Abstract
Aspergillus fumigatus is the most pathogenic species among the Aspergilli, and the major fungal agent of human pulmonary infection. To prosper in diverse ecological niches, Aspergilli have evolved numerous mechanisms for adaptive gene regulation, some of which are also crucial for mammalian infection. Among the molecules which govern such responses, integral membrane receptors are thought to be the most amenable to therapeutic modulation. This is due to the localization of these molecular sensors at the periphery of the fungal cell, and to the prevalence of small molecules and licensed drugs which target receptor-mediated signaling in higher eukaryotic cells. In this review we highlight the progress made in characterizing receptor-mediated environmental adaptation in A. fumigatus and its relevance for pathogenicity in mammals. By presenting a first genomic survey of integral membrane proteins in this organism, we highlight an abundance of putative seven transmembrane domain (7TMD) receptors, the majority of which remain uncharacterized. Given the dependency of A. fumigatus upon stress adaptation for colonization and infection of mammalian hosts, and the merits of targeting receptor-mediated signaling as an antifungal strategy, a closer scrutiny of sensory perception and signal transduction in this organism is warranted.
Keywords: Aspergillus fumigatus, stress, virulence, signaling
Introduction
The genus Aspergillus is comprised of environmental filamentous mold fungi which utilize decaying organic matter for metabolic energy and nutrition. Aspergillus fumigatus is the most pathogenic, and is commonly isolated as an agent of human pulmonary infections (Dagenais and Keller, 2009). In healthy individuals, mucociliary clearance and pulmonary immune defences clear the hundreds of conidia inhaled daily (Balloy and Chignard, 2009). However, medical advances in transplantation and anticancer therapies have expanded the immunosuppressed patient population, and the number of individuals infected by opportunistic organisms, such as A. fumigatus, has drastically increased (McNeil et al., 2001; Chamilos et al., 2006). For opportunistic fungal pathogens, the phenomenon of “ready-made” virulence has been postulated, whereby traits which evolved for survival in ecological niches also govern survival in susceptible immuno-compromised hosts (Casadevall et al., 2003; Rhodes, 2006).
Beyond residual host immune responses, there are additional obstacles to successful colonization of the mammalian lung, including tolerance of host-facilitated stresses, such as iron starvation (Schrettl et al., 2004, 2007) and alkaline pH (Peñalva and Arst, 2004; Bignell et al., 2005; Peñalva et al., 2008). The requirement for infecting fungi to detect and respond to such extracellular cues is often essential for infectious growth, and in A. fumigatus the fungal receptors through which the extracellular environment is sensed remain largely unknown. This review discusses current knowledge on receptor-mediated signaling in A. fumigatus (Figure 1) and catalogues all of the putative seven transmembrane domain (7TMD) sensors encoded by the A. fumigatus genome (Table A1). Our analysis exposes the vast numbers of uncharacterized A. fumigatus receptor-like proteins.
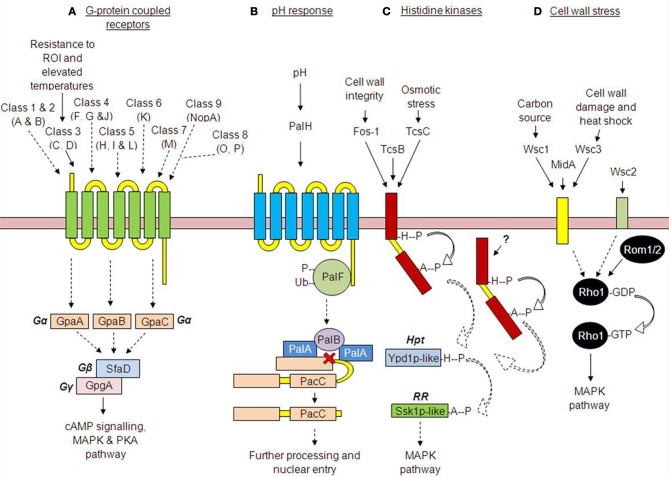
Figure 1.
Receptor-mediated signaling in Aspergillus fumigatus. (A) G-protein coupled receptor pathways—So far 15 GPCRs have been identified in A. fumigatus, though only two have been characterized (GprC and D). (B) The pH-response pathway—A shift from acidic to alkaline environmental pH is thought to be detected by the receptor PalH, mediating the phosphorylation, and subsequent ubiquitination of the C-terminally bound arrestin, PalF. This stimulates the proteolytic cleavage of the transcription factor PacC. PacC then undergoes further pH independent cleavage, before translocating to the nucleus. (C) The histidine kinase receptor pathway—The phosphorelay histidine kinases mediate the transduction of specific external or cytosolic stimuli such as cell wall integrity (Fos-1) and osmotic stress (TcsC), stimulating autophosphorylation upon a conserved histidine residue. The activating stimulus for TcsB remains elusive. (D) The cell wall stress pathway—Cell wall receptors detect environmental stress such as cell wall damage/heat shock (MidA) and alternative carbon sources (Wsc1); however, the specific stimuli for receptors Wsc2 and 3 remain elusive. (Dotted lines indicate predictions based on studies in other fungi).
G protein coupled receptors (GPCRs) in A. fumigatus
In silico analyses of fungal genome sequences have identified genes encoding putative GPCR proteins. In the phytopathogenic fungus Magnaporthe grisea, a screen of the predicted proteome using all GPCR sequences at the time available in the GPCR Database (GPCRDB) (Horn et al., 2003) yielded 14 GPCR-like sequences (Kulkarni et al., 2005). A similar exercise applied to A. fumigatus identified 15 putative GPCRs (Lafon et al., 2006).
In Aspergilli, putative GPCRs are classified by homology, and according to a convention established by Lafon et al. (2006) in A. nidulans, into nine groupings. In A. fumigatus, Classes 1 and 2 are comprised, respectively, of two putative pheromone receptors GprA (AFUA_3G14330) and GprB (AFUA_5G07880); Class 3 is comprised of two putative carbon sensors GprC (AFUA_7G04800), GprD (AFUA_2G12640); Class 4 is comprised of three putative nitrogen sensors GprF (AFUA_5G04100), GprG (AFUA_1G11900), and GprJ (AFUA_1G06840); Class 5 of three putative cAMP receptors GprH (AFUA_5G04140), GprI (AFUA_3G00780), and GprL (AFUA_ 3G01750), the latter being unique to A. fumigatus; Class 6 is comprised of a single putative GPCR, GprK (AFUA_4G01350) having a regulator of G-protein signaling (RGS) domain, unique to filamentous fungi; Class 7 includes two putative GPCRs with homology to rat growth hormone-releasing factor receptors (Miller et al., 1999) only one of which is found in A. fumigatus, GprM (AFUA_7G05300); Class 8 is comprised of three putative GPCRs with identity to yeast Izh zinc regulators (Karpichev et al., 2002; Lyons et al., 2004), two of which are found in A. fumigatus GprO (AFUA_ 3G10570) and GprP (AFUA_6G07160), and Class 9 is comprised of a single putative GPCR, NopA (AFUA_7g01430) having identity to bacterial opsins. The roles of some of these receptors have been identified in other species though in A. fumigatus little is known (Figure 1).
Among the 15 predicted GPCR-like proteins in A. fumigatus, only two, GprC (AFUA_7G04800) and GprD (AFUA_2G12640), have been characterized (Gehrke et al., 2010). GprC and GprD have been noted as having homology to Gpr1p of Saccharomyces cerevisiae which activates the cAMP pathway in response to glucose, as demonstrated by cAMP enzyme immunoassay (Yun et al., 1998; Kraakman et al., 1999). Furthermore, the A. nidulans GprD homologue mediates increase of intracellular cAMP in response to oxygenated polyunsaturated fatty acids (oxylipins), which act as autocrine and paracrine mediators in eukaryotic organisms (Affeldt et al., 2012). Deletion of A. fumigatus GprC and GprD resulted in significant growth impairment under all tested growth conditions and analysis of virulence revealed significant attenuation of virulence for ΔgprD and delayed mortality for ΔgprC in a murine model of aspergillosis (Gehrke et al., 2010). The remainder of the putative A. fumigatus GPCRs remain to be investigated and nothing is known about their molecular linkages to multi-subunit G-proteins. Unlike most Aspergillus spp. where four predicted Gα subunits occur, only three (GpaA, AFUA_1G13140, GpaB, AFUA_1G12930, and GpaC, AFUA_3G12400) have been identified for A. fumigatus (Liebmann et al., 2003), which presumably act via interaction with the Gβ and Gγ subunits (SfaD, AFUA_5G12210 and GpgA, AFUA_1G05210). In the current absence of other identified G protein subunits, or similar proteins, it is thought that the aforementioned five proteins service the entire A. fumigatus GPCR repertoire (Figure 1). Undoubtedly the relevance of A. fumigatus Gβ and Gγ subunits for viability and vegetative growth is significant as ΔsfaD and ΔgpgA gene deletion mutants are extremely impaired for germination and vegetative growth (Shin et al., 2009).
Genome-wide in silico predictions of A. fumigatus integral membrane proteins
Kulkarni et al. (2005) noted, based upon membrane topology, that the number of putative GPCR-like proteins encoded by the M. grisea genome rose to 76 when the criteria were relaxed to include homologs of the Pth11 receptor (DeZwaan et al., 1999) which is required for M. grisea pathogenicity in rice. Applying a more universal approach to A. fumigatus, we used the published genome sequence (Nierman et al., 2005) to catalogue all A. fumigatus proteins having predicted TMDs (Figure 1). To implement this, we used the TMPRED (Hofmann and Stoffel, 1993) predictive tool to perform an analysis of all 9497 A. fumigatus proteins encoded by the reference genome Af293 (Nierman et al., 2005) http://www.cadre-genomes.org.uk/Aspergillus_fumigatus/Info/Index. In total we identified 6496 proteins having putative TMDs. Among them, 161 proteins were found to encode seven predicted TMDs (Tables 1 and A1). The majority of the predicted 7TMD proteins are of hypothetical function (Table A1).
Table 1.
Numbers of predicted A. fumigatus TMD proteins, by chromosome.
| TMDs | Chromosome number | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 0 | 557 | 519 | 424 | 398 | 428 | 358 | 179 | 138 |
| 1 | 311 | 404 | 378 | 251 | 288 | 282 | 179 | 161 |
| 2 | 267 | 257 | 228 | 199 | 206 | 194 | 106 | 114 |
| 3 | 127 | 233 | 100 | 100 | 141 | 116 | 51 | 57 |
| 4 | 81 | 85 | 59 | 60 | 56 | 57 | 25 | 40 |
| 5 | 132 | 38 | 23 | 3 | 22 | 31 | 18 | 17 |
| 6 | 22 | 26 | 17 | 21 | 16 | 21 | 14 | 8 |
| 7 | 18 | 22 | 23 | 19 | 28 | 27 | 7 | 16 |
| 8 | 9 | 10 | 16 | 9 | 10 | 5 | 5 | 4 |
| 9 | 19 | 13 | 12 | 13 | 15 | 15 | 4 | 9 |
| 10 | 15 | 23 | 20 | 13 | 16 | 7 | 7 | 12 |
| 11 | 22 | 22 | 23 | 29 | 21 | 16 | 9 | 18 |
| 12 | 23 | 25 | 24 | 17 | 18 | 19 | 12 | 9 |
| 13 | 7 | 6 | 9 | 8 | 7 | 8 | 2 | 1 |
| 14 | 3 | 2 | 6 | 3 | 5 | 6 | 2 | 3 |
| 15 | 3 | 1 | 4 | 1 | 2 | 2 | 1 | 1 |
| 16 | 0 | 1 | 2 | 0 | 2 | 4 | 0 | 1 |
| 17 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| 18 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
PalH: a putative 7TMD pH sensor
During colonization of the mammalian lung A. fumigatus is exposed to a range of microenvironments, of likely differing pHs, not only within the pulmonary niche but also following phagocytosis by macrophages or ingestion by neutrophils and exposure to their vacuole contents (Levitz et al., 1999; Newman, 1999; Reeves et al., 2002; Ibrahim-Granet et al., 2003). Versatility of metabolism and physiology is required to survive such extremes, including appropriate pH-responsive gene expression for nutrient acquisition and survival (Bignell et al., 2005). In the model ascomycete and occasional pathogen A. nidulans, the PacC transcription factor governs gene expression in response to extracellular pH (Tilburn et al., 1995; Diez et al., 2002) and is vital for mammalian pathogenicity (Peñalva and Arst, 2004; Bignell et al., 2005; Peñalva et al., 2008). Under alkaline conditions, a signaling cascade involving seven proteins is involved in activation of PacC. A putative pH sensor, PalH, has 7TMDs and a cytoplasmic C-terminus (Negrete-Urtasun et al., 1997, 1999), which interacts with a cognate arrestin encoded by palF (Herranz et al., 2005; Hervas-Aguilar et al., 2010). Unlike canonical GPCR receptors, PalH is not thought to act via interaction with G-protein subunits (Kroeze et al., 2003). When an alkaline response is triggered, PalF is phosphorylated and subsequently ubiquitinated in a PalH-dependent manner (Herranz et al., 2005), leading to PalB-mediated, signal dependent, proteolytic cleavage of the pH-responsive transcription factor PacC (Penas et al., 2007; Rodriguez-Galan et al., 2009). Subsequent translocation of the truncated PacC protein, from cytoplasm to nucleus, permits alkaline adaptation via differential expression of genes required to enable growth under alkaline extracellular conditions (Tilburn et al., 1995; Mingot et al., 1999, 2001; Espeso and Arst, 2000; Espeso et al., 2000). In A. fumigatus the amino acid residues crucial for PalH and PalF interaction are conserved, and in split-ubiquitin analyses the proteins enter into close proximity (Bertuzzi and Bignell, 2011; Bignell, 2012). We have also recently demonstrated the requirement for A. fumigatus PalH for murine infection (Bertuzzi et al., in preparation).
Histidine kinase sensors in A. fumigatus
Histidine kinases (HK) are phosphorelay protein sensors which transduce extracellular signals. HKs are common in the fungal kingdom, and apparently absent in humans (West and Stock, 2001). Amongst archaea, bacteria and fungi, two classes of HK (two-component and hybrid) are found. The former class of two-component receptor systems predominate in bacteria and archaea, whereby autophosphorylation of the HK protein precedes transfer of the phosphoryl group to a conserved aspartate residue in a second protein, termed the response regulator (RR)(Li et al., 2012). HK activities have been associated with both the osmo- and peroxide-regulatory pathways in multiple fungi, and have been most extensively characterized in S. cerevisiae (Santos and Shiozaki, 2001). However, RR proteins are not abundantly encoded by fungal genomes; Skn7 and Ssk1 are two examples of such proteins, which in S. cerevisiae and C. albicans, account for the entire RR cohort of these species (Kaserer et al., 2009; Oide et al., 2010). The fungal phosphotransfer relay can involve three proteins, as exemplified by the S. cerevisiae HOG1 MAPK phosphorelay, where an HK (Sln1), a histidine phosphointermediate (Ypd1) and an RR (Ssk1) collectively mediate a multistep phosphotransfer (Kaserer et al., 2009).
Fungal HKs most commonly fall into the hybrid class of regulators which utilize a single polypeptide. This protein possesses both a Histidine kinase A (HiskA) and a receiver domain (REC) containing a conserved aspartate residue (Li et al., 2012). Other domains, such as the ATP-binding HATPase_c domain (Dago et al., 2012) are also found; however, as these proteins are largely uncharacterized for A. fumigatus, the functional relevance of domain organization is unknown. The composition, and/or relative positioning, of additional domains provides the basis for sub-classification of HKs (Catlett et al., 2003), presented for 12 A. fumigatus HKs in Table A2. Amongst these, only three have been studied: the two-component system proteins A, B, and C (TcsA/Fos-1 AFUA_5G10240, TcsB AFUA_2G00660 and TcsC AFUA_2G03560).
Despite the significance of the HK Fos-1 for detection of extracellular stresses, this hybrid HK has been previously predicted as possessing no TMDs, implying a cytosolic presence (Pott et al., 2000). However, our TMPRED analyses predicted TMDs for all of the three HKs, with Fos-1 possessing a single TMD (Table A2). Deletion of the fos-1 gene leads to a ~66% reduction in conidiation after 48 h in liquid YG medium, as well as heightened resistance to the cell wall-degrading enzyme mix novozym 234, suggesting the role of fos-1 in cell wall assembly (Pott et al., 2000). Δfos-1 mutants were found to have normal morphology, germination, osmotic and oxidative stress tolerance, and antifungal susceptibilities. Subsequent transcriptional analyses found a significant increase in fos-1 expression, relative to in vitro growth, during the first 72 h of infection in a murine model of pulmonary aspergillosis (Zhang et al., 2005), and reduced virulence of A. fumigatus in a systemic murine model of infection (Clemons et al., 2002).
In a study addressing the role of oxidative stress in A. fumigatus pathogenicity, Du et al. (2006) characterized the A. fumigatus TcsB protein, a putative homolog of Sln1 in S. cerevisiae. In A. nidulans, TMPRED analysis predicted 2 TMDs for TcsB at the N-terminus (Furukawa et al., 2002), though in A. fumigatus, our prediction extends this to 4TMDs (Table A1). Unlike S. cerevisiae where deletion of sln1 is lethal (Maeda et al., 1994), an A. fumigatus ΔtcsB mutant is viable, demonstrates normal morphology, and is as tolerant as the wild type to increased temperatures, various cell wall damaging agents, and poor nitrogen/carbon sources. The only phenotype discernable for the mutant was a minor sensitivity to SDS (Du et al., 2006). This data suggests a non-essential role for TcsB, or redundancy of function with other, as yet uncharacterized protein(s).
It is believed that group III HK mediate resistance to high osmotic pressure via the high osmolarity glycerol pathway (HOG). For this reason, the characterization of the sole A. fumigatus group III hybrid HK TcsC, classified as such on the basis of putative HAMP (HK, adenyl cyclase, methyl-accepting chemotaxis protein, phosphatase) domains, was investigated (McCormick et al., 2012). The significance of the HAMP domains, based upon studies of other sensor proteins and signaling is postulated as providing the means to couple input and output since HAMP domains of integral membrane hybrid HKs are found in close proximity to the membrane-spanning segment (Parkinson, 2010). It is speculated that in response to extracellular signals, such as altered osmolarity, a conformational rearrangement is triggered which prompts activation of an output domain (Parkinson, 2010). In A. fumigatus, deletion of the tcsC gene resulted in an extended white colony rim and a reduced number of extending hyphae. However, unlike the A. nidulans homologue nikA (Hagiwara et al., 2009), no detrimental effects on sporulation and conidial growth were observed. In the presence of nitrate as a nitrogen source a significant reduction in radial growth was detectable, and furthermore, compared to the control strain, growth of ΔtcsC at 2% O2 abolished sporulation and prompted a dome-shaped morphology indicative of oxygen starvation. A strong inhibition of growth resulted from exposure to hyperosmotic stress (1.2 M sorbitol, 1 M KCl, or 1 M NaCl) but sensitivity to calcofluor white, amphotericin B, posaconazole and caspofungin, extremes of pH/temperature, or oxidative stress were reportedly normal.
In a comparative analysis of wild type and ΔtcsC virulence, no discernable differences in pathogenicity analysis in a murine model of invasive aspergillosis were detected (McCormick et al., 2012).
Cell wall receptors
The fungal cell wall is essential for viability and an important target of antifungal drugs (Latgé et al., 2005; Latgé, 2007; Walker et al., 2010). In fungi a conserved MAPK signaling module is responsible for maintaining cellular integrity, shape and resilience to environmental stresses (Lee and Levin, 1992; Levin, 2005; Lesage and Bussey, 2006). In S. cerevisiae, such MAPK signaling (Chen and Thorner, 2007) is initiated through stress detection at five integral membrane receptors Wsc1-3, Mid2, and Mtl1 (Lodder et al., 1999). This promotes guanine nucleotide exchange factor (GEFs—Rom1 and Rom2)-mediated nucleotide exchange upon the GTPase Rho1, facilitating the regulation of numerous downstream effectors (Zu et al., 2001). In a quest to find equivalent cell wall sensors in A. fumigatus, Dichtl et al. (2012) performed BLAST analyses to reveal three previously uncharacterized open reading frames with domain structures similar to those of Wsc1-3 (Af Wsc1, AFUA_4G13670, Af Wsc2, AFUA_3G07050, and Af Wsc3, AFUA_5G09020 respectively). Bioinformatic analyses predicted the presence of characteristic cell wall integrity (CWI) sensor N-terminal WSC domains with downstream, though truncated, ser/thr rich regions, and (with the exception of Wsc2) transmembrane domains. In common with the S. cerevisiae sensors a short cytosolic C-terminus was also predicted for two of the sensors (Dichtl et al., 2012).
To discern subcellular localization, ectopically integrating vectors were applied to generate four putative CWI sensor-GFP fusions, Wsc1-3, and MidA. From these, localization of all C-terminally tagged sensors was observed at the fungal surface. Additionally a strong presence was observed in vacuoles, though this was dismissed as a by-product of over expression or misfolding. Phenotypic analyses of single and double mutants identified a significant impairment of radial growth in the case of a Δwsc1Δwsc3 double mutant. These findings were further exacerbated in a triple mutant Δwsc1Δwsc3ΔmidA. Furthermore, in all mutants lacking wsc1, provision of glycerol as carbon source lead to a significant reduction in radial growth on minimal media (Dichtl et al., 2012).
Previously, mutants lacking members of the CWI MAPK pathway have demonstrated a clear sensitivity to echinocandins and azole antifungals (Fujioka et al., 2007; Dirr et al., 2010). Extending this analysis to the A. fumigatus mutant phenotypes revealed a single relevant phenotype, namely the heightened sensitivity of the Δwsc1 mutant to the echinocandin, caspofungin (Dichtl et al., 2012).
To study stress-induced activation of intracellular signaling, effects on growth and MpkA phosphorylation were analyzed in the presence of the cell wall perturbing agent, calcoflour white, or following heat shock (48°C). None of the Wsc mutants were found to be sensitive to cell wall perturbation or heat shock, however, ΔmidA was highly sensitive to all of these stresses. In agreement with phenotypic data, calcofluor white-induced MpkA phosphorylation was significantly reduced in the ΔmidA mutant compared with wild type, while phosphorylation of MpkA was not diminished in mutants lacking wsc1 or wsc1 and wsc3 (Dichtl et al., 2012). In S. cerevisiae, the Wsc1 cell wall sensor mediates signaling of alkaline stress via the CWI MAPK module (Serrano et al., 2006); no evidence for such a role in A. fumigatus was obtained. Thus, while Mkk2 null mutants are sensitive to alkalinization of the medium (Dirr et al., 2010), the identity of the activating cell wall sensor remains unknown.
Taken together these findings suggest that MidA is the sole cell wall perturbation sensor, while Wsc1 is required for glycerol carbon source assimilation. Furthermore, a compensatory role between Wsc1 and Wsc3 with regards to efficient growth and conidiation has been demonstrated. Despite these observations, a role for Wsc2 has yet to be identified, while the putative CWI pH sensor remains elusive.
Receptor-mediated signaling during A. fumigatus infection: relevance for therapeutic strategy
Drugs which target GPCR function account for >50% of currently licensed drugs (Davies et al., 2008). It therefore follows that fungal GPCRs are likely to be similarly responsive to chemical perturbations. This fact, coupled with the absolute requirement for some GPCRs in fungal growth make a compelling case for these proteins as antifungal drug targets. Although the pharmaceutical market is dominated by GPCR-active molecules, the discovery of most of these agents was made on the basis of functional activity in high throughput screens, only later were the targets and modes of action clarified (Filmore, 2004). In the post-genomic era, with confidence in the pharmaceutical relevance of such proteins, drug discovery can become target-driven. An expanding repertoire of technologies to probe 7TMD protein activities provides the basis upon which to confront functional studies of the uncharacterized receptors in A. fumigatus and to screen for inhibitory molecules. It has recently been suggested that considering seven transmembrane receptors as disordered proteins able to allosterically respond to a number of binding partners, is useful in understanding the plasticity of function exhibited by such proteins (Kenakin and Miller, 2010). Conformational changes which occur in response to extracellular ligands and/or stimuli expose cytosolic signaling domains and present three distinct arenas open to perturbation: extracellular sensing/ligand binding, cytosolic surfaces, and intramembrane domains. In order to prioritise the most promising candidates for drug development, a crucial experiment will be to assess the requirement of such receptors, via regulatable promoters, for sustained viability of established fungal mass in murine models of infection (Gossen and Bujard, 1992).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the assistance of Arshad Khan in automating the genome-wide TMPRED analyses. This work was assisted by funding from the Medical Research Council [G0501164 to E. M. Bignell], the Biotechnology and Biological Sciences Research Council [PhD studentship to C. M. Grice, BB/F016239/1], and the Wellcome Trust [WT093596MA to E. M. Bignell].
Appendix
Table A1.
Identity and annotations of A. fumigatus proteins having seven predicted transmembrane domains.
| Annotation | ORF | Accession number |
|---|---|---|
| CHROMOSOME 1 | ||
| Polyketide synthase | AFUA_1G01010 | XP_749851.1 |
| Conserved hypothetical protein | AFUA_1G01190 | XP_749869.1 |
| High affinity zinc ion transporter/membrane zinc transporter | AFUA_1G01550 | XP_749905.1 |
| Conserved hypothetical protein | AFUA_1G01620 | XP_749912.1 |
| MFS alpha-glucoside transporter | AFUA_1G03280 | XP_750078.1 |
| Peroxisomal ABC transporter (PXA1) | AFUA_1G04780 | XP_750227.1 |
| Vacuolar membrane PQ loop repeat protein | AFUA_1G06840 | XP_750433.1 |
| Phosphatidate cytidylytransferase | AFUA_1G07010 | XP_750449.1 |
| Export control protein CHS7-Like | AFUA_1G07110 | XP_750458.1 |
| Rhomboid family protein | AFUA_1G09150 | XP_752282.1 |
| Conserved hypothetical protein | AFUA_1G10160 | XP_752382.1 |
| COP11-coated vesicle protein SurF4/Erv29 | AFUA_1G11770 | XP_752543.1 |
| PQ loop repeat protein | AFUA_1G11900 | XP_752556.1 |
| Integral membrane protein Pth11-like | AFUA_1G14080 | XP_752778.1 |
| DUF409 domain protein | AFUA_1G14140 | XP_752784.1 |
| DUF803 domain membrane protein | AFUA_1G15880 | XP_752954.1 |
| Potassium transporter | AFUA_1G16340 | XP_753000.1 |
| Fatty acid elongase (Gig30) | AFUA_1G16710 | XP_753038.1 |
| Conserved hypothetical protein | AFUA_1G16720 | XP_753039.1 |
| CHROMOSOME 2 | ||
| RTA1 domain protein | AFUA_2G00420 | XP_749178.1 |
| DUF1275 domain protein | AFUA_2G00530 | XP_749189.1 |
| Alpha-amylase | AFUA_2G00710 | XP_749208.1 |
| Cellobiose dehydrogenase | AFUA_2G01180 | XP_749254.1 |
| Bax inhibitor family protein | AFUA_2G03220 | XP_749457.1 |
| Extracellular threonine rice protein | AFUA_2G03540 | XP_749488.1 |
| ZIP metal ion transporter | AFUA_2G08740 | XP_755208.1 |
| Nickel transport protein | AFUA_2G08830 | XP_755217.1 |
| Serine/threonine protein kinase | AFUA_2G09570 | XP_755291.1 |
| ZIP metal ion transporter | AFUA_2G12050 | XP_755537.1 |
| Midasin | AFUA_2G12150 | XP_755547.1 |
| Integral membrane protein | AFUA_2G12640 | XP_755596.1 |
| HEAT repeat protein | AFUA_2G14180 | XP_755751.2 |
| Conserved hypothetical protein | AFUA_2G15100 | XP_755844.2 |
| Integral membrane protein | AFUA_2G15440 | XP_755879.2 |
| DUF92 domain protein | AFUA_2G15640 | XP_755899.1 |
| Rhomboid family membrane protein | AFUA_2G16490 | XP_755986.1 |
| Integral membrane protein | AFUA_2G16985 | XP_001481687.1 |
| Sulfatase domain protein | AFUA_2G17610 | XP_756096.1 |
| Integral membrane protein | AFUA_2G17760 | XP_756111.1 |
| RTA1 domain protein | AFUA_2G17810 | XP_756116.1 |
| RTA1 domain protein | AFUA_2G17890 | XP_756125.1 |
| CHROMOSOME 3 | ||
| RTA1 domain protein | AFUA_3G00480 | XP_748368.1 |
| DUF1275 domain protein | AFUA_3G00670 | XP_748388.1 |
| cAMP receptor (Car4) | AFUA_3G00780 | XP_748399.1 |
| Hypothetical protein | AFUA_3G00850 | XP_748406.2 |
| RTA1 domain protein | AFUA_3G00920 | XP_748413.1 |
| Integral membrane protein Pth11-like | AFUA_3G01200 | XP_748441.2 |
| RTA1 domain protein | AFUA_3G01630 | XP_748484.1 |
| G protein coupled receptor family protein | AFUA_3G01750 | XP_748496.1 |
| Conserved hypothetical protein | AFUA_3G02450 | XP_748568.1 |
| RTA1 domain protein | AFUA_3G03310 | XP_748650.1 |
| PKS-like enzyme | AFUA_3G03540 | XP_748674.1 |
| UPF0016 domain protein | AFUA_3G07080 | XP_754898.1 |
| Phosphatidylinositol: UDP-GlcNAc transferase PIG-C | AFUA_3G07170 | XP_754889.1 |
| Conserved hypothetical protein | AFUA_3G07420 | XP_754867.2 |
| DUF1275 domain protein | AFUA_3G07550 | XP_754856.1 |
| Sucrose transporter | AFUA_3G08480 | XP_754766.1 |
| Conserved hypothetical protein | AFUA_3G09650 | XP_754654.1 |
| PQ loop repeat protein | AFUA_3G10470 | XP_754572.1 |
| RTA1 domain protein | AFUA_3G10770 | XP_754542.1 |
| RTA1 domain protein | AFUA_3G12830 | XP_754338.2 |
| Nonribosomal peptide synthase | AFUA_3G13730 | XP_754251.1 |
| Mating-type alpha-pheromone receptor PreB | AFUA_3G14330 | XP_754193.1 |
| Conserved hypothetical protein | AFUA_3G14870 | XP_754138.1 |
| Integral membrane protein | AFUA_3G15100 | XP_754114.2 |
| CHROMOSOME 4 | ||
| Polyketide synthase | AFUA_4G00210 | XP_746435.1 |
| Hypothetical protein | AFUA_4G00580 | XP_746398.1 |
| Hypothetical protein | AFUA_4G01242 | XP_746333.2 |
| Conserved hypothetical protein | AFUA_4G01350 | XP_746323.2 |
| Patatin-like serine hydrolase | AFUA_4G03000 | XP_746486.1 |
| Aquaporin | AFUA_4G03390 | XP_746526.2 |
| Integral membrane protein | AFUA_4G03540 | XP_746541.1 |
| C4-dicarboxylate transporter/malic acid transport protein, putative | AFUA_4G04540 | XP_746640.1 |
| Para-hydroxybenzoate-polyprenyltransferase Coq2 | AFUA_4G05970 | XP_752227.1 |
| Longevity-assurance protein (LAC1) | AFUA_4G06290 | XP_752195.1 |
| RNA polymerase II mediator complex subunit Nut1 | AFUA_4G06600 | XP_752166.2 |
| CaaX prenyl protease Ste24 | AFUA_4G07590 | XP_752066.2 |
| Conserved hypothetical protein | AFUA_4G07680 | XP_752057.1 |
| 26S proteasome regulatory subunit Rpn2 | AFUA_4G08480 | XP_751978.1 |
| Conserved hypothetical protein | AFUA_4G10080 | XP_751818.2 |
| Endosomal peripheral membrane protein (Mon2) | AFUA_4G12070 | XP_751624.1 |
| Potassium uptake transporter | AFUA_4G13540 | XP_751477.1 |
| Conserved hypothetical protein | AFUA_4G14210 | XP_751411.1 |
| Low affinity iron transporter | AFUA_4G14640 | XP_751369.1 |
| CHROMOSOME 5 | ||
| Integral membrane protein | AFUA_5G00100 | XP_748330.2 |
| RTA1 domain protein | AFUA_5G01230 | XP_748219.1 |
| RTA1 domain protein | AFUA_5G01310 | XP_748211.1 |
| Phosphate permease | AFUA_5G01320 | XP_748210.1 |
| Histone acetylase complex subunit Paf400 | AFUA_5G02570 | XP_748085.1 |
| Integral membrane protein | AFUA_5G02860 | XP_748057.2 |
| PQ loop repeat protein | AFUA_5G04100 | XP_747934.1 |
| cAMP receptor-like protein | AFUA_5G04135 | XP_001481495.1 |
| Conserved hypothetical protein | AFUA_5G06570 | XP_753976.1 |
| Integral membrane protein | AFUA_5G06670 | XP_753966.1 |
| DUF300 domain protein | AFUA_5G07250 | XP_753909.1 |
| a-pheromone receptor PreA | AFUA_5G07880 | XP_753848.1 |
| PQ loop repeat protein | AFUA_5G08410 | XP_753796.1 |
| Spermine/spermidine synthase family protein | AFUA_5G08500 | XP_753787.1 |
| Beige/BEACH domain protein | AFUA_5G09220 | XP_753717.1 |
| Bax Inhibitor family protein | AFUA_5G09310 | XP_753708.2 |
| RTA1 domain protein | AFUA_5G09900 | XP_753650.1 |
| MFS multidrug transporter | AFUA_5G10140 | XP_753627.1 |
| MHYT domain signaling protein | AFUA_5G11310 | XP_753518.2 |
| 26S proteasome regulatory subunit Mts4 | AFUA_5G11720 | XP_753478.1 |
| Guanine nucleotide exchange factor (Gea2) | AFUA_5G11900 | XP_753461.1 |
| Integral membrane protein (Ptm1) | AFUA_5G12390 | XP_753413.1 |
| Integral membrane protein TmpA | AFUA_5G12520 | XP_753400.1 |
| DUF1275 domain protein | AFUA_5G13060 | XP_753348.1 |
| pH signal transduction protein PalH | AFUA_5G13270 | XP_753327.1 |
| Integral membrane protein | AFUA_5G13725 | XP_753282.2 |
| Integral membrane protein | AFUA_5G14600 | XP_753197.1 |
| CHROMOSOME 6 | ||
| Integral membrane protein | AFUA_6G00320 | XP_731523.1 |
| Cation diffusion facilitator | AFUA_6G00440 | XP_731511.1 |
| Hypothetical protein | AFUA_6G00460 | XP_731509.1 |
| Integral membrane protein | AFUA_6G00640 | XP_731492.1 |
| Signal peptide peptidase | AFUA_6G02150 | XP_747862.1 |
| Hypothetical protein | AFUA_6G03180 | XP_747759.1 |
| Conserved hypothetical protein | AFUA_6G03380 | XP_747738.2 |
| Nonribosomal peptide synthase | AFUA_6G03480 | XP_747729.1 |
| Integral membrane protein (Pth11) | AFUA_6G03600 | XP_747717.1 |
| GTPase activating protein (Tsc2) | AFUA_6G04000 | XP_747677.1 |
| Conserved hypothetical protein | AFUA_6G06950 | XP_750588.2 |
| IZH family channel protein (Izh3) | AFUA_6G07160 | XP_750609.1 |
| 4-hydroxybenzoate polyprenyl transferase | AFUA_6G07240 | XP_750617.1 |
| Integral membrane protein | AFUA_6G07820 | XP_750673.2 |
| Aquaglyceroporin | AFUA_6G08480 | XP_750737.1 |
| RTA1 domain protein | AFUA_6G09550 | XP_750844.1 |
| Ceramide synthase membrane component (LAG1) | AFUA_6G10460 | XP_750934.1 |
| Cell morphogenesis protein (PAG1) | AFUA_6G11010 | XP_750987.1 |
| Integral membrane protein | AFUA_6G11560 | XP_751039.1 |
| RTA1 domain protein | AFUA_6G11800 | XP_751062.1 |
| GPI transamidase component (GAA1) | AFUA_6G12760 | XP_751154.1 |
| ABC iron exporter Atm1 | AFUA_6G12870 | XP_751165.1 |
| UDP-galactose transporter | AFUA_6G13070 | XP_751184.1 |
| Ferric-chelate reductase | AFUA_6G13750 | XP_751251.1 |
| Integral membrane protein Pth11-like | AFUA_6G13800 | XP_751256.1 |
| Integral membrane protein | AFUA_6G13950 | XP_751270.1 |
| RTA1 domain protein | AFUA_6G14140 | XP_751288.1 |
| CHROMOSOME 7 | ||
| Plasma membrane hexose transporter | AFUA_7G00220 | XP_746907.1 |
| Conserved hypothetical protein | AFUA_7G00280 | XP_746901.1 |
| Squalene-hopene-cyclase | AFUA_7G00300 | XP_746899.1 |
| Conserved hypothetical protein | AFUA_7G04800 | XP_749030.2 |
| Plasma membrane protein Pth11-like | AFUA_7G06130 | XP_748897.2 |
| Conserved hypothetical protein | AFUA_7G06660 | XP_748845.2 |
| Metalloreductase | AFUA_7G07120 | XP_748799.1 |
| CHROMOSOME 8 | ||
| Solute transporter | AFUA_8G00660 | XP_747139.1 |
| Glycosyl transferase | AFUA_8G00680 | XP_747137.1 |
| Conserved hypothetical protein | AFUA_8G01300 | XP_747076.1 |
| GABA permease | AFUA_8G01450 | XP_747061.1 |
| NRPS-like enzyme | AFUA_8G01640 | XP_747042.1 |
| Conserved hypothetical protein | AFUA_8G01840 | XP_747022.2 |
| Conserved hypothetical protein | AFUA_8G02390 | XP_746967.1 |
| ZIP family zinc transporter | AFUA_8G04010 | XP_747208.2 |
| Integral membrane protein | AFUA_8G04560 | XP_747263.1 |
| Integral membrane protein | AFUA_8G05510 | XP_747353.1 |
| Chitin synthase F | AFUA_8G05630 | XP_747364.1 |
| RTA1 domain protein | AFUA_8G05740 | XP_747375.1 |
| Cellobiose dehydrogenase | AFUA_8G05805 | XP_747382.1 |
| DUF1295 domain protein | AFUA_8G05810 | XP_747383.2 |
| Toxin biosynthesis protein (Tri7) | AFUA_8G05970 | XP_747399.1 |
| Metalloreductase transmembrane component | AFUA_8G06210 | XP_747422.2 |
Table A2.
Identity and annotations of A. fumigatus histidine kinase receptors.
| Annotation | ORF | Accession number | No. of transmembrane domains | Putative conserved domains | Putative group no. |
|---|---|---|---|---|---|
| CHROMOSOME 2 | |||||
| Sensor histidine kinase/response regulator TcsB/Sln1 | AFUA_2G00660 | XP_001481640.1 | 4 |
|
6 |
| Two-component osmosensing histidine kinase (Bos1)-TcsC | AFUA_2G03560 | XP_749489.1 | 1 |
|
3 |
| CHROMOSOME 3 | |||||
| Sensor histidine kinase/response regulator | AFUA_3G07130 | XP_754893.1 | 3 |
|
7 |
| Sensor histidine kinase/response regulator | AFUA_3G12530 | XP_754368.1 | 2 |
|
5 |
| Sensor histidine kinase/response regulator | AFUA_3G12550 | XP_754366.1 | 9 |
|
10 |
| CHROMOSOME 4 | |||||
| Sensor histidine kinase/response regulator | AFUA_4G00320 | XP_746424.2 | 2 |
|
2 |
| Sensor histidine kinase/response regulator | AFUA_4G00660 | XP_746390.1 | 1 |
|
2 |
| Sensor histidine kinase/response regulator | AFUA_4G02900 | XP_746476.1 | 2 |
|
8 |
| Sensor histidine kinase/response regulator | AFUA_4G01020 | XP_746355.1 | 3 |
|
2 |
| Sensor histidine kinase/response regulator | AFUA_4G07400 | XP_752086.2 | 0 |
|
7 |
| CHROMOSOME 6 | |||||
| Sensor histidine kinase/response regulator Fos-1/TcsA | AFUA_6G10240 | XP_750913.1 | 1 |
|
5 |
| CHROMOSOME 8 | |||||
| Sensor histidine kinase/response regulator | AFUA_8G06140 | XP_747415.2 | 2 |
|
7 |
HiskA, Histidine kinase A; HAMP, Histidine kinases, adenyl cyclases, methyl-accepting chemotaxis protein, phosphatase; PAS, Per—period circadian protein, Arnt—Al receptor nuclear translocator protein, Sim—single minded protein; GAF, presence in cGMP-regulated cyclic nucleotides PDEs, certain adenyl cyclases and the bacterial transcription factor FhlA; AAA, ATPases associated with diverse cellular activities.
References
- Affeldt K. J., Brodhagen M., Keller N. P. (2012). Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins (Basel) 4, 695–717 10.3390/toxins4090695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloy V., Chignard M. (2009). The innate immune response to Aspergillus fumigatus. Microbes Infect. 11, 919–927 10.1016/j.micinf.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Bertuzzi M., Bignell E. M. (2011). Sensory perception in fungal pathogens: applications of the split-ubiquitin Membrane Yeast Two-Hybrid (MYTH) technique. Fungal Biol. Rev. 25, 7–7 [Google Scholar]
- Bignell E. (2012). Conservation of seven transmembrane domain receptors and arrestins required for pH signalling in Aspergillus fumigatus, and implications for drug discovery. Ann. N.Y. Acad. Sci. 1273, 35–43 10.1111/j.1749-6632.2012.06814.x [DOI] [PubMed] [Google Scholar]
- Bignell E., Negrete-Urtasun S., Calcagno A. M., Haynes K., Arst H. N., Jr., Rogers T. (2005). The Aspergillus pH-responsive transcription factor PacC regulates virulence. Mol. Microbiol. 55, 1072–1084 10.1111/j.1365-2958.2004.04472.x [DOI] [PubMed] [Google Scholar]
- Casadevall A., Steenbergen J. N., Nosanchuk J. D. (2003). ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi–the Cryptococcus neoformans paradigm. Curr. Opin. Microbiol. 6, 332–337 10.1016/S1369-5274(03)00082-1 [DOI] [PubMed] [Google Scholar]
- Catlett N. L., Yoder O. C., Turgeon B. G. (2003). Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2, 1151–1161 10.1128/EC.2.6.1151-1161.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos G., Luna M., Lewis R. E., Bodey G. P., Chemaly R., Tarrand J. J., et al. (2006). Invasive fungal infections in patients with hematologic malignancies in a tertiary care cancer center: an autopsy study over a 15-year period (1989–2003). Haematologica 91, 986–989 [PubMed] [Google Scholar]
- Chen R. E., Thorner J. (2007). Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773, 1311–1340 10.1016/j.bbamcr.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons K. V., Miller T. K., Selitrennikoff C. P., Stevens D. A. (2002). fos-1, a putative histidine kinase as a virulence factor for systemic aspergillosis. Med. Mycol. 40, 259–262 [DOI] [PubMed] [Google Scholar]
- Dagenais T. R., Keller N. P. (2009). Pathogenesis of Aspergillus fumigatus in invasive Aspergillosis. Clin. Microbiol. Rev. 22, 447–465 10.1128/CMR.00055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dago A. E., Schug A., Procaccini A., Hoch J. A., Weigt M., Szurmant H. (2012). Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 109, E1733–E1742 10.1073/pnas.1201301109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. N., Secker A., Halling-Brown M., Moss D. S., Freitas A. A., Timmis J., et al. (2008). GPCRTree: online hierarchical classification of GPCR function. BMC Res. Notes 1:67 10.1186/1756-0500-1-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T. M., Carroll A. M., Valent B., Sweigard J. A. (1999). Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11, 2013–2030 10.1105/tpc.11.10.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl K., Helmschrott C., Dirr F., Wagener J. (2012). Deciphering cell wall integrity signalling in Aspergillus fumigatus: identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol. Microbiol. 83, 506–519 10.1111/j.1365-2958.2011.07946.x [DOI] [PubMed] [Google Scholar]
- Diez E., Alvaro J., Espeso E. A., Rainbow L., Suarez T., Tilburn J., et al. (2002). Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 21, 1350–1359 10.1093/emboj/21.6.1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirr F., Echtenacher B., Heesemann J., Hoffmann P., Ebel F., Wagener J. (2010). AfMkk2 is required for cell wall integrity signaling, adhesion, and full virulence of the human pathogen Aspergillus fumigatus. Int. J. Med. Microbiol. 300, 496–502 10.1016/j.ijmm.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Du C., Sarfati J., Latge J. P., Calderone R. (2006). The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 44, 211–218 10.1080/13693780500338886 [DOI] [PubMed] [Google Scholar]
- Espeso E., Arst H. N., Jr. (2000). On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol. Cell. Biol. 20, 3355 10.1128/MCB.20.10.3355-3363.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeso E., Roncal T., Díez E., Rainbow L., Bignell E., Álvaro J., et al. (2000). On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 19,Vol. 7 719–728. 10.1093/emboj/19.4.719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmore D. (2004). It's a GPCR World. Mod. Drug Discov. 7, 24–28 [Google Scholar]
- Fujioka T., Mizutani O., Furukawa K., Sato N., Yoshimi A., Yamagata Y., et al. (2007). MpkA-Dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510 10.1128/EC.00281-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Katsuno Y., Urao T., Yabe T., Yamada-Okabe Y., Yamada-Okabe H., et al. (2002). Isolation and functional analysis of a gene, tcsB, encoding a transmembrane hybrid-type histidine kinase from Aspergillus nidulans. Appl. Environ. Microbiol. 68, 5304–5310 10.1128/AEM.68.11.5304-5310.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke A., Heinekamp T., Jacobsen I. D., Brakhage A. A. (2010). Heptahelical receptors GprC and GprD of Aspergillus fumigatus are essential regulators of colony growth, hyphal morphogenesis, and virulence. Appl. Environ. Microbiol. 76, 3989–3998 10.1128/AEM.00052-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Bujard H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Mizuno T., Abe K. (2009). Characterization of NikA histidine kinase and two response regulators with special reference to osmotic adaptation and asexual development in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 73, 1566–1571 [DOI] [PubMed] [Google Scholar]
- Herranz S., Rodriguez J. M., Bussink H. J., Sanchez-Ferrero J. C., Arst H. N., Jr., Penalva M. A., et al. (2005). Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U.S.A. 102, 12141–12146 10.1073/pnas.0504776102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas-Aguilar A., Galindo A., Penalva M. A. (2010). Receptor-independent Ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J. Biol. Chem. 285, 18095–18102 10.1074/jbc.M110.114371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. (1993). TMbase - a database of membrane spanning protein segments. Biol. Chem. 374, 166 [Google Scholar]
- Horn F., Bettler E., Oliveira L., Campagne F., Cohen F. E., Vriend G. (2003). GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 31, 294–297 10.1093/nar/gkg103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim-Granet O., Philippe B., Boleti H., Boisvieux-Ulrich E., Grenet D., Stern M., et al. (2003). Phagocytosis and intracellular fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect. Immun. 71, 891–903 10.1128/IAI.71.2.891-903.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpichev I. V., Cornivelli L., Small G. M. (2002). Multiple regulatory roles of a novel Saccharomyces cerevisiae protein, encoded by YOL002c, in lipid and phosphate metabolism. J. Biol. Chem. 277, 19609–19617 10.1074/jbc.M202045200 [DOI] [PubMed] [Google Scholar]
- Kaserer A. O., Andi B., Cook P. F., West A. H. (2009). Effects of osmolytes on the SLN1-YPD1-SSK1 phosphorelay system from Saccharomyces cerevisiae. Biochemistry 48, 8044–8050 10.1021/bi900886g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T., Miller L. J. (2010). Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol. Rev. 62, 265–304 10.1124/pr.108.000992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman L., Lemaire K., Ma P., Teunissen A. W., Donaton M. C., Van Dijck P., et al. (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002–1012 10.1046/j.1365-2958.1999.01413.x [DOI] [PubMed] [Google Scholar]
- Kroeze W. K., Sheffler D. J., Roth B. L. (2003). G-protein-coupled receptors at a glance. J. Cell Sci. 116, 4867–4869 10.1242/jcs.00902 [DOI] [PubMed] [Google Scholar]
- Kulkarni R. D., Thon M. R., Pan H., Dean R. A. (2005). Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 6:R24 10.1186/gb-2005-6-3-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A., Han K. H., Seo J. A., Yu J. H., d'Enfert C. (2006). G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet. Biol. 43, 490–502 10.1016/j.fgb.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Latgé J.-P. (2007). The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66, 279–290 10.1111/j.1365-2958.2007.05872.x [DOI] [PubMed] [Google Scholar]
- Latgé J. P., Mouyna I., Tekaia F., Beauvais A., Debeaupuis J. P., Nierman W. (2005). Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med. Mycol. 43Suppl. 1,15–22 [DOI] [PubMed] [Google Scholar]
- Lee K. S., Levin D. E. (1992). Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12, 172–182 10.1128/MCB.12.1.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Bussey H. (2006). Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343 10.1128/MMBR.00038-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291 10.1128/MMBR.69.2.262-291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz S. M., Nong S. H., Seetoo K. F., Harrison T. S., Speizer R. A., Simons E. R. (1999). Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Agrellos O. A., Calderone R. (2012). Histidine kinases keep fungi safe and vigorous. Curr. Opin. Microbiol. 13, 424–430 10.1016/j.mib.2010.04.007 [DOI] [PubMed] [Google Scholar]
- Liebmann B., Gattung S., Jahn B., Brakhage A. A. (2003). cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269, 420–435 10.1007/s00438-003-0852-0 [DOI] [PubMed] [Google Scholar]
- Lodder A. L., Lee T. K., Ballester R. (1999). Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T. J., Villa N. Y., Regalla L. M., Kupchak B. R., Vagstad A., Eide D. J. (2004). Metalloregulation of yeast membrane steroid receptor homologs. Proc. Natl. Acad. Sci. U.S.A. 101, 5506–5511 10.1073/pnas.0306324101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S. M., Saito H. (1994). A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369, 242–245 10.1038/369242a0 [DOI] [PubMed] [Google Scholar]
- McCormick A., Jacobsen I. D., Broniszewska M., Beck J., Heesemann J., Ebel F. (2012). The two-component sensor kinase TcsC and its role in stress resistance of the human-pathogenic mold Aspergillus fumigatus. PLoS ONE 7:e38262 10.1371/journal.pone.0038262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M. M., Nash S. L., Hajjeh R. A., Phelan M. A., Conn L. A., Plikaytis B. D., et al. (2001). Trends in mortality due to invasive mycotic diseases in the United States, 1980–1997. Clin. Infect. Dis. 33, 641–647 10.1086/322606 [DOI] [PubMed] [Google Scholar]
- Miller T. L., Godfrey P. A., Dealmeida V. I., Mayo K. E. (1999). The rat growth hormone-releasing hormone receptor gene: structure, regulation, and generation of receptor isoforms with different signaling properties. Endocrinology 140, 4152–4165 10.1210/en.140.9.4152 [DOI] [PubMed] [Google Scholar]
- Mingot J. M., Espeso E. A., Diez E., Penalva M. A. (2001). Ambient pH signaling regulates nuclear localization of the Aspergillus nidulans PacC transcription factor. Mol. Cell. Biol. 21, 1688–1699 10.1128/MCB.21.5.1688-1699.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot J. M., Tilburn J., Diez E., Bignell E., Orejas M., Widdick D. A., et al. (1999). Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol. Cell. Biol. 19, 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete-Urtasun S., Denison S. H., Arst H. N., Jr. (1997). Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J. Bacteriol. 179, 1832–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete-Urtasun S., Reiter W., Diez E., Denison S. H., Tilburn J., Espeso E. A., et al. (1999). Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol. Microbiol. 33, 994–1003 10.1046/j.1365-2958.1999.01540.x [DOI] [PubMed] [Google Scholar]
- Newman S. L. (1999). Macrophages in host defense against Histoplasma capsulatum. Trends Microbiol. 7, 67–71 10.1016/S0966-842X(98)01431-0 [DOI] [PubMed] [Google Scholar]
- Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S., Arroyo J., et al. (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438, 1151–1156 10.1038/nature04332 [DOI] [PubMed] [Google Scholar]
- Oide S., Liu J. Y., Yun S. H., Wu D. L., Michev A., Choi M. Y., et al. (2010). Histidine kinase two-component response regulator proteins regulate reproductive development, virulence, and stress responses of the fungal cereal pathogens Cochliobolus heterostrophus and Gibberella zeae. Eukaryot. Cell 9, 1867–1880 10.1128/EC.00150-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. (2010). Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64, 101–122 10.1146/annurev.micro.112408.134215 [DOI] [PubMed] [Google Scholar]
- Peñalva M., Arst H. N., Jr. (2004). Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58, 425–451 10.1146/annurev.micro.58.030603.123715 [DOI] [PubMed] [Google Scholar]
- Peñalva M., Tilburn J., Bignell E., Arst H. N., Jr. (2008). Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16, 291–300 10.1016/j.tim.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Penas M. M., Hervas-Aguilar A., Munera-Huertas T., Reoyo E., Penalva M. A., Arst H. N., Jr., et al. (2007). Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot. Cell 6, 960–970 10.1128/EC.00047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott G. B., Miller T. K., Bartlett J. A., Palas J. S., Selitrennikoff C. P. (2000). The isolation of FOS-1, a gene encoding a putative two-component histidine kinase from Aspergillus fumigatus. Fungal Genet. Biol. 31, 55–67 10.1006/fgbi.2000.1225 [DOI] [PubMed] [Google Scholar]
- Reeves E. P., Lu H., Jacobs H. L., Messina C. G., Bolsover S., Gabella G., et al. (2002). Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416, 291–297 10.1038/416291a [DOI] [PubMed] [Google Scholar]
- Rhodes J. C. (2006). Aspergillus fumigatus: growth and virulence. Med. Mycol. 44Suppl. 1, S77–S81 10.1080/13693780600779419 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Galan O., Galindo A., Hervas-Aguilar A., Arst H. N., Jr., Penalva M. A. (2009). Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J. Biol. Chem. 284, 4404–4412 10.1074/jbc.M808645200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. L., Shiozaki K. (2001). Fungal histidine kinases. Sci. STKE 2001:re1 10.1126/stke.2001.98.re1 [DOI] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Joechl C., Rogers T., Arst H. N., Jr., et al. (2004). Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200, 1213–1219 10.1084/jem.20041242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M., et al. (2007). Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3:1195–1207 10.1371/journal.ppat.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R., Martin H., Casamayor A., Arino J. (2006). Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 281, 39785–39795 10.1074/jbc.M604497200 [DOI] [PubMed] [Google Scholar]
- Shin K. S., Kwon N. J., Yu J. H. (2009). Gbg-mediated growth and developmental control in Aspergillus fumigatus. Curr. Genet. 55, 631–641 10.1007/s00294-009-0276-4 [DOI] [PubMed] [Google Scholar]
- Tilburn J., Sarkar S., Widdick D. A., Espeso E. A., Orejas M., Mungroo J., et al. (1995). The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14, 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. A., Gow N. A. R., Munro C. A. (2010). Fungal echinocandin resistance. Fungal Genet. Biol. 47, 117–126 10.1016/j.fgb.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. H., Stock A. M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376 [DOI] [PubMed] [Google Scholar]
- Yun C. W., Tamaki H., Nakayama R., Yamamoto K., Kumagai H. (1998). Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252, 29–33 10.1006/bbrc.1998.9600 [DOI] [PubMed] [Google Scholar]
- Zhang L. J., Wang M. Y., Li R. Y., Calderone R. (2005). Expression of Aspergillus fumigatus virulence-related genes detected in vitro and in vivo with competitive RT-PCR. Mycopathologia 160, 201–206 10.1007/s11046-005-0141-z [DOI] [PubMed] [Google Scholar]
- Zu T., Verna J., Ballester R. (2001). Mutations in WSC genes for putative stress receptors result in sensitivity to multiple stress conditions and impairment of Rlm1-dependent gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 266, 142–155 10.1007/s004380100537 [DOI] [PubMed] [Google Scholar]