Abstract
Activation of the polycyclic polyketide prenyltransferases (pcPTase)-containing silent clusters in Aspergillus fumigatus and Neosartorya fischeri led to isolation of a new metabolite neosartoricin (3). The structure of 3 was solved by X-ray crystallography and NMR to be a prenylated anthracenone. 3 Exhibits T-cell antiproliferative activity with an IC50 of 3 μM, suggestive of a physiological role as an immunosuppressive agent.
Genome mining of sequenced microorganisms has yielded new natural products with interesting activities.1 Mining of pathogenic species may yield new virulence factors or compounds active against human targets that are not produced in routine laboratory culturing conditions. As part of our efforts to understand fungal polyketide biosynthesis, we recently identified a group of polycyclic prenyltransferases (pcPTases) that catalyze the Friedel-Crafts transfer of C5 and C10 prenyl groups to aromatic polyketides. Phylogenetic analyses showed that the homologous pcPTase genes formed a distinct clade from the common indole prenyltransferase. 2 The pcPTases are found mostly in the aspergillosis- and keratitits-causing Aspergillus fumigatus and Neosartorya fischeri, and the seven genome-sequenced arthrodermataceous dermatophytes.3
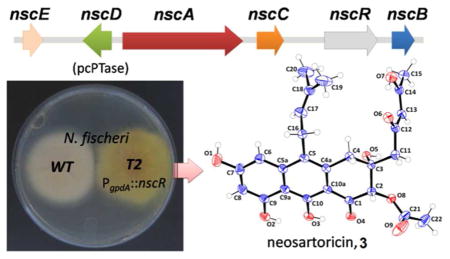
Interestingly, all these pcPTase genes are associated with a three-gene ensemble that makes polycyclic aromatic polyketides such as anthracenones (e.g. asperthecin) and napthacenediones (e.g. TAN-1612 1, viridicatumtoxin). 4, 5 The ensemble includes genes that encode a non-reducing polyketide synthase (NR-PKS), a flavin-dependent monooxygenase (FMO) and a metallo-β-lactamase-like thioesterase (MβL-TE) (Figure 1). The prevalence of these pcPTase-encoding gene clusters among human and animal-associated fungi has invited the speculation that they may encode for production of a group of structurally-related prenylated polycyclic polyketides that have a common ecological role. When cloned and assayed in vitro, pcPTases from N. fischeri, M. canis and T. tonsurans were confirmed to be dimethylallyltransferases that can utilize naphthacenedione substrates including 1 (Scheme 1), supporting the hypothesis that prenylated polycyclic compounds may be produced by these organisms.
Figure 1.
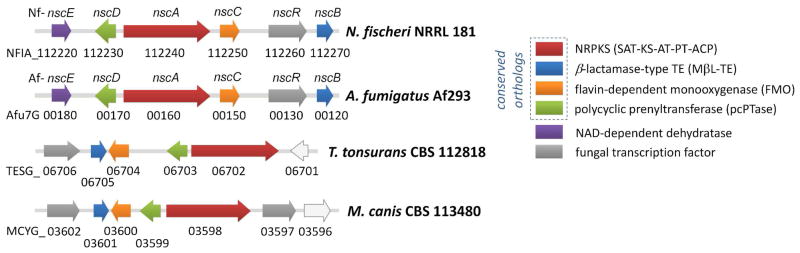
Evolutionary conserved pcPTase-containing gene clusters in N. fisheri, A. fumigatus, T. tonsurans and M. canis.
Scheme 1.
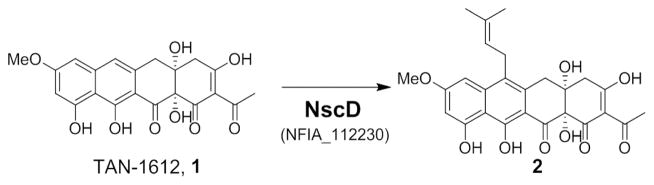
Conversion of 1 to 2 by pcPTase NscD.
Using the N. fischeri pcPTase (NFIA_112230, Nf-NscD) as a lead, we aim to identify the compound(s) produced by the gene cluster (designated as nsc gene cluster). We expect the highly homologous and syntenic nsc cluster (Afu7g00120-170) in A. fumigatus to encode for identical metabolite(s) as in N. fischeri (>90% protein identities, Table S2). Under our laboratory culturing conditions, neither N. fischeri NRRL 181 nor A. fumigatus Af293 produced any detectable prenylated polyaromatics, consistent with the lack of such compounds associated with strains in literature.
Reverse transcriptase-PCR (RT-PCR) from the total RNA of N. fischeri did not detect transcription of the nsc cluster genes (Figure S1A), suggesting that the cluster is transcriptionally silent. To activate the pathway, we overexpressed a putative pathway-specific Zn(II)2Cys6 transcriptional factor (FTF) encoded by NFIA_112260 (Nf-nscR), a strategy that has been successfully used in various fungi recently.4,6 Transformation of N. fischeri with pBGP-Nf112260, which places nscR under the regulation of A. nidulans gpdA promoter (Pgpda) and contains the bar resistance gene, resulted in several glufosinate-resistant colonies that exhibit yellow pigmentation on the reverse side on agar plates (Figure S1B). Diagnostic PCR confirmed the integration of nscR-overexpression cassette in transformant T2, while RT-PCR analysis showed that the nsc genes (nscA-E) were indeed upregulated (Figure S1A).
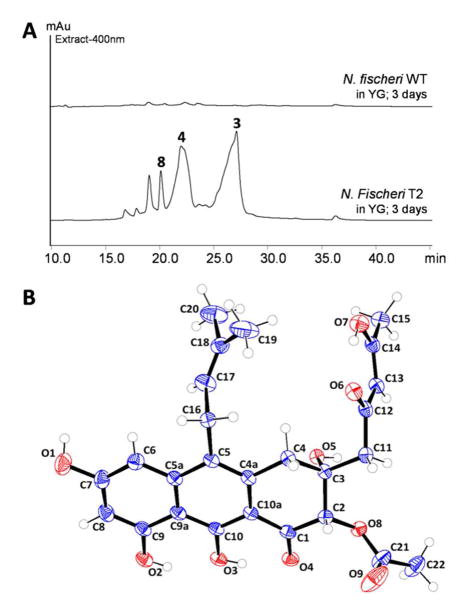
The transformant T2 cultured in stationary liquid GMM culture was extracted after 3 days. LC/MS analysis of the extract indicates production of multiple new compounds that are not found in the wild type N. fischeri (Figure 2A). All the new compounds exhibit similar UV spectra and λmax between 395–410 nm, while the MW range from 358 to 484. We reasoned that the higher MW peaks, including 3 (m/z 485 [M+H]+) and 7 (m/z 425 [M+H]+) at later RT are likely to be prenylated, while the compounds at earlier RT are unprenylated intermediates and shunt products, i.e. 4 (m/z = 359 [M+H]+) and 8 (m/z = 317 [M+H]+) (Figure 2A and Figure S2). To investigate if the same set of metabolites is produced by A. fumigatus upon transcriptional activation of the syntenic nsc cluster (Figure 1), an overexpression cassette encoding the putative FTF Afu7G00170 (p402-Af0170) with the resistance marker phleoR for selection was randomly integrated into the genome. In identical fashion to N. fischeri, A. fumigatus incorporating the cassette exhibits yellow pigmentation. The metabolite profile of A. fumigatus T1 is indeed identical to N. fischeri T2 based on UV spectra, retention time and m/z values of major compounds 3, 4, 7 and 8 during LC-MS analysis (Figure S3). Thus, both clusters in N. fischeri and A. fumigatus have been activated to produce the same set of new metabolites.
Figure 2.
Activation of a silent pathway in N. fischeri. (A) Metabolic profile of N. fisheri T2 (nscR overexpression) compared to WT. (B) Perspective drawing of the molecular confirmation of neosartoricin 3.
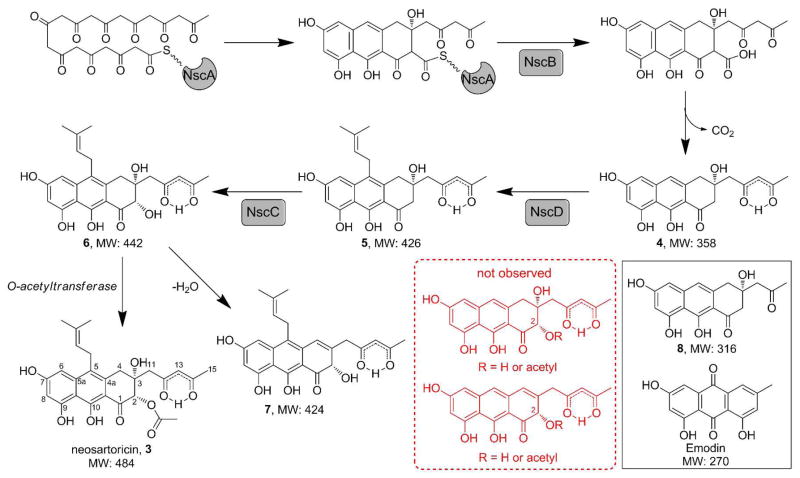
About 35 mg of 3 (as bright yellow, amorphous powder) was successfully purified from 2 L of the N. fischeri T2 culture. Orange platelet-like crystals were obtained in an EtOAc/n-hexane solvent system and the atom connectivity of 3 was resolved by X-ray diffraction (Figure 2B). 3 Apparently co-crystalized with residual DMSO-d6 from NMR analyses. Anomalous scattering of the X-rays by the sulfur atom in DMSO allowed elucidation of the absolute three-dimensional structure of 3. The X-ray structure assisted in the assignment of the NMR spectra (Table S2). The structure of 3 consists of a tricyclic 3,4-dihydroanthracen-1(2H)-one core with a 2,4-keto-enol pentyl side chain extending from a decaketide (C20) backbone, a dimethylallyl side chain at C5, and a C2-acetoxy substitution syn to a C3-hydroxyl. The 2,4-keto-enol pentyl chain tautomerizes in organic solvent, where the β-keto-enol form is dominant over the β-diketo form as observed in the 1H, 13C and 2D NMR spectra (Table S2 and Figure S5–S8). 3 was named neosartoricin, based on the common distribution in both N. fisheri and A. fumigatus (given the Teleomorph name Neosartorya fumigata). 7 Besides 3, the desacetyl derivative 7 (overlapped with 3 in Figure 2A) was isolated in small quantity from the N. fisheri T2. Due to low titer (<1 mg/L) and instability upon purification, only 1H-NMR and LCMS data can be obtained for 7. Spectra anaysis and comparison to 3 led to assignment of the structure in Scheme 2 (Table S3). 4 and 8 are non-prenylated anthracenones derived from a decaketide and a nonaketide (C18) respectively. Both compounds were isolated and characterized from yeast expressing the NR-PKS and FMO pair from the A. niger ada pathway for 1 and A. nidulans apt pathway for asperthecin, previously.4
Scheme 2.
Proposed pathway for biosynthesis of 3
See Figure S2 and Supplementary Discussion for more details.
A pathway for biosynthesis of 3 was proposed based on analysis of the intermediates (Scheme 2 and Figure S2). As NscA-C are highly homologous to AdaA-C in A. niger, the biosynthesis of the anthracenones portion of 3 is likely to follow a parallel pathway for that of 1: NscA synthesizes and cyclizes the decaketide backbone; NscB mediates the product release through hydrolysis followed by spontaneous decarboxylation to afford 4; and NscC is responsible for the stereospecfic hydroxylation at C2. The NscD catalyzes the addition of dimethylallyl group to the aromatic C5, consistent with the previously determined regioselectivity (Scheme 1). The timing of NscC, which was proposed to occur after prenylation of 4 to yield 5, likely directs the formation of the anthracenone present in 6 instead of the naphthacenedione in 1, although an alternative pathway via a Baeyer-Villiger cleavage of the fourth ring, such as that observed in mithramycin pathway, 8 cannot be completely ruled out (See Supplementary Discussion). There is no gene encoding O-acetyltransferase in the nsc gene cluster; thus, the 2-O-acetylation of 6 to 3 maybe catalyzed by an unidentified O-acetyltransferase in N. fisheri, while off-pathway dehydration of 6 affords 7.
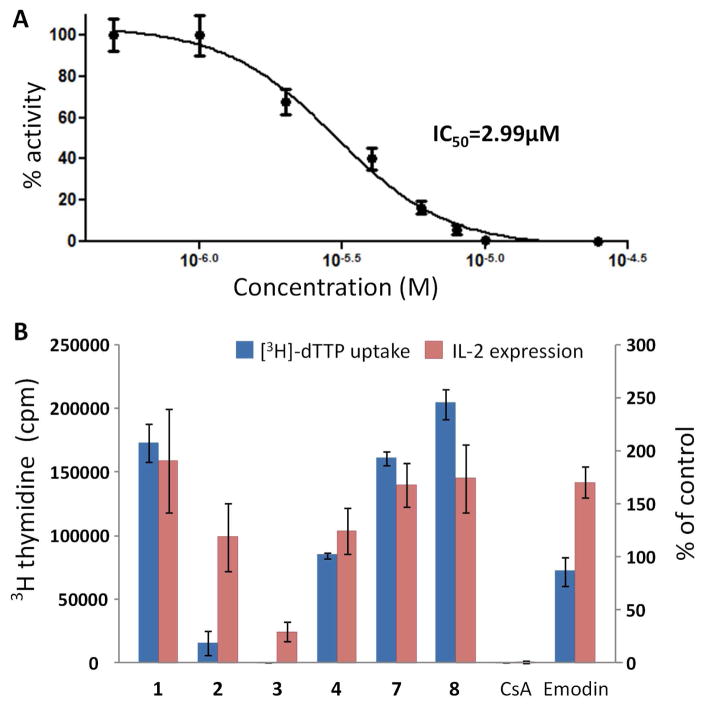
3 Does not exhibit significant inhibitory activity against either Gram positive or Gram negative bacteria, nor the yeasts Saccharomyces cerevisiae and Candida albicans (> 64 μg/mL). We tested 3 for immunosuppressive activity in a cell-based in vitro assay, which showed that 3 exhibits antiproliferative activity on anti-CD3/CD28-activated murine spleenic T-cells with an IC50 of 2.99 μM (1.45 Vg/mL) (Figure 3A). The effect of 3 on [3H]-thymidine uptake by murine T-cells is dosage-dependent. Cyclosporin A (CsA) was used as a control in the assay and was shown to exhibit an IC50 of 26 nM, which is comparable to the value reported previously.9 Next, we included compound 1, 2, 4, 7, and 8 in a structural-activity relationship studies along with emodin, which has been previously shown to exhibit moderate immunosuppressive activity. 10 The data show that 3 exhibits a higher inhibitory activity compared to all the analogs tested including emodin (Figure 3B) and that the dimethylallyl group and O-acetyl is required for its activity. 3 Was then assayed for cytotoxicity against HeLa and HFF cells, which shows that 3 is less toxic to the two cell lines compared to 1 and 2 at up to 50 μM concentration, suggesting that the antiproliferateive activity of 3 is relatively specific to T-cells (Figure S9).
Figure 3.
Effect of 3 on murine T-cell proliferation. (A) Dose-response curve of 3 on [3H]-thymidine uptake by murine T-cells (% of DMSO control). (B) Comparison of the activity of 3 with 1, 2, 4, 7, 8, cyclosporine A (CsA) and emodin at 10 μM concentration.
We next examined the possible role of 3 in establishing primary infection in human hosts with a 51Cr release cell damage assay on A549 lung epithelial cells and RAW 264.7 macrophage cells. 3 Does not appear to cause any damage to both cell types up to 10 μM (< 5% specific release). The extent of A549 cell damage caused by the nsc pathway-overexpressing A. fumigatus T1 in vitro was also investigated, but no statiscally significant differences in cell damage was observed compared to wild type A. fumigatus (Figure S10). Thus, we concluded that the nsc pathway compounds including 3 are not involved in primary virulence but may facilitate infection through suppressing the host adaptive immunity.11
Supplementary Material
Acknowledgments
We thank Dr. Saeed Khan at the UCLA crystallography facility for solving the X-ray structures. Anu Biswas and Muxun Zhao of UCLA are thanked for their assistance in cytotoxicity assays. This work is supported by NIH 1R01GM085128 and 1DP1GM106413 to YT, and R01AI073829 to SGF.
Footnotes
Supporting Information Available NMR and MS characterization of compounds and additional experimental information and discussion. CIF Crystallographic Information File of compound 3 (CCDC 866028).
References
- 1.Winter JM, Behnken S, Hertweck C. Curr Opin Chem Biol. 2011;15:22. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Chooi YH, Wang P, Fang J, Li Y, Wu K, Tang Y. J Am Chem Soc. 2012;134:9428. doi: 10.1021/ja3028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, Monod M, Shelest E, Barton RC, Birch E, Brakhage AA, Chen Z, Gurr SJ, Heiman D, Heitman J, Kosti I, Rossi A, Saif S, Samalova M, Saunders CW, Shea T, Summerbell RC, Xu J, Young S, Zeng Q, Birren BW, Cuomo CA, White TC. MBio. 2012;3:e00259. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Chooi Y-H, Sheng Y, Valentine JS, Tang Y. J Am Chem Soc. 2011;133:15773. doi: 10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chooi YH, Cacho R, Tang Y. Chem Biol. 2010;17:483. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Nat Chem Biol. 2007;3:213. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 7.O’Gorman CM, Fuller HT, Dyer PS. Nature. 2008;457:471. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 8.Gibson M, Nur-e-alam Mohammad, Lipata F, Oliveira MA, Rohr J. J Am Chem Soc. 2005;127:17594. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 9.Zenke G, Strittmatter U, Fuchs S, Quesniaux VFJ, Brinkmann V, Schuler W, Zurini M, Enz A, Billich A, Sanglier JJ. J Immunol. 2001;166:7165. doi: 10.4049/jimmunol.166.12.7165. [DOI] [PubMed] [Google Scholar]
- 10.Huang HC, Chu SH, Chao PDL. Eur J Pharmacol. 1991;198:211. doi: 10.1016/0014-2999(91)90624-y. [DOI] [PubMed] [Google Scholar]
- 11.Blanco JL, Garcia ME. Vet Immunol Immunopathol. 2008;125:47. doi: 10.1016/j.vetimm.2008.04.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.