional conservation of crucifer building blocks between the A, B, and C subgenomes of Brassica species as revealed by comparative QTL mapping
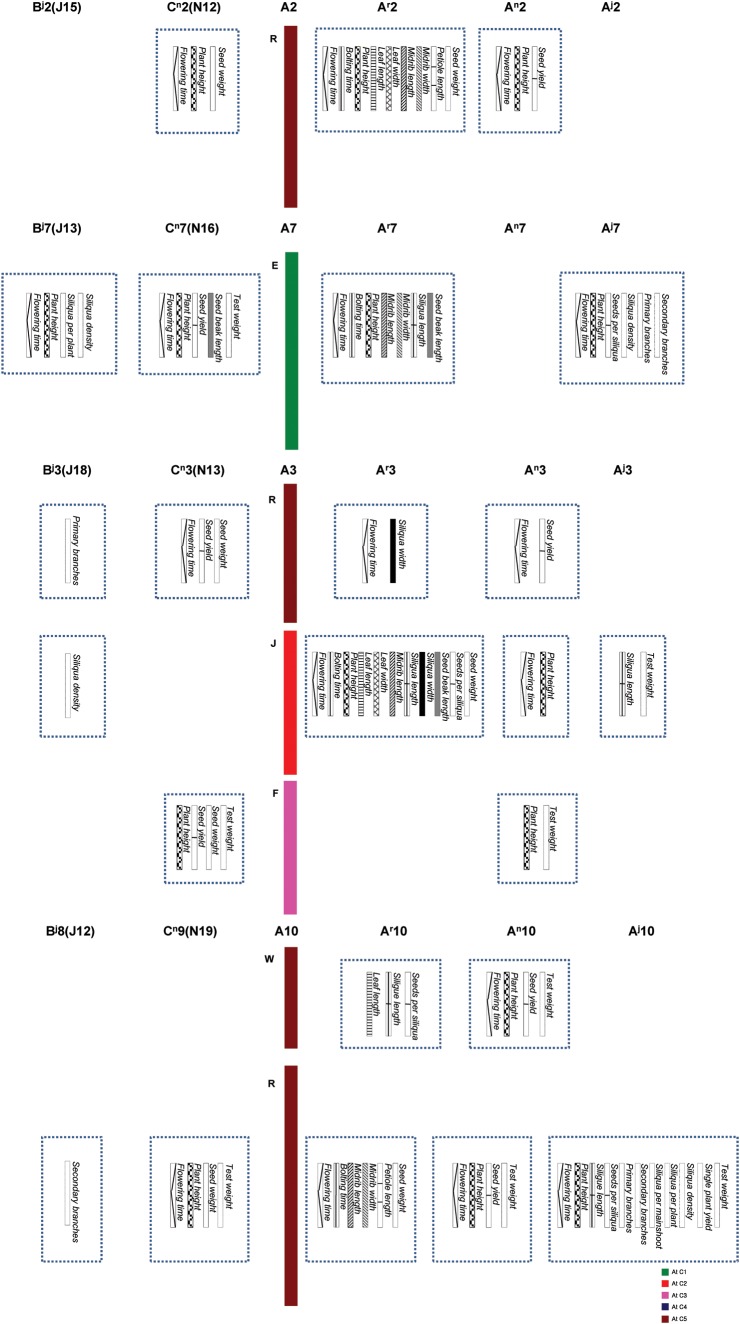
We aligned the QTL map of B. rapa from this study with the B. juncea map of Ramchiary et al.20 and B. napus QTL maps of Quijada et al.21 and Udall et al.22 to identify the structural and functional conservation of QTL blocks for important morphological and yield component traits. Comparative alignment of the QTL maps between these three species identified a total of four homologous/homoeologous groups (HGs) containing QTL for agronomic and yield component traits (Fig. 5). The first group was the R block of Ar2, Aj2, An2, Cn2, and Bj2 (r, j, and n stand for B. rapa, B. juncea, and B. napus, respectively). Although many major QTL for different traits were detected in the B. rapa A subgenome, in comparison with B. napus, three traits, namely, plant height, flowering time, and seed yield, were commonly detected in the A subgenome of the two species and the homoeologous region of the same block in the C subgenome of B. napus. We did not detect any QTL in the Aj2 or Bj2 subgenomes of B. juncea (Fig. 3). The second HG was the A3, B3, and C3 chromosomes of the A, B, and C subgenomes of Brassica species. Three blocks, namely, R, J, and F, harboured common QTL between the three subgenomes. The R block of this HG contained common QTL for seed weight, yield, and test weight (hereafter referred to collectively as SW) and flowering time QTL between the A and C subgenomes of B. napus and the A subgenome of B. rapa. In addition, the homoeologous region of this block contained QTL for a number of branches of the B subgenome of B. juncea. The J block of this group contained many QTL in Ar3, but few in An3, Aj3, and Bj3. The common loci were for SW and flowering time in addition to siliqua traits, e.g. siliqua length, density, and number, and plant height. The last common block, i.e. the F block of this group, harboured common trait QTL mainly for SW and plant height between An3 and Cn3. The most important common block of HG7 was the E block. This block contained common QTL for plant height, SW, flowering time, siliqua number and density, and seed number per siliqua in Ar7, Aj7, Cn7, and Bj7. In addition, some leaf traits were detected in Ar7. The last common group of the A, B, and C Brassica subgenomes was HG10 that contained the W and R blocks. The W block contained common QTL for siliqua length, seed number per siliqua, and SW between Ar10, An10, and Aj10. In addition, QTL for flowering, plant height, and leaf traits were detected in Ar10 and An10, respectively. However, in this block, no single or corresponding common QTL were detected in Bj10 and Cn10. The most important block of HG10 was the R block. This block harboured common major QTL for plant height, branch number, flowering time, siliqua traits, and SW in Ar10, Aj10, An10, Bj10, and Cn10, respectively. Similarly, the middle region of Ar6 (X and Q block) in the present study detected QTL for SW, leaf, and flowering traits while the same chromosomal region in B. juncea detected major QTL for siliqua length and density.20 We also detected common QTL for flowering and SW, both in Ar6 and Cn6 subgenomes.21
Figure 5.
Comparative QTL mapping between B. rapa, B. juncea, and B. napus revealed conservation of crucifer building blocks governing morphological and yield component traits. Superscripts ‘r’, ‘n’, and ‘j’ after each A, B, and C LGs denote chromosomes that were derived from B. rapa, B. napus, and B. juncea, respectively. QTL in B genome of B. juncea were taken from Ramchiary et al.,20 and the C genome of B. napus from Quijada et al.,21 and Udall et al.22
4. Discussion
4.1. QTL mapping revealed some crucifer blocks that were functionally more active than duplicated paralogous blocks in the B. rapa genome
Although in many studies, immortal mapping populations such as doubled haploid and recombinant inbred lines are used, in our present study, we used F3 families for three consecutive years to detect QTL, as F3 families represent the comprehensive genetic information of heterozygous and homozygous plants and their interacting effects on phenotype. As the parental lines Chiifu and RCBr were very different with respect to all the traits under study, it was easy to detect many QTL in B. rapa using the segregating F3 population. Correlation coefficient analysis revealed significant correlations between leaf and yield-related traits that were further supported by the co-mapping of several QTL governing different traits in a few chromosomal blocks of the B. rapa genome. The co-localization of QTL governing different traits in the same genetic interval has been suggested to be due to either pleiotropy exhibited by a single gene or tight linkage of genetic loci governing different traits in the same genetic interval.8,20 The R block of A2 contained QTL for eight traits while the same paralogous blocks on A10 and A3 contained QTL for five and four traits, respectively (Fig. 1). Prominent QTL clusters were observed for seven traits related to leaf, flowering, bolting, and siliqua traits on the E block of A7 while the same block on A2 contained only one QTL for siliqua length. A similar case was observed for the J blocks, where the same block on A3 harboured QTL for 10 traits, but only harboured QTL for 2 traits on A4 and A5. These findings suggest that the R block of A2, E block of A7, and J block of A3 contain more active genes that govern leaf traits, plant morphology, and seed traits. Lou et al.8 and Li et al.9 also observed QTL clusters in these LGs for days to flowering, bolting, and leaf traits. We presumed that the chromosomal blocks of Lou et al.8 were the R block on A2 and E block on A7. The W block on A2 contained QTL clusters for days to flowering and bolting and seeds per siliqua, whereas A10 contained QTL for siliqua and leaf traits. This kind of observation suggests that only those crucifer blocks harbouring QTL might possess functionally active genes for the traits under study, as reported by Wang et al.16 and Tang et al.35 for the B. rapa genome. They reported that, of the triplicate duplicated subgenomes, fractionation and deletion of genes were more frequent in some subgenomes when compared with other paralogs. We also observed that some candidate genes were missing in major QTL blocks, such as BraGASA4 from A10 and BraCKX5 from A3, which both belong to the R block that is homologous to A. thaliana chromosome 5. Other reasons for not finding equal QTL/genes in all paralogous blocks may be that the genes are not functionally active or the absence of polymorphisms in the causal genes between the parental lines.
4.2. Gene family members were detected in different QTL blocks of the B. rapa genome
Many candidate genes in QTL blocks for flowering, leaf, seed, and siliqua traits were identified through synteny analysis of the B. rapa and A. thaliana genomes. The R block, homologous to A. thaliana chromosome 5, harbours many flowering genes, including FLC and COL1. We identified BraFLC and BraCOL1 paralogs in those blocks containing QTL for flowering time, as reported by earlier studies.8,36 Recent studies have shown that members of cytokinin oxidase/dehydrogenases (CKX), GA-stimulated Arabidopsis (GASA), and GA oxidase (GA2OX, GA20OXs, etc.) gene families are involved in the regulation of flower and inflorescence development, thereby increasing the seed size and yield.28–31CKX genes are important for agricultural productivity as mutations in these genes cause accumulation of high levels of cytokinin in the inflorescence meristem and increased reproductive organs and larger flowers and seeds.29,30 These genes co-localized in the QTL blocks of B. rapa LGs A2, A3, A4, A7, A9, and A10. GASA genes are active in the shoot apical meristem, developing flowers, and embryos. GASA mutants and GASA over-expressing plants demonstrated that GASA genes are involved in floral meristem identity and affect seed size and yield in A. thaliana.29 We observed two paralogs of BraGASA4, one on each R block of A2 and A3, but not on the R block of A10, suggesting that the paralogs on A10 may have been lost over time, although all blocks harboured QTL for flowering, siliqua, and seed traits. GA oxidases were observed in the QTL regions of the R, J, and other blocks containing prominent QTL for leaf and flowering traits, and a possible involvement of this gene family in leaf traits was reported by Li et al.9 The BraGRF members found on A1 and A5 are also candidates for leaf and flowering traits, as supported by their co-localization with these QTL regions. Several members of the BraCOL and BraFT families were also found in different LGs in the QTL regions, suggesting an involvement in trait variation.
SNPs analysis revealed some amino acid changes between the parental lines in many genes belonging to constans like 1(COL1), cytokinin oxidases/dehydrogenases, growth-regulating factor, and GA oxidases gene families. Further expression analysis showed higher expression of genes that facilitate early flowering such as BraCOL1a, BraFT, BraAP2 in RCBr, but minimal expression in Chiifu, whereas the flowering repressor gene BraFLCa was highly expressed in Chiifu suggesting an association between flowering trait and expression of these genes. The co-mapping of these genes with flowering QTL in different LGs further supported this notion. Members of growth-regulating factor, GA oxidase, and cytokinin oxidase/dehydrogenases gene families that co-localized within the QTL region showed higher expression in Chiifu, suggesting their role in better growth and development of leaf, plants, and reproductive organs when compared with RCBr. Either non-synonymous SNPs or differential genes expression or a combination of these factors may underlie phenotypic differences between the parental lines and in the segregating populations. However, this should be further assessed in different plant tissues.
4.3. QTL blocks are evolutionarily conserved between the A, B, and C subgenomes of Brassica species
Our comparative QTL map alignment between B. rapa, B. napus, and B. juncea revealed conservation of QTL blocks that contain several genetic loci influencing plant height, flowering, siliqua, and seed traits between the A, B, and C subgenomes of Brassica species (Fig. 5). Although many earlier studies reported the conservation of chromosomal blocks at the gross level,10,11,14 a comparison of QTL location has not been performed for yield component traits between different Brassica subgenomes. In our present study, we identified conserved QTL blocks in 4 of the 10 HGs of the A, B, and C subgenomes reported by Panjabi et al.11 As plant height, flowering time, siliqua, and seed traits were the only common traits studied in the three species, we could study the conservation of these traits in the four HG QTL blocks. The R blocks of HG2, HG3, and HG10, E block of HG7, W block of HG10, and J and F blocks of HG2 were major QTL blocks harbouring common QTL for SW, plant height, flowering, and siliqua traits between the A, B, and C subgenomes of these three species. This suggest that the genes governing these traits are structurally and functionally conserved not only between the A, B, and C subgenomes of Brassica species but also between A. thaliana and Brassica genomes, even though they diverged from their common ancestor a long time ago.12 We previously found the conservation of QTL for seed oil, total seed glucosinolates, and seed protein content between the A subgenomes of B. rapa and B. napus.5 Studies on B. napus21,22 and B. juncea20 detected QTL in homoeologous LGs of the A and C and A and B subgenomes, respectively, but detailed information with respect to specific crucifer blocks was not provided. We found more common QTL between the A and C subgenomes when compared with the B subgenome with either the A or C subgenomes. Although we believe that more comparisons using QTL maps from different mapping populations are needed, the present study suggests that A and C (diverged 8 MYA) are more close than the A/B and C subgenomes, as reported earlier.12 We also observed many QTL in one species that had no counterparts controlling the same traits in the corresponding locations of other species, which might have been the result of a lack of genetic polymorphisms between the parental lines or diversified molecular polymorphisms between the species due to the presence of genomic rearrangements/structural changes causing differences in functional expression between different Brassica species. We believe that adding more markers to generate a high-density map and a detailed comparison of QTL for common traits using different mapping populations of more Brassica species would give more detailed information about the conservation and diversification of genetic loci governing morphological and yield component traits.
4.4. Breeding opportunities in Brassica species using candidate gene information from B. rapa
The present QTL mapping study identified some interesting findings regarding the presence of candidate genes for important traits in a few specific crucifer building blocks of the B. rapa genome (Fig. 1). Although, the crucifer blocks demonstrated the co-localization of important QTL and candidate genes, association analysis using large segregating population with different trait phenotypes and functional validation of candidate genes by transformation should be done because each QTL block contains several genes governing overlapping traits. We observed few gene paralogs within the QTL interval in one duplicated block but not in others, suggesting the need for further validation. However, we believe that some of the SNPs identified in candidate genes such as BraCKXs, BraGASA4, BraGA20OXs, BraGRFs, BraFLC, BraCOLs, BraMAF, and BraAP2 would be helpful in breeding leaf, flowering, bolting, siliqua, and seed traits of B. rapa, as their differential expressions in parental lines and previous studies suggest their role in creating phenotypic differences9. However, additional expression studies using different tissues are needed. Although genome sequencing of other cultivated Brassica species is ongoing, gene sequence information of B. rapa could be used to isolate and develop molecular markers for breeding other Brassica species, especially B. juncea and B. napus, as B. rapa is one of the progenitor species of both species and there still exists QTL block conservation for important agronomic and yield component traits between these three species.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This research was partially supported by Chinese Cabbage Molecular Marker Research Center (Grant No. 607003-05) and Agriculture Research Center (Grant No. 06362010-0010) from the Technology Development Program for Agriculture and Forestry, Ministry of Agriculture, Forestry and Fisheries, as well as a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ007992), Rural Development Administration, Republic of Korea.
Supplementary Material
Acknowledgements
We are thankful to the Korea Brassica Genome Resource Bank for providing Chiifu and CRF3 mapping population, Prof. Deepak Pental and Prof. Akshay K. Pradhan, Department of Genetics, Delhi University South Campus, India for the updated B. juncea map.
Footnotes
Edited by Dr Masahiro Yano
References
- 1.Suwabe K., Tsukazaki H., Iketani H., et al. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 2006;173:309–19. doi: 10.1534/genetics.104.038968. doi:10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X., Ramchiary N., Choi S.R., et al. Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome. 2010;53:939–47. doi: 10.1139/G10-054. doi:10.1139/G10-054. [DOI] [PubMed] [Google Scholar]
- 3.Tanhuanpaa P.K., Vilkke J.P., Vikki H.J. Mapping a QTL for oleic acid concentration in spring turnip rape (Brassica rapa ssp. oleifera) Theor. Appl. Genet. 1996;92:952–6. doi: 10.1007/BF00224034. doi:10.1007/BF00224034. [DOI] [PubMed] [Google Scholar]
- 4.Lou P., Zhao J., He H., et al. Quantitative trait loci for glucosinolate accumulation in Brassica rapa leaves. New Phytol. 2008;179:1017–32. doi: 10.1111/j.1469-8137.2008.02530.x. doi:10.1111/j.1469-8137.2008.02530.x. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C., Ramchiary N., Ma Y., et al. Structural and functional comparative mapping between the Brassica A genomes in allotetraploid Brassica napus and diploid Brassica rapa. Theor. Appl. Genet. 2011;123:927–41. doi: 10.1007/s00122-011-1637-1. doi:10.1007/s00122-011-1637-1. [DOI] [PubMed] [Google Scholar]
- 6.Saito M., Kubo N., Matsumoto S., Suwabe K., Tsukada M., Hirai M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 2006;114:81–91. doi: 10.1007/s00122-006-0412-1. doi:10.1007/s00122-006-0412-1. [DOI] [PubMed] [Google Scholar]
- 7.Soengas P., Hand P., Vicente J.G., Pole J.M., Pink D.A. Identification of quantitative trait loci for resistance to Xanthomonas campestris pv.campestris in Brassica rapa. Theor. Appl. Genet. 2007;114:637–45. doi: 10.1007/s00122-006-0464-2. doi:10.1007/s00122-006-0464-2. [DOI] [PubMed] [Google Scholar]
- 8.Lou P., Zhao J.J., Kim J.S., et al. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 2007;58:4005–16. doi: 10.1093/jxb/erm255. doi:10.1093/jxb/erm255. [DOI] [PubMed] [Google Scholar]
- 9.Li F., Kitashiba H., Inaba K., Nishio T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009;16:311–23. doi: 10.1093/dnares/dsp020. doi:10.1093/dnares/dsp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkin I.A.P., Gulden S.M., Sharpe A.G., et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–81. doi: 10.1534/genetics.105.042093. doi:10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panjabi P., Jagannath A., Bisht N., et al. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. 2008;9:113. doi: 10.1186/1471-2164-9-113. doi:10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mun J.H., Kwon S.J., Yang T.J., et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. doi:10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagercrantz U., Lydiate D.J. Comparative genome mapping in Brassica. Genetics. 1996;144:1903–10. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schranz M.E., Lysak M.A., Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–42. doi: 10.1016/j.tplants.2006.09.002. doi:10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Yang T.J., Kim J.S., Lim K.B., et al. The Korea Brassica Genome Project: a glimpse of the Brassica genome based on comparative genome analysis with Arabidopsis. Comp. Funct. Genomics. 2005;6:138–46. doi: 10.1002/cfg.465. doi:10.1002/cfg.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Wang H., Wang J., et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–9. doi: 10.1038/ng.919. doi:10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J., Lu Y., Yuan Y., et al. Map based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol. Biol. 2009;69:553–63. doi: 10.1007/s11103-008-9437-y. doi:10.1007/s11103-008-9437-y. [DOI] [PubMed] [Google Scholar]
- 18.Ramchiary N., Nguyen D.V., Li X., et al. Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res. 2011;18:305–20. doi: 10.1093/dnares/dsr017. doi:10.1093/dnares/dsr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Basten C.J., Zeng Z.B. Windows QTL Cartographer 2.5. Raleigh, NC: North Carolina State University; 2005. [Google Scholar]
- 20.Ramchiary N., Padmaja K.L., Sharma S., et al. Mapping of yield influencing QTL in Brassica juncea: implications for breeding of a major oilseed crop of dryland areas. Theor. Appl. Genet. 2007;115:807–17. doi: 10.1007/s00122-007-0610-5. doi:10.1007/s00122-007-0610-5. [DOI] [PubMed] [Google Scholar]
- 21.Quijada P.A., Udall J.A., Lambert B., Osborn T.C. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor. Appl. Genet. 2006;113:549–61. doi: 10.1007/s00122-006-0323-1. doi:10.1007/s00122-006-0323-1. [DOI] [PubMed] [Google Scholar]
- 22.Udall J.A., Quijada P.A., Lambert B., Osborn T.C. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor. Appl. Genet. 2006;113:597–609. doi: 10.1007/s00122-006-0324-0. doi:10.1007/s00122-006-0324-0. [DOI] [PubMed] [Google Scholar]
- 23.Stanke M., Tzvetkova A., Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006;7:S11. doi: 10.1186/gb-2006-7-s1-s11. doi:10.1186/gb-2006-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R., Li Y., Kristiansen K., Wang J. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. doi:10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Koeyer D., Douglass K., Murphy A., et al. Application of high-resolution DNA melting for genotyping and variant scanning of diploid and autotetraploid potato. Mol. Breed. 2010;25:67–90. doi:10.1007/s11032-009-9309-4. [Google Scholar]
- 27.Lee S.C., Lim M.H., Kim J.A., et al. Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K oligo microarray. Mol. Cells. 2008;26:595–605. [PubMed] [Google Scholar]
- 28.Plackett A.R.G., Powers S.J., Fernandez-Garcia N., et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–60. doi: 10.1105/tpc.111.095109. doi:10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashikari M., Sakakibara H., Lin S., et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–5. doi: 10.1126/science.1113373. doi:10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 30.Bartrina I., Otto E., Strnad M., Werner T., Schmulling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23:69–80. doi: 10.1105/tpc.110.079079. doi:10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roxrud I., Lid S.E., Fletcher J.C., Schmidt E.D., Opsahl-Sorteberg H.G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007;48:471–83. doi: 10.1093/pcp/pcm016. doi:10.1093/pcp/pcm016. [DOI] [PubMed] [Google Scholar]
- 32.Kim J.H., Choi D., Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313x.2003.01862.x. doi:10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 33.Hay A., Barkoulas M., Tsiantis M. Asymmetric leaves1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development. 2006;133:3955–61. doi: 10.1242/dev.02545. doi:10.1242/dev.02545. [DOI] [PubMed] [Google Scholar]
- 34.Rieu I., Eriksson S., Powers S.J., et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20:2420–36. doi: 10.1105/tpc.108.058818. doi:10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang H., Woodhouse M.R., Cheng F., et al. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics. 2012;190:1563–74. doi: 10.1534/genetics.111.137349. doi:10.1534/genetics.111.137349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schranz M.E., Quijada P., Sung S-B., Lukens L., Amasino R., Osborn T.C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162:1457–68. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.