Abstract
In this study, we developed the first genetic linkage map for the major rice insect pest, the brown planthopper (BPH, Nilaparvata lugens). The linkage map was constructed by integrating linkage data from two backcross populations derived from three inbred BPH strains. The consensus map consists of 474 simple sequence repeats, 43 single-nucleotide polymorphisms, and 1 sequence-tagged site, for a total of 518 markers at 472 unique positions in 17 linkage groups. The linkage groups cover 1093.9 cM, with an average distance of 2.3 cM between loci. The average number of marker loci per linkage group was 27.8. The sex-linkage group was identified by exploiting X-linked and Y-specific markers. Our linkage map and the newly developed markers used to create it constitute an essential resource and a useful framework for future genetic analyses in BPH.
Keywords: Nilaparvata lugens, brown planthopper, genetic linkage map, SSR, SNP
1. Introduction
The brown planthopper (BPH), Nilaparvata lugens Stål (Homoptera: Delphacidae), is one of the most economically important insect pests of rice (Oryza sativa L.) throughout Asia. The widespread damage caused by BPH leads to substantial and unpredictable decreases in rice yield. Continuous rice cultivation, accompanied by extensive use and frequent misuse of insecticides along with a high rate of nitrogen fertilizer application, often causes BPH outbreaks in rice fields.1 More than 50 million ha of rice fields throughout Asia have been heavily affected by this insect in the past decade.2–4 In addition to directly feeding on rice plants, BPH also causes indirect damage by transmitting viruses that cause the ragged stunt and grassy stunt diseases of rice.5
Host-plant resistance offers a practical approach for BPH control, and a succession of resistant rice varieties has been bred and cultivated widely in Asia. Nevertheless, the appearance of BPH with new virulence characteristics can overcome their resistance soon after the varieties have been released.4,6 The virulence, which refers to the ability to adapt to resistant rice varieties, has been reported to be under genetic control and is inherited as a quantitative trait.7 Understanding the genetic basis of the virulence against rice cultivars will provide significant information for future BPH management strategies. A genetic linkage map would provide an important framework for identifying quantitative trait loci (QTL) related to the virulence characteristics. Unfortunately, no detailed genetic linkage map based on molecular markers has yet been developed for BPH or for other closely related delphacid species.
BPH has a diploid chromosome number of 30 (28 autosomes combined with the sex determination system XY and XX in males and females, respectively).8–10 The genome size of BPH is estimated to be 1.2 Gbp.11 Recently, a number of genomic resources for BPH have been made available, and they provide highly useful information to support experimental genetics studies.12–16 More than 350 expressed sequence tag (EST)-derived simple sequence repeat (e-SSR) markers for BPH have been successfully developed subsequent to the availability of large EST datasets.17–19 In addition, a total of 136 SSRs have been isolated from BPH genomic DNA.20 These newly developed e-SSRs and genomic SSR (g-SSR) markers have been used to assess the genetic diversity of BPH populations.17,20 However, the exact genomic locations of the newly developed SSR markers have not yet been determined by genetic mapping.
Genetic maps are valuable genomic resources and represent a powerful research tool for identifying the genetic basis of phenotypic variation. The purpose of the present study was to construct a baseline genetic linkage map for BPH using both new and existing SSRs and single-nucleotide polymorphisms (SNPs). G-SSR markers were generated from the sequence information of repeat regions derived from high-throughput microsatellite isolation procedures. We also developed SNP markers using next-generation sequencing technologies. The newly developed and public markers were successfully applied to construct a consensus linkage map. This map will provide initial information for identifying the genetic basis of BPH virulence and other important traits, including insecticide resistance, virus transmission, and wing polymorphisms. In addition, the newly developed markers will support studies of the morphology, physiology, evolution, and ecology of this significant insect pest.
2. Materials and methods
2.1. Insect strains and crosses
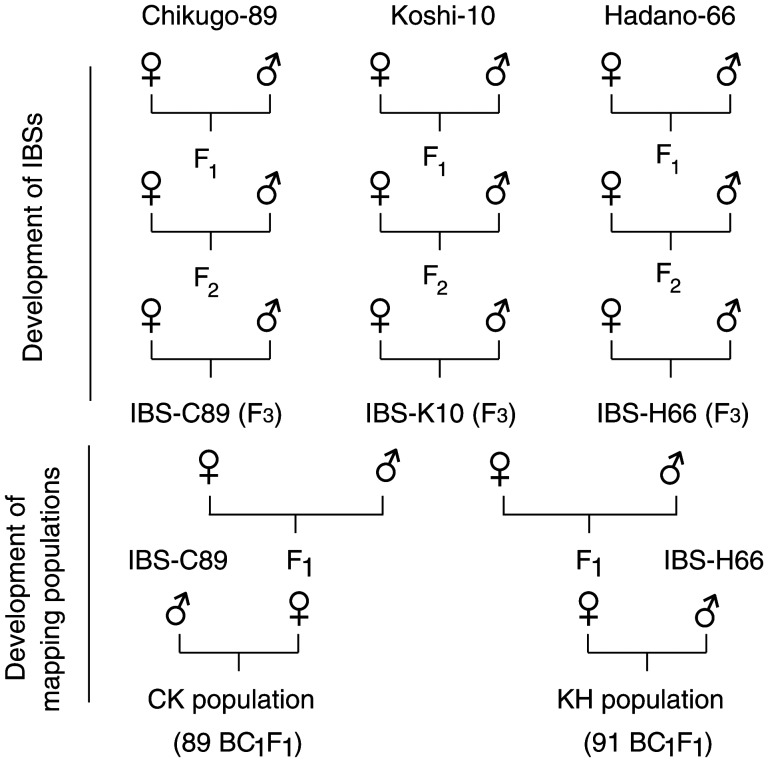
Four BPH strains collected in Japan were used in this study. (i) Hadano-66 was collected in Hadano, Kanagawa Prefecture, in 1966 and was maintained in a laboratory colony for ∼500 generations, (ii) Chikugo-89 was collected in Chikugo, Fukuoka Prefecture, in 1989 and was maintained in a laboratory colony for more than 250 generations, (iii) Koshi-10 was collected in Koshi, Kumamoto Prefecture, in 2010 and was maintained for ∼20 generations in a laboratory colony and (iv) Izumo-87 was collected in Izumo, Shimane Prefecture, in 1987 and was maintained in a laboratory colony for ∼270 generations. These BPH strains immigrated into Japan from East Asian regions.21,22 The Izumo-87 strain was used for whole-genome sequencing to generate a reference sequence12 and detect SNPs. Hadano-66, Chikugo-89, and Koshi-10 strains were used to generate the inbred BPH strains (IBSs) and the mapping populations. One pair of insects from each BPH strain was selected for crossing to generate the IBSs H66 for Hadano-66, C89 for Chikugo-89, and K10 for Koshi-10. The IBSs were prepared without outcrossing through three generations of single-pair, full-sibling mating (Fig. 1). All BPH strains were reared on seedlings of cv. ‘Reiho’, a japonica rice variety, and maintained in the laboratory at 25 ± 2°C under a 16L:8D photoperiod. To generate the mapping populations, a pair of insects from two different IBSs was selected and crossed using single-pair mating. Two backcross populations (BC1F1) were generated from crosses (♀ × ♂) of K10 × H66 with backcrossing with H66 (referred to as the KH population) and C89 × K10 with backcrossing with C89 (referred to as the CK population).
Figure 1.
Mating scheme of an experiment perform to obtain IBSs and mapping populations.
The crosses were performed by placing a virgin female and a male from each IBS on ‘Reiho’ seedlings in a glass tube. After mating and oviposition, the parental insects were collected and stored in 95% ethanol. The F1 offspring from each cross was cultured in separate containers. Newly hatched nymphs of F1 progeny from each cross were transferred into a plastic 800-ml container (dimensions: 8.5 cm wide by 16 cm tall) containing 8–10 seedlings and reared up to the fifth-instar nymph stage. An individual fifth-instar nymph of each sex was selected and reared in a separate glass tube to obtain unmated adults. F1 females were backcrossed with their parent family to generate the BC1F1 generation (Fig. 1). BC1F1 offspring from each cross were reared up to the second- and third-instar nymph stages. Although it is not possible to sex the insects at this stage, using early instars greatly accelerates the analytical process and avoids viability problems that may occur at later stages. All offspring were then collected and stored in 95% ethanol. We randomly selected 91 KH offspring and 89 CK offspring for the linkage analysis. All crossing experiments were conducted in the insectary at the Kyushu Okinawa Agricultural Research Center, Kumamoto, Japan.
2.2. Total DNA extraction and whole-genome amplification
Genomic DNA of the preserved samples (grandparents, parents, and offspring) was isolated individually using a modified version of the potassium acetate procedure of Dellaporta et al.23 For the adult insects, only the head and thorax were used for DNA extraction to reduce the possibility of contamination by fertilized eggs and bacterial symbionts.24 The whole-genome amplification technique25 was used to produce large amounts of DNA from individual insects for large-scale genotyping.
2.3. Microsatellite isolation, sequencing, and primer design
Two SSR-enrichment genomic libraries were prepared from Hadano-66 genomic DNA that was extracted from ∼50 insects, including nymphs, adults, males, and females. The first library (referred to as the SREL library) was produced by the Savannah River Ecology Laboratory (University of Georgia), following the method of Glenn and Schable.26 Briefly, 2 μg of BPH genomic DNA was digested with RsaI. The digested DNA was ligated with double-stranded SuperSNX linkers (5′–GTTTAAGGCCTAGCTAGCAGAATC–3′/5′–GATTCTGCTAGCTAGGC CTTAAACAAAA–3′). The linker-ligated DNA was hybridized with seven biotinylated oligo probes: (AG)12, (TG)12, (AAC)6, (AAG)8, (AAT)12, (ACT)12, and (ATC)8. The DNA fragments with a microsatellite repeat were captured using Dynabeads (Dynal, Oslo, Norway) under a magnetic field. The amount of eluted DNA was increased by using the polymerase chain reaction (PCR) to recover enriched DNA fragments using the SuperSNX primer pairs.
The fast isolation by amplified fragment length polymorphism of sequences containing repeats (FIASCO) procedure described by Zane et al.27 was used to isolate the second library (referred to as the KU library) at the Plant Breeding Laboratory (Kyushu University). Briefly, genomic DNA was digested with MseI and ligated to MseI adaptors (5′–TACTCAGGACTCAT–3/5–GACGATGAGTCCTGAG–3′). The mixture was amplified with an MseI adaptor primer (5′–GATGAGTCCTGAGTAAN–3′). The PCR product was then hybridized with six biotinylated probes: (CT)10, (CA)10, (CAC)7, (AAG)7, (TCC)7, and (GACA)5. The DNA molecules that were hybridized to biotinylated probes were captured using streptavidin magnetic particles (Roche Applied Science, Mannheim, Germany) using a magnetic field. PCR was performed to recover enriched DNA fragments using the MseI adaptor primer.
The SREL and KU libraries were sequenced using the Roche 454 GS FLX Titanium system (454 Life Sciences, Branford, CT, USA). SSRs in the read data were detected using a Perl script in the Simple Sequence Repeat Identification Tool (SSRIT),28 with minimum thresholds of nine, six, five, five, five, and five repeat units for di-, tri-, tetra-, penta-, hexa-, and heptanucleotide repeats, respectively. The terminal sequences derived from the ligated adapters were eliminated in both directions. SSR regions were masked by continuous N letters with the same length as the detected SSRs. The masked sequences were then de novo assembled using the Newbler software (454 Life Sciences). A flanking sequence of an appropriate length for primer design was calculated using a Perl script and Primer3.29
To develop e-SSR markers, we obtained the EST sequences contributed by Noda et al.12 from the NCBI database (http://www.ncbi.nlm.nih.gov/). The EST sequences were analysed for SSR discovery using SSRIT with minimum thresholds of six, five, five, three, and three repeat units for di-, tri-, tetra-, penta-, and hexanucleotide SSRs, respectively. All EST sequences that contained SSRs were assembled using the CodonCode Aligner software (CodonCode Corporation, http://www.codoncode.com/). Primer3 was used to design the primers. We designed 341 primer pairs, but only 50 primer pairs were randomly selected and used for the polymorphism test.
The newly developed g-SSR and e-SSR markers were named using the prefixes NLGS (Nilaparvata lugens g-SSR) and NLES (Nilaparvata lugens e-SSR), respectively, followed by the number of the marker. For the primers from Jing et al.,17 we retained the original published names. Primer functionality and polymorphism were tested on the parents of the backcross populations. Only clearly polymorphic markers were employed to genotype individuals from the mapping populations. The PCR products were electrophoresed on 4% agarose gels in 0.5× Tris–borate–EDTA buffer, stained with ethidium bromide, and visualized under ultraviolet light. All PCR conditions are summarized in Supplementary Tables S1–S3.
2.4. SNP discovery and analysis
SNPs were discovered by comparing whole-genome sequence data from two BPH strains, Izumo-87 (I87)12 and IBS-C89. The reference genomic sequences of I87 were acquired using the Roche 454 GS FLX pyrosequencing platform (H. Noda, National Institute of Agrobiological Sciences, unpublished data). To obtain reference genomic contigs, the short-read next-generation sequencing data were subjected to de novo assembly using the Celera Assembler software (v6.1).30 The genome sequence of IBS-C89 was performed using the Illumina HiSeq 2000 platform, and a total of about 77.3 million reads were generated. The read data were mapped to reference genomic contigs using the BWA software (v0.6.1).31 SNPs between I87 and IBS-C89 were called using the SAMtools/BCFtools software (v0.1.12a)32 and visualized using the Tablet software (v1.12).33 We randomly selected 234 SNPs that were homogeneous within the strain. PCR primers were designed to amplify 500- to 600-bp fragments carrying each potential SNP. The fragments were sequenced using a 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA) to screen for polymorphic SNPs between the reference strains (I87 and IBS-C89) and two of the parents of the KH mapping populations (K10 and H66). The selected polymorphic SNPs were used to genotype individual offspring of the KH population. The developed SNP markers were named using the prefixes NLSP (Nilaparvata lugens SNP), followed by the number of the marker. The primers and the PCR conditions are summarized in Supplementary Table S4.
2.5. Genetic linkage analysis and map construction
A genetic linkage map of each mapping population was analysed using the JoinMap software (v4.1).34 All markers were tested for significant deviation from a normal Mendelian segregation ratio to assess the goodness-of-fit to the expected ratios of 1:1, 1:1:1:1, and 1:2:1 for each marker at a 5% significance level. The ‘outcross pollinated’ (CP) option was used as the population type in this analysis. Grouping of markers was performed using a minimum-independence logarithm of odds (LOD) threshold of 5.0. Groups were converted into maps using the regression algorithm provided by JoinMap with a recombination frequency smaller than 0.45 used for the linkages, and then Kosambi's mapping function35 was used to calculate map distances. Distorted markers were retained for mapping, if their presence did not alter the surrounding marker order in a given linkage group (LG).
On completion of the linkage maps for KH and CK, the individual maps were integrated into a consensus map by applying the ‘combine groups for map integration’ module of JoinMap. An integrated map was created by merging bridging-SSR markers in homologous LGs into a single consensus map. Linkage maps were drawn using the MapChart software for Windows.36 The expected genome coverage was estimated according to the method of Chakravarti et al.,37 in which the length of each LG was multiplied by (m + 1)/(m − 1), where m is the marker number on each LG.
3. Results
3.1. SSR development and genotyping
SSR development and primer design: High-throughput sequencing by the 454 GS FLX Titanium generated ∼23.6 Mbp of read data with 87 068 reads from the SREL library. The sequence data were de novo assembled into 1906 contigs. After the assembly, the contigs had an average length of 303 bp. Among the 1906 unique sequences, 970 had flanking regions that were large enough for primer design. For the KU library, ∼24.1 Mbp of read data were generated. From 83 630 reads, 4490 unique contigs were produced, with an average length of 314 bp after the assembly. Of the 4490 contigs, 2556 had flanking regions that were large enough for primer design. Finally, 860 and 2300 primer pairs were designed from the read data for the SREL and KU libraries, respectively.
SSR polymorphism: A total of 3160 g-SSR primer pairs from the SSR enrichments, 50 newly developed e-SSRs, and 351 publicly available e-SSRs were used to screen for polymorphisms in the parents of the KH and CK populations. Of the 3160 g-SSR markers, 2048 (64.8%) were successfully amplified by PCR. Of these 2048 SSR primer pairs, 922 (45.0%) amplified fragments were polymorphic between the parents. From these polymorphic g-SSRs, only 418 (45.3%) were informative in the mapping populations. The remaining 504 (54.7%) polymorphic markers could not be scored, mostly due to banding patterns that were too difficult to interpret, including multiple-banding patterns, bands that amplified poorly in the offspring, and bands that did not show clear polymorphism on the 4% agarose gel. Of the 50 newly developed e-SSR markers, 15 (30.0%) amplified fragments were polymorphic between the parents. Of the 351 published e-SSR markers that we tested, 317 (90.3%) were successfully amplified by PCR and 100 of the 317 (31.5%) showed polymorphism. Of the total of 1037 polymorphic markers, including g-SSRs, public e-SSRs, and newly developed e-SSRs, only 408 and 213 primer pairs were selected to analyse offspring of the KH and CK populations, respectively. Details of the polymorphic SSR markers are summarized in Supplementary Tables S1–S3.
3.2. SNP discovery and genotyping
Of the 234 primer pairs designed to amplify SNP target sequences, 164 (70.1%) were successfully used to amplify a single amplicon of the expected size. Among them, 79 loci (48.2%) were polymorphic between the survey strains (I87 and IBS-C89), and 48 loci (29.3%) were polymorphic between the parents of the KH population. Six SNP markers that were not heterozygous in the offspring of the KH population were excluded from the analysis, leaving 42 loci. After adding 4 SNPs from a preliminary experiment, 46 SNPs were selected and used to genotype the offspring of the KH mapping population. The SNP markers and PCR conditions are summarized in Supplementary Table S4.
3.3. Linkage analysis and map construction
Individual genetic linkage maps: The polymorphic markers used for linkage map construction consisted of e-SSR, g-SSR, SNP, and sequence-tagged site (STS) markers. Based on the results of the LG analysis, we constructed individual linkage maps for the KH and CK populations. The KH linkage map was developed from g-SSR, e-SSR, and SNP markers with an LOD score of 5.0. The map consisted of 18 LGs with 357 unique positions (excluding duplicated markers), derived from 348 SSR and 44 SNP markers and covered 870.2 cM. The average distance between the markers was 2.8 cM (Supplementary Table S5, Fig. S1). The LGs ranged in length from 3.8 to 83.2 cM. Of these markers, 21 were unlinked or unsuccessfully positioned. Of the 21 unmapped markers, 13 were not linked to another marker and the remaining 8 markers were assigned to LGs, but no map positions were determined. Chi-square analysis indicated that more than 10% of the markers deviated from Mendelian segregation at P < 0.01.
Only 180 g-SSR markers, 32 e-SSR markers, and 1 STS marker that showed polymorphism among the parents (for a total of 213 markers) were selected to analyse the offspring of the CK population. A linkage map was constructed with a recombination threshold of 0.45, and the minimum number of markers per group was set to two. Linkage analysis was performed at a minimum LOD of 5.0. The linkage map consisted of 207 marker loci in 16 LGs that covered 770.7 cM. (Supplementary Table S6, Fig. S1). The LGs ranged in length from 1.4 to 85.9 cM, with an average distance of 4.4 cM between markers. We positioned 43 distorted loci (20.8%) in LGs 1 to 15. The majority was clustered in LG16. We found four unlinked markers and excluded two other distorted markers from the analysis. The Y chromosomal marker PMn3 was mapped at the end of the LG arm next to NLGS201 in LG16 of the CK population.
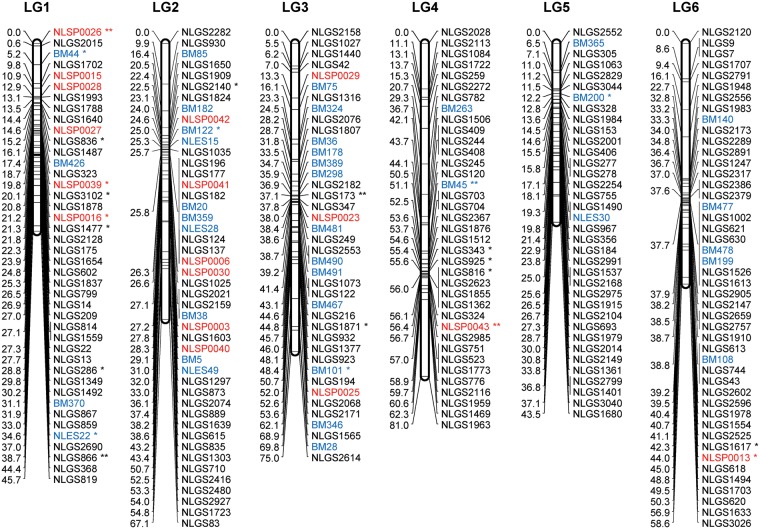
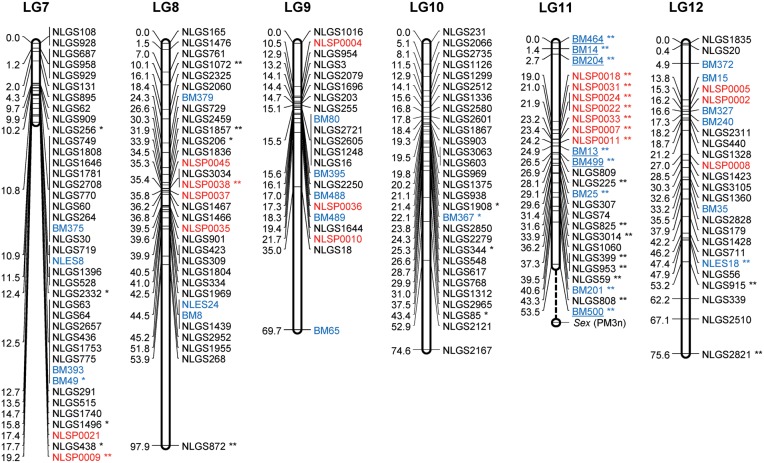
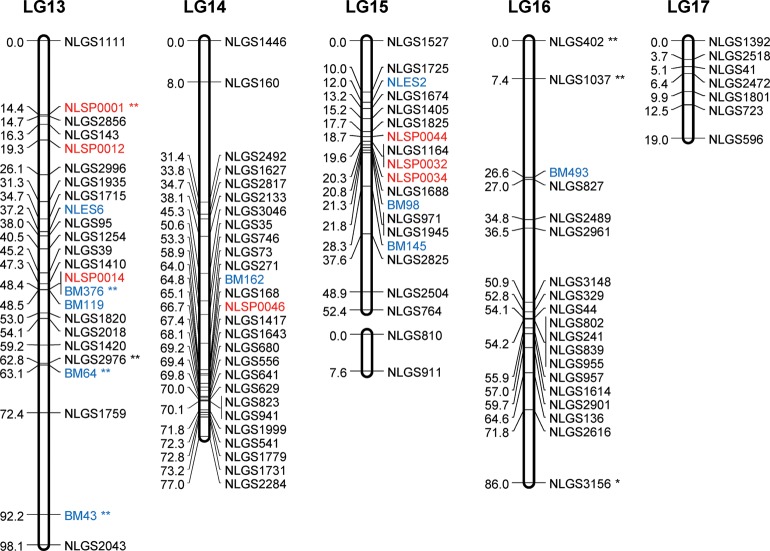
Consensus linkage map: The consensus map was constructed by integrating linkage data from the two backcross populations (KH and CK). The ‘combine groups for map integration’ module of JoinMap was used to integrate the two individual maps into a consensus map. A total of 73 common SSR markers between the homologous LGs were used as bridges to integrate the individual maps. The order of the bridged markers was consistent between the two individual maps, except for 16 marker inversions in LG2 to LG8, in LG10, and in LG12 to LG13 (Supplementary Fig. S1). The final consensus map contained 400 g-SSR, 74 e-SSR, 43 SNP, and 1 STS markers (for a total of 518), with 472 unique positions in 17 LGs. Table 1 summarizes the statistics for the consensus LGs, and Supplementary Table S7 reports the locus name and its position for all loci in the 17 LGs in the consensus map. The length of the 17 LGs ranged from 19.0 cM (LG17) to 98.1 cM (LG13). The average genetic distance between loci was 2.3 cM. Gaps larger than 20 cM were found in LG 8 to LG11 and in LG14 (Fig. 2). The largest gap in the framework map was 43.9 cM and occurred in LG8. The large gaps observed in the individual maps were reduced in the consensus map. In total, 30 markers remained unlinked. The number of integrated (consensus) LGs was close to the actual number of haploid chromosomes in BPH (n = 15). The 17 LGs contained 518 loci that encompassed 1093.9 cM of the total map distance (excluding the distance from PM3n to the distal end of LG11) and covered 92.3% of the estimated genome size. According to the estimated genome size of BPH (1200 Mbp),11 the recombination rate is ∼0.91 cM/Mbp.
Table 1.
Description of the integrated consensus map
| LG | Total length (cM) | No. of loci | No. of markers | Average distance (cM) | No. of distorted markers (P < 0.01) | Estimated genome size (cM) | Genome coverage (%) | Largest interval (cM) | No. of bridging markers | DNA markers |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| e-SSR | g-SSR | SNP | STS | ||||||||||
| LG1 | 45.7 | 42 | 42 | 1.1 | 8 | 48.0 | 95.3 | 5.7 | 3 | 4 | 32 | 6 | 0 |
| LG2 | 67.1 | 37 | 46 | 1.8 | 2 | 70.1 | 95.7 | 12.3 | 7 | 10 | 30 | 6 | 0 |
| LG3 | 75.0 | 39 | 40 | 1.9 | 3 | 78.8 | 95.1 | 8.5 | 6 | 13 | 24 | 3 | 0 |
| LG4 | 81.0 | 32 | 37 | 2.5 | 3 | 85.5 | 94.7 | 18.7 | 7 | 2 | 34 | 1 | 0 |
| LG5 | 43.5 | 35 | 36 | 1.2 | 1 | 46.0 | 94.6 | 6.5 | 10 | 3 | 33 | 0 | 0 |
| LG6 | 58.6 | 42 | 46 | 1.4 | 2 | 61.2 | 95.7 | 10.1 | 5 | 5 | 40 | 1 | 0 |
| LG7 | 19.2 | 24 | 40 | 0.8 | 6 | 20.2 | 95.1 | 5.4 | 6 | 4 | 34 | 2 | 0 |
| LG8 | 97.9 | 29 | 32 | 3.4 | 5 | 104.2 | 93.9 | 44.0 | 2 | 3 | 25 | 4 | 0 |
| LG9 | 69.7 | 20 | 22 | 3.5 | 0 | 76.4 | 91.3 | 34.8 | 0 | 5 | 14 | 3 | 0 |
| LG10 | 74.6 | 29 | 29 | 2.6 | 4 | 79.9 | 93.3 | 21.7 | 8 | 1 | 28 | 0 | 0 |
| LG11 | 53.5 | 27 | 27 | 2.0 | 22 | 57.8 | 92.6 | 16.3 | 3 | 8 | 11 | 7 | 1 |
| LG12 | 75.6 | 26 | 26 | 2.9 | 3 | 81.6 | 92.6 | 9.0 | 5 | 6 | 17 | 3 | 0 |
| LG13 | 98.1 | 23 | 24 | 4.3 | 5 | 106.6 | 92.0 | 19.8 | 3 | 5 | 16 | 3 | 0 |
| LG14 | 77.0 | 26 | 27 | 3.0 | 0 | 82.9 | 92.9 | 23.4 | 3 | 1 | 25 | 1 | 0 |
| LG15 | 52.4 | 18 | 18 | 2.9 | 0 | 58.5 | 89.5 | 11.4 | 3 | 3 | 12 | 3 | 0 |
| LG16 | 86.0 | 16 | 19 | 5.4 | 3 | 95.6 | 90.0 | 19.2 | 2 | 1 | 18 | 0 | 0 |
| LG17 | 19.0 | 7 | 7 | 2.7 | 0 | 25.3 | 75.0 | 6.5 | 0 | 0 | 7 | 0 | 0 |
| Total | 1093.9 | 472 | 518 | 67.0 | 1178.6 | 273.0 | 73 | 74 | 400 | 43 | 1 | ||
| Average | 64.3 | 27.8 | 30.5 | 2.3 | 3.9 | 69.3 | 92.3 | 16.1 | 4.3 | 4.4 | 23.5 | 2.5 | 0.1 |
Figure 2.
A genetic linkage map of BPH. The LGs were constructed by integrating linkage data from two backcross populations (KH and CK). Grouping of markers was performed using an independence LOD threshold of five. Groups were converted to maps using the regression algorithm provided by JoinMap, with a recombination frequency smaller than 0.45 and Kosambi's mapping function applied for calculation of map distances. The LGs have been arbitrarily numbered LG1–LG17 based on the order generated by JoinMap (v4.1). Distances in centimorgans are indicated at the left of each LG. Marker names starting with NLGS (black) represent g-SSRs, those starting with NLES and BM represent e-SSRs (blue), and those starting with NLSP (red) represent SNP markers. PM3n is an STS marker located on the Y chromosome. A gap between the marker BM500 and PM3n on the LG11 is expected to include the differentiated region of the Y chromosome. e-SSR markers BM13, BM14, BM204, BM464, BM499, and BM500 are for vitellogenin genes that are genes with female-specific expression located on the X chromosome. Segregation-distorted loci indicate different significant levels; *P < 0.01 and **P < 0.001.
3.4. Identification of markers linked with the sex chromosomes
We found that most SSR and SNP markers in LG16 of the CK population deviated significantly from the expected Mendelian segregation ratio of 1:2:1 (homozygous parental: heterozygous: homozygous parental). The segregation of all marker loci in LG16 was exploited to distinguish the X chromosome from the autosomes. If the genotypes of the F1 maternal and BC1 paternal parents are designated as X1X2 and X1Y1, respectively, there would be four equal proportions of BC1F1 genotypes in the offspring in which the females are X1X1 or X1X2 and the males are X2Y1 or X1Y1. In addition, a ratio of 2:1:1 [homozygous/hemizygous paternal bands (X1X1/X1Y1): heterozygous (X1X2): homozygous maternal band (X2Y1)] would be expected from the amplified DNA bands. The Y-specific dominant marker PM3n, which was informative in the CK population segregated in a 1:1 ratio, suggested that the CK population had an equal number of males (43) and females (44) in the BC1F1 generation. The observed ratio for all SSR markers in LG16 of CK population matched the expected ratio of 2:1:1, suggesting that these markers are located on the X chromosome (Table 2).
Table 2.
Segregation of marker loci in the X-LGs of the CK and KH populations
| Marker locus | Banding pattern |
No. of insects |
P-value |
|||
|---|---|---|---|---|---|---|
| X1X1/X1X2 | X2Y1 | X1Y1 | 1:2:1a | 2:1:1b | ||
| CK population (codominant marker) | ||||||
| NLGS225 | 39 | 26 | 21 | 86 | <0.001** | 0.52c |
| BM13 | 40 | 28 | 20 | 88 | <0.001** | 0.34c |
| BM25 | 38 | 28 | 22 | 88 | <0.001** | 0.29c |
| NLGS3014 | 40 | 24 | 23 | 87 | <0.001** | 0.75c |
| NLGS953 | 35 | 29 | 25 | 89 | <0.001** | 0.11c |
| NLGS825 | 38 | 28 | 23 | 89 | <0.001** | 0.29c |
| BM201 | 37 | 27 | 25 | 89 | <0.001** | 0.27c |
| KH population (codominant marker) | ||||||
| NLSP0007 | 45 | 18 | 24 | 87 | <0.001** | 0.63c |
| NLSP0011 | 45 | 21 | 24 | 90 | <0.001** | 0.90c |
| NLSP0018 | 50 | 14 | 24 | 88 | <0.001** | 0.14c |
| NLSP0022 | 48 | 19 | 22 | 89 | <0.001** | 0.69c |
| NLSP0024 | 45 | 20 | 23 | 88 | <0.001** | 0.88c |
| NLSP0031 | 49 | 18 | 24 | 91 | <0.001** | 0.51c |
| NLSP0033 | 43 | 19 | 23 | 85 | <0.001** | 0.82c |
| BM464 | 50 | 20 | 21 | 91 | <0.001** | 0.63c |
| BM204 | 49 | 20 | 22 | 91 | <0.001** | 0.73c |
| BM499 | 47 | 18 | 26 | 91 | <0.001** | 0.47c |
| BM500 | 55 | 17 | 18 | 90 | <0.001** | 0.11c |
| BM13 | 45 | 20 | 24 | 89 | <0.001** | 0.83c |
| KH population (dominant marker) | 3:1a | 3:1b | ||||
| NLGS74 | 68 | 23 | 91 | 1.00c | 1.00c | |
| NLGS225 | 66 | 25 | 91 | 0.96c | 0.96c | |
| NLGS307 | 64 | 27 | 91 | 0.97c | 0.97c | |
| NLGS399 | 64 | 27 | 91 | 0.94c | 0.94c | |
| NLGS809 | 66 | 22 | 88 | 0.98c | 0.98c | |
| NLGS1060 | 67 | 23 | 90 | 1.00c | 1.00c | |
| NLGS3014 | 62 | 27 | 89 | 0.80c | 0.80c | |
| NLGS59 | 60 | 30 | 90 | 0.90c | 0.90c | |
| NLGS808 | 61 | 29 | 90 | 0.73c | 0.73c | |
| BM14 | 50 | 35 | 85 | 0.23c | 0.23c | |
aExpected ratio for autosomes.
bExpected ratio for the X chromosome.
cNon-significant deviation from the expected ratio.
**Significant at the 0.1% level.
Based on LG11 in the integrated map, the bridged markers in LG16 of the CK population were located in LG11 of the KH population. Of the 22 markers in LG11 of the KH population, 12 loci were codominant markers. Because the PM3n marker was not informative for the KH population, the sexes of BC1F1 were not determined. If we assume that the KH population had an equal number of males and females in the BC1F1 generation, the ratio of 2:1:1 would be expected for codominant markers. The codominant markers, including seven SNPs and five SSRs, segregated in a 2:1:1 ratio of homozygous/hemizygous paternal bands: heterozygous: homozygous maternal band in the KH offspring. Using 4% agarose gel to visualize the amplified fragments, we could not distinguish between heterozygous and homozygous genotypes for the paternal genotype of the 10 SSR markers in LG11. These markers were dominant markers, and segregated in a 3:1 ratio for all paternal band types: homozygous maternal band types. These loci also segregated in the ratio expected for a sex-linked marker (Table 2).
4. Discussion
4.1. Development and analysis of SSR markers
Early research focused on the development of SSR markers from BPH genomic DNA.38 Because of a lack of appropriate strategies at that time, few markers were developed. Later, after an enormous number of BPH ESTs became available,12 more than 350 e-SSR markers were developed to support genetic studies.17–19 Recently, 30 new g-SSR markers were developed via the FIASCO method.20 In the present study, we combined enrichment-based SSR isolation with next-generation sequencing approaches and demonstrated that this approach was a powerful tool for developing SSR markers from genomic DNA. Through these strategies, we developed 3160 g-SSR markers, of which 922 (29.2%) were successfully amplified and showed polymorphism among the BPH strains used as parents for the mapping. As the polymorphisms were detected on a simple agarose gel, only 418 SSR markers (45.3%) were informative. The remaining 54.7% were not informative, mostly due to banding patterns that were too difficult to interpret or bands that were too close together. The number of informative markers could be increased using more powerful fragment separation systems such as fluorescently labelled-fragment capillary arrays.
The newly developed e-SSR primers in this study were less polymorphic (30.0%) when compared with the g-SSRs (45.0%). Likewise, the polymorphisms of the publicly available e-SSRs also were <32%. It might be because of greater DNA sequence conservation in transcribed regions. However, e-SSR markers produced a high proportion of high-quality markers with stronger and clearer amplified bands.
4.2. Development and analysis of SNP markers
We found a large number of candidate SNPs (>100 000) between the two survey strains in the next-generation sequencing data. However, the frequency of valid SNPs for linkage analysis was low because of several difficulties in developing informative markers using the strategies described in this paper. The major reason for these difficulties was the lack of a reliable reference genome sequence for BPH. The draft assembly of the BPH genome contains a considerable number of intraspecies sequence polymorphisms, leading to insufficiently precise calling of SNPs. Although generating pure IBSs is an effective strategy for reducing heterogeneity, repetitive inbreeding is not easy because it decreases the fecundity and fitness of the BPH populations (T. Kobayashi, unpublished data). Differences between the strains used for the SNP survey (I87 vs. IBS-C89) and the linkage analysis (K10 vs. H66) also reduced the number of valid SNPs. To increase SNP markers in this species, high-quality reference genome sequences and a better strategy to evaluate potential SNPs will be necessary.39
4.3. Genetic linkage map
Based on the results of our study, we have constructed an SSR- and SNP-based linkage map for BPH. The genetic map consists of 518 markers that cover 1093.9 cM, with an average marker spacing of 2.3 cM. The majority of the mapped e-SSR and SNP markers were widely distributed among several LGs. Disagreement in the number of LGs between the two populations (KH and CK) might have resulted from the large portion of markers in some LGs (LG8 and LG11) and the existence of one small LG (LG17), possibly due to the relatively small number of BC1F1 individuals (<100) used for mapping and the lack of informative markers among LGs belonging to the same chromosome.40,41 The quality and accuracy of marker order in such maps depend on numerous factors, including segregation distortion, the population size, and scoring errors. In some cases, the quality can be improved and the problem can be solved.42 Numerous examples of a high level of deviations from Mendelian segregation ratios have been reported in previous studies (Bombyx mori,43 Chlamys farreri,44 and Crassostrea gigas45). In our study, segregation distortion (P < 0.01) was observed for 8.7% of the total marker loci that we analysed. This may have several possible explanations, including inbreeding depression,46 sex-biased ratio distortion,47 erroneous scoring, or linkage between molecular markers and distorting factors such as recessive lethal genes or incompatible alleles.48
Population size is one of the factors associated with the accuracy of detecting recombination events, and it can affect the efficiency of the mapping process.49 Because we used small population sizes, inaccurate marker order may have occurred due to inversions and marker distance. However, numerous linkage maps have been successfully produced from small mapping populations (50–94 individuals) in several species.40,50–52 To obtain accurate estimates of marker positions in the map, we only selected clearly polymorphic markers and carefully scored them to reduce errors and ensure the accuracy of the linkage map. Nonetheless, inversions of some markers were observed in several LGs of the consensus map (Supplementary Fig. S1). A more accurate marker order can be acquired by means of map validation, when a draft genome sequence for BPH becomes available.
Recombination frequencies in mapping populations of some vertebrate and arthropod species show distinct differences between males and females.53–57 Although the mechanism responsible for these differences in recombination rates between the sexes is not well understood, the phenomenon is common. The conventional way to construct a linkage map is usually to analyse the two sexes separately. Because we collected the offspring of the mapping populations during their early growth stages, the sex of the offspring could not be identified. In addition, the number of females and males in the populations was too small to enable mapping separately within each sex. Consequently, we included both sexes in our linkage analysis to produce a single map. Developing separate linkage maps for the two sexes in future mapping experiments will provide a better understanding of differences in the recombination rates between the sexes of BPH.
Mapping with multiple populations is advantageous because a larger number of markers can be placed on a single map, thereby providing greater genomic coverage. Integrated linkage maps have been constructed in various plant and animal species, including insects.52,58–60 We combined two genetic linkage maps from different genetic backgrounds derived from three IBSs that have different virulence against rice differential varieties to BPH,61 into one integrated map. The numbers of markers and the genomic coverage were increased by this approach, and the gaps between markers were reduced in the integrated map. However, the marker density of the final linkage map was still low when compared with the denser linkage maps of other model insect species (e.g. B. mori57,62 and Apis mellifera59). The construction of a highly saturated linkage map with full genome coverage will be possible in future research with more SNP markers. Nevertheless, the new SSR markers and the consensus map developed in the present study provide useful genetic and genomic tools for molecular analysis of the BPH genome and will facilitate the construction of a higher density second-generation linkage map.
4.4. Determination of the sex-LG
As we could not identify the sex of individual nymphs in our mapping populations, we used the Y-specific marker PM3n63 as a sex-determining locus (Sex). In the consensus map, the putative location of PM3n was mapped at the distal end of LG11. Additional evidence to confirm the sex-LG would require X-linked loci. Because the e-SSRs were developed from the transcribed regions of the genome, mapping e-SSR markers directly shows the location of the corresponding genes within the linkage map. Vitellogenin genes exhibit female-specific expression and are generally located on the X chromosome in various organisms, including insects.64–68 An interesting feature of the present map is that six e-SSRs derived from vitellogenin genes17 were gathered in only LG11 of the consensus map. We tested the segregation of all marker loci on LG11 and found that all the markers fitted the segregation ratio expected for genes located on the X chromosome. The results indicated that LG11 corresponds to the X-LG in BPH. The X-linked markers would be useful in future genetic analysis of the sex chromosome.
4.5. Conclusions and future perspectives
In this paper, we report the development of g-SSR, e-SSR, and SNP markers and present a preliminary genetic map for BPH based on these markers. Two individual linkage maps from the KH and CK populations were integrated into a consensus map to increase locus density and provide more dense coverage of the genome. The consensus map consisted of 518 markers that covered a total of 1093.9 cM. Denser coverage is needed to reduce the gaps and group the smallest LGs so that the consensus map matches the actual number (15) of chromosomes. Additional SNP identification and mapping should be performed in future research. It should be possible to extend the genome coverage in this manner. The mean marker distance of 2.3 cM in the consensus map may offer enough resolution for the genetic dissection of QTLs for agriculturally important traits, and particularly the virulence of BPH against the host plant. An understanding of the genetics responsible for virulence is a crucial aspect for future management of BPH. In addition, the newly developed markers can be applied in various studies of the genetics and ecology of this species at a molecular level (e.g. population genetics, migration sources, and the evolution of BPH).
The primer information is available at http://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?class=unka. All the sequence data used for the generation of the SSR markers and the whole-genome sequence data were submitted to the DDBJ BioProject Database under ID PRJDB683.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Grant-in-Aid for fellows of the Japan Society for the Promotion of Science (No. P10400) and the NIAS Strategic Research Fund. This work was also partially supported by the Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 23580009). A Postdoctoral Fellowship for Foreign Researchers was awarded to J.J.
Supplementary Material
Acknowledgements
We thank the Savannah River Ecology laboratory (The University of Georgia) for preparing the enriched genomic DNA. We thank Dr S. Fukuoka and Ms T. Ando (National Institute of Agrobiological Sciences) for technical support and SSR genotyping. We are grateful to Prof. A. Yoshimura (Kyushu University) for his highly valuable suggestions on linkage map construction. Many thanks to our colleagues at the Plant Breeding laboratory (Kyushu University), the staff of the Kyushu Okinawa Agricultural Research Center, and the National Institute of Agrobiological Sciences for their assistance and encouragement.
Footnotes
Edited by Dr Satoshi Tabata
References
- 1.Bottrell D.G., Schoenly K.G. Resurrecting the ghost of green revolutions past: the brown planthopper as a recurring threat to high-yielding rice production in tropical Asia. J. Asia-Pacific Entomol. 2012;15:122–40. [Google Scholar]
- 2.Catindig J.L.A., Arida G.S., Baehaki S.E., et al. In: Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia. Heong K.L., Hardy B., editors. Los Baños, Philippines: IRRI; 2009. pp. 191–220. [Google Scholar]
- 3.Heong K.L. Are planthopper problems caused by a breakdown in ecosystem services? In: Heong K.L., Hardy B., editors. Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia. Los Baños, Philippines: IRRI; 2009. pp. 221–31. [Google Scholar]
- 4.Gallagher K.D., Kenmore P.E., Sogawa K. Judicial use of insecticides deter planthopper outbreaks and extend the life of resistant varieties in Southwest Asian rice. In: Denno R.F., Perfect T.J., editors. Planthoppers: Their Ecology and Management. New York: Chapman and Hall; 1994. pp. 599–614. [Google Scholar]
- 5.Cabauatan P.Q., Cabunagan R.C., Choi I.R. Rice viruses transmitted by the brown planthopper Nilaparvata lugens Stål. In: Heong K.L., Hardy B., editors. Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia. Los Baños, Philippines: IRRI; 2009. pp. 357–68. [Google Scholar]
- 6.Hare J.D. Status and prospects for an integrated approach to the control of rice planthoppers. In: Denno R.F., Perfect T.J., editors. Planthoppers: Their Ecology and Management. New York: Chapman and Hall; 1994. pp. 615–32. [Google Scholar]
- 7.Tanaka K. Quantitative genetic analysis of biotypes of the brown planthopper Nilaparvata lugens: heritability of virulence to resistant rice varieties. Entomologia Experimentalis et Applicata. 1999;90:279–87. [Google Scholar]
- 8.Den Hollander J. The chromosomes of Nilaparvata lugens Stål. and some other Auchenorrhyncha. Cytologia. 1982;47:227–36. [Google Scholar]
- 9.Saxena R.C., Barrion A.A. Brain cells and chromosomes of the brown planthopper Nilaparvata lugens (Stål) Cytologia. 1987;52:859–65. [Google Scholar]
- 10.Noda H., Tatewaki R. Re-examination of chromosomes of three species of rice planthoppers (Homoptera: Delphacidae) Appl. Entomol. Zool. 1990;4:538–40. [Google Scholar]
- 11.Wang Y., Fan H.W., Huang H.J., et al. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae) Insect Biochem. Mol. Biol. 2012;42:637–46. doi: 10.1016/j.ibmb.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Noda H., Kawai S., Koizumi Y., et al. Annotated ESTs from various tissues of the brown planthopper Nilaparvata lugens: a genomic resource for studying agricultural pests. BMC Genomics. 2008;9:117. doi: 10.1186/1471-2164-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue J., Bao Y.Y., Li B., et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS One. 2010;5:e14233. doi: 10.1371/journal.pone.0014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass C., Hebsgaard M.B., Hughes J. Genomic resources for the brown planthopper, Nilaparvata lugens: transcriptome pyrosequencing and microarray design. Insect Sci. 2012;9:1–12. [Google Scholar]
- 15.Peng X., Zha W., He R., et al. Pyrosequencing the midgut transcriptome of the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2011;20:745–62. doi: 10.1111/j.1365-2583.2011.01104.x. [DOI] [PubMed] [Google Scholar]
- 16.Bao Y., Wang Y., Wu W., et al. De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics. 2012;99:256–64. doi: 10.1016/j.ygeno.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Jing S., Liu B., Peng L., et al. Development and use of e-SSR markers for assessing genetic diversity in the brown planthopper (Nilaparvata lugens Stål) Bull. Entomol. Res. 2011;102:113–22. doi: 10.1017/S0007485311000435. [DOI] [PubMed] [Google Scholar]
- 18.Abercrombie L.G., Anderson C.M., et al. Molecular Ecology Resources Primer Development Consortium. Permanent genetic resources added to molecular ecology resources database 1 January 2009–30 April 2009. Mol. Ecol. Resources. 2009;9:1375–429. doi: 10.1111/j.1755-0998.2009.02746.x. [DOI] [PubMed] [Google Scholar]
- 19.Sun J.T., Zhang Y.K., Ge C., Hong X.Y. Mining and characterization of sequence tagged microsatellites from the brown planthopper Nilaparvata lugens. J. Insect Sci. 2011;11:134. doi: 10.1673/031.011.13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jing S., Zhou X., Yu H., et al. Isolation and characterization of microsatellite markers in brown planthopper (Nilaparvata lugens Stål) Int. J. Mol. Sci. 2012;13:9527–33. doi: 10.3390/ijms13089527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K., Matsumura M. Development of virulence to resistant rice varieties in the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae), immigrating into Japan. Appl. Entomol. Zool. 2000;35:529–33. [Google Scholar]
- 22.Otuka A., Watanabe T., Suzuki Y., Matsumura M., Furuno A., Chino M. A migration analysis of the rice planthopper Nilaparvata lugens from the Philippines to East Asia with three-dimensional computer simulations. Popul. Ecol. 2005;47:143–50. [Google Scholar]
- 23.Dellaporta S.L., Wood J., Hicks J.R. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1983;1:19–21. [Google Scholar]
- 24.Tang M., Lv L., Jing S., Zhu L., He G. Bacterial symbionts of the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae) Appl. Environ. Microbiol. 2010;76:1740–5. doi: 10.1128/AEM.02240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S. Polymerase and multiply-primed rolling circle amplification rapid amplification of plasmid and phage DNA using Phi29 DNA. Genome Res. 2001;11:1095–9. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glenn T.C., Schable N.A. Isolating microsatellite DNA loci. In: Zimmer E.A., Roalson E.H., editors. Methods in Enzymology. vol. 395. San Diego, CA: Academic Press; 2005. pp. 202–22. [DOI] [PubMed] [Google Scholar]
- 27.Zane L., Bargelloni L., Patarnello T. Strategies for microsatellite isolation: a review. Mol. Ecol. 2002;11:1–16. doi: 10.1046/j.0962-1083.2001.01418.x. [DOI] [PubMed] [Google Scholar]
- 28.Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S., McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–52. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 30.Miller J.R., Delcher A.L., Koren S., et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–24. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., et al. The sequence alignment/map (SAM) format and SAM tools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milne I., Bayer M., Cardle L., et al. Tablet–next generation sequence assembly visualization. Bioinformatics. 2010;26:401–2. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Ooijen J.W. JoinMap®4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen, Netherlands: Kyazma, B.V; 2006. [Google Scholar]
- 35.Kosambi D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944;12:172–5. [Google Scholar]
- 36.Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 37.Chakravarti A., Lasher L.K., Reefer J.E. A maximum likelihood method for estimating genome length using genetic linkage data. Genetics. 1991;128:175–82. doi: 10.1093/genetics/128.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mun J., Song Y.H., Roderick G.K. Isolation and characterization of microsatellites in the brown planthopper, Nilaparvata lugenes Stål. Korea J. Appl. Entomol. 1994;43:311–5. [Google Scholar]
- 39.Nielsen R., Paul J.S., Albrechtsen A., Song Y.S. Genotype and SNP calling from next generation sequencing data. Nature Rev. 2011;12:443–51. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chistiakov D.A., Hellemans B., Haley C.S., et al. A microsatellite linkage map of the European sea bass Dicentrarchus labrax L. Genetics. 2005;170:1821–6. doi: 10.1534/genetics.104.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C.M., Zhu Z.Y., Lo L.C., et al. A microsatellite linkage map of barramundi, Lates calcarifer. Genetics. 2007;175:907–15. doi: 10.1534/genetics.106.059972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronin Y., Mester D., Minkov D., et al. Two-phase analysis in consensus genetic mapping. G3. 2012;2:537–49. doi: 10.1534/g3.112.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan Y.D., Wan C., Zhu Y., et al. An amplified fragment length polymorphism map of the silkworm. Genetics. 2001;157:1277–84. doi: 10.1093/genetics/157.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L., Xiang J.H., Liu X., Zhang Y., Dong B., Zhang X. Construction of AFLP-based genetic linkage map for Zhikong scallop, Chlamys farreri and mapping of sex-linked markers. Aquaculture. 2005;245:63–73. [Google Scholar]
- 45.Guo X., Li Q., Wang Q.Z., Kong L.F. Genetic mapping and QTL analysis of growth-related traits in the Pacific oyster. Mar. Biotechnol. 2012;14:218–26. doi: 10.1007/s10126-011-9405-4. [DOI] [PubMed] [Google Scholar]
- 46.McColdrick D.J., Hedgecock D. Fixation, segregation and linkage of allozyme loci in inbred families of the Pacific oyster Crassostrea giga (Thunberg): implications for the causes of inbreeding depression. Genetics. 1997;146:321–34. doi: 10.1093/genetics/146.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao Y., Hart D.L., Laurie C.C. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Nat. Acad. Sci. USA. 2001;98:13183–8. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyttle T.W. Cheaters sometimes prosper: distortion of mendelian segregation by meiotic drive. Trends Genet. 1993;9:205–10. doi: 10.1016/0168-9525(93)90120-7. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira A., da Silva M.F., da Costa e Silva L., Cruz C.D. Estimating the effects of population size and type on the accuracy of genetic maps. Genet. Mol. Biol. 2006;29:182–92. [Google Scholar]
- 50.Hawthorne D.J. AFLP-based genetic linkage map of the Colorado potato beetle Leptinotarsa decemlineata: sex chromosomes and a pyrethroid-resistance candidate gene. Genetics. 2001;158:695–700. doi: 10.1093/genetics/158.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z., Karsi A., Li P., Cao D., Dunham R. An AFLP-based genetic linkage map of channel catfish (Ictalurus punctatus) constructed by using an interspecific hybrid resource family. Genetics. 2003;165:687–94. doi: 10.1093/genetics/165.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobler A., Kapan D., Flanagan N.S., et al. First-generation linkage map of the warningly colored butterfly Heliconius erato. Heredity. 2005;94:408–17. doi: 10.1038/sj.hdy.6800619. [DOI] [PubMed] [Google Scholar]
- 53.Feldmeyer B., Pen I., Beukeboom L.W. A microsatellite marker linkage map of the housefly, Musca domestica: evidence for male recombination. Insect Mol. Biol. 2010;19:575–81. doi: 10.1111/j.1365-2583.2010.01016.x. [DOI] [PubMed] [Google Scholar]
- 54.Sakamoto T., Danzmann R.G., Gharbi K., et al. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics. 2000;155:1331–45. doi: 10.1093/genetics/155.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lien S., Gidskehaug L., Moen T., et al. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homologies and striking differences in sex-specific recombination patterns. BMC Genomics. 2011;12:615. doi: 10.1186/1471-2164-12-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhan X., Fan F., You W., Yu J., Ke C. Construction of an integrated map of Haliotis diversicolor using microsatellite markers. Mar. Biotechnol. 2012;14:79–86. doi: 10.1007/s10126-011-9390-7. [DOI] [PubMed] [Google Scholar]
- 57.Miao X.X., Xu S.J., Li M.H., et al. Simple sequence repeat-based consensus linkage map of Bombyx mori. Proc. Nat. Acad. Sci. USA. 2005;102:16303–8. doi: 10.1073/pnas.0507794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schafer M.A., Mazzi D., Klappert K., et al. A microsatellite linkage map for Drosophila montana shows large variation in recombination rates, and a courtship song trait maps to an area of low recombination. J. Evol. Biol. 2010;23:518–27. doi: 10.1111/j.1420-9101.2009.01916.x. [DOI] [PubMed] [Google Scholar]
- 59.Solignac M., Vautrin D., Baudry E., Mougel F., Loiseau A., Cornuet J.M. A microsatellite-based linkage map of the honeybee, Apis mellifera L. Genetics. 2004;167:253–62. doi: 10.1534/genetics.167.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong D., Pai A., Yan G. AFLP-based genetic linkage map for the red flour beetle (Tribolium castaneum) J. Heredity. 2004;95:53–61. doi: 10.1093/jhered/esh012. [DOI] [PubMed] [Google Scholar]
- 61.Myint K.K.M., Yasui H., Takagi M., Matsumura M. Virulence of long-term laboratory populations of the brown planthopper, Nilaparvata lugens (Stål), and whitebacked planthopper, Sogatella furcifera (Horváth) (Homoptera: Delphacidae), on rice differential varieties. Appl. Entomol. Zool. 2009;44:149–53. [Google Scholar]
- 62.Yamamoto K., Narukawa J., Kadono-Okuda K., et al. Construction of a single nucleotide polymorphism linkage map for the silkworm, Bombyx mori, based on bacterial artificial chromosome end sequences. Genetics. 2006;173:151–61. doi: 10.1534/genetics.105.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kobayashi T., Noda H. Identification of Y chromosomal PCR marker and production of a selected strain for molecular sexing in the brown planthopper, Nilaparvata lugens. Arch. Insect Biochem. Physiol. 2007;65:1–10. doi: 10.1002/arch.20173. [DOI] [PubMed] [Google Scholar]
- 64.Barnett T., Pachl C., Gergen J.P., Wensink P.C. The isolation and characterization of Drosophila yolk protein genes. Cell. 1980;21:729–38. doi: 10.1016/0092-8674(80)90436-5. [DOI] [PubMed] [Google Scholar]
- 65.Higgins M.J., Walker V.K., Holden J.A., White B.N. Isolation of two Drosophila melanogaster genes abundantly expressed in the ovary during vitelline membrane synthesis. Dev. Biol. 1984;105:155–65. doi: 10.1016/0012-1606(84)90271-9. [DOI] [PubMed] [Google Scholar]
- 66.Riddell D.C., Higgins M.J., McMillan B.J., White B.N. Structural analysis of the three vitellogenin genes in Drosophila melanogaster. Nucl. Acids Res. 1981;9:1323–8. doi: 10.1093/nar/9.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradfield J.Y., Wyatt G.R. X-linkage of a vitellogenin gene in Locusta migratoria. Chromosoma. 1983;88:190–3. [Google Scholar]
- 68.Heine U., Blumenthal T. Characterization of regions of the Caenorhabditis elegans X chromosome containing vitellogenin genes. J. Mol. Biol. 1986;188:301–12. doi: 10.1016/0022-2836(86)90156-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.